Abstract
Acetylcholine (ACh) is the neurotransmitter used by cholinergic neurons at the neuromuscular junction and in parasympathetic nerve terminals in the periphery, as well as important memory-related circuits in the brain and also takes part in several critical functions. ACh is synthesized from choline and acetyl coenzyme-A by the enzyme choline acetyltransferase (ChAT). The formation of acetylcholine in cholinergic nerve terminals requires both the transport of choline into the cells from the extracellular space, and the activity of ChAT. High affinity choline uptake (HACU) represents the majority of choline uptake into the nerve terminal, and is the acutely regulated, rate-limiting step in ACh synthesis. The HACU component of choline uptake can be differentiated from non-specific choline uptake by inhibition of the choline transporter with hemicholinium. Several methods have been described previously to measure HACU and ChAT simultaneously in synaptosomes, but a well-documented protocol for cultured cells is lacking. We describe a procedure to simultaneously measure HACU and ChAT in cultured cells by simple radionuclide-based techniques. In this procedure we have quantitatively determined HACU and ChAT activity in cholinergically differentiated human neuroblastoma (SK-N-SH) cells. These simple methods can be used for neurochemical and drug discovery studies relevant to several disorders including Alzheimer’s disease, myasthenia gravis, and cardiovascular disease.
Keywords: brain enzyme, cholinesterase, CNS, synapse, neuronal differentiation, enzymatic activity, cell culture
INTRODUCTION
Acetylcholine is the chemical neurotransmitter secreted by cholinergic neurons at the neuromuscular junction, at parasympathetic nerve terminals, and in certain memory-related circuits in the brain. It is synthesized in a single step chemical reaction by the enzyme choline acetyltransferase (ChAT) (Tucek, 1982). The choline transporter (ChT) transports choline into the cell by a sodium-dependent mechanism (Apparsundaram et al., 2000), where it is combined with the acetyl group of acetyl CoA generated in the mitochondria in a reaction catalyzed by the ChAT enzyme to form acetylcholine (ACh). Choline uptake into cholinergic cells through this transporter can be blocked by hemicholinium-3 (HC-3), and this HC-3 sensitive transport is referred to as high affinity choline uptake (HACU) (Simon et al., 1975). We have previously reported successful application of a radioisotope-based method to measure HACU and ChAT activity in the same population of cultured cells (Ray et al., 2009). In the current manuscript, we describe these methods in the same in vitro system of retinoic acid- (RA) differentiated human neuroblastoma (SK-N-SH) cells.
Human neuroblastoma (NB) cells display characteristics of multipotent embryonic precursor cells of neural crest origin (Pizzi et al., 2002). Upon treatment with retinoic acid (RA), human NB cells undergo mitotic arrest and differentiate to cholinergic neurons (Wainwright et al., 2001) (see Basic Protocol 1). Human neuroblastoma cells differentiated in the presence of RA for 7 days are incubated in buffer containing [3H] choline chloride and the amount of transported choline is determined by measuring [3H] in the cell lysate by liquid scintillation counting (see Basic Protocols 1 and 4). The entire uptake procedure is performed in live cells attached to cell culture plates (see Basic Protocol 2), and values for HACU are corrected for variation in cell number by Cell Titer Glo (CTG) or other suitable assays of cell number and cell viability (see Basic Protocol 3). The CTG assay measures the ATP content of the cells in culture, and highly correlates with total cell number (Crouch et al., 1993). HACU is determined by subtracting non-specific choline uptake, which is determined in the presence of HC-3, from total choline uptake in the absence of HC-3 (see Basic Protocol 2). Lysates produced in the HACU assay are subsequently used to determine the activity of ChAT in the same population of cells.
Choline acetyltransferase activity is determined from the same population of cells by extracting the soluble cytosolic enzyme followed by incubation with choline and [14C]-acetyl coenzyme A (AcCoA) (see Basic Protocol 5). The assay is based on the transfer of the radiolabeled acetyl moiety from acetyl CoA to choline, and separation of the radiolabeled substrate, [14C] acetyl CoA, from the radiolabeled product, [14C]-ACh. This latter step is achieved by a simple organic extraction of the aqueous reaction mixture with 3-heptanone containing tetraphenylboron, and separation of the resulting layers by low speed centrifugation (Fonnum, 1969). Workflows of both HACU and ChAT procedures are outlined in Figure 1.
Figure 1.
Human neuroblastoma cells were cultured and differentiated with ATRA as described in the text. Approximately 100,000 cells were plated onto 24-well cell culture plate and subjected to HACU and ChAT assay. Different steps and incubation conditions are briefly described in this flow sheet diagram.
These simple protocols not only accurately measure HACU in cell culture models, but at the same time assess ChAT activity. Concurrent measurement of HACU and ChAT in the same cells provides complementary information on ACh production. HACU provides a measure of ACh terminal activity, because it is acutely regulated with ACh release, and ChAT activity may provide an indication of ACh synthetic capacity, as it is not acutely regulated by activity (Simon et al. 1975).
BASIC PROTOCOL 1: CHOLINERGIC DIFFERENTIATION OF HUMAN SK-N-SH CELLS
Human SK-N-SH cells were originally derived from metastatic bone marrow of a female neuroblastoma patient (Biedler et al. 1973); they exhibit epithelial morphology and contain 47 modal chromosomes (for details refer to www.atcc.org). One of the important features of this line is its ability to differentiate into neuronal cells upon treatment with retinoic acid (RA). Pizzi et al. differentiated SK-N-SH cells with RA for two weeks and demonstrated the presence of neuronal markers, including NMDA receptor subunits and cholinergic terminal markers, which were observed at very low or undetectable levels in undifferentiated SK-N-SH cells (Pizzi et al., 2002).
Materials
Minimum Essential Medium (MEM) (Sigma, St. Louis, MO, USA; cat. no. M4655)
MEM-complete (see Reagents and Solutions)
MEM-complete/ATRA (see Reagents and Solutions)
Fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA; cat. no. S11050H)
Penicillin-Streptomycin-Amphotericin B (Cellgro/Mediatech; cat. no. 30-004-Cl)
All-trans Retinoic acid (ATRA) (Sigma; cat. no. R2625; See Reagents and Solutions).
Trypsin-EDTA (Cellgro/Mediatech; cat. no. 25-051-Cl)
Trypan blue (Sigma; cat. no. T-8154)
100 mm tissue culture-treated plates (Corning Incorporated, Corning, NY, USA; cat. no.430167)
24-well tissue culture-treated plates (Corning; cat. no.3526) (6-well and 48-wells can also be used)
50 and 15 ml polyethylene (PE) tubes. (Corning, cat. no. 430828; 430052, respectively)
Pasteur pipettes (Fisher Scientific, Pittsburgh, PA, USA; cat. no. 13-678-30)
Improved Neubauer hemocytometer
Standard cell culture equipment (humidified incubator with 37ºC temperature and 5% CO2, laminar flow hood).
Cell culture and differentiation
Unless otherwise noted, all media, solutions, and reagents added to cells should be sterile and pre-warmed to 37°C prior to use.
Obtain cells from American Type Culture Collection (ATCC) and grow them in 100 mm plates in MEM-complete as described previously (Lahiri et al., 1999).
- When cells are approximately 50% confluent, replace the medium with MEM-complete/ATRA to differentiate SK-N-SH cells into cholinergic neurons. Replace the medium (with MEM-complete/ATRA) after an additional 72 hours of culture.ATRA is light sensitive and should be stored in the dark.
- After 5 days of differentiation (i.e. five days after cells are first treated with MEM-complete/ATRA), remove the medium by gentle aspiration and wash the cells once with PBS to remove all serum containing media. Add 5ml of trypsin-EDTA into the culture plate; return plate to 37°C incubator. Once cells have detached (periodically gently tap the plate to speed the process up, but keep cells at 37°C in-between checks), transfer them into a 15 ml polyethylene (PE) tube and centrifuge at 300×g at room temperature for 5 minutes.Trypsin-EDTA detaches the cells from the plate so that they can be collected. After addition of trypsin-EDTA, do not pipette vigorously to detach the cells, as this can damage them. Always use a mixture of trypsin-EDTA instead of trypsin alone.
- Carefully remove the supernatant by gentle aspiration and add 1ml of MEM-complete/ATRA in the PE tube. Serum present in the complete MEM will neutralize the trypsinAspirate the medium very gently and cautiously to avoid accidental removal of the cell pellet.
Resuspend the cells by gently (but thoroughly) pipetting up and down using a fire-polished Pasteur pipette. Take care to avoid excessive shearing force.
Count cells using the trypan blue exclusion method. Gently mix cells and remove a 20 μL aliquot and transfer to a fresh microcentrifuge tube. Add 20 μL of stock (0.4%) trypan blue to aforementioned tube, mix well by pipetting, and incubate for 1 minute. Dispense 20 μL of the mix into the hemocytometer chambers. Count only viable cells and more than 90% cells are expected to be viable.
Dilute cells to a concentration of 1million viable cells/mL. In a 24-well plate, add 400 μL of MEM-complete/ATRA into each well and aliquot 100 μL of the cell dilution into each well for a final concentration of 100,000 viable cells/well, and a final volume of 500 μL/well. (Cell number can be increased or decreased according to the desired multiwell culture plate format. For example, the equivalent cell density for 48-well plates would be approximately 40,000 cells per well). Typically, cells settle down within 5–6 hrs after plating and any treatment of the cells can be carried out after 36–48 hrs of plating; at that time, wells will be approximately 70–80% confluent (if 100,000 cells are plated in each well of a 24-well plate).
HACU and ChAT assays may be carried out in these cells 36–48 hrs after plating; the total differentiation time for SK-N-SH cells is therefore approximately 7 days (5 days for differentiation + 2 days for culture post-differentiation). One can perform HACU or ChAT assay in more differentiated cells (i.e. 15 days of ATRA treatment). A minimum of 7 days treatment with ATRA is recommended for visible differentiation of the cells.
ALTERNATE PROTOCOL: HACU AND CHAT ASSAYS IN SYNAPTOSOMES
HACU and ChAT activities can also be measured ex vivo in synaptosomal preparations taken from brain tissues that contain cholinergic terminals. For example, this procedure has been used to measure the effects of drug treatments (Richter et al., 1982; PMID 7150428) and electrical stimulation on HACU as a surrogate measurement for ACh release (Simon et al., 1975, PMID 1271069). Alternatively, post-mortem measurements of ChAT activity measured by this procedure have been used to determine the viability of cholinergic cells implanted into the hippocampus of lesioned animals (Tarricone et al., 1993, PMID8149244). Such measurements require different protocols for sample preparation; however, the subsequent steps of these procedures are similar.
Materials
0.32 M sucrose solution
Other materials used in Basic Protocol 1
Synaptosome preparation for the HACU assay
-
1
Homogenize brain tissue in 30 – 50 volumes of 0.32 M sucrose. Homogenize using about 10 – 12 up-and-down strokes using an appropriately sized Potter Elvehjem homogenizer with a Teflon coated pestle, at approximately 800 rpm. A 50ml homogenizer can be used for large (e.g., whole brain), or a 3ml homogenizer for smaller samples (e.g., hippocampus). Transfer homogenate to a centrifuge tube, maintaining the
-
2
The homogenate is then centrifuged at 1000 × g for 10 minutes at 4°C. This precipitates a crude nuclear pellet. Transfer the supernatant to a fresh tube and discard the nuclear pellet.
-
3
Centrifuge the supernatant at 17,000 × g for 15 minutes. This produces the crude mitochondrial pellet which contains the synaptosomes. Discard the supernatant and resuspend the pellet in 0.32 M sucrose, using the same volume as in step 1. This suspension can be used for the HACU assay by adding 50 μl to a microcentrifuge tube with the 500 μl HACU buffer (see Step 4 of Basic Protocol 2). The synaptosomes are recovered after the incubation step by centrifugation, washed once with 750 μl DPBS and recovered again by centrifugation. Synaptosomes are then lysed and uptake is determined by scintillation counting (see Basic Protocol 4). In a typical experiment, a 100 mg tissue sample is suspended in 30 volumes (3 ml) of 0.32M sucrose. Using 50 μl per assay, this is sufficient for 60 assays.
Sample preparation for the ChAT assay
-
4
Fresh or frozen tissue from the brain region of interest can be used. Isolate the tissue (e.g. cortex, hippocampus, or striatum all have high levels of ChAT activity) and place it in a microcentrifuge tube. A small amount of tissue, such as a single hippocampus or ~100 mg of cortical tissue would be sufficient for this assay.
-
5
Homogenize the tissue by sonication in lysis buffer (e.g., M-PER) with protease inhibitors. Typically, 10 volumes of M-PER buffer added to the tissue is sufficient for lysis. This homogenate can be used for the ChAT assay.
BASIC PROTOCOL 2: THE HIGH AFFINITY CHOLINE UPTAKE (HACU) ASSAY
In this protocol, the process of determining high affinity choline uptake by means of a subtractive radiolabel-based assay is described. Uptake by the high-affinity transport mechanism (mediated by CHT1) is sensitive to inhibition by the competitive choline (re)uptake inhibitor, HC-3; on the other hand, low-affinity/nonspecific uptake is mediated by an HC-3-insensitive transporter or family of transporters (Apparsundaram et al., 2000; Okuda and Haga, 2000). Therefore, by determining total choline uptake along with uptake in the presence of maximally effective concentrations of HC-3, the portion of uptake attributable to HACU (via CHT1) can be calculated by simple subtraction.
Details for executing the assay in plated, differentiated SK-N-SH cells are provided in this protocol. However, the underlying principles are applicable to other in vitro systems such as other neuron cultures including primary neurons and synaptosomes.
Materials
MEM-complete/ATRA (see Reagents and Solutions)
HACU buffer (see Reagents and Solutions)
HACU/HC-3 buffer (see Reagents and Solutions)
[3H] choline chloride solution, 66.7 Ci/mmol or 1 mCi/ml (see Reagents and Solutions)
1X Dulbecco’s Phosphate Buffered Saline (DPBS)
Potassium chloride (KCl), 1.15% solution
Sodium chloride (NaCl), 2M solution
Normal saline (0.9% NaCl solution)
Calcium chloride (CaCl2), 1.22% solution
Sodium hydrogen phosphate, dibasic (Na2HPO4), 0.1 M solution
Magnesium sulfate (MgSO4) heptahydrate, 3.8% solution
D-glucose (Sigma; cat. no. G-5767)
Hemicholinium-3 (HC-3) (Sigma; cat. no. H108)
[3H] choline chloride (Perkin Elmer, Boston, MA, USA; cat. no. NET109250UC)
Mammalian protein extraction buffer (M-PER) (Pierce, Rockford, IL, USA; product no.78505)
Plastic liquid scintillation counting vials (Research Products, International Corp, Mount Prospect, IL, USA; cat. no. 125501)
Water bath with heater and shaker (Precision Scientific Company; Chicago, IL, USA. cat. no. 66802)
Liquid scintillation spectrometer (Beckman model LS 3801)
Inverted phase contrast microscope (Leica DMIL HC) (Leica Microsystems GmbH, Wetzler, Germany)
HACU assay setup
Note: All volumes assume the use of 24-well plates
-
1
Plan the experimental layout, taking into consideration that half of each plate must be used for measurement of low-affinity choline transport, and the other for total choline uptake. Figure 2 shows a schematic representation of total uptake and HC-3 negative control wells (i.e., “blanks,” which are used to determine nonspecific activity because HC-3 inhibits high affinity choline uptake) in a 24-well format.
-
2Prepare HACU and HACU/HC-3 buffers, as well as [3H] choline chloride solution prior to starting the assay (see Reagents and Solutions).Tritium waste must be disposed of properly and handled only by a trained researcher.
-
3Gently aspirate the MEM-complete/ATRA media from each well as described in earlier protocol, and wash each well by slowly adding 500 μL of room temperature DPBS down the side-wall of the well, followed by careful aspiration.DPBS wash is required for complete removal of the growth medium from the wells. Alternatively, 0.9% NaCl can be used for this wash step.
Figure 2.
A schematic showing cells in a 24-well plate for HACU experiment. Four replicate sets can be performed in a 24-well format. Three wells of each row are allocated for total HACU and the other three wells for hemicholinium (HC-3) blank (i.e. for each set n=3). Number of replicate sets can be decreased or increased by using 12-well or 48-well plates, respectively.
Initiation of assay
-
4
Add 500 μL of HACU buffer to each well designated for total uptake, and add the same volume (500 μL) of HACU/HC-3 buffer to each well designated for HC-3 blank uptake.
-
5
Add 20 μL of previously prepared [3H] choline chloride to the center of each well. Replace the lid on the plate, and place the plate in a 37ºC water bath oscillating at 50 oscillation/minute. Incubate for one hour.
At this time, also pipette 20 μL of the [3H] choline chloride directly into a scintillation vial. This is referred to as the “direct count”. The “direct count” is used to calculate the final concentration of choline present in the incubation media. This is based on the number of DPM per ml, and the specific radioactivity of the [3H]-choline being used. The aliquot of [3H]-choline chloride should be taken using the same pipette and type of tip as was used to dispense the radiolabel into the experimental plate.
While pipetting [3H] choline, try to keep the tip of the pipette close to the surface of the incubation buffer so that radioactive choline does not touch the wall of the wells. Carefully adjust the water level in the water bath such that the culture plate is evenly heated, but not so high that water from the bath can flood or splash into the wells. Add a small weight on the lid of the plate if necessary to prevent sliding. It is possible to use smaller volumes to cut down on radioactive waste (e.g. 250μl), although this may require re-establishing experimental conditions (i.e. cell density and incubation time) to ensure linearity within the target data collection time frame.
Termination of assay
-
6
Following incubation, terminate the uptake by carefully returning the plate to ice, and aspirating the radioactive incubation media from each well. Follow this by gently adding ice cold DPBS (750 μL) down the side wall of each well in order to wash the cells. Remove the wash buffer by careful aspiration.
The aspirated fluid contains radioactive choline and should be disposed of appropriately. -
7
While cells are in cold DPBS, quickly view the plate under an inverted microscope to assess any cell loss and/or morphological changes that might have occurred (Figure 3).
Cells should appear intact and there should not be any gross change in cell morphology before and after the uptake. Once cold buffer is added to the cells, there is still a possibility of efflux of 3H-choline over time, so these steps should be done as quickly as possible. -
8
After washing is complete, if cell morphology appears normal, aspirate the DPBS and add 100 μL of M-PER buffer to each well and shake the plate at 75 oscillations/minute for 7–10 minutes to lyse all the cells. The plate should be kept on a flat ice container while oscillating. After 7–10 minutes, examine the wells under microscope to ensure that the cells were lysed.
-
9
To assay cell viability, proceed to Basic Protocol 3. This is best done immediately after cell lysis, however, it is possible to perform this assay at a later time. If storage is necessary, transfer the samples from ice to a −80°C freezer as quickly as possible. When ready, thaw samples on ice and proceed with Basic Protocol 3. Scintillation counting can also be performed after a period of cold storage.
Figure 3.

Phase contrast images of 7 days differentiated human SK-N-SH cells before and after HACU experiment. A representative area of a well was imaged at 10x magnification. Cell morphology and number, both remain unchanged after the uptake of [3H] choline.
BASIC PROTOCOL 3: CELL TITER GLO (CTG) ASSAY
Cell Titer Glo (CTG) is a luminescence-based assay, which measures the total ATP content of the cells. Luminescence results from the activity of firefly luciferase, which is included in the assay buffer. This enzyme catalyzes the reaction of ATP in the sample with ambient O2 and luciferin in the assay. Relative ATP content of cell lysates corresponds to the number of cells; hence CTG can be a good indicator of overall viable cell number (Ray et al., 2011). The relative values obtained by this method agree with cell density as determined by standard protein assays (unpublished observations).
Materials
Cell Titer Glo (CTG) reagent (Promega, Fitchburg, WI, USA; cat. no.G7572)
96-well flat bottom white polystyrene plates (Corning; cat. no.3688)
−80ºC freezer
Glowmax luminometer (Turner Biosystems, Sunnyvale, CA, USA)
ATP measurement using lysates from Basic Protocol 2
-
1Transfer 30 μL of the cell lysate from each well to the corresponding well of a 96-well flat bottom white polystyrene plate kept on ice. Add an equal volume (30μl) of CTG reagent to each well of the 96-well plate. Include at least one well containing 30μl lysis buffer as a blank.For new kits, aliquot CTG reagent and store at −80°C for future use.
-
2
Shake the plate at room temperature for 10 minutes.
-
3Obtain relative luminescence signals using a luminometer.For a Glowmax luminometer, integration time is set to 1 second. Take multiple readings at 1 to 5 minute intervals and select the highest reading set.
Data interpretation
-
4
Subtract the value of the blank well(s) from the values obtained from the lysates for the raw luminescence signal.
-
5
Luminescence values represent relative ATP content, which in turn is reflective of total viable cells (assuming that any treatments performed were without effect on total ATP). Therefore, values from the CTG assay may be used as a reference point for adjusting other readings from the same sample fraction (i.e. HACU values)
CTG values are one of the adjustment parameters which may be used to determine net choline uptake. Alternatively or additionally, 5 μL of the lysate can be used to determine protein concentration. The most appropriate adjustment parameter must be empirically determined.
BASIC PROTOCOL 4: DETERMINATION OF [3H] RADIOACTIVITY IN THE LYSATE
Determination of choline uptake by differentiated SK-N-SH cells is carried out by measuring total tritium levels in the cellular lysates by scintillation counting. Since we used radioactive [3H]-choline chloride, the DPM value for [3H] will indicate total effective uptake. In the assay that follows (measuring ChAT activity, Basic Protocol 5), 14C is used to label the enzymatic product. While 3H and 14C are both β particle emitters, these signals can be separated by most liquid scintillation spectrophotometers based on the energy of the β particles emitted. In the first liquid scintillation detection step, only 3H will be present, however, in the detection step for Basic Protocol 5, both 3H and 14C, respectively will be present. Separation of these signals will depend on the nearly 10-fold difference in energy of the β particles emitted by these two isotopes, which are 18.6keV for 3H and 156keV for 14C. Setting the upper and lower energy thresholds (the “window”) of the instrument will allow the two isotopes to be measured independently from the same sample. Details on the settings used for the instrument used to optimize this assay will be given here, but these may need to be adjusted for different instruments.
Materials and instruments
Liquid biodegradable counting cocktail
Plastic counting vials
Scintillation counter
Liquid scintillation spectrophotometer setup
-
1
To detect 3H, use a lower window setting of 0, and an upper window setting of 1000 keV
-
2
To detect Carbon-14, use a lower window setting of 400, and an upper window setting of 1000.
-
3
Disintegrations per minute are determined automatically by the instrument based on standard quench curves for [3H] and [14C]. The average efficiency for counting [3H] was approximately 37% and the average efficiency for counting [14C] was about 91% for our instrument. These values are likely to differ between instruments and based on other materials used, such as the scintillation cocktail and scintillation vials used.
Determination of radioactivity, using lysates from Basic Protocol 2
Note: Measurement of ChAT activity can be performed on the same day as the HACU assay; otherwise store the lysate at −80ºC and use within one month.
-
4
Transfer 50 μL of cell lysate from each well (from Basic Protocol 2) to a correspondingly labeled scintillation vial. At this time, also transfer 15 μL of the remaining lysates from all the wells into labeled 1.5 ml plastic microcentrifuge tubes for subsequent ChAT activity measurement and store on ice or at −80°C until ready.
Properly number or label all scintillation vials before taking them to the scintillation counter to avoid errors. -
5
Add 5 ml of biodegradable liquid scintillation cocktail to each scintillation vial and determine radioactivity by liquid scintillation spectrometry.
-
6
To calculate the relative net [3H]-choline uptake, divide the DPM values of all wells by the corresponding CTG values obtained in Basic Protocol 3. This approximates the DPM per cell number, and adjusts for well-to-well differences in cell number. Then, subtract the CTG-adjusted DPM value of the HC-3 blank wells from the CTG-adjusted DPM value of the corresponding experimental wells to calculate the relative net uptake.
Optional: these values can be further adjusted as, e.g., percent control or percent maximum uptake if desired.Please refer to Figure 4 for an overview of the calculation workflow for determining net HACU activity with CTG (viability) adjustment.
Figure 4.
Schematic shows the calculations involved in determining net HACU activity in neuronal cells. Raw [3H] choline uptake (in DPM) of both ‘total uptake’ and ‘blank’ wells is adjusted by the relative CTG value to obtain adjusted DPM. First, cells were counted by cell viability assay (CTG) as shown here. The relative CTG value is determined by dividing the raw CTG value (in Relative Luminescence Unit or RLU) of all wells by the lowest CTG value. Net HACU is calculated by subtracting the adjusted DPM values from each ‘total uptake’ well by the average of the adjusted DPM value of the three ‘blank’ wells.
BASIC PROTOCOL 5: MEASUREMENT OF CHAT ACTIVITY
The ChAT enzyme catalyzes the synthesis of acetylcholine (ACh) from acetyl coenzyme-A and choline. It is not rate-limiting in ACh production, and is not acutely regulated by cholinergic terminal activity to the same extent as HACU. Thus, this assay can be used to detect the presence of cholinergic terminals in a sample without regard to neuron activity. This assay is highly sensitive and can be performed using a small aliquot of lysate generated during the HACU assay (Basic Protocol 3).
Materials
ChAT buffer (see Reagents and Solutions).
[14C] acetyl CoA (Perkin Elmer; Boston, MA, USA; cat. no. NEC313010UC)
Eserine hemisulfate, 6M (Sigma; cat. no.E8625)
Choline chloride, 100 mM (Sigma; cat. no. C7527)
EDTA, 100 mM (See Reagents and Solutions) (Sigma; cat. no. ED2SS)
Hydrochloric acid (HCl), 0.06 M (Sigma; cat. no. 84435):
Sodium tetraphenylboron (Strem Chemicals, Newburyport, MA, USA; cat. no. 93-057)
3-Heptanone (MP Biomedicals LLC, Solon, OH, USA; cat. no. 195217)
Biodegradable counting cocktail (Econo-Safe) (Research Products, International Corp; cat. no.111175)
ChAT assay setup
-
1Label a sufficient number of 1.5 mL microcentrifuge tubes for the number of anticipated samples, with provision for replicate samples (at least duplicates).To preserve the activity of ChAT, samples must be kept on ice at all times. Therefore, it is strongly recommended that the prepared microcentrifuge tubes be pre-chilled.
-
2
Thaw the frozen cell lysates obtained after radioactivity measurement in the HACU procedure (see BASIC PROTOCOL 4 step #1) on ice before the experiment
-
3Transfer 4 μL of the thawed lysates to the labeled 1.5 ml microcentrifuge tubes. Include one tube containing 4ul lysis buffer to use as a blank.If the procedure is to be performed on the same day as HACU, 4μl of the lysate can be transferred to labeled 1.5 ml microcentrifuge tubes kept on ice in lieu of freezing the 15 μL aliquots set aside at the beginning Basic Protocol 4, otherwise freeze the 15 μl aliquots as soon as possible
Start the ChAT reaction
-
4
Add 10 μL of ChAT buffer to each tube (including the blank) and vortex gently.
-
5
Incubate the samples at 37ºC and shake at 50 oscillations/minute for 60 minutes.
-
6
While samples are incubating, add 10 μl of ChAT buffer to a scintillation vial for a direct count.
Stop the ChAT reaction and process samples -
7
Return the tubes to ice after 60 minutes and add 25 μL of 0.05 M HCl to each tube to stop the reaction.
-
8
Add 100 μL of sodium tetraphenylboron/3-heptanone (30 mg/ml) solution to each tube and vortex the tubes for 10 seconds
-
9
Centrifuge tubes for 2 minutes at 1,200 × g at room temperature.
Centrifugation separates liquid phase into two layers; top organic (clear) and bottom inorganic (turbid) which is illustrated in Figure 5. -
10
Transfer 90 μL of the top organic layer from the microcentrifuge tube to scintillation vials. Take care to avoid contamination of the top organic layer with the bottom aqueous layer. If necessary, a smaller volume (e.g., 75ul) of the organic layer can be used to avoid drawing from the aqueous layer.
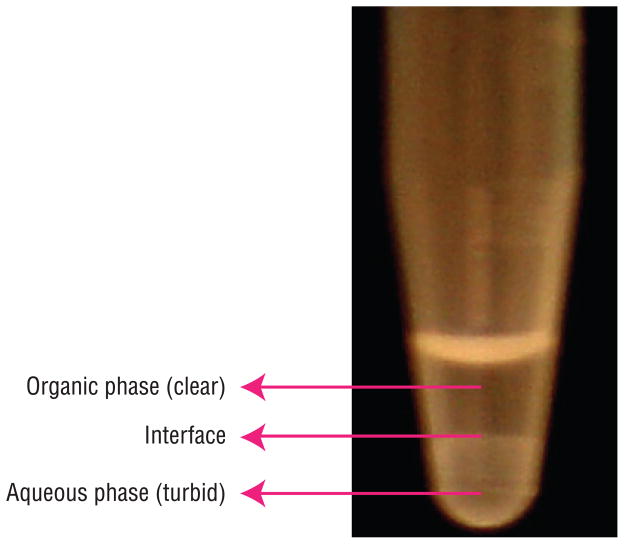
Figure 5.
Separation of [14C]-Ach from the ChAT assay reaction mixture is shown here. Eppendorf tube shows the bi-phasic liquid obtained in the ChAT assay after the addition of sodium tetraphenylboron/3-heptanone solution followed by centrifugation. The clear organic layer contains [14C]-ACh.
Collect and analyze data
-
11
Add 5 ml of bio-degradable liquid scintillation cocktail to each vial and measure radioactivity by liquid scintillation spectrometry.
-
12
Subtract DPM value obtained from the blank tube from the experimental tubes to obtain the raw DPM value. Divide this value by the raw CTG value (see Basic Protocol 3). These data can be further calculated, for example, as percent control or percent maximum if desired.
REAGENTS AND SOLUTIONS
MEM-complete
To one 500 ml bottle of MEM, add 50 ml heat-inactivated fetal bovine serum and 5 ml of a standard 100x antibiotic-antifungal-antimycotic cocktail. Store at 4 °C and warm to 37 °C immediately prior to use.
Serum must be heat-inactivated. Serum that has not been heat-inactivated is highly toxic to SK-N-SH cells.
MEM-complete/ATRA
All-trans retinoic acid (ATRA): Mix 50 mg of ATRA with 16.7 ml of 200 proof ethanol to make 10 mM (1000x) stock solution. Store at −20 °C. Some of the ATRA will precipitate at −20 °C. Be sure ATRA is dissolved completely at room temperature or 37 °C before use. Add 1 μl of the 1000x stock per 1 ml of MEM-complete immediately prior to use.
HACU buffer
HACU buffer is an isotonic mixture of normal saline, KCl, CaCl2, Na2HPO4, MgSO4 and dextrose and is a modified form of Krebs buffer initially described by H. Krebs (Krebs, 1951). Please refer to table 1 for additional details.
Always prepare HACU buffer immediately before the experiment.
Table 1.
Composition of HACU buffer
| Stock solution | Volume (mL) | Concentration in individual well (mM) |
|---|---|---|
| 0.9% normal saline | 100.0 | 122.0 |
| 1.15% KCl solution | 4.0 | 4.9 |
| 1.22% CaCl2 solution | 1.5 | 4.3 |
| 1.42% Na2HPO4 solution | 20.0 | 15.8 |
| 3.8% MgSO4 solution | 1.0 | 1.2 |
Add 2 mg/mL of glucose in the above mixture so that the final concentration of glucose in the assay wells equals 11 mM
HACU/HC-3 buffer
To make HACU/HC-3 buffer, dissolve 6 mg of HC-3 in sterile distilled water to make a stock solution of 10 mM HC-3. Add 80 μL of the 10 mM stock solution to 7.92 ml of normal HACU buffer (1:100 dilution) to yield HACU buffer containing 100 μM HC-3. The concentration of HC-3 used here is several orders of magnitude above the IC50 of 18nM determined previously in rat synaptosomal preparations (Chatterjee et al., 1988, PMID 3409952), although we have observed that 100 μM HC-3 produces near complete inhibition of HACU and is not toxic to any cells tested to date.
HC-3 stock solution (10 mM) can be stored at −20ºC but should be used within 7 days.
[3H] choline solution
Mix 12μl of the stock solution (66.7 Ci/mmole; 1 mCi/ml) with 588 μL of normal HACU buffer (1:50 dilution) for an uptake experiment requiring 24 wells.
Since this procedure requires radioactive materials, only individuals with radiation safety training should perform the procedure.
ChAT buffer
Composition of the ChAT buffer is shown in Table2. Aliquot ChAT buffer and store at −20ºC or −80 º C for future use.
Table 2.
Composition of ChAT buffer
| Stock solution | Volume (μL) | Final concentration in buffer (mM) | Final concentration in assay tube (mM) |
|---|---|---|---|
| 500 mM sodium phosphate buffer (pH 7.4) | 120 | 60.000 | 42.900 |
| 100 mM EDTA (pH 7.5) | 2 | 0.200 | 0.140 |
| 100 mM choline chloride | 120 | 12.000 | 8.600 |
| 6 mM eserine hemisulfate | 20 | 0.120 | 0.086 |
| 2 M sodium chloride | 120 | 240.000 | 171.000 |
| 1.53 M [14C]-acetyl-CoA* | 100 | 0.153 | 0.110 |
| H2O | 518 | - | - |
[14C]-acetyl-CoA should be prepared by mixing appropriate amounts of labeled and unlabeled acetyl coenzyme A, and this will depend on the specific radioactivity of the labeled material.
Ingredients of ChAT buffer in ‘stock’ solution and their final concentration in the ChAT enzyme assay reaction is shown here.
Sodium tetraphenylboron/3-heptanone solution
Dissolve sodium tetraphenylboron in 3-heptanone at a concentration of 30 mg/ml. For example, if there are 12 samples and 100 μL of sodium tetraphenylboron/3-heptanone is to be added to each tube, one should make 1500μl or 1.5 ml of the solution. To do this, 30 mg/ml × 1.5 ml = 45 mg of sodium tetraphenylboron is dissolved in 1.5 ml of 3-heptanone. Since 3-heptanone is an eye and skin irritant, it should be handled with caution (wear gloves and work in a fume hood).
Store [14C] acetyl CoA at −20°C and use within one year.
Choline chloride is hygroscopic and should be stored in a dry place
EDTA, 100 mM
To make a 100 mM solution, dissolve 150 mg EDTA in 5 ml of distilled water and titrate with 10 M NaOH to facilitate dissolution (if not using the Na salt).
COMMENTARY
The individual HACU and ChAT protocols were originally proposed by Simon and Kuhar (Simon and Kuhar, 1975) and Fonnum (Fonnum, 1969), respectively. However, both original protocols measured HACU and ChAT activity in synaptosomes obtained from rodent brain tissue. We have recently validated these protocols in cultured cells, and different uptake kinetic parameters including Km and Vmax were evaluated (Ray et al., 2009). Furthermore, we also evaluated the differences in choline uptake and ChAT activity between naïve and differentiated human neuroblastoma cells. As expected, we observe more choline uptake and ChAT activity in differentiated SK-N-SH cells compared to naïve cells. Both HACU and ChAT were found to be linear as a function of cell number, at least over the range of 100,000 to 200,000 differentiated SK-N-SH cells. Apart from differentiated SK-N-SH cells, we also applied this method to study neurodegeneration under different insults. Cholinergic neuronal loss in the presence of reactive oxygen species (ROS) is one of the hallmarks of several neurodegenerative diseases including Alzheimer’s disease (AD) (Lourenssen et al., 2009). We tested the effect of ROS-mediated insult, as induced by H2O2 treatment, on choline uptake in differentiated SK-N-SH cells. When 100 μM H2O2 was used as a pretreatment of the cells, a significant reduction in HACU was observed. Interestingly, our preliminary data also showed that several drugs including some cholinesterase inhibitors (ChEI) can restore HACU activity when present with the ROS. Taken together, these protocols can serve as powerful tools in the areas of neurochemical and drug developmental research in different neurodegenerative disorders.
Troubleshooting
For a summary of troubleshooting and commonly encountered issues, please refer to Table 3.
Table 3.
Troubleshooting summary
| Step | Problem | Troubleshooting |
|---|---|---|
| Basic Protocol 2; step 2 | HACU buffer is cloudy | HACU buffer can turn cloudy if CaCl2 and/or phosphate buffer is incorrectly prepared or added incorrectly. This results in insoluble calcium salt precipitation. Prepare fresh CaCl2 and phosphate buffer, and discard the old HACU buffer. |
| Basic Protocol 2; step 5 | Gross morphological changes are seen in the cells after uptake | Discard the old HACU and HACU/HC3 buffer. Make fresh buffer and carefully check pH. |
| Basic Protocol 3; step 2 | CTG reading is low | Cells are not properly lysed. After adding M-PER buffer, shake the plate on ice for at least 10 minutes and look under microscope to ensure that all cells are lysed. |
| Basic Protocol 4; step 3 | Uptake and blank readings are low because cells washed from the plate during DPBS washing | Add cold DPBS carefully by pipetting along the side of the well and look under the microscope to ensure that all cells are attached in wells. |
| Basic Protocol 4; step 3 | Sample:Blank ratio declines over time | Check purity of the [3H] choline chloride and the integrity of the HC-3 solution |
| Basic Protocol 5; step 6 | The top layer becomes contaminated with the bottom layer | Take < 90 μL, e.g. remove 50 μL and take this volume into account during final calculations. |
| Basic Protocol 5; step 7 | Radioactivity signal is low | Check purity of the [14C] acetyl-CoA and avoid repeated freeze-thaw of the ChAT buffer. |
| Basic Protocol 5; step 8 | Inconsistent radioactive signals between assays | Adjust windows setting of the scintillation counter. Tritium is present during [14C] measurement as well, and this may influence readings. Set the lower window at a higher energy level to omit tritium signals. |
Anticipated problems in various steps and possible troubleshooting steps are indicated.
Basic Protocol 1
Variable amounts of retinoic acid and other signaling molecules (e.g., hormones) may be present in animal sera. If inconsistent results are obtained, the use of dialyzed or charcoal-dextran stripped sera may improve the results.
If robust differentiation and neurite sprouting is not observed in the initial 5 days of differentiation, this can be due to cells too densely populating the culture dish. To correct this, start with a lower number of cells.
Basic Protocol 2
The DPBS wash following termination of uptake is necessary to remove all traces of [3H] choline from the wells after the one hour incubation. It is important to add a volume of DPBS (e.g., 700–750 μl) greater than that of the [3H] choline-containing buffer (500μl).
While shaking, place the plate on an ice-containing tray to minimize degradation of ATP, which will be measured later as an index of cell number. After lysis, examine the cell culture plate under the microscope to ensure that all cells are lysed.
Low DPM readings in test samples could indicate low specific activity of the [3H]-choline. Check that the correct amount was added to the buffer. The direct count should produce a DPM value between 200,000 and 300,000.
Low DPM readings could also result from low temperature during incubation with the assay buffer. Check the temperature of the water bath.
DPM values that are undesirably high in the test sample (e.g., >5% of the direct count) can result if the number of cells is too high and exceeds the linear range. The upper limit of 5% was selected to limit the change in extracellular choline concentration caused by excessive uptake. A different upper limit may be chosen by the individual investigator, although it is important to ensure the assay maintains linearity at higher uptake levels. Following the protocol outlined here, the final choline concentration in the HACU buffer of 60nM produces results in the linear range. If modifications to the protocol (e.g., different cell types) cause uptake values that are in excess of a predetermined limit or above the linear range for the assay, reducing the number of cells should correct this problem.
High DPM values in the HC-3 blank wells could indicate inadequate washing. Increase the incubation time in the wash buffer (e.g., 30–60s), or add an additional wash to the procedure. Could also indicate a problem with the HC-3 solution. If not prepared properly of if too old, could get high blank values.
Basic Protocol 3
A low or highly variable signal could indicate either inadequate lysis or uneven cell seeding among wells. Insufficient lysis should be detectable by observing the cells under a microscope. To resolve this, increase the incubation time in the lysis buffer. Uneven seeding of wells should also be observable under a microscope, and may be resolved by more rigorously disbursing the pellet before counting to avoid clumping of cells.
Substantial luminescence (e.g., >200 relative luminescence units) detected in the blank well(s) indicates cross-talk between wells. For example, a very high luminescent signal in one well can produce sufficient light to interfere with the detector in the instrument as it reads an adjacent well. Resolve this by spacing out the samples such that there are empty wells between samples.
Basic Protocol 4
Low DPM values could indicate low specific activity of the [3H] stock solution. Verify the activity of the stock solution with the “direct count” mentioned above. These DPM values are typically between 200,000 and 300,000.
Low DPM values could also indicate a problem with the HACU assay, such as incorrect water bath temperature, too few cells per well, or poorly differentiated cells. Double-check the parameters used in earlier steps of the assay.
High DPM values, with DPM values in the total HACU wells above 5% of the direct count, can be corrected by reducing the number of cells in each well, or by reducing the incubation time.
High DPM values in the HC-3 wells relative to the total count of the experimental wells (HC-3 reading should not be more than ~6–7% of the experimental wells) indicate ineffective HC-3 or inadequate washing between the incubation and detection steps. Make sure the HC-3 stock is fresh, and ensure careful washing and complete as possible removal of the wash buffer before lysis.
Basic Protocol 5
Unexpectedly high DPM values in experimental samples or in blank could be caused by accidental aspiration of some of the aqueous layer during the organic extraction step. Pipette very slowly and use a smaller volume if necessary.
Low DPM values in the experimental samples may result from tissue degradation due to improper storage of the samples, or from errors in mixing the buffer solutions. Test the ChAT assay buffer with a positive control tissue, such as fresh homogenate from rat striatum or other structure known to be rich in cholinergic terminals.
A low signal:noise ratio (e.g., < 2:1) can be addressed in two ways. First, increase the read time in the scintillation counter. This will not change the signal:noise ratio, but rather it will increase the precision of the instrument and give the results statistical validity even with a low signal:noise ratio. Second, repeat the assay with a more concentrated lysate sample. This can be obtained by using a greater number of cells in each well.
Anticipated Results
Basic Protocol 1
After 3 days of RA treatment, SK-N-SH cells should show signs of differentiation, including growth arrest and neurite extension. After 7 days of RA treatment, these cells should be maximally differentiated.
Basic Protocol 2
Uptake is typically up to 5% of the total [3H]-choline in the individual wells, e.g., 10,000 DPM if the direct count is 200,000. This upper limit of 5% uptake was chosen in order to prevent significant changes in extracellular substrate concentrations and can be adjusted as long as linearity is maintained. Uptake can vary between cell lines, and if uptake is determined to be in excess of the linear range of the assay, the protocol should be adjusted to reduce total (e.g., by using a smaller number of cells, or shorter incubation time) so that substrate does not become limiting.
Uptake in the experimental wells should also have DPM values that are at least double the values of the corresponding HC-3 blank wells, i.e., the signal:noise ratio should be at least 2:1.
Basic Protocol 3
Signal intensity depends on cell density, but if performed as described here, this assay should produce results in the 100,000’s of relative luminescent units. This value should not be affected by the radioactive signal or HC-3 treatment.
Basic Protocol 4
Ideal DPM values in the experimental wells will represent 1–5% of the direct count value in order to limit changes in extracellular substrate concentrations. If analysis of uptake kinetics is not required, higher uptake values (i.e., > 5%) can be tolerated as long as uptake values are still within the linear range of the assay. In this situation, linearity would need to be determined experimentally.
Basic Protocol 5
Similar to the HACU assay, ideal DPM values in the experimental samples will represent ≤ 5% of the direct count value, while DPM values in the blank samples should be no more than half the DPM value of the experimental wells, and are typically much lower. The direct count values are typically between 10,000 and 20,000 DPM, which will result in a range of DPM values in the samples of 100 to 1000 and a minimum signal:noise ratio of 3:1, assuming a typical background reading of about 30DPM.
ChAT enzyme activity is less dependent on the neurotransmitter release activity of the neuron than the HACU assay, and thus, the two assays may not co-vary with all treatments. ChAT activity will provide an estimation of the ACh generating capacity of the tissue.
Time Considerations
Basic Protocol 1
Upon receipt of the cells from ATCC (or upon removing them from frozen storage), cells may require a 2–3 week recovery period in order to reach log-phase growth and provide sufficient cells for these experiments.
Treating cells, including warming medium and adding RA, should require approximately 30 minutes. Counting and seeding multiwell culture plates should require approximately 1 hour.
Basic Protocol 2
75–90 minutes
Basic Protocol 3
25–30 minutes
Basic Protocol 4
65–90 minutes.
Basic Protocol 5
90–120 minutes
Acknowledgments
This work was supported by grants from Alzheimer’s Associations (Zenith Award), and the National Institutes of Health (AG18379 and AG18884) to DKL.
References
- Apparsundaram S, Ferguson SM, George AL, Jr, Blakely RD. Molecular cloning of a human, hemicholinium-3-sensitive choline transporter. Biochem Biophys Res Commun. 2000;276:862–867. doi: 10.1006/bbrc.2000.3561. [DOI] [PubMed] [Google Scholar]
- Crouch SP, Kozlowski R, Slater KJ, Fletcher J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods. 1993;160:81–88. doi: 10.1016/0022-1759(93)90011-u. [DOI] [PubMed] [Google Scholar]
- Fonnum F. Radiochemical micro assays for the determination of choline acetyltransferase and acetylcholinesterase activities. Biochem J. 1969;115:465–472. doi: 10.1042/bj1150465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA. The use of ‘CO2 buffers’ in manometric measurements of cell metabolism. Biochem J. 1951;48:349–359. doi: 10.1042/bj0480349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Nall C, Ge YW. Promoter activity of the beta-amyloid precursor protein gene is negatively modulated by an upstream regulatory element. Brain Res Mol Brain Res. 1999;71:32–41. doi: 10.1016/s0169-328x(99)00150-3. [DOI] [PubMed] [Google Scholar]
- Lourenssen S, Miller KG, Blennerhassett MG. Discrete responses of myenteric neurons to structural and functional damage by neurotoxins in vitro. Am J Physiol Gastrointest Liver Physiol. 2009;297:G228–239. doi: 10.1152/ajpgi.90705.2008. [DOI] [PubMed] [Google Scholar]
- Okuda T, Haga T. Functional characterization of the human high-affinity choline transporter. FEBS Lett. 2000;484:92–97. doi: 10.1016/s0014-5793(00)02134-7. [DOI] [PubMed] [Google Scholar]
- Pizzi M, Boroni F, Bianchetti A, Moraitis C, Sarnico I, Benarese M, Goffi F, Valerio A, Spano P. Expression of functional NR1/NR2B-type NMDA receptors in neuronally differentiated SK-N-SH human cell line. Eur J Neurosci. 2002;16:2342–2350. doi: 10.1046/j.1460-9568.2002.02403.x. [DOI] [PubMed] [Google Scholar]
- Ray B, Bisht S, Maitra A, Lahiri DK. Neuroprotective and Neurorescue Effects of a Novel Polymeric Nanoparticle Formulation of Curcumin (NanoCurc) in the Neuronal Cell Culture and Animal Model: Implications for Alzheimer’s disease. J Alzheimers Dis. 2011;23:61–77. doi: 10.3233/JAD-2010-101374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B, Simon JR, Lahiri DK. Determination of high-affinity choline uptake (HACU) and choline acetyltransferase (ChAT) activity in the same population of cultured cells. Brain Res. 2009 doi: 10.1016/j.brainres.2009.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JR, Kuhar MG. Impulse-flow regulation of high affinity choline uptake in brain cholinergic nerve terminals. Nature. 1975;255:162–163. doi: 10.1038/255162a0. [DOI] [PubMed] [Google Scholar]
- Simon JR, Mittag TW, Kuhar JM. Inhibition of synaptosomal uptake of choline by various choline analogs. Biochem Pharmacol. 1975;24:1139–1142. doi: 10.1016/0006-2952(75)90208-7. [DOI] [PubMed] [Google Scholar]
- Tucek S. The synthesis of acetylcholine in skeletal muscles of the rat. J Physiol. 1982;322:53–69. doi: 10.1113/jphysiol.1982.sp014022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright LJ, Lasorella A, Iavarone A. Distinct mechanisms of cell cycle arrest control the decision between differentiation and senescence in human neuroblastoma cells. Proc Natl Acad Sci U S A. 2001;98:9396–9400. doi: 10.1073/pnas.161288698. [DOI] [PMC free article] [PubMed] [Google Scholar]