Abstract
Tissue-nonspecific alkaline phosphatase (TNAP) plays a major role in maintaining a ratio of phosphate to inorganic pyrophosphate (Pi/PPi) in biological fluids that is conducive to controlled skeletal mineralization while preventing inappropriate ectopic calcification. Medial calcification associated with Enpp1 or Ank deficiency or with end–stage renal disease is associated with an increase in TNAP activity in arteries that leads to reduced levels of PPi and increased vascular calcification. Here, we describe in detail a high-throughput screening (HTS) campaign to identify inhibitors of TNAP, performed within the Molecular Library Screening Center Network (MLSCN). A homogeneous luminescent TNAP assay was developed and optimized for identification of compounds with diverse mechanism of action (MOA). The MLSCN compound collection, containing 64,394 molecules at the time of screening, was tested in the assay. Several novel inhibitory scaffold classes were identified and demonstrated to have diverse selectivity and mode of inhibition (MOI) profiles. Representatives of the novel scaffolds exhibited nanomolar potency surpassing the inhibitors known to date.
This paper sets a successful example in which pharmacologically active compounds, with outstanding selectivity in a panel of more than 200 assays, are identified from high throughput screening. Integral to the success of the project were a well-designed compound collection, an industrial-level screening facility and a deep knowledge of target biology that were brought together through the NIH-sponsored Roadmap Initiative.
Keywords: NIH Roadmap Initiatives, MLSCN, TNAP inhibitors, diverse MOA, compound selectivity
INTRODUCTION
Alkaline phosphatases (E.C.3.1.3.1) (APs) are dimeric enzymes, present in most organisms (McComb et al., 1979), where they catalyze the hydrolysis of phosphomonoesters. In humans, three of the four isozymes are tissue-specific, i.e., the intestinal (IAP), placental (PLAP), and germ cell (GCAP) APs; the fourth AP is tissue-nonspecific (TNAP) and is expressed in bone, liver and kidney (Millán, 2006). Recent studies have provided compelling proof that a major role for TNAP in bone tissue is to hydrolyze extracellular inorganic pyrophosphate, PPi, to avoid accumulation of this mineralization inhibitor, thus ensuring normal bone mineralization. PPi is a potent inhibitor of hydroxyapatite formation at concentrations normally found in plasma (Russell et al., 1969; Meyer et al., 1984; Francis et al., 1969). PPi prevents calcification of rat aortas in culture (Lomashvili et al., 2004) and in vitamin D-toxic rats in vivo (Schibler et al., 1968). Humans with low levels of PPi due to the absence of the PPi-producing enzyme ecto-nucleotide pyrophosphatase/phosphodiesterases-1 (NPP1, a.k.a PC-1) develop severe, fatal arterial calcification (Garg et al, 2005; Terkeltaub, 2001; Rutsch et al., 2001; Rutsch et al., 2003). Humans undergoing chronic hemodialysis, in whom arterial calcification is common, have reduced plasma levels of PPi (Lomashvili et al., 2005). Thus, there are compelling data that PPi is an important endogenous inhibitor of medial vascular calcification. Normalization of PPi levels in NPP1 null and ANK-deficient mice improves their soft-tissue ossification abnormalities (Hessle et al., 2002; Harmey et al., 2004). Importantly, these studies have suggested that TNAP may be a useful therapeutic target for the treatment of arterial calcification. Indeed, substantial evidence points to the presence of TNAP-rich vesicles at sites of mineralization in human arteries. The presence of TNAP-enriched matrix vesicles (MVs) in human atherosclerotic lesions suggests an active role in the promotion of the accompanying vascular calcification (Hsu and Camacho, 1999; Hui et al., 1997; 1998; Tanimura et al., 1986). Increased expression of TNAP accelerates calcification by bovine vascular smooth muscle cells (VSMCs) (Shioi et al., 1995), and macrophages can induce a calcifying phenotype in human VSMCs by activating TNAP in the presence of IFNγ and 1,25(OH)2D3 (Shioi et al., 2002). Recently we have shown upregulation of TNAP activity in Enpp1−/− and ank/ank VSMCs (Narisawa et al., 2007) and in the aortas of uremic rats (Lomashvili et al., 2008) and we have shown that the pharmacological downregulation of this upregulated TNAP activity suppresses VSMC-dependent calcification (Narisawa et al., 2007). Thus, there is ample evidence warranting exploration of the therapeutic potential of TNAP inhibition at sites of arterial calcification to increase local concentration of PPi thereby reducing inappropriate mineralization. Discovery of potent and selective TNAP inhibitors would facilitate these explorations.
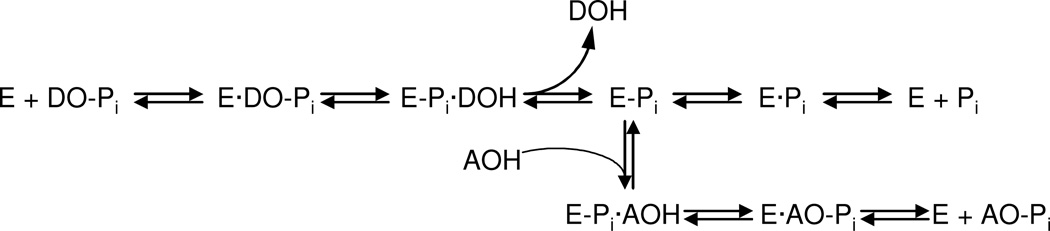
The molecular mechanism of the AP catalytic reaction is common to the enzyme from various species and tissues and is depicted in Scheme 1 (Holtz et al. 1999). The initial AP (designated as E in the scheme) catalyzed reaction consists of a substrate (DO-Pi) binding step, phosphate-moiety transfer to the active site Ser and product alcohol (DOH) release. In the second part of the reaction, phosphate is released through hydrolysis of the covalent intermediate (E-Pi) and dissociation of inorganic phosphate from the non-covalent complex (E·Pi). Depending on the origin of the enzyme and the exact conditions of the reaction, either hydrolysis of E-Pi or release of the phosphate from E·Pi is rate-limiting leading to the elevated relative concentration of E-Pi and E·Pi comparing with other enzyme-substrate species. In the presence of alcohol molecules (AOH), phosphate is also released via a faster transphosphorylation reaction mechanism. AP assays commonly utilized in clinical practice (Stinson, 1993; WHO Guidelines on Standard Operating Procedures for Clinical Chemistry, Section B) are based on dephosphorylation of p-nitrophenol phosphate (pNPP) in the presence of high concentration of amino-containing buffers, such as 2-amino-2-methyl-1-propanol and diethanolamine (DEA). Besides maintaining an alkaline pH, the buffer also provides saturating levels of substrate for the AP transphosphorylation reaction. Interestingly, the biological significance of AOH is still unknown, and it is unclear if any biological molecule is capable of performing the function of AOH in vivo.
Scheme 1.
Catalytic mechanism of alkaline phosphatase reaction (Holtz et al, 1999). Abbreviations: E - alkaline phosphatase enzyme molecule; DO-Pi - substrate molecule; E-Pi – phosphorenzyme (enzyme phosphorylated on Ser-93 in the TNAP sequence of its active site); DOH - product alcohol; E·Pi - non-covalent complex of inorganic phosphate in the active site; AOH – an alcohol molecule, acceptor substrate of transphosphorylation reaction acceptor; AO-Pi – product of transphosphorylation reaction.
TNAP, like all mammalian APs, is inhibited uncompetitively by a number of small molecule compounds. They include L-homoarginine (Fishman and Sie, 1971), as well as some unrelated compounds, such as levamisole (Van Belle, 1976) and theophylline (Farley et al., 1980). However, these known inhibitors of TNAP are not entirely specific for this AP isozyme, have low affinity and are not particularly effective at inhibiting the pyrophosphatase activity of TNAP. For these reasons, we initiated a comprehensive effort to screen for and optimize, via structure-activity and medicinal chemistry efforts, novel potent TNAP inhibitors that are capable of specifically inhibiting pyrophosphatase activity of TNAP at physiological pH.
The current manuscript documents a successful screening campaign that led to the identification of potent small molecule TNAP inhibitors. In the scope of this campaign a novel TNAP assay was developed and optimized to ensure maximum sensitivity in detection of ligands with different modes of action (MOA). Subsequent HTS and follow-up studies led to identification of several potent and selective inhibitory scaffolds targeting TNAP but not related enzymes, and exhibiting diverse biochemical properties.
MATERIALS AND METHODS
Reagents
The CDP-star substrate (2-chloro-5-(4-methoxyspiro{1,2-dioxetane-3,2´(5´-chloro)-tricyclo [3.3.1.13.7]decan}-4-yl)-1-phenyl phosphate disodium salt; CAS 160081-61-9) was obtained from New England Biolabs. All other chemicals were of an analytical grade.
Compound collection utilized in HTS
The compound library was supplied by the NIH Molecular Libraries Small Molecule Repository (MLSMR, http://www.mli.nih.gov/mlsmr). The MLSMR, funded by the NIH, is responsible for the selection of small molecules for HTS screening, their purchase and QC analysis, library maintenance and distribution within the NIH Molecular Libraries Screening Center Network (MLSCN, http://www.mli.nih.gov/mlscn). Both MLSMR and MLSCN are parts of the Molecular Libraries Initiatives (MLI, http://nihroadmap.nih.gov/molecularlibraries) under the NIH Roadmap Initiative (www.nihroadmap.nih.gov). MLSMR compounds are acquired from commercial, and in part from academic and government sources and are selected based on the following criteria: samples are available for re-supply in 10 mg quantity, are at least 90% pure, have acceptable physicochemical properties and contain no functional groups or moieties which are known to generate artifacts in HTS (http://mlsmr.glpg.com/). Compounds are selected to represent diversified chemical space with clusters of closely related analogs around them to aid in the HTS-based SAR analysis.
Expression and preparation of test enzymes
Expression plasmids containing a secreted epitope-tagged TNAP were transfected into COS-1 cells for transient expression (Kozlenkov et al., 2004). The medium was replaced with Opti-MEM 24 hrs later, and the serum free media containing secreted proteins were collected 60 hrs after electroporation. The conditioned medium was dialyzed against TBS containing 1 mM MgCl2 and 20 mM ZnCl2 (to remove phosphate) and filtered through a 0.22 µm cellulose acetate filter.
High throughput screening assays
Development of the novel luminescent TNAP assay using CDP-star substrate and detailed protocol of its application for HTS is described in more details elsewhere (ES&JLM, 2008). Briefly, the assay was performed in 384-well white plates (784075, Greiner) at 50 µM CDP-star substrate in assay buffer containing 100 mM DEA-HCl, pH 9.8, 1 mM MgCl2 and 20 uM ZnCl2. The luminescence signal was measured after 30 min incubation at room temperature on an EnVision plate reader (Perkin Elmer, Inc.). Analogous luminescent assays were optimized and utilized for PLAP and IAP with the CDP-star concentration adjusted to their respective Km values, 85 µM and 177 uM respectively.
The colorimetric TNAP assay was performed as previously described (Narisawa et al., 2007) with slight modifications. TNAP in assay buffer containing 1 M DEA-HCl, pH 9.8, 1 mM MgCl2 and 20 uM ZnCl2 was added with 0.5-mM pNPP substrate and after 1h incubation, the reaction was terminated by adding 2,3-dimercapto-1-propanesulfonate to 100 µM. TNAP activity was measured using absorbance at 405 nm (OD405 assay) or at 620 nm after addition of malachite green-based phosphate detection reagent (MG assay) prepared according to Cogan et al., 1999.
Compound solubility was measured on a NEPHELOstar plate reader (BMG Labtech, Inc.) that measures the intensity of light scattered by compound solution to detect potential compound precipitation. Compound concentrations where the signal in the well was higher than an average value of the control wells by >3-fold the standard deviation was excluded from dose-response data analysis.
High throughput screening and hit confirmation
TNAP screening was carried out with 64,394 compounds from the MLSMR. Primary screening was performed using the luminescent assay at a single concentration of 20 µM. Levamisole at 1 mM concentration was utilized as positive control. Z’-factor was calculated according to Zhang et al (1999).
Compound efficacy was estimated after data conversion to percent inhibition using Eq. (1):
| Eq. (1), |
where NC and PC represent average values of negative and positive control wells respectively, Y is a value observed in a well with the tested compound. Compounds exhibiting greater than 50% inhibition of TNAP at this concentration were defined as primary positives and confirmed in single-concentration and dose-response inhibition studies. The data was analyzed using Cbis software (ChemInnovation, Inc.). Dose-response data was fitted to the Hill equation (Eq. (2)) according non-linear regression:
| Eq. (2), |
where [C] is compound concentration in micromolar units, IC50 is the concentration value of half-maximal inhibition, nH is the Hill coefficient of the curve. For mode of inhibition studies, the compounds efficacy was tested in the presence of several concentrations of CDP-star or DEA and analyzed using Prism 4.03 (GraphPad Software Inc.).
Hit clustering
A relocation method based on maximal dissimilarity partitioning provided in the Accelrys Pipeline Pilot’s Chemistry Component was used for clustering of the hit molecules (http://accelrys.com/products/scitegic/component-collections/chemistry.html).
RESULTS
High throughput Screening
The original assay protocol called for utilization of a colorimetric assay that used pNPP as the substrate and employed saturating concentration of DEA. The assay demonstrated robust performance and reproducibility during HTS implementation stage. However, after screening 20.5K MLSCN compounds in the assay (see PubChem AID 615) or with a modified version that employed malachite green-based phosphate detection (see PubChem AID 614) we obtained no hits at all. Taking this into consideration, we decided to switch to the luminescent assay that we recently developed for confirmation purposes and utilize it as the primary HTS assay.
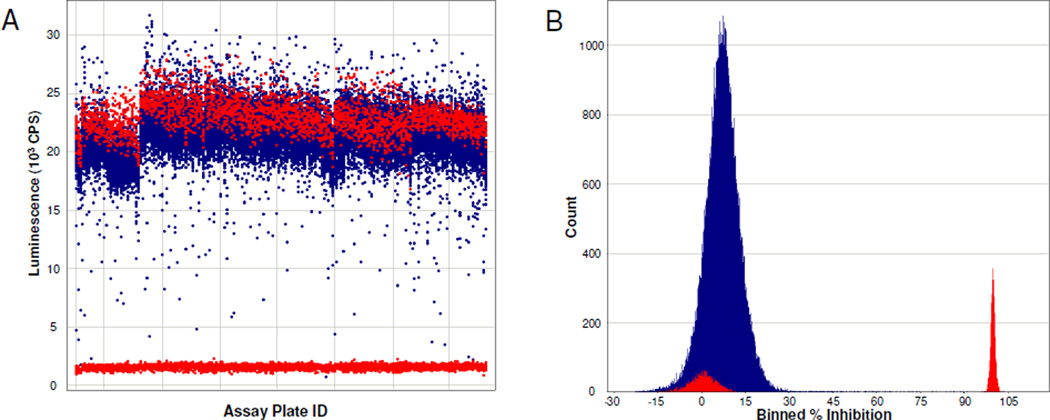
A detailed description of the development and utilization of the novel luminescent HTS assay for TNAP is described elsewhere (ES&JLM, 2008). Briefly, the assay was optimized to enable screening TNAP in the presence of phosphate-donor and phosphate-acceptor substrates present at their respective Km values. TNAP was screened against the MLSMR collection containing 64,394 compounds. The average Z’-factor for the full screen was equal 0.82 (range 0.75–0.89). A histogram and scatter-plots of the primary screening data presented in Figure 1.
Figure 1.
HTS data for the luminescence-based TNAP assay. MLSMR compound collection was screened at 20 uM compound concentration in the assay. The actual experimental data from screening 191 assay plates on seven different days is displayed in panel A without further modifications. Compounds inhibition calculated according to Eq. 1 is summarized in panel B in a histogram format. Tested compounds and control wells are depicted in blue and red color, respectively. Graphs are prepared using Spotfire software (TIBCO).
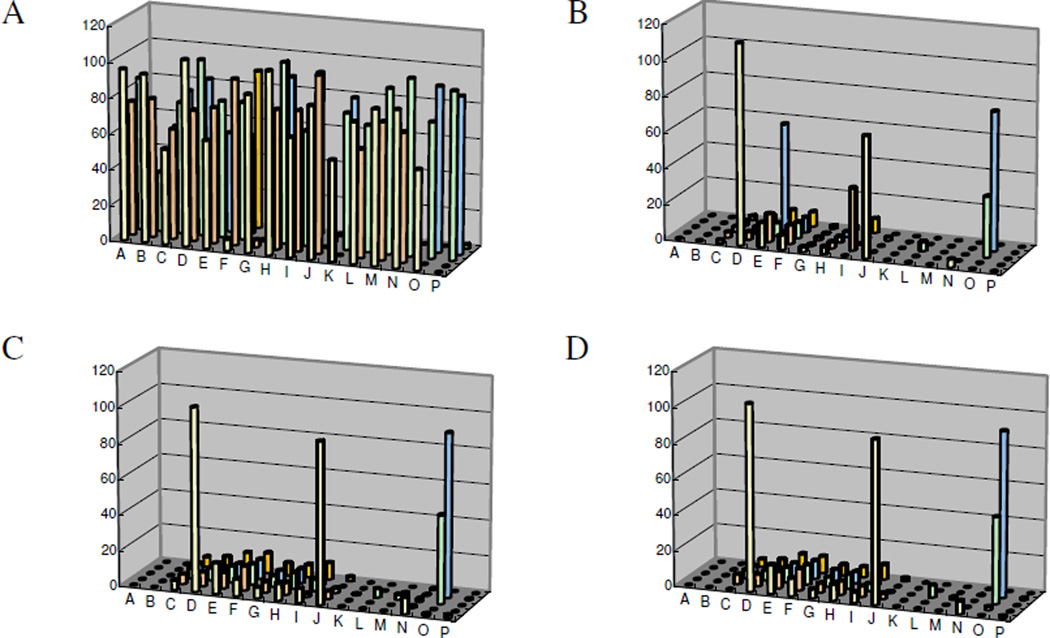
From the primary screen, 73 primary positives, compounds causing >50% inhibition, were identified resulting in 0.11% hit rate. For initial hit confirmation, aliquots for all the primary positives were obtained from the MLSMR and the compounds were tested in single concentration mode. Each compound was tested in quadruplicates at 20 µM, the concentration utilized in primary screening. Hits were similarly tested in the two colorimetric assays, OD405 and MG. Surprisingly, only four hits were confirmed in either of the colorimetric assay (Figure 2 C and D); whereas 55 of the primary hits reconfirmed in the primary HTS luminescent assay (Figure 2A). To address the apparent discrepancy, the compounds were also tested in the luminescent assay performed in the presence of 1M DEA-based buffer similar to the colorimetric assays. As can be seen in Figure 2B, the number of hits significantly decreased and matched the hit rate of the colorimetric assays, suggesting that a high DEA concentration interferes with the binding of most of hits, perhaps, through a direct competition for the binding site.
Figure 2.
Confirmation of primary TNAP hits. Cherry-picked hits were tested at 20 uM concentration in the luminescent (A and B) and colorimetric (C and D) assays. The concentration of DEA was 100 mM (A) or 1M (B, C, and D). Activity of the enzyme in the colorimetric assays was measured using absorbance of either p-nitrophenyl (C) or malachite green-phosphomolybdate complex (D). Experimental data was converted to % inhibition according to Eq. 1 using control values of the assay. Average values of each set of quadruplicates were calculated and utilized for each compound.
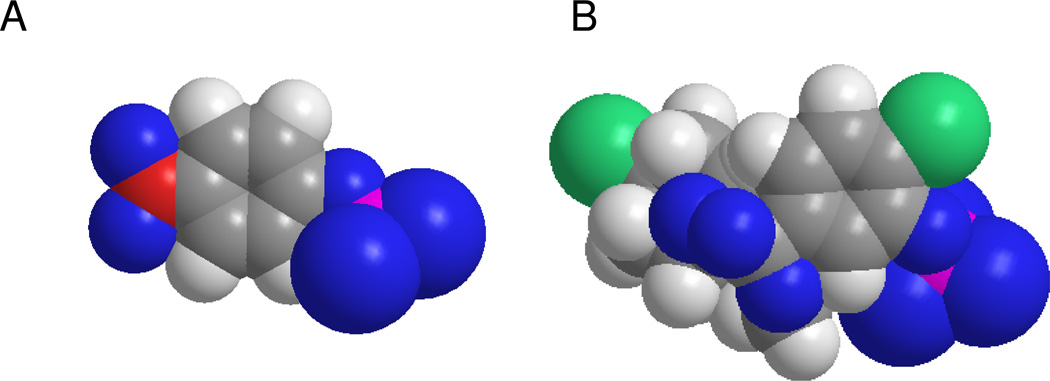
An additional observation from Figure 2B–D, two compounds display higher activity in the high-DEA luminescent assay compared with their activity in the colorimetric assays. It is hard to imagine that utilization of a bulkier substrate molecule (see Figure 3 for comparison of the two substrate molecules) would improve inhibitor binding unless it nonspecifically interacts with the non-catalytic part of the CDP-star molecule or is an artifact of the detection method utilized in the luminescent assay. In either case, those types of molecules would not be valuable for further development since they would be unlikely to inhibit the pyrophosphatase activity of TNAP. Early on in the project we developed and implemented a luminescent counterscreen utilizing PLAP. This assay served two purposes: 1) it detects compounds that have the potential to interfere with the assay detection system; 2) provides a measure of the specificity of the compounds in the family of APs.
Figure 3.
Spatial models of TNAP substrates. Space-fill rendering of the molecules of pNPP (A) and CDP-star (B) was generated using ChemBio3D software (CambridgeSoft).
The primary HTS hits were tested in a dose-response mode using the luminescent-based assay. Of the 73 primary positive compounds, 53 compounds were confirmed to have apparent inhibition and IC50 values below 20 µM in the luminescent assay with potency ranging from micromolar to submicromolar. The results of the HTS campaign are summarized in Table 1. A detailed protocol and experimental data for TNAP luminescent-based screening is described in PubChem AID 518.
Table 1.
Summary of HTS campaign for screening TNAP using luminescent assay
| Compounds screened | Hits identified | Hit criteria | Hit rate | |
|---|---|---|---|---|
| Primary HTS | 64,394 | 73 | >50% inhibition | 0.11% |
| Single-concentration confirmation | 73 | 55 | >50% inhibition | 75.3% |
| Dose-response confirmation | 55 | 53 | IC50 < 20 uM | 96.4% |
Primary HTS data analysis
The 53 confirmed hit molecules were clustered into 4 scaffold groups (see Figure 4) and several singletons using Chemistry Component Pipeline Pilot Software (Accelrys). Expanded HTS data analysis in relation to the hits was performed on 3 scaffolds (I, II and III in Fig. 4) each possessing members with submicromolar potency.
Figure 4.
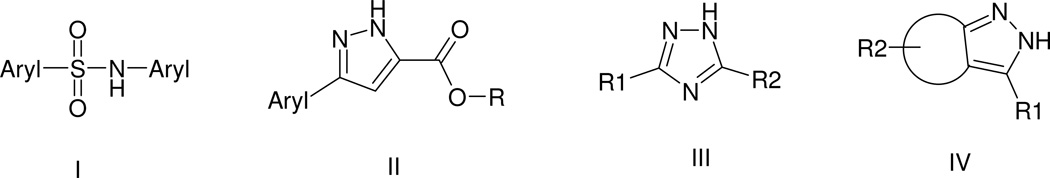
Scaffolds identified in the HTS. TNAP positives were clustered using Accelrys Pipeline Pilot’s Chemistry Component. The following clusters were identified: I – biarylsulfonamide; II – pyrazole; III – triazole; IV - fused pyrazole.
Biarylsulfonamides
For the biarylsulfonamide scaffold (I in Figure 4) series, the 3 most active compounds shown in Table 2, share high similarity to each other: all have a pyridine moiety and an alkoxy group at the ortho position of the sulfonylbenzene. The weaker compound lacked the pyridine moiety but contained the o-alkoxy group. A logical conclusion that could be immediately drawn from these observations is that pyridine and o-alkoxy-substituted sulfonylbenzene are necessary for the activity of this scaffold. HTS data was further “mined” to evaluate the validity of the above conclusion.
Table 2.
Biaryl sulfanilamide scaffold positive compounds
| Compound ID | PubChem SID |
PubChem CID |
Lumi HTS IC50 (µM) |
|
|---|---|---|---|---|
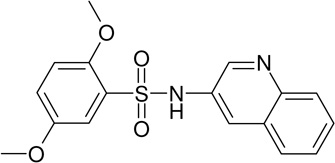 |
MLS-0038949 | 4261842 | 2931238 | 0.155 |
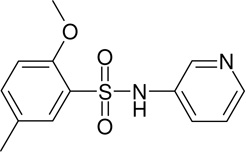 |
MLS-0010847 | 843550 | 645853 | 1.78 |
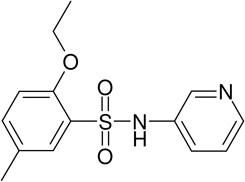 |
MLS-0043344 | 4260922 | 934901 | 1.93 |
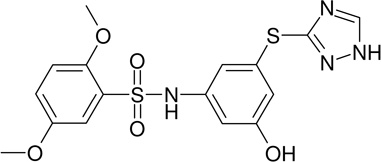 |
MLS-0043574 | 4262468 | 1589729 | 8.45 |
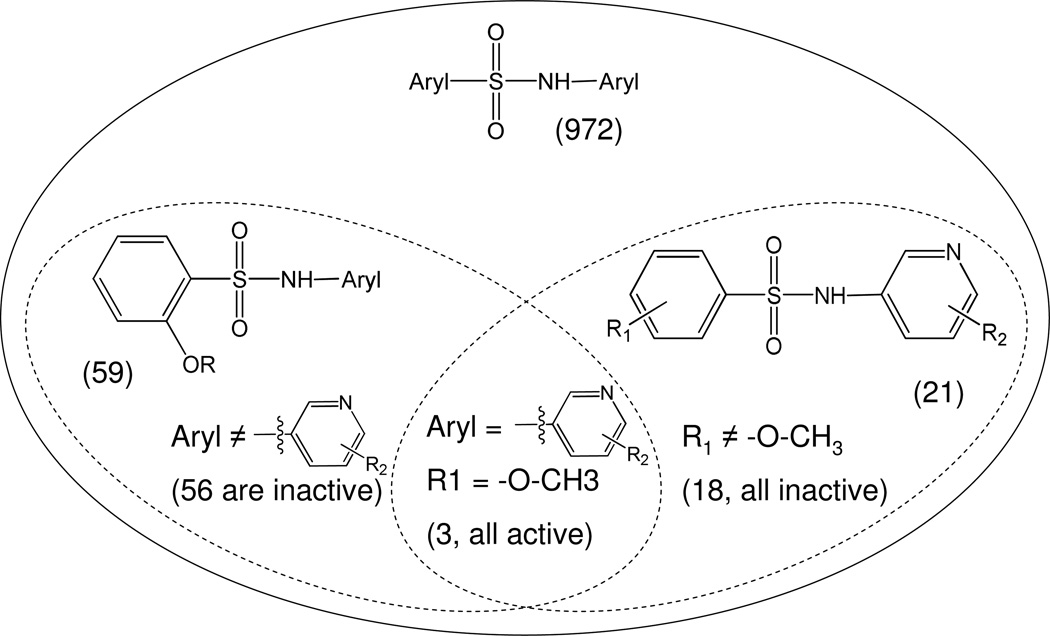
There are 972 molecules belonging to the biarylsulfonamide scaffold in the library used for the primary screen (Fig. 5). Further analysis showed that there are 59 molecules containing o-alkoxy-substituted sulfonylbenzene. Only 4 of these 59 compounds were active; three of them contained pyridine moiety. Among the 972 biarylsulfonamide derivatives 21 contained a pyridine moiety. Only three of them contained an o-alkoxy-substituted sulfonylbenzene and were active in the assay (Fig. 5). Therefore, this supports our conclusion that both a pyridine moiety and an o-alkoxy-sulfonylbenzene are required for biarylsulfonamide activity.
Figure 5.
HTS data analysis for biarylsulfonamide series
Pyrazole series
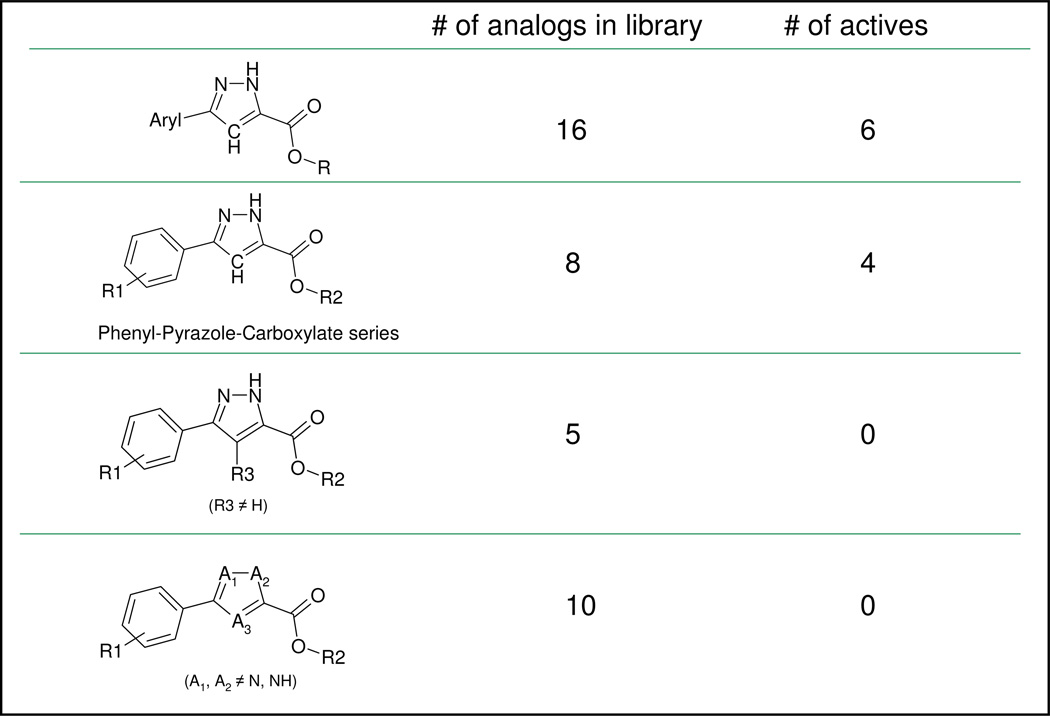
The available data did not suggest immediately which aryl group or R group is preferred for scaffold II (Figure 4). However, a similar HTS data analysis was performed to investigate the role of the N and NH pair of the pyrazole ring and the effect of substitution at position 4 of the pyrazole ring in scaffold II (Fig. 6). There were 5 molecules with a substituent at position 4 and none of them were active in the HTS assay, suggesting that an unsubstituted position 4 of the pyrazole ring is preferred for TNAP inhibition. Other 5-membered ring molecules that lack N-NH pair did not substitute for the pyrazole ring since 10 molecules in this category were all inactive.
Figure 6.
HTS data analysis for bi-substituted pyrazole series
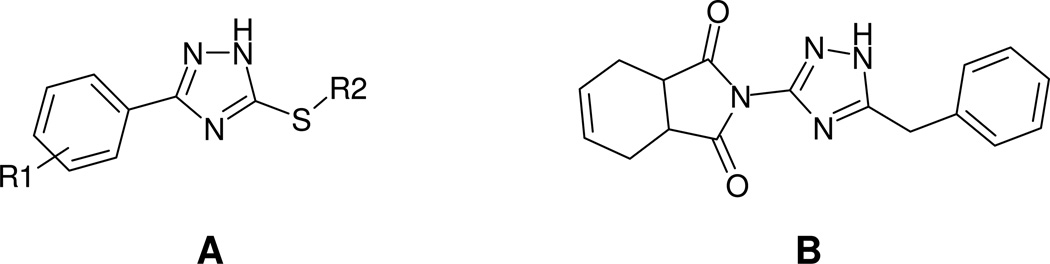
Triazole series
Most of the triazole derivatives (scaffold III in Figure 4) identified in primary screening belong to a more specific sub-series, thio-phenyl-triazole (Fig. 7A) resembling the ones identified in a previous screening effort (Narisawa et al., 2007). Another triazole derivative (Fig. 7B) was the most potent in the series and was identified as a singleton. Interesting observation, the only biarylsulfonamide compound that did not contain a pyridine moiety yet displayed some activity (MLS-0043574 in Table 2) also contained a triazole moiety. It is possible that its activity is a product of contribution from structural elements of these two scaffolds.
Figure 7.
Representatives of triazole series identified in HTS.
Hit confirmation
All 53 compounds identified and confirmed as positive inhibitors of TNAP originated from commercial sources. This significantly simplified compound structure verification and potency confirmation performed on dry powder compounds. The representatives of the major scaffolds were purchased from their original vendors. Table 3 summarizes the results of the reconfirmation stage. IC50 values obtained for dry powder compounds, designated SAR IC50 in Table 3, closely match the ones obtained for compounds provided by the MLSMR (HTS IC50 values in the Table 3).
Table 3.
Confirmation of major HTS scaffolds from dry powders
| Compound ID | PubChem SID |
PubChem CID |
Structure | IC50 (uM) | ||||
|---|---|---|---|---|---|---|---|---|
| HTS- Lumi |
HTS- OD405 |
SAR- Lumi |
SAR- OD405 |
SAR- MG |
||||
| MLS-0010847 | 26514189 | 645853 | 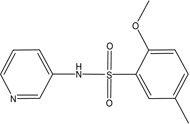 |
1.78 | 0.708 | 0.926 | 0.825 | 0.945 |
| MLS-0038949 | 26514170 | 2931238 | 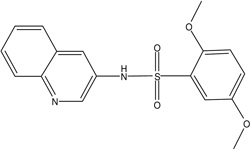 |
0.155 | 0.043 | 0.193 | 0.158 | 0.192 |
| MLS-0005718 | 843997 | 646303 | 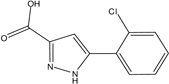 |
0.982 | >100 | 0.743 | >100 | >100 |
| MLS-0039961 | 4257586 | 610864 | 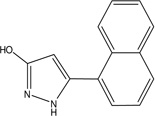 |
3.24 | 21 | 4.62 | 45.8 | 73.6 |
| MLS-0067142 | 4260165 | 2975791 | 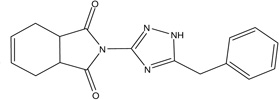 |
0.236 | >100 | 0.186 | >100 | >100 |
We purchased and tested 46, 29 and 10 commercially available analogs of the biarylsulfonamide, pyrazole and triazole series, respectively. These analogs offered confirmation for the SAR conclusions drawn from HTS. Extended SAR studies for these scaffolds will be described elsewhere (Ardecky et al, 2008; Dahl et al, in preparation; Sergienko et al, in preparation).
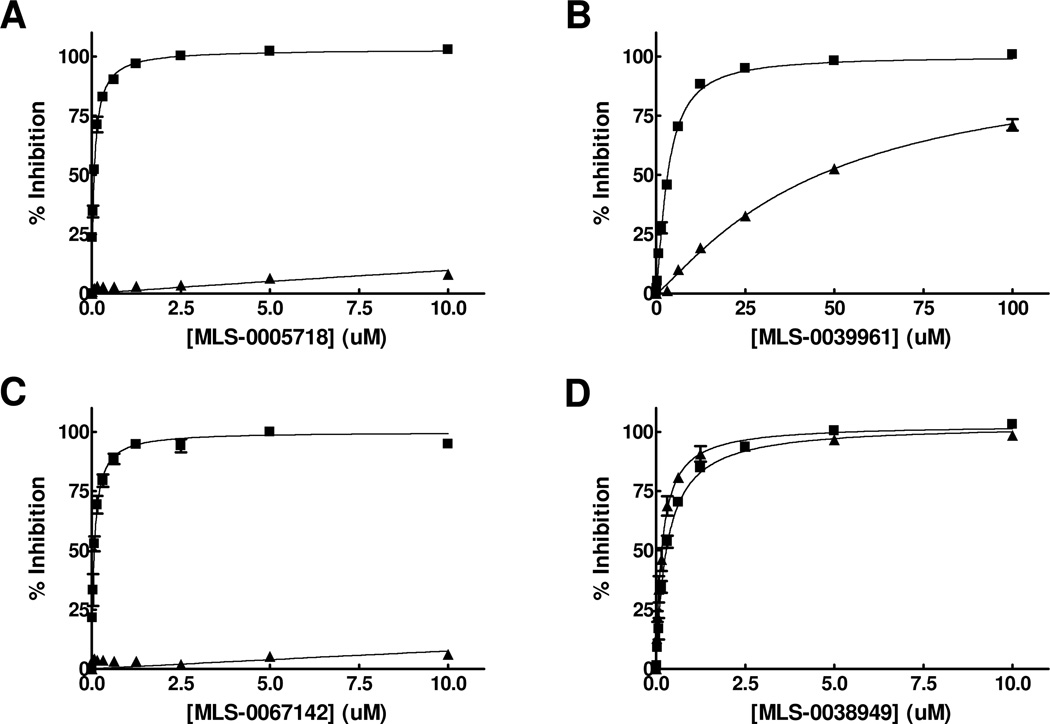
Similarly to the single-concentration confirmation stage during HTS, all the hits were tested in the luminescent and the colorimetric assays. Three biarylsulfonamides and a single representative of the pyrazole scaffold, MLS-0039961 (Table 3 and Figure 8), were the only compounds that demonstrated similar activity in both assays; all other primary positives were inactive in the colorimetric assay.
Figure 8.
Dose-response curves for representatives of three scaffolds luminescent and colorimetric assays. Compounds MLS-0005718 (A), MLS-0039961 (B), MLS-0067142 (C), MLS-0038949 (D) were tested the HTS luminescent assay containing 100 mM DEA (squares) and colorimetric OD405 (triangles) assays.
Exploratory Studies on the Mechanism of Inhibition of the Leading Scaffolds
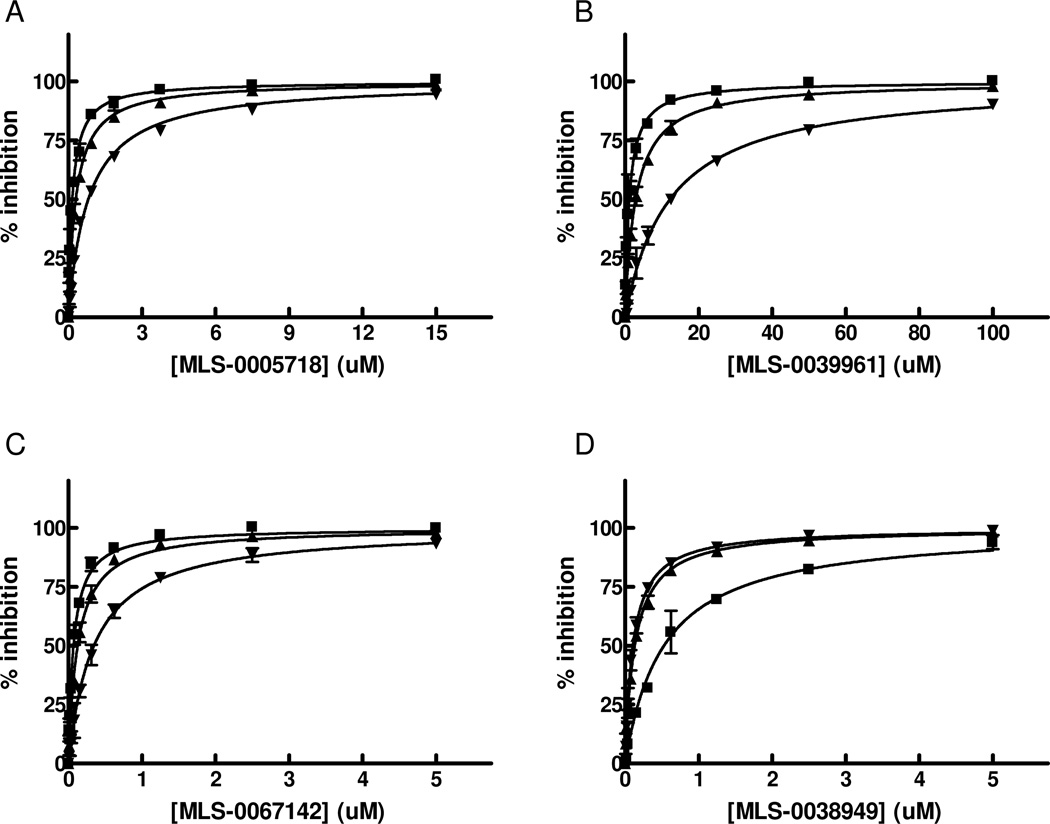
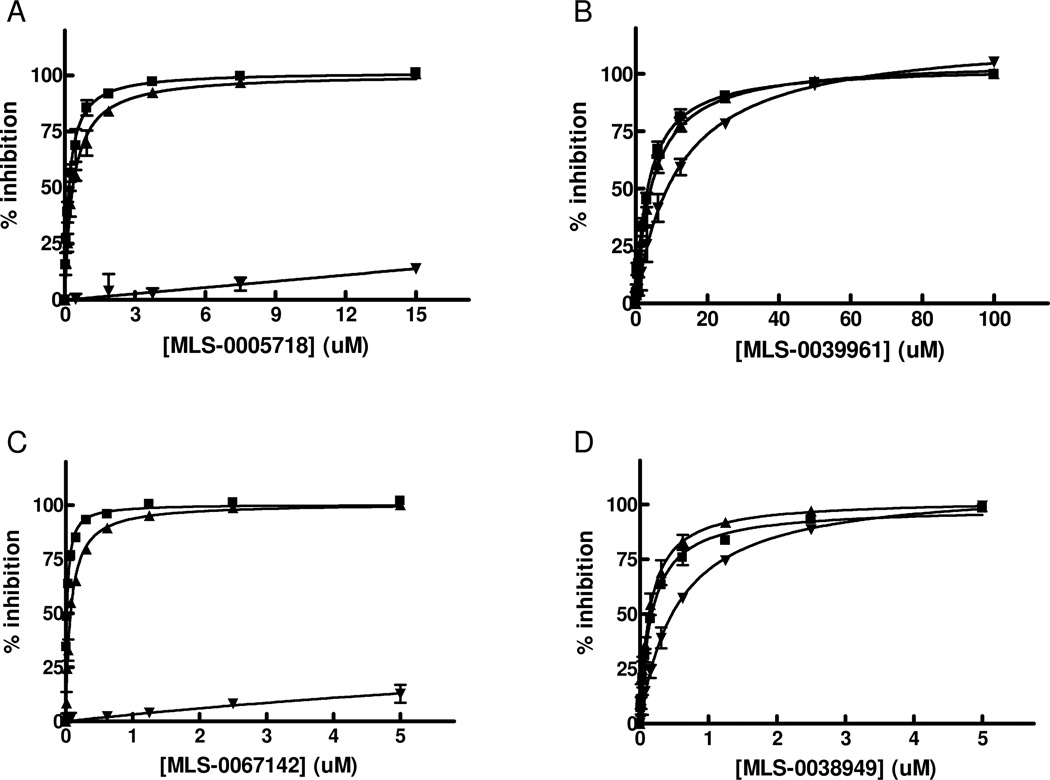
We performed exploratory mechanistic substrate-competition studies on the three leading scaffolds in order to shed the light on their dissimilar behavior in the luminescent and colorimetric assays. For these studies we selected the most potent compounds from each series, MLS-0038949, MLS-0005718 and MLS-0067142, and also added MLS-0039961, the pyrazole compound active in the colorimetric assay. The data are summarized in Figures 9 and 10 and Table 4.
Figure 9.
Mode of inhibition studies for primary HTS hits in respect to CDP-star substrate. TNAP inhibition with MLS-0005718 (A), MLS-0039961 (B), MLS-0067142 (C), and MLS-0038949 (D) was measured in the presence of 1 uM (squares), 50 uM (triangles) or 300 uM (inversed triangles) CDP-star. The concentration of DEA was equal 100 mM.
Figure 10.
Mode of inhibition studies for primary HTS hits in respect to DEA substrate. TNAP inhibition with MLS-0005718 (A), MLS-0039961 (B), MLS-0067142 (C), and MLS-0038949 (D) was measured in the presence of 0 M (squares), 0.1M (triangles) or 1M (inversed triangles) DEA. The concentration of CDP-star was equal 50 uM.
Table 4.
Parameters of MOI experiment
| Compound ID | PubChem SID |
PubChem CID |
Structure | IC50 (uM) at different [CDP*] and fixed [DEA] |
MOI vs CDP * |
IC50 (uM) at different [DEA] and fixed [CDP*] |
MOI vs DEA |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 uM CDP* |
50 uM CDP* |
300 uM CDP* |
0M DEA |
0.1M DEA |
1M DEA |
||||||
| MLS-0038949 | 26514170 | 2931238 | 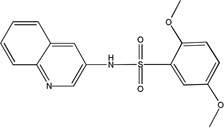 |
0.527 ± 0.039 | 0.132 ± 0.004 | 0.107 ± 0.002 | UC | 0.177 ± 0.007 | 0.134 ± 0.005 | 0.442 ± 0.028 | NC |
| MLS-0005718 | 843997 | 646303 | 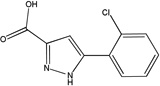 |
0.160 ± 0.008 | 0.303 ± 0.009 | 0.81 ± 0.032 | C | 0.178 ± 0.007 | 0.339 ± 0.022 | 95.31 ± 14.13 | C |
| MLS-0039961 | 4257586 | 610864 | 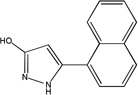 |
1.148 ± 0.058 | 2.883 ± 0.086 | 12.2 ± 0.383 | C | 3.212 ± 0.168 | 4.008 ± 0.223 | 7.869 ± 0.853 | NC |
| MLS-0067142 | 4260165 | 2975791 | 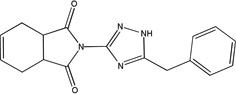 |
0.07 ± 0.003 | 0.126 ± 0.005 | 0.346 ± 0.007 | C | 0.021 ± 0.001 | 0.073 ± 0.003 | 32.63 ± 3.553 | C |
Competition studies with respect to CDP-star (Figure 9) demonstrated that compounds MLS-0005718, MLS-0039961, and MLS-0067142 are competitive with CDP-star. Their potency decreased in the presence of increasing concentrations of CDP-star. Compound MLS-0038949 was the only one that demonstrated an inverse behavior - increasing potency at higher substrate concentrations, indicative of uncompetitive inhibition.
Experiments with DEA demonstrated that inhibition with MLS-0005718 and MLS-0067142 was strongly effected by DEA, consistent with a competitive mechanism (Figure 10 and Table 4). On the other hand, inhibition by the other two compounds, MLS-0038949 and MLS-0039961, was virtually the same at the three DEA concentrations tested suggesting the non-competitive character of their inhibition.
Primary Hits Selectivity
A PLAP selectivity assay was implemented as an integral part of the hit optimization process. For the lead characterization stage we added to the panel a luminescent assay for IAP. The selectivity data for the best representatives of the scaffolds obtained at SDCCG and extracted from PubChem database are summarized in Table 5. Biarylsulfonamide scaffold compounds demonstrated no inhibition of PLAP and IAP and were inactive in more than 200 PubChem assays. Both representatives of the pyrazole scaffold demonstrated no inhibition of PLAP and reasonable selectivity versus IAP, whereas the triazole representative was equally potent in both the TNAP and IAP assays and slightly less potent against PLAP.
Table 5.
Selectivity of primary TNAP scaffolds
| Compound ID | PubChem SID |
PubChem CID |
Structure | SDCCG panel (IC50, uM) |
PubChem panel (# of assays) |
|||
|---|---|---|---|---|---|---|---|---|
| TNAP | PLAP | IAP | Active | Total | ||||
| MLS-0010847 | 26514189 | 645853 | 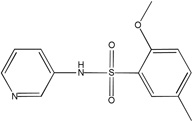 |
0.926 | >100 | >100 | 1 (TNAP) | 234 |
| MLS-0038949 | 26514170 | 2931238 | 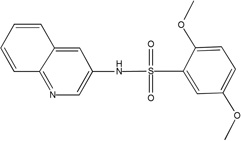 |
0.193 | >100 | >100 | 2 (TNAP, CYP2C19) |
214 |
| MLS-0005718 | 843997 | 646303 | 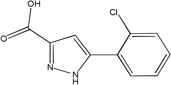 |
0.743 | >100 | 43.5 | 1 (TNAP) | 242 |
| MLS-0039961 | 4257586 | 610864 | 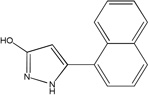 |
4.33 | >100 | >100 | 1 (TNAP) | 213 |
| MLS-0067142 | 4260165 | 2975791 | 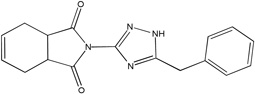 |
0.186 | 3.14 | 0.539 | 2 (TNAP, IAP) |
221 |
DISCUSSION
MLSCN project for screening TNAP
As mentioned above, the TNAP screening project was performed within the Molecular Library Screening Center Network (MLSCN) funded through the NIH Roadmap Initiatives (http://nihroadmap.nih.gov) for accelerated discovery of small molecule tools for basic research (Austin et al, 2004). The project was carried out by the Burnham Screening Center (http://sdccg.burnham.org), part of the Network, in collaboration with the laboratory specializing in TNAP-related research. The main goal of the project was to identify small-molecule chemical probes that could be utilized as research tools for studying TNAP’s physiological and pathological roles and significance. In addition, compounds active in the pyrophosphatase reaction of TNAP were to be tested for their efficacy in biologically relevant model systems and to serve as starting points for the development of therapeutic agents to treat soft-tissue calcification.
Although the TNAP enzyme has been known for many decades, its participation in diverse biological processes are still being actively investigated (Millán, 2006). Many phosphate derivatives have been documented as substrates of TNAP, i.e., pNPP, α-naphthylphosphate, β-glycerophosphate, phosphoserine, PPi, pyridoxal-5’-phosphate (PLP), phosphothreonine, phosphotyrosine, phosphatidates, polyphosphates, ATP, ADP, AMP, glucose-1-phosphate, glucose-6-phosphate, fructose-6-phosphate and others (Moss, 1967; Millán, 2006). However, only two, PPi and PLP, have been unequivocally confirmed as physiological substrates of TNAP, as abnormalities in their metabolism explain the phenotypic changes observed in hypophosphatasia, a genetic disease resulting from TNAP deficiency (Whyte et al., 1988; Fedde et al., 1999; Millán et al., 2008). Furthermore, the role of TNAP in maintaining proper concentrations of extracellular PPi are further documented by the association of low PPi and high TNAP in the vasculature in diverse models of arterial calcification (Narisawa et al., 2007; Lomashvili et al., 2008).
The in vitro reaction of TNAP exhibits a significant response to amino-containing alcohols acting as phosphate-acceptor substrates in the transphosphorylation reaction (ES&JLM, 2008). The physiological role of this reaction is unknown as it is also unclear what metabolite if any could be acting as a natural phosphate-acceptor substrate. Thus, there is an unmet need for small molecule compounds capable of probing TNAP binding sites and deciphering their functions. To be able to interrogate the diversity of TNAP functional features a broad assortment of chemical probes is required. To aid in the identification of the chemical probes with diverse MOA, we developed a principally new homogeneous assay for TNAP and configured it to be as unrestrictive as possible.
Luminescent assay for TNAP
The development of a novel assay (described in ES&JLM, 2008) was a critical step in the successful HTS campaign for the identification of TNAP inhibitors. It provided a more sensitive and automation-friendly approach than the one employed earlier (Narisawa et al., 2007). The readout signal (light intensity) of the chemiluminescent reaction is directly proportional to the steady-state rate of the TNAP reaction and is stable for more than 5h; therefore, the measurement of activity of the enzyme does not require termination of the reaction and can be reliably performed in real-time. The reagents are stable for several days at room temperature enabling flexible scheduling and automation of the screening. The developed assay was very robust and reproducible. The average Z’-factor value obtained from the screening of 64,394 compounds was greater than 0.8 (see Figure 1). High fidelity of the HTS data resulted in a confirmation rate of 75% either in single-concentration or dose-response mode.
The luminescent assay is very sensitive and could tolerate >100-fold decrease in TNAP concentration compared to the colorimetric assays. The assay conditions were optimized for maximal sensitivity. The concentrations of CDP-star and DEA substrates were kept equal to their respective Km values to ensure optimal response to inhibitors with different MOA, competitive and uncompetitive against the substrates molecules. In contrast, utilization of a saturating concentration of phosphoacceptor/buffer is common in the clinical tests for the determination of AP activity in different tissue specimens (Stinson, 1993; WHO SOP).
Generation of diverse HTS hit classes
MOA with respect to phosphate-donating substrate
Both competitive and uncompetitive inhibitors with respect to CDP-star substrate were identified. Of note, competitive inhibitors of APs have never been reported prior to this work. The best representatives of pyrazole and triazole scaffolds with this type of MOA inhibit TNAP with nanomolar potency. The competition with a bulky CDP-star substrate does not necessarily mean that the compounds would be competitive with a much smaller inorganic pyrophosphate substrate. Thus further experimentation is required for clarification of the behavior of pyrazole and triazole compounds in the pyrophosphatase reaction. Nevertheless, it seems safe to assume that the compounds are much more likely to be competitive with bulkier substrates of TNAP, such as AMP and PLP, than with the smaller PPi. It is also possible that molecules with narrow specificities for targeting diverse natural reactions of TNAP could be identified and/or designed via medicinal chemistry approaches within the CDP-star-competitive series, such as pyrazoles and triazoles. This supposition would require additional experimental confirmation, but if found to be true, it would enable differential inhibition of the unique biological activities of TNAP by the aforementioned inhibitors.
Inhibitors competitive with a phosphate-donor substrate are expected to be ineffective in the presence of elevated concentrations (relative to Km) of the substrate. Thus they could also act as sensors of the local concentration of natural substrates of TNAP in a specific biological environment. If employed as therapeutic agents they have the potential to be selective with respect to specific physiological functions of TNAP.
All the inhibitors known prior to this publication acted through an uncompetitive mechanism with respect to phosphate-donating substrates. Biarylsulfonamides identified in the current project were the only scaffold with this common MOA. They represent significant improvement of the existing state of the art: unlike the earlier inhibitors, the potency of the best compound in the biarylsulfonamide series, MLS-0038949, is in the mid-nanomolar range, thus exceeding levamisole (Van Belle, 1976) and ChemBridge 5804079 (identified in our earlier screening efforts, Narisawa et al., 2007) by 2 orders and 1 order of magnitude, respectively.
Compounds uncompetitive with the phosphate-donor substrate, especially as potent as MLS-0038949, would be useful in experiments of chemical knockdown of TNAP and expected to act irrespective of the actual substrate molecule involved in the catalysis. This mode of inhibition relies on the kinetic mechanism of the AP reaction (Holtz et al, 1999): the rate-limiting step of the TNAP reaction is downstream of the active site serine phosphorylation and alcohol product release (DOH in Scheme 1) thus leading to elevated concentration of phosphoenzyme species in the steady state independent of the nature of substrate. Consequently, uncompetitive inhibitors of TNAP are expected to sustain their mode of action irrespective of the actual phosphate-donating substrates providing they are present at sufficiently high concentration. Thus, these inhibitors could also act as sensors of the TNAP catalytical commitment specific to a particular environment.
MOA with respect to phosphate-accepting substrate
The majority of compounds that we identified in the HTS luminescent assay did not inhibit TNAP in the presence of high DEA concentration. Competition with DEA, a phosphate-acceptor substrate, is proposed to explain these observations. For two of the scaffolds (pyrazoles and triazoles) that were further characterized, their competition with DEA was confirmed in the MOA studies. TNAP inhibitors competing with DEA were unknown prior to this study. A possible explanation is that most AP assays performed to date employed a saturating concentration of phosphate-acceptor substrate (Narisawa et al., 2007, Stinson, 1993; WHO SOP).
Inhibitors competitive with the phosphate-acceptor substrate are poised to serve as valuable research tools to study the transphosphorylation reaction in vivo and to help in the identification of its potential substrates. DEA molecule is relatively small in size; thus, the compounds competitive with DEA are expected to compete with any other phosphate-acceptor substrates (AOH in Scheme 1). It is also possible that careful characterization and further MedChem optimization of the scaffolds identified in this study could furnish the molecules with a spectrum of selectivity in respect to phosphate-acceptor substrates, and thus would result in probes selective to different classes of transphosphorylation substrates.
On the other hand, compounds non-competitive with DEA are not expected to interfere with the transphosphorylation reaction. They could serve as valuable reference compounds to differentiate between the effects on phosphate-donor- and phosphate–acceptor-driven stages of the reaction. Two different types of DEA-competitive inhibitors were identified in this study. For example, MLS-0039961 is competitive with the phosphate-donor substrate similar to pyrazoles and triazoles, but non-competitive with the phosphate-acceptor substrate. In turn, the biarylsulfonamide compounds are uncompetitive with the phosphate-donor and non-competitive with the phosphate-acceptor substrates.
As mentioned above, only four primary hits demonstrated activity in the colorimetric assay. One of them was MLS-0039961.Three others were representatives of the biarylsulfonamide scaffold and all three possessed pyridine and o-alkoxybenzene groups connected with a sulfonamide linker (first three compounds in Table 2). The fourth biarylsulfonamide that lacked the pyridine but contained a triazole moiety instead (compound MLS-0043574 in Table 2) is inactive in the colorimetric assay and thus potentially competes with DEA for binding. Perhaps the triazole moiety in this case defines the binding site and thus the MOI of the compound. As a more general conclusion, the triazole moiety of the name-sake scaffold may be targeting the binding site occupied by the nitrogen atom of DEA. It would be interesting to see if MLS-0043574 sustained the uncompetitive MOA against CDP-star innate to other biarylsulfonamides or if it adopted the competitive character of other triazoles.
Concluding remarks: TNAP screening campaign as an MLSCN project
The specifics of performing a project within the MLSCN was outlined elsewhere (Austin, 2004; Lazo et al, 2007). Carrying out the TNAP screening through the MLSCN Roadmap Initiative brought several essential benefits that were not available otherwise, yet were critical for the success of the project. To start with, the cost of a chemical probe identification project including the assay development, screening, SAR studies and medicinal chemistry hit optimization is estimated at several hundreds of thousands of dollars, and is prohibitive to an ordinary research laboratory. All the expenses related to the project were covered by the MLSCN U54 grant supporting the operations of the Burnham Center for Chemical Genomics.
The X01 grant mechanism allowed free access to the services and resources available within the MLSCN at no cost to the research lab. Among other resources, the MLSCN compound library is available to the projects performed within the Network. The library is assembled, maintained and supplied by the MLSMR for primary HTS and hit confirmation. As mentioned in Materials and Methods, compounds are selected to represent diversified chemical space with clusters of closely related analogs. Incorporation of multiple analogs within the compound collection provides the basis for rapid HTS-based SAR analysis. As can be seen in this paper, this type of HTS data analysis served as a valuable tool for preliminary SAR studies, especially for the biarylsulfonamide scaffold which contained large number of compounds.
As mentioned above, the MLSMR supplies compound solutions for both primary HTS and hit confirmation. The fact that most of the compounds in the MLSMR collection originate from commercial sources is a key factor for rapid confirmation of hit efficacy and structures based on commercial compounds. Outstanding reproducibility of the results between the library and the dry-powder commercial compounds (apparent from Table 3) attests to the high integrity of the screening compound collection.
The commercial origin of the compounds in the MLSMR collection is a key factor not only for rapid compound confirmation but also for expanding the preliminary SAR. According to the common practice of the chemical supplies industry, most commercial compounds are produced in a series of multiple analogues. Maximizing the number of analogs in a series allows manufacturers to make the best use of their initial investment in the development of novel synthetic routes. This sets a foundation for the commonly used approach of “analoging by cataloging” (ABC) which we successfully utilized for the confirmation of HTS SAR models and the optimization of the primary hits as will be reported elsewhere (Ardecky et al, 2008; Dahl et al, in preparation; Sergienko et al, in preparation).
Another benefit of performing the assay within the MLSCN is the unprecedented amount of selectivity data available for hundreds of thousands of MLSCN compounds. The MLSMR compound collection was supplied to 10 MLSCN centers. The data produced in hundreds of MLSCN-based HTS campaigns is released into the public domain and can be accessed through the PubChem portal maintained by NCBI. For example, the representatives of the three scaffolds identified in this study were screened in >220 assays (in addition to our own AP isoforms selectivity panels) each providing an immense selectivity profiling.
This project exemplifies a highly successful HTS campaign in which a diverse set of chemical probes was identified and developed. To summarize, three major scaffolds of TNAP inhibitors were identified. They were characterized with high potency, diverse mode of actions and extraordinary level of specificity to TNAP. These chemical probes will provide tools for the characterization of the physiological functions and the catalytic mechanism of TNAP. Several hits demonstrate high potency in the TNAP pyrophosphatase activity and are utilized as starting points for future intervention of soft-tissue ossification, as will be reported elsewhere.
ACKNOWLEDGMENTS
This work was supported by NIH Roadmap Initiative grant U54HG003916 and performed at the Burnham Center for Chemical Genomics, part of MLSCN. S.N. and J.L.M. were supported by NIH grants DE12889 and 1X01MH077602-01.
We wish to thank Drs. Stefan Vasile, Thomas “T.C.” Chung and Nicholas D. P. Cosford for helpful comments on the manuscript.
Abbreviations
- ABC
analog by catalog
- ANK
Ankylosis protein
- AP
alkaline phosphatase
- DEA
diethanolamine
- GCAP
germ cell alkaline phosphatase
- IAP
intestinal alkaline phosphatase
- MG assay
colorimetric assay for TNAP based on malachite green-based detection of phosphate release from pNPP substrate
- MLSCN
Molecular Library Screening Center Network
- MOA (MOI)
mode of action (inhibition)
- MV
matrix vesicle
- OD405 assay
colorimetric assay for TNAP based on absorbance detection of p-nitrophenol release from pNPP substrate
- NPP1
ecto-nucleotide pyrophosphatase/phosphodiesterases-1
- pNPP
p-nitrophenol phosphate
- PLAP
placental alkaline phosphatase
- PLP
pyridoxal-5’-phosphate
- Pi
inorganic phosphate
- PPi
inorganic pyrophosphate
- TNAP
tissue-non-specific alkaline phosphatase
- VSMC
vascular smooth muscle cell
REFERENCES
- Ardecky R, Sidique S, Su Y, Narisawa S, Brown B, Millan JL, Sergienko EA, Cosford N. Design and synthesis of pyrazole derivatives as potent and selective inhibitors of tissue-nonspecific alkaline phosphatase (TNAP) 2008 doi: 10.1016/j.bmcl.2008.10.107. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Brady LS, Insel TR, Collins FS. Molecular Biology: NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- Cogan EB, Birrell GB, Griffith OH. A robotics-based automated assay for inorganic and organic phosphates. Analytical Biochemistry. 1999;271:29–35. doi: 10.1006/abio.1999.4100. [DOI] [PubMed] [Google Scholar]
- Farley JR, Ivey JL, Baylink DJ. Human skeletal alkaline phosphatase - Kinetic studies including pH dependence and inhibition by theophylline. J Biol Chem. 1980;255:4680–4686. [PubMed] [Google Scholar]
- Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millán JL, MacGregor GR, Whyte MP. Alkaline phosphatase knock-out mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J Bone Miner Res. 1999;14:2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman WH, Sie H-G. Organ-specific inhibition of human alkaline phosphatase isoenzymes of liver, bone, intestine and placenta; L-phenylalanine, L-tryptophan and L-homoarginine. Enzymologia. 1971;41:140–167. [PubMed] [Google Scholar]
- Francis MD, Russell RGG, Fleisch H. Diphosphonates inhibit formation of calcium phosphate crystals in vitro and pathologic calcification in vivo. Science. 1969;165:1264–1266. doi: 10.1126/science.165.3899.1264. [DOI] [PubMed] [Google Scholar]
- Garg P, Lomashvili KA, O'Neill WC. Uremic vascular calcification: role of pyrophosphate deficiency and prevention by bisphosphonate therapy. J Am Soc Nephrol. 2005;16:53. [Google Scholar]
- Harmey D, Hessle L, Narisawa S, Johnson KA, Terkeltaub RA, Millán JL. Concerted regulation of inorganic pyrophosphate and osteopontin by Akp2, Enpp1 and Ank: An integrated model of the pathogenesis of mineralization disorders. Am J Pathol. 2004;164:1199–1209. doi: 10.1016/S0002-9440(10)63208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessle L, Johnson KA, Anderson HC, Narisawa S, Sali A, Goding JW, Terkeltaub R, Millán JL. Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc Natl Acad Sci USA. 2002;99:9445–9449. doi: 10.1073/pnas.142063399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz KM, Stec B, Kantrowitz ER. A model of the transition state in the alkaline phosphatase reaction. J Biol Chem. 1999;274:8351–8354. doi: 10.1074/jbc.274.13.8351. [DOI] [PubMed] [Google Scholar]
- Hsu HH, Camacho NP. Isolation of calcifiable versicles from human atherosclerotic aortas. Atherosclerosis. 1999;143:353–362. doi: 10.1016/s0021-9150(98)00322-0. [DOI] [PubMed] [Google Scholar]
- Hui M, Li SQ, Holmyard D, Cheng P. Stable transfection of nonosteogenic cell lines with tissue nonspecific alkaline phosphatase enhances mineral deposition both in the presence and absence of beta-glycerophosphate: possible role for alkaline phosphatase in pathological mineralization. Calcified Tissue International. 1997;60:467–472. doi: 10.1007/s002239900264. [DOI] [PubMed] [Google Scholar]
- Hui M, Tenenbaum HC. New face of an old enzyme: alkaline phosphatase may contribute to human tissue aging by inducing tissue hardening and calcification. Anatomical Record. 1998;253:91–94. doi: 10.1002/(SICI)1097-0185(199806)253:3<91::AID-AR5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Kozlenkov A, Hoylaerts MF, Ny T, Le Du MH, Millán JL. Residues determining the binding specificity of uncompetitive inhibitors to tissue-nonspecific alkaline phosphatase. J. Bone Min Res. 2004;19:1862–1872. doi: 10.1359/JBMR.040608. [DOI] [PubMed] [Google Scholar]
- Lazo JS, Brady LS, Dingledine R. Building a Pharmacological Lexicon: Small Molecule Discovery in Academia. Mol. Pharmacol. 2007;72:1–7. doi: 10.1124/mol.107.035113. [DOI] [PubMed] [Google Scholar]
- Lomashvili KA, Cobbs S, Hennigar RA, Hardcas’le KI, O'Neill WC. Phosphate-induced vascular calcification: role of pyrophosphate and osteopontin. J Am Soc Nephrol. 2004;15:1392–1401. doi: 10.1097/01.asn.0000128955.83129.9c. [DOI] [PubMed] [Google Scholar]
- Lomashvili KA, Khawandi W, O'Neill WC. Reduced plasma pyrophosphate levels in hemodialysis patients. J Am Soc Nephrol. 2005;16:2495–2500. doi: 10.1681/ASN.2004080694. [DOI] [PubMed] [Google Scholar]
- Lomashvili KA, Garg P, Narisawa S, Millán JL, O’Neill WC. Upregulation of Alkaline Phosphatase and Pyrophosphate Hydrolysis: Potential Mechanism for Uremic Vascular Calcification. Kidney Int. 2008;73:1024–1030. doi: 10.1038/ki.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComb RB, Bowers GN, Jr, Posen S. Alkaline phosphatase. New York: Plenum; 1979. [Google Scholar]
- Meyer JL. Can biological calcification occur in the presence of pyrophosphate? Arch Biochem Biophys. 1984;231:1–8. doi: 10.1016/0003-9861(84)90356-4. [DOI] [PubMed] [Google Scholar]
- Millán JL. From biology to applications in medicine and biotechnology. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co; 2006. Mammalian alkaline phosphatases; pp. 1–322. [Google Scholar]
- Millán JL, Narisawa S, Lemire I, Loisel TP, Boileau G, Leonard P, Gramatikova S, Terkeltaub R, Pleshko Camacho N, McKee MD, Crine P, Whyte MP. Enzyme replacement therapy for murine hypophosphatasia. J Bone Miner Res. 2008;23:777–787. doi: 10.1359/JBMR.071213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss DW, Eaton RH, Smith JK, Whitby LG. Association of inorganic pyrophosphatase activity with human alkaline phosphatase preparations. Biochem J. 1967;102:53–57. doi: 10.1042/bj1020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa S, Harmey D, Yadav MC, O’Neill WC, Hoylaerts MF, Millán JL. High throughput screening and identification of novel alkaline phosphatase inhibitors - A druggable target for the treatment of vascular calcification. J Bone Miner Res. 2007;22:1700–1710. doi: 10.1359/jbmr.070714. [DOI] [PubMed] [Google Scholar]
- Russell RGG, Bisaz S, Fleisch H. Pyrophosphate and Diphosphates in Calcium Metabolism and Their Possible Role in Renal Failure. Arch Intern Med. 1969;124:571–575. [PubMed] [Google Scholar]
- Rutsch F, Vaingankar S, Johnson K, Goldfine I, Maddux B, Schauerte P, Kalhoff H, Sano K, Boisvert WA, Superti-Furga A, Terkeltaub R. PC-1 nucleoside triphosphate pyrophosphohydrolase deficiency in idiopathic infantile arterial calcification. Am J Pathol. 2001;158:543–554. doi: 10.1016/S0002-9440(10)63996-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, Ruf N, Vaingankar S, Toliat MR, Suk A, Hohne W, Schauer G, Lehmann M, Roscioli T, Schnabel D, Epplen JT, Knisely A, Superti-Furga A, McGill J, Filippone M, Sinaiko AR, Vallance H, Hinrichs B, Smith W, Ferre M, Terkeltaub R, Nurnberg P. Mutations in ENPP1 a‘e associate’d with 'idiopathic' infantile arterial calcification. Nat Genet. 2003;34:379–381. doi: 10.1038/ng1221. [DOI] [PubMed] [Google Scholar]
- Schibler D, Russell GG, Fleisch H. Inhibition by pyrophosphate and polyphosphate of aortic calcification induced by vitamin D3 in rats. Clin Sci. 1968;35:363–372. [PubMed] [Google Scholar]
- Sergienko EA, Millán JL. Screening for novel inhibitors of alkaline phosphatase of diverse modes of action. Nature Protocols. 2008 doi: 10.1038/nprot.2010.86. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi A, Nishizawa Y, Jono S, Koyama H, Hosoi M, Morii H. Beta-glycerophosphate accelerates calcification in cultured bovine vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 1995;15:2003–2009. doi: 10.1161/01.atv.15.11.2003. [DOI] [PubMed] [Google Scholar]
- Shioi A, Katagi M, Okuno Y, Mori K, Jono S, Koyama H, Nishizawa Y. Induction of bone-type alkaline phosphatase in human vascular smooth muscle cells: roles of tumor necrosis factor-alpha and oncostatin M derived from macrophages. Circ Res. 2002;91:9–16. doi: 10.1161/01.res.0000026421.61398.f2. [DOI] [PubMed] [Google Scholar]
- Stinson RA. Kinetic Parameters for the Cleaved Substrate, and Enzyme and Substrate Stability, Vary with the Phosphoacceptor in Alkaline Phosphatase Catalysis. Clin Chem. 1993;39:2293–2297. [PubMed] [Google Scholar]
- Tanimura A, McGregor DH, Anderson HC. Calcification in atherosclerosis. I. Human studies. J Exp Pathol. 1986;2:261–273. [PubMed] [Google Scholar]
- Tanimura A, McGregor DH, Anderson HC. Calcification in atherosclerosis. II. Animal studies. J Exp Pathol. 1986;2:275–297. [PubMed] [Google Scholar]
- Terkeltaub RA. Inorganic pyrophosphate generation and disposition in pathology. Am J Physiol : Cell Physiol. 2001;281:C1–C11. doi: 10.1152/ajpcell.2001.281.1.C1. [DOI] [PubMed] [Google Scholar]
- Van Belle H. Alkaline phosphatase. I. Kinetics and inhibition by levamisole of purified isoenzymes from humans. Clin Chem. 1976;22:972–976. [PubMed] [Google Scholar]
- WHO Guidelines on Standard Operating Procedures for Clinical Chemistry, Section B. ( http://www.searo.who.int/EN/Section10/Section17/Section53/Section481_1761.htm)
- Whyte MP, Mahuren JD, Fedde KN, Cole FS, McCabe ER, Coburn SP. Perinatal hypophosphatasia: Tissue levels of vitamin B6 are unremarkable despite markedly increased circulating concentrations of pyridoxal-5/-phosphate. Evidence for an ectoenzyme role for tissue-nonspecific alkaline phosphatase. J Clin Invest. 1988;81:1234–1239. doi: 10.1172/JCI113440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]