Abstract
Imaging techniques have greatly improved our understanding of lymphocyte activation. Technical advances in spatial and temporal resolution and new labelling tools have enabled researchers to directly observe the activation process. Consequently, research using imaging approaches to study lymphocyte activation has expanded, providing an unprecedented level of cellular and molecular detail in the field. As a result, certain models of lymphocyte activation have been verified, others have been revised and yet others have been replaced with new concepts. In this article, we review the current imaging techniques that are used to assess lymphocyte activation in different contexts, from whole animals to single molecules, and discuss the advantages and potential limitations of these methods.
Lymphocytes are a central component of immune defence mechanisms and have a pivotal role in our battle against pathogens. During adaptive immune responses, lymphocytes bearing antigen receptors identify and respond to rare pathogen-derived antigens without responding to self antigens. These cells continuously patrol the body, each in search of its cognate antigen. In many cases, such as T cell activation, physical contact between an antigen-presenting cell (APC) and a lymphocyte is required for the antigen-specific receptor to recognize and bind antigen. This initial binding event must be translated into a productive signal in the lymphocyte to generate a successful immune response. The consequences of inappropriate activation in this system are significant. Autoimmunity could result from inappropriate recognition of self, whereas a compromised immune response could lead to infection and death.
Information on the events that are triggered by the binding of an antigen receptor to its ligand was initially obtained by biochemical studies, which successfully identified a large number of signalling molecules (including receptors, enzymes, adaptors and second messengers) that are required for lymphocyte activation1-3. Genetic manipulations have confirmed the role of many of these proteins and have aided in understanding the functional hierarchy of molecules in these signalling cascades4. These techniques provide very limited temporal and spatial information at the level of a single cell or molecule. Imaging approaches are unique in providing the ability to monitor individual events and to follow these events in time, thus allowing the investigator to determine heterogeneity in the immune response and to understand the dynamics of lymphocyte signalling. Consequently, imaging studies have led to unexpected observations of the diversity and dynamics of lymphocyte–APC contacts, the spatial organization of the contact zone between the two cells and the intracellular molecular events.
Although imaging of the immune system began more than 100 years ago with Elie Metchnikoff’s early work on phagocytosis5, in the past three decades rapid advances in light microscopy have revolutionized our understanding of immune processes. Electron and advanced light microscopy techniques have been used to produce high-resolution images of lymphocytes in vitro. The advent of two-photon microscopy in the past decade has also made available data from in vivo settings. Most recently, high-resolution methods have broken the diffraction limit of light to probe subcellular features as small as single molecules. Thus, advances in imaging techniques have enabled the visualization of signalling events in lymphocytes with progressively greater spatial and temporal precision.
In this Review, we provide an overview of the imaging toolbox that is used for visualizing lymphocytes during activation. We start out at the whole animal or tissue level, then zoom in to the cellular and subcellular levels and finally discuss techniques for imaging cells at molecular resolution (FIG. 1). Whole body imaging methods such as positron emission tomography (PET), magnetic resonance imaging (MRI) and bioluminescence are not covered and in vivo imaging is discussed only briefly. Instead, we focus on microscopy techniques that have been used to visualize the subcellular details of lymphocyte activation, with a focus on T cells. We discuss each of the techniques in light of their advantages and limitations regarding resolution, sensitivity and physiological relevance. Although we highlight how these studies have offered unique insights into the molecular and cellular mechanisms that underlie lymphocyte activation, for an in-depth discussion of the biological implications of the data readers are referred to other reviews. Our goal is to provide an up-to-date assessment of the methodologies for researchers who are considering the application of imaging techniques to the study of lymphocyte activation.
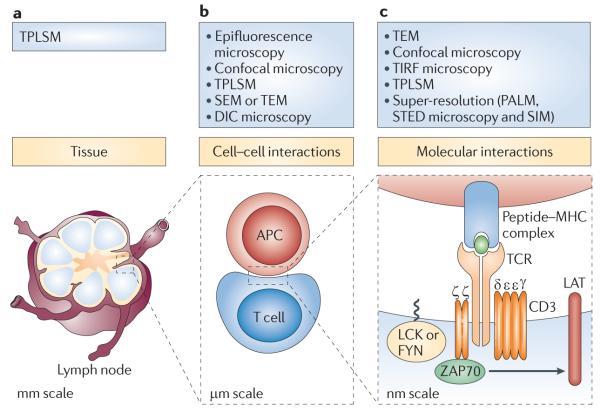
Figure 1. Imaging techniques: hierarchy of scale.
The study of lymphocyte activation requires observation of samples that vary in size over six orders of magnitude. This figure shows the T cell receptor (TCR)-mediated signalling pathway and microscopy techniques used at three levels of sample size. a | In whole organisms or intact tissues, encounters between lymphocytes and antigen-presenting cells (APCs) can be studied in situ using two-photon laser scanning microscopy (TPLSM). The specimen size is tens of millimetres. b | Cell–cell interactions and many subcellular details can be monitored using a large number of conventional microscopy techniques. These samples are typically several micrometres in size, but some details are at the diffraction limit of light scale. c | To observe molecular detail, high- and super-resolution imaging techniques are required. The samples here are individual molecules, only a few nanometres in size. DIC, differential interference contrast; LAT, linker for activation of T cells; PALM, photoactivated localization microscopy; SEM, scanning electron microscopy; SIM, structured illumination microscopy; STED, stimulated emission depletion; TEM, transmission electron microscopy; TIRF, total internal reflection fluorescence; ZAP70, ζ-chain-associated protein kinase of 70kDa.
In vivo imaging
The visualization of lymphocyte activation in vivo has always been a major goal of immune system imaging. After decades of inferring the in situ behaviour of lymphocytes from static tissue sections, three studies were published in the early 2000s that showed the interactions between live T cells and APCs in thymic organ cultures or explanted lymph nodes. One of these studies used conventional confocal microscopy6, whereas the others used two-photon laser scanning microscopy (TPLSM)7,8. Two-photon systems are advantageous because they use long-wavelength infrared lasers that allow observation deep into tissues, while their small excitation volume decreases photobleaching and phototoxicity. These systems were then used for true in situ imaging of exposed lymphoid organs in living animals9,10.
The first TPLSM imaging studies generated contradictory data on the motility of naive T cells as well as on the incidence, duration and stability of interactions between T cells and APCs. Further experiments showed that these parameters varied both with the strength of the T cell receptor (TCR) signal and with the location of the cells within the lymph node, information that could only be gained by observing individual cells in an intact organ11,12. Intravital TPLSM studies have continued, generating insights into the regulation of cytotoxic T lymphocytes (CTLs)13, CTL-mediated killing of tumour cells14 and autoimmune interactions of T cells in the central nervous system15. Recently, TPLSM has been used to monitor the correlation between interferon-γ gene (IFNG) expression and the diversification of T cell responses16.
TPLSM has also been applied to the in vivo visualization of B cells. An early report showed an important role for chemokines in B cell responses17. Other studies revealed that B cells contact antigens not just in solution, but also on the surface of dendritic cells (DCs) and macrophages18-21. In addition, TPLSM has been used to view a wide range of other processes, including germinal centre formation, antigen delivery by macrophages and calcium signalling in T and B cells12.
In many in vivo imaging studies, lymphocytes are labelled with dyes or fluorescent proteins that are expressed throughout the cytoplasm (for TPLSM methods, refer to REF. 22). This generates an image of lymphocytes in a dark tissue volume, but offers little insight into how other components of the lymphoid organs affect lymphocyte activation. However, visualization of the extracellular matrix, either by second-harmonic generation signals23 or by labelling of stromal cells with green fluorescent protein (GFP)24, showed that lymphocytes migrate along reticular networks rather than wandering randomly through empty space. The generation of additional mouse strains that express fluorescently tagged molecules in non-lymphoid cells, as well as mice that express combinations of several proteins labelled with different fluorophores, will undoubtedly aid future studies.
There are technical limitations that accompany the benefits that are gained by using TPLSM, including scattering of the emitted light and tissue heating by the infrared excitation light. Furthermore, two-photon systems are generally used at lower magnification and have lower axial resolution than confocal microscopy, limiting the view of subcellular details. In fact, the use of a confocal microscope in the study by Stoll et al.6 allowed the authors to observe molecular details such as CD43 exclusion from the T cell–APC interface, providing early evidence for immunological synapse formation in vivo. Exciting recent work has extended in vivo imaging to the molecular level, with experiments in lymph nodes showing the localization of two fluorescent fusion proteins (linker for activation of T cells (LAT)–enhanced GFP25 and TCR–GFP26) during T cell activation.
The performance and applicability of in vivo imaging improve with each technical advance. Better detectors lead to better images, and high-sensitivity gallium arsenide phosphide photomultipliers are already being used in conjunction with TPLSM26. In situations in which the increased penetration of TPLSM is not needed, the sensitivity of confocal systems can be improved with electron-multiplying charge-coupled-device cameras, and this technology has been used to detect fungal infection of the brain in vivo27. Furthermore, the recent combination of two-photon excitation with fluorescence lifetime microscopy allows more precise measurement of intracellular fluorescent resonance energy transfer (FRET) signals, thus enabling the use of FRET sensors in situ28,29. Adding an optical parametric oscillator to an infrared laser can extend the range of usable wavelengths to 1,600 nm, allowing imaging of deeper regions of tissues as well as imaging of red fluorescent proteins30. This technology has been used for intravital brain imaging of three fluorescent proteins31. Finally, other techniques are available for in vivo imaging, including optical-resolution photoacoustic microscopy, which has been used for imaging vascular beds, cancer cells and amyloid plaques32,33. So, although TPLSM has markedly improved our understanding of lymphocyte behaviour, the future will bring even higher resolution and more physiological imaging of the immune system.
Imaging of cell–cell interactions
T cells and APCs have to find each other and engage in direct cell–cell contact to initiate an immune response. Although the in vivo studies described above examine the behaviour of cells in their natural or near-natural environment, current technologies do not have sufficient resolution to detect dynamic molecular details of activation in vivo. The in vitro studies described below have provided information about the activities of single cells at the subcellular and molecular levels and proved to be particularly fruitful in the study of lymphocyte activation.
Morphological changes on initial contact
Migrating lymphocytes have a polarized morphology with a leading edge that is rich in actin and a trailing edge or uropod in which the microtubule-organizing centre and the Golgi apparatus are localized34. In the early 1980s, when the fluorescence microscope came into routine use, transmission light microscopy combined with fluorescent antibody detection techniques revealed that on contact with APCs, T cells rapidly polarize their cytoskeleton and Golgi apparatus towards the APC35,36. In one of the earliest imaging studies of immune cell activation, Geiger et al.37 combined electron and fluorescence microscopy to demonstrate the polarization of CTLs during the directional killing of target cells by monitoring the reorientation of their microtubule-organizing centres towards the contact area between the cells. Since then, immunologists have used increasingly advanced imaging techniques to visualize the APC–lymphocyte junction.
Imaging of the increase in intracellular calcium levels on TCR engagement was one of the first real-time observations of lymphocyte activation in live cells38. Simultaneous differential interference contrast microscopy (DIC microscopy; also known as Nomarski microscopy) and epifluorescence microscopy imaging of chemical calcium indicators revealed morphological changes and sustained global calcium increases in T cells following ligand engagement39,40.
As the resolution of light microscopy is limited, immunologists turned to other classic imaging techniques such as transmission electron microscopy and scanning electron microscopy to image the morphological changes induced by antigen recognition. The spatial resolution achieved by transmission electron microscopy (~0.1 nm) is far greater than that of light microscopy (~200 nm) and has thus allowed for imaging of the fine structure at the T cell–APC interface41,42. The resolution of scanning electron microscopy is lower than that of transmission electron microscopy but provides a good three-dimensional representation of cell surface morphology, producing striking images of the spreading response of T and B cells on activation43,44.
Supramolecular changes on cell–cell contact
In concert with cell shape changes during antigen recognition, spatial reorganization of membrane proteins occurs at the junction of the T cell and the APC. The result is a clearly organized interface, several micrometres in diameter, termed the immunological synapse. The first detailed view of the immunological synapse was provided by the Kupfer laboratory more than a decade ago45 using optical deconvolution and digital reconstruction. This study revealed a ‘bull’s eye’ synapse with discrete concentric domains. TCRs and CD3 molecules occupy the central region termed the central supramolecular activation cluster (cSMAC), which is surrounded by an outer ring of adhesion molecules in the peripheral SMAC (pSMAC)45. Subsequently, a third SMAC termed the distal SMAC (dSMAC), which is enriched with the phosphatase CD45, was detected in a ring outside the pSMAC46. The immunological synapse has been identified at the stimulated interface of CD8+ T cells47, B cells44, natural killer (NK) cells48 and mast cells49. Over the past decade it has become clear that there is not just one type of immunological synapse50,51. For example: some T cell lines, thymocytes and weakly stimulated T cells do not show SMAC formation52-54; in vitro T cell–DC conjugates tend to have multifocal synapses41; and mobile T cell–APC junctions known as kinapses55 have been observed. At the same time, our understanding of immunological synapse function has evolved considerably, and this subject has been recently reviewed56.
The Dustin laboratory provided the first dynamic pictures of the stimulated T cell interface in a study in which the APC was replaced by planar lipid bilayers57 (discussed further below). Other investigators probed the dynamics of activation in the context of two interacting cells. These studies used three-dimensional video microscopy to capture the T cell–APC interface and revealed that small TCR clusters and increased calcium concentrations preceded the consolidation of the cSMAC58,59. The recruitment of downstream signalling molecules into small, peripheral TCR clusters was also observed before cSMAC consolidation in subsequent studies46,60.
Other supramolecular structures have also been observed using imaging techniques. Away from the immunological synapse, on the opposite side of the cell, an accumulation of proteins forms the distal pole complex61,62. Interestingly, in addition to being localized to the immunological synapse as discrete puncta, the calcium-release-activated calcium (CRAC) channel components stromal interaction molecule 1 (STIM1) and CRAC channel protein 1 (ORAI1) accumulate in cap-like structures opposite the immunological synapse at the distal pole of the cell63.
Visualization of the various SMACs, the distal pole complex and protein clusters highlights an important contribution that imaging studies have made to the understanding of T cell activation: specifically, how the partitioning of different molecules in different cellular compartments contributes to T cell responses. Moreover, the changing molecular composition of distinct domains over time indicates the importance of intracellular kinetics.
However, imaging of the immune cell interface is fraught with challenges. As it is impossible to predict where the conjugates and synapses will form, it is extremely difficult to catch the earliest events of T cell activation. The complex topology of the T cell and the APC at the contact site requires the acquisition of large z stacks to capture the interaction between the cells. This results in low temporal resolution, while spatial resolution is reduced to minimize acquisition time. Several current approaches address these concerns. Optical trapping can be used to orient cells during synapse formation, allowing the direct visualization of the contact interface between the cells at markedly increased temporal and spatial resolution64. Another approach that has proved to be productive in the study of the stimulated lymphocyte surface is the use of surrogate stimuli on planar substrates.
Modelling the stimulated cell interface
To experimentally manipulate lymphocyte activation, researchers have reduced the complexity of the system by substituting non-cellular surrogates for the APC. Ligand-coated beads are good substitutes for APCs and allow precise control of the activating ligands (FIG. 2a). Bead-induced activation has been used to address many issues, including T cell migration, calcium fluxes and the role of mitochondrial polarization during lymphocyte activation. However, it is still difficult to capture the dynamics of activation because the locations of the first contacts are unpredictable and it is still necessary to image a large three-dimensional volume. Instead, model systems in which the APC is replaced with a planar activation surface (such as an antigen-containing mobile lipid bilayer (FIG. 2b) or antibody-coated coverslip (FIG. 2c)) have produced the best high-resolution images of T cell activation. Planar substrates improve resolution because in lens-based microscopy the resolution in the horizontal x–y plane is substantially better than that in the axial or z plane. As only a thin z-stack is required to capture the entire contact surface and the plane of activation is known beforehand, confocal microscopy (FIG. 3) can be used to view the rapid changes that are induced by activation. Moreover, since the activating surface is mounted on a coverslip, two additional high-resolution imaging techniques are available. The first, total internal reflection fluorescence microscopy (TIRF microscopy)65 (FIG. 3), improves z-plane resolution, and the second, interference reflection microscopy (IRM)66, produces an image that contains only the regions of close contact between the cell and the activating surface (0–200 nm from the surface).
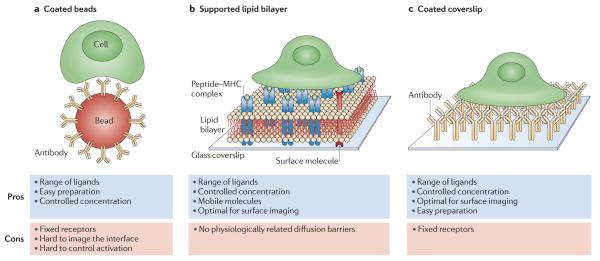
Figure 2. Antigen-presenting cell substitutes used for lymphocyte activation.
Several model systems have been developed to enable lymphocyte imaging at improved resolution. These models use a surrogate substrate for activation instead of an antigen-presenting cell (APC). a | Beads, typically coated with stimulatory antibodies, can be easily prepared and customized but do not allow rapid imaging of cell activation. b | Supported lipid bilayers that incorporate peptide–MHC molecules and other molecules such as integrins provide a better surface for imaging. Although the molecules are mobile, the bilayer lacks diffusion barriers that may be present in real cells. c | Coverslips that are coated with stimulatory molecules, including peptide–MHC molecules and antibodies, also allow controlled concentrations of a wide range of ligands and optimal surface imaging. However, the bound molecules are immobile.
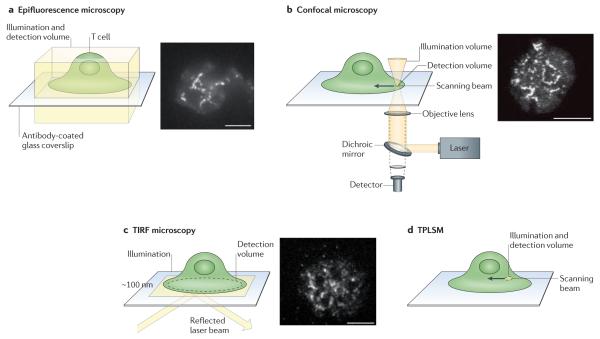
Figure 3. Light-microscopy techniques that are widely used for cell imaging.
a | Epifluorescence microscopy. b | Confocal microscopy. c | Total internal reflection fluorescence (TIRF) microscopy. d | Two-photon laser scanning microscopy (TPLSM). Illumination is depicted in yellow and the detection volume is labelled. The images show fluorescent clusters of linker for activation of T cells (LAT) in Jurkat T cells activated on antibody-coated coverslips.
Morphological changes on planar substrates
IRM imaging of T cells on both bilayers and immobilized ligands demonstrated that dramatic morphological changes occur rapidly following T cell activation. Tight contacts containing polymerized actin were seen as the T cell touched the activating surface. On lipid bilayers, these tight contacts were made on the leading edge of migrating T cells57, whereas on immobilized ligands, the areas of first contact resembled filopodia67. Initial contact was immediately followed by a rapid burst of cell expansion that involves lamellipodia and the formation of an actin-rich ring67,68. On immobile ligands, the cells maintain maximal spreading, whereas on mobile substrates, the spreading phase was followed by contraction involving retrograde actin flow and myosin II-generated forces68,69. Experiments on planar substrates have shown that the polarization of the microtubule-organizing centre towards the immunological synapse depends on signalling through ζ-chain-associated protein kinase of 70 kDa (ZAP70), LAT and SH2 domain-containing leukocyte protein of 76 kDa (SLP76) to activate phospholipase Cγ1 (PLCγ1), creating local changes in diacylglycerol levels70.
Microclusters as sites of activation
In 1999, Grakoui et al.57 visualized the changes in the localization of TCRs and lymphocyte function-associated antigen 1 (LFA1) following activation of T cells on a lipid bilayer. Following engagement with activating ligand, small peripheral clusters of TCRs were seen within 30 seconds of contact. These TCR-enriched discrete structures have been termed microclusters. In 2002, studies from the Samelson laboratory using immobilized TCR-specific antibodies on coverslips and confocal microscopy showed that several signalling molecules were rapidly recruited to microclusters that were generated at the edge of the spreading T cells71. Studies using immobilized ligands have determined that the initial complexes contain many kinases, scaffold proteins and effector molecules. These proteins include the TCR; the protein tyrosine kinases LCK and ZAP70; the adaptors LAT, growth factor receptor-bound protein 2 (GRB2), GRB2-related adaptor protein 2 (GRAP2; also known as GADS), SLP76, non-catalytic region of tyrosine kinase (NCK) and Wiskott–Aldrich syndrome protein (WASP); and the enzymes PLCγ1, Casitas B-lineage lymphoma (CBL) and VAV171-73. Larger glycoproteins, such as CD43 and the tyrosine phosphatase CD45, are excluded71. Huse et al.74 further refined the timing of these events using immobilized and photoactivatable peptide–MHC complexes, and showed that adaptor proteins are recruited to signalling microclusters within 4 seconds of activation.
Investigators using TIRF microscopy of antigen–MHC ligands on lipid bilayers observed analogous TCR microclusters into which ZAP70, LCK, LAT and SLP76 were rapidly recruited75. Importantly, in both coverslip and bilayer systems, peripheral microclusters are predominant sites of tyrosine phosphorylation, and cytosolic calcium fluxes coincide with the onset of microcluster formation, indicating that they are sites of signal initiation. Moreover, defects in cluster assembly and persistence are associated with defects in T cell activation76,77.
Planar activation has now also been used to study B cells, NK cells and mast cells. When activated by mobile ligands in a bilayer, the B cell receptor (BCR) forms microclusters that are necessary for early signalling events and for the subsequent translocation of the BCR microclusters to the centre of the contact interface. The recruitment of LYN, PLCγ2, BLNK (B cell linker protein) and VAV1 to BCR microclusters has been demonstrated to date78.
Microcluster dynamics and signal termination
Real-time imaging studies have demonstrated the dynamic nature and changing composition of microclusters. On antibody-coated coverslips, the composition of the microclusters changes quickly as LAT and associated molecules separate from the immobilized TCR and ZAP7071. On lipid bilayers, the TCR is mobile and all of these proteins move towards the forming cSMAC. However, signalling proteins, including kinases and adaptors, dissociate from the TCR as they translocate radially75. Therefore, in both systems, although signalling molecules are initially recruited to TCR microclusters, a subset of molecules rapidly dissociates and can form separate signalling clusters.
Studies on lipid bilayers have also investigated the dynamics of the co-stimulatory receptors CD2 and CD28. Although both of these co-receptors initially form microclusters that contain TCRs, in the cSMAC they localize to discrete regions away from the TCRs and support sustained signalling79,80.
Microcluster movement towards the centre of the contact interface involves the cytoskeleton, as radial movement requires actin retrograde flow, myosin II and an intact microtubule network68,69,81.This movement of microclusters has been linked with signal termination. Tyrosine phosphorylation occurs predominantly in peripheral clusters, whereas central clusters are thought to be sites where signalling is terminated by endocytosis and degradation82. Multiple studies have reported that clusters of signalling molecules undergo endocytosis as they move towards the centre of the synapse and that this endocytosis depends on the cellular ubiquitin system81,83-86. Inhibition of the movement or internalization of clusters by mechanical trapping, inhibition of endocytosis or co-stimulation leads to enhanced signalling83,85,87,88.
An evaluation of planar models
Antibody-coated coverslips immobilize the TCR, and as a result individual microclusters can be visualized as they form, mature and evolve in composition over time. In addition, the lifetime of each constituent in a fixed cluster can be accurately determined. Furthermore, visualization of signalling complexes after they dissociate from the initiating receptor is possible. By comparison, in lipid bilayer studies the molecules can move laterally and physiological ligands can be incorporated into the bilayer. Importantly, the mobility of the ligands allows the tracking of receptor microclusters as they migrate radially inwards to form the cSMAC.
However, neither of these model systems accurately reproduces the forces exerted on microclusters following activation by an APC. Immobilized ligands fail to support TCR movement and immunological synapse formation, whereas the ligands in lipid bilayers are much more mobile than those presented by an APC. Moreover, the APC itself is important for activation, as perturbation of the DC cytoskeleton decreases T cell activation and WASP-deficient DCs fail to form a stable immunological synapse89. Nonetheless, experiments on planar substrates are responsible for the discovery and characterization of the dynamic microclusters that are the first sites of TCR activation. The existence and importance of microclusters have since been confirmed in cell–cell conjugates90. Thus, the investigation of model systems that can be imaged at high resolution can generate important observations, and these findings can then be verified with APC–lymphocyte conjugates.
Molecular interactions and dynamics
The composition and dynamics of activation-induced microclusters have been largely studied by live-cell fluorescence imaging, as described above. We now discuss fluorescence microscopy techniques that have helped to refine the spatial and temporal resolution in the study of the motility and interactions of molecular signalling assemblies with submicrometre dimensions (FIG. 4).
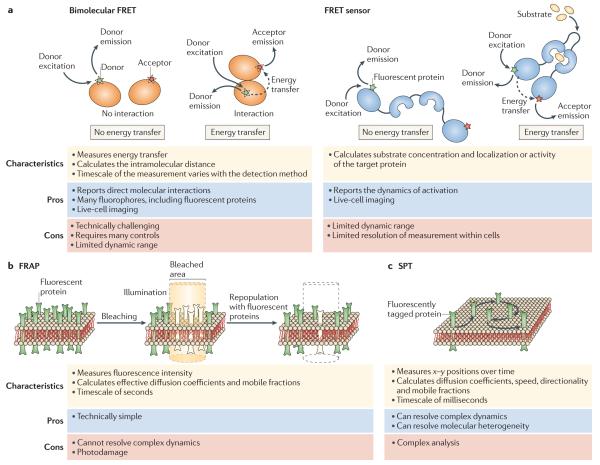
Figure 4. Techniques for imaging molecular dynamics and interactions.
a | Fluorescent resonance energy transfer (FRET) occurs when a suitable donor is in close proximity to an acceptor fluorophore; the process is illustrated here by arrows showing donor excitation, donor emission, energy transfer and acceptor emission. For bimolecular FRET, synthetic fluorescent probes (green and red stars) or fluorescent proteins can be used, whereas FRET sensors primarily incorporate fluorescent proteins. See Supplementary Information S1 (box) for further details on FRET measurements. b | Fluorescence recovery after photobleaching (FRAP), showing fluorescent proteins in a region of membrane undergoing selective photobleaching, followed by the recovery of fluorescence in this area as a result of repopulation with unbleached mobile molecules. c | Single-particle tracking (SPT), showing a single fluorescently tagged molecule as it is tracked over time.
Early receptor activation events revealed by FRET
FRET has been used to look at ligand-induced conformational changes in antigen receptors, protein–protein interactions and early signalling events in lymphocytes. FRET is the non-radiative transfer of excited-state energy from an excited molecule, the donor, to a non-excited molecule, the acceptor. For energy transfer to occur, the emission spectrum of the donor molecule must overlap with the excitation spectrum of the acceptor, and the donor and acceptor must be in a favourable orientation, no more than 10 nm away from each other. On conducting careful control measurements, energy transfer can be reported in several different ways (see Supplementary information S1 (box)).
The immune recognition receptors can undergo significant clustering on binding to antigen, and receptor clustering is considered to be important for the activation of signalling cascades that are associated with lymphocyte activation. However, the molecular events that initiate clustering remain undefined. To assess the earliest events that follow receptor binding, the Pierce laboratory used live-cell FRET imaging to probe the interactions between the different BCR subunits. They provided FRET-based evidence for monomeric BCRs on the surface of resting cells, and found that these BCRs then clustered on antigen binding. In addition, they showed that whereas antigen binding induced a conformational change in the membrane-proximal Fc portion of the BCR that promoted BCR clustering, the cytoplasmic domains of the BCR moved apart91,92. Thus, initiation of BCR signalling involves conformational changes in both the ectodomain and cytoplasmic domains of antigen-engaged BCRs.
Conformational changes have also been invoked to explain TCR activation. However, this process has been poorly understood despite the efforts of several groups. Recently, Xu et al.93 used live-cell FRET imaging to show a close interaction between the CD3ε cytoplasmic domain and the plasma membrane. The FRET data, in combination with striking nuclear magnetic resonance measurements, led to the conclusion that CD3ε binds to the inner leaflet of the plasma membrane, burying tyrosine residues that are crucial for activation in the lipid bilayer. FRET has also been used to look at interactions between the TCR and co-receptors94.
Measuring TCR–peptide–MHC binding with single-molecule FRET
The relationship between the strength of TCR–peptide–MHC interactions and the final functional activation of a T cell remains an important unknown issue. Recently, Huppa et al.95 used FRET between single TCRs (on live cells) bound to peptide–MHC complexes on planar lipid bilayers to measure on-rates and to calculate off-rates from estimates of inter-probe distances. Strikingly, they found dissociation rates in situ that were significantly (4–12-fold) faster than the rates obtained from measurements made in solution. However, an even larger (100-fold) increase in association rates resulted in an apparent increase in net affinity compared with solution measurements. In these experiments the ligands could freely diffuse away from the TCR following dissociation, but it is unclear whether the same mobility is allowed in a cellular context.
FRET between proximal signalling molecules
FRET has been used to study interactions between various molecules in the signalling cascade that colocalize in microclusters. In addition to FRET between the TCR and the kinase ZAP70 (REF. 96), FRET was detected between PLCγ1 and LAT, CBL, VAV1 or SLP76, and between SLP76 and NCK within microclusters, indicating that there are direct interactions between these proteins when clustered72,73. By comparison, lower FRET was observed between LAT and either SLP76 or NCK, and this is consistent with previous data showing that these molecules interact indirectly. FRET was also used to follow the interactions between NCK and WASP that lead to actin polymerization72. In addition, when the levels of proteins in multimolecular complexes were manipulated through gene silencing, FRET measurements were used to probe the cooperative nature of multimolecular interactions97. Further downstream in the signalling cascade, positive FRET between the calcium channel proteins STIM1 (which is resident in the endoplasmic reticulum) and ORAI1 (which is localized in the plasma membrane) indicated a direct interaction between the calcium sensor and channel63,128,129.
Mapping protein activation across the cell
A wide range of FRET sensors have been developed and extensively used to study protein–protein interactions, enzymatic activity and local concentrations of second messengers in live cells. Of specific interest to the study of lymphocyte activation are the sensors for monitoring membrane-restricted lipid molecules and intracellular levels of Ca2+, and activity reporters for proteins such as PLCγ, WASP and protein kinase C98. Importantly, two recently developed sensors for monitoring the activity of LCK99 and ZAP70 (REF. 100) showed unexpected results when used in activated T cells. The LCK sensor indicated that neither significant conformational change nor phosphorylation on a key regulatory tyrosine is required for LCK to phosphorylate the TCRζ and CD3 chains. In experiments using the second sensor, ZAP70 activity was detected not just at the contact area but also at the distal pole of the cell following TCR activation.
Highly sensitive FLIM-FRET (see Supplementary information S1 (box)) has also been used to look directly at early signalling events in lymphocytes. Using FRET between a fluorescently tagged inhibitory NK cell receptor and a fluorescently tagged phosphotyrosine-specific antibody, the authors showed that phosphorylation was localized to discrete regions within receptor clusters101.
Characterizing protein–lipid interactions
The first steps in antigen recognition and activation of lymphocytes take place at the plasma membrane, where protein–lipid interactions can have an important role. It has been suggested that lipid rafts, which are ordered sphingolipid- and cholesterol-rich domains in the plasma membrane, can coalesce on receptor activation into larger signalling domains that have an important role in lymphocyte activation102. However, the role of lipid rafts has come under considerable scrutiny103-105 and imaging methods have provided evidence on both sides of the debate.
Dynamic studies using dyes that are sensitive to plasma membrane ordering have indicated that raft-like domains arise during T cell activation at the contact site between T cells and APCs106-108. Studies using transmission electron microscopy showed that ordered lipid phases were associated with signalling complexes109-111. Furthermore, FRET-based studies in B cells showed that clustered BCR–lipid interactions exist, although they are weak and transient112. By contrast, studies in T cells failed to show significant association of raft markers and microclusters, either by colocalization or by FRET analysis71,96,113. Furthermore, studies using LAT mutants showed that protein interactions, not lipid raft recruitment, were required for LAT localization to microclusters and confinement to signalling domains114. Further study will be needed to understand the role of lipid domains and to determine the forces that are involved in the formation of signalling complexes.
Tracking of molecular dynamics using FRAP and SPT
The mobility of signalling molecules affects how signalling is translated into activation. Fluorescence recovery after photobleaching (FRAP) methods allow the evaluation of the mobility and diffusion of populations of molecules. Studies of the dynamics of microclusters using this technique revealed the fast exchange of molecules at the clusters, although the clusters themselves were static71,114,115. In contrast to the unexpectedly fast dynamics of interactions between TCRs and peptide–MHC complexes that were revealed by single-molecule FRET95, as discussed above, FRAP measurements showed unexpectedly slow off-rates for the interactions between the cell surface molecules CD2 and CD58 (REF. 116), suggesting that this interaction is longer-lived in the contact area than in solution. These FRAP measurements emphasize once again the need for detailed imaging measurements of molecular interaction rates in the contact areas between T cells and model APC interfaces or intact APCs, as the results are difficult to predict.
By contrast, single-particle tracking (SPT) using TIRF microscopy tracks single fluorescent molecules in space and time, and is a powerful tool for following the dynamics of individual plasma membrane constituents. For example, previous FRAP and anisotropy studies suggested that the high-affinity Fc receptor for IgE (FcεRI) must be immobilized to induce signalling. However, in a recent study, single quantum dot tracking showed that small, antigen-induced oligomers of IgE and FcεRI remained competent for signalling while being mobile, and became immobile only at elevated levels of multivalent antigen117.
SPT studies have provided detailed insights into the mobility of proteins in TCR microclusters. SPT of the adaptor LAT revealed that the mobility of LAT molecules at the T cell plasma membrane can change abruptly between immobility and rapid diffusion. Transient immobilization of LAT correlated strongly with encounter with clusters, indicating that LAT molecules diffuse between clusters and are occasionally trapped within clusters114. In a more recent study, the Vale laboratory used SPT to gain insight into the mechanism of formation of the ‘bull’s eye’ immunological synapse68. Tracking of single intercellular adhesion molecule 1 (ICAM1) molecules showed that they rarely penetrated into the TCR-rich cSMAC and were deflected when they encountered the edge of this region. These observations led to the suggestion that a diffusion barrier around the mature cSMAC may help to exclude adhesion molecules from this region68.
A closer look at signalling domains
The ability to study intact live cells in physiologically relevant conditions is a great advantage of light microscopy. However, the resolution of light microscopy is generally subject to the diffraction limit of light. Recent developments in light microscopy have been able to break the diffraction limit of light in multiple ways118 and currently pose new opportunities for studying the subcellular and molecular events that lead to the activation of lymphocytes.
Super-resolution imaging of signalling microclusters
Several techniques have been used to generate high-resolution images of the microclusters that are formed following lymphocyte activation. Transmission electron microscopy images of the plasma membrane sheets of activated mast cells showed the formation of ‘primary signalling domains’ that include the receptor FcεRI, spleen tyrosine kinase (SYK) and PLCγ2, and ‘secondary signalling domains’ that include LAT and PLCγ1 but not the Fc receptor109. These submicrometre-sized domains have been shown to exist before receptor activation. However, on receptor activation, these domains come into close contact to form larger patches that are probably equivalent to the microclusters observed by diffraction-limited light microscopy. More recent work from the Davis group has harnessed photoactivated localization microscopy (PALM) to give us the first direct look at the nanoscale organization of TCRs and LAT molecules at the plasma membrane of live T cells that are spread on lipid bilayers119. PALM is a super-resolution technique that is capable of resolving single fluorescent molecules. Stochastic photoactivation of a few well-spaced molecules allows these individual emitters to be imaged until they are photobleached, and this sequence of events is repeated to build up an image120. Related techniques include fluorescence PALM (FPALM), stochastic optical reconstruction microscopy (STORM; which uses sets of photoactivatable fluorophores) and a considerable list of other variants (FIG. 5). Using rapid PALM imaging of live cells, Lillemeier et al.119 could identify the existence of discrete TCR and LAT domains (with dimensions in the order of 60–130 nm) in naive unactivated cells. In stimulated T cells, these ‘islands’ seemed to coalesce to form larger structures that were comparable in size to the microclusters observed by TIRF and confocal microscopy. Moreover, using dual-colour fluorescence cross-correlation spectroscopy (FCCS), they could further show that these domains migrated together only on TCR activation119.
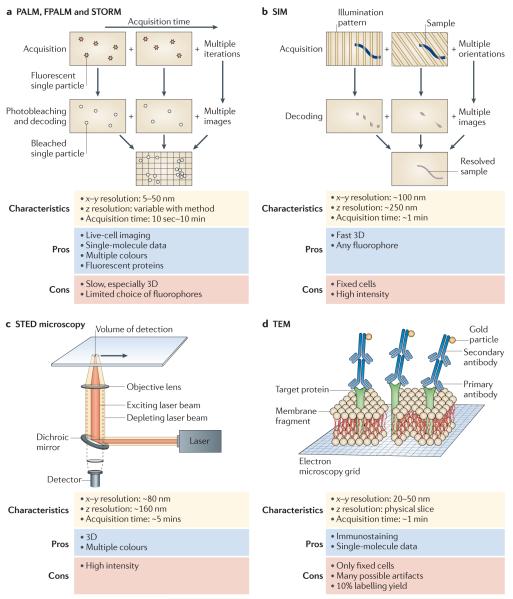
Figure 5. High-resolution imaging techniques.
These techniques are needed to image cellular structures beyond the diffraction limit of light. a | Super-resolution light microscopy techniques — photoactivated localization microscopy (PALM), fluorescence PALM (FPALM) and stochastic optical reconstruction microscopy (STORM) — are based on stochastic detection of single particles (red stars) and their subsequent bleaching and localization (white circles). Localized molecules are summed over time to reveal sub-diffraction-sized features. b | Structured illumination microscopy (SIM) uses a sequence of illumination patterns to decode cellular features (blue line) with different components of spatial orientation. Summing all of the resolved spatial components results in a super-resolved image of the cellular feature (solid grey line). c | Stimulated emission depletion (STED) microscopy uses two concentric focused illumination beams (yellow and orange cones) to reduce the effective volume of detection. The illumination beams are scanned across the cell to resolve small cellular features. d | Transmission electron microscopy (TEM) uses a beam of high-voltage electrons to image features within thin layers or membrane sheets of cells.
Perspectives
Current imaging technologies have generated considerable advances in our understanding of lymphocyte activation. Intravital imaging has provided new insights into lymphocyte behaviour in a physiological environment. Imaging of cell–cell contacts has led to an appreciation of the dramatic supramolecular changes that take place at the site of antigen engagement. Imaging at molecular resolution in cells has given us an understanding of the dynamic and heterogeneous nature of complexes and clusters as they evolve over time. These are just some examples of the observations from imaging studies that have substantially influenced our thinking about immune cell function.
When considering the developments in imaging technologies and their effect on the study of lymphocyte activation, it is clear that one must carefully consider the strengths and limitations of the different techniques in light of the specific research question at hand (TABLE 1). No single current technology provides a simple solution for studying the molecular details of lymphocyte activation in vivo in real time. Physiological context may have to be sacrificed to some degree for higher resolution, or vice versa. Given these trade-offs, a combination of low-resolution in vivo imaging and high-resolution in vitro experiments will allow advances in the study of where, when and how lymphocytes become activated.
Table 1.
Applications of imaging techniques
| Research question | Live-cell imaging | Fixed-cell imaging |
|---|---|---|
| Molecular structure | No | Crystallography, electron microscopy |
| Conformational changes | FRET, single-molecule FRET | Crystallography, electron microscopy |
| Mobility of bound species | FRAP, FCCS, SPT | No |
| Intracellular activity of proteins | FRET sensors, FRET | No |
| Intracellular localization | Confocal microscopy, STED microscopy | Confocal microscopy, STED microscopy, SIM, PALM |
| Aggregation state of receptors | Anisotropy, FRET, PALM, STORM, FCCS and related analyses of molecular brightness |
TEM, PALM, STORM, FRET |
| Mobility at the plasma membrane |
TIRF microscopy, FRAP, SPT and sptPALM, confocal microscopy, STED microscopy |
No |
| Cell morphology | Confocal microscopy, epifluorescence microscopy, TIRF microscopy, DIC microscopy |
Confocal microscopy, epifluorescence microscopy, TIRF microscopy, DIC microscopy |
| Cell adherence to a surface | TIRF microscopy, DIC microscopy, IRM | TIRF microscopy, DIC microscopy, IRM, TEM |
DIC, differential interference contrast; FCCS, fluorescence cross-correlation spectroscopy; FRAP, fluorescence recovery after photobleaching; FRET, fluorescent resonance energy transfer; IRM, interference reflection microscopy; PALM, photoactivated localization microscopy; SIM, structured illumination microscopy; SPT, single-particle tracking; STED, stimulated emission depletion; STORM, stochastic optical reconstruction microscopy; TEM, transmission electron microscopy; TIRF, total internal reflection fluorescence.
Nevertheless, there have been constant improvements in both in vivo and in vitro imaging systems. Advances in TPLSM technology121 and the generation of various fluorescent reporter mice will take us a long way towards the long-term goal of imaging the molecular details of cells in their natural environment. At the same time, the addition of better planar models of the cellular interface122 and the use of optical tweezers or other techniques to optimally visualize cell–cell interactions64 will improve the high-resolution imaging of activated lymphocytes. Another key issue related to cell imaging is the diversity and heterogeneity of the cells themselves, and the choice of samples may be subject to the bias of the observer. As imaging is generally a low-throughput process, the recent introduction of automated instruments for high-speed sequential imaging of cells (such as imaging flow cytometry systems123,124 and high-throughput screening systems with imaging capabilities) will allow users to collect an immense number of unbiased images with minimal compromise on resolution.
Looking ahead, recent developments in advanced imaging techniques and their integration into commercially available systems are likely to greatly improve our capability to image lymphocytes. Many turn-key microscopes can now integrate some kind of super-resolution technique, including structured illumination microscopy (SIM), PALM or direct STORM (dSTORM), STORM and stimulated emission depletion (STED) microscopy117 (FIG. 5). Furthermore, the continuous introduction of new labelling reagents and modalities125-127 promises a bright and colourful future for imaging of molecular details. Given the rapid rate of technological advances, we expect significant progress in the near future towards our ultimate wish of real-time, targeted probing of molecular dynamics and interactions in live cells in vivo (BOX 1 ).
Box 1. Desired and novel imaging-related capabilities.
Combination of super-resolution and automated live-cell imaging
Automated high-throughput imaging systems are currently used for multiparametric screening of cells. In principle, such systems are technically compatible with super-resolution imaging techniques, with the understanding that super-resolution will slow down the throughput of the system.
High-resolution molecular imaging in situ
Two-photon laser scanning microscopy (TPLSM) is able to activate several genetically encoded fluorescent proteins, thus enabling molecular imaging in situ. However, high-resolution imaging is limited by the low numerical aperture of long-working-distance objective lenses and the significant loss of signal owing to tissue scattering and absorption. Using optimized, long-wavelength-emitting fluorescent proteins and getting closer to the imaged cells, for example using minimally invasive endoscopes, could substantially enhance in situ imaging capabilities.
Real-time multiple-colour three-dimensional imaging
The incorporation of highly sensitive imaging sensors, such as electron-multiplying charge-coupled devices, into the fastest three-dimensional imaging systems (for example, spinning-disk confocal microscopes or structured illumination microscopy systems with no moving parts) is expected to significantly accelerate the imaging of cells, while maintaining multiple-colour imaging capabilities (for example, by spectral multiplexing).
Electron microscopy with genetic tagging
Genetically encoded probes could be created for electron microscopy imaging that would serve as analogues to fluorescent proteins in light microscopy, but with the enhanced resolution provided by electron microscopy.
Supplementary Material
Acknowledgements
We thank R. Kortum for critically reading the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research (CCR).
Glossary
- Diffraction limit of light
This refers to the physical impossibility of focusing light that is emitted from a point source into a single point owing to diffraction, which limits optical resolution to a distance of about half of the light wavelength (~200 nm for green light).
- Confocal microscopy
A technique in which light that is emitted by fluorescent targets is passed through a pinhole, thus removing out-of-focus light and allowing accurate volume observation by the sequential acquisition of x-y images along the z axis.
- Two-photon laser scanning microscopy
A technique in which an image is formed by scanning a sample with a high-power pulsed laser. A spot of excitation is produced where the combined energy from the simultaneous absorption of two low-energy photons is sufficient to excite a fluorophore.
- Transmission light microscopy
A technique that uses light to enlarge and image objects by passing the light through a set of lenses and subsequently detecting it by eye or with a detector.
- Differential interference contrast microscopy
A phase-imaging technique that produces contrast from differences in refractive indices at various parts of the sample.
- Epifluorescence microscopy
A technique that captures the fluorescence coming from the entire emitting volume of the sample.
- Transmission electron microscopy
A technique that produces an image from a beam of electrons that are transmitted through a thin specimen containing electron-dense material to create an image with a very high resolution of several Angstroms.
- Scanning electron microscopy
A technique that images the surface of a solid sample with high-energy electrons and detects features on its surface with a resolution of several nanometres.
- Deconvolution
A computational image restoration technique that removes the out-of-focus blur that is typical of epifluorescence images and improves both lateral and axial resolution.
- Optical trapping
A technique that uses a focused laser beam to exert small mechanical forces to trap cells or other microscopic objects in suspension, thus restricting or directing their motion and orientation and allowing their subsequent study by light microscopy.
- Total internal reflection fluorescence microscopy
A technique that uses an evanescent wave, which is generated when the excitation beam is completely reflected from the coverslip, to excite fluorescent molecules in a thin layer within about one hundred nanometres of the coverslip.
- Interference reflection microscopy
A technique that uses the interference of reflected rays of light to produce an image that contains only the regions of close contact between the cell and the contact surface (0–200 nm).
- Mechanical trapping
The use of nanometre-scale structures built into lipid bilayers that act as barriers and inhibit the movement of T cell receptor microclusters.
- Lipid raft
An ordered sphingolipid- and cholesterol-rich membrane domain. These domains are thought to reside within the more diffusive and unordered pool of lipids of the plasma membrane.
- Fluorescence recovery after photobleaching
A technique that involves photobleaching fluorescent molecules in a region of a cell and then measuring the recovery of fluorescence that is due to the repopulation of the bleached area by diffusion of unbleached molecules.
- Anisotropy
A method that measures the loss of correlation in polarization between the polarized excitation light and the light emitted from a rotating probe; this can be used to indicate changes in rotation speed caused by binding of the labelled molecule.
- Plasma membrane sheets
The part of the plasma membrane of an adherent cell that remains on the adhering surface after the rest of the cell is removed during preparation for subsequent electron microscopy imaging.
- Photoactivatable fluorophores
Fluorophores (fluorescent proteins or synthetic fluorophores) that change their spectral properties on the absorption of light, providing a unique method for the optical labelling and tracking of molecules.
- Fluorescence cross-correlation spectroscopy
A spectroscopy method that correlates the fluctuations in intensity of two types of probes that diffuse through a small illumination volume, thus reporting on their binding.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
SUPPLEMENTARY INFORMATION
See online article: S1 (box)
References
- 1.Huse M. The T-cell-receptor signaling network. J. Cell Sci. 2009;122:1269–1273. doi: 10.1242/jcs.042762. [DOI] [PubMed] [Google Scholar]
- 2.Lin J, Weiss A. T cell receptor signalling. J. Cell Sci. 2001;114:243–244. doi: 10.1242/jcs.114.2.243. [DOI] [PubMed] [Google Scholar]
- 3.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 4.Schwartzberg PL. Genetic approaches to tyrosine kinase signaling pathways in the immune system. Immunol. Res. 2003;27:481–488. doi: 10.1385/IR:27:2-3:481. [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann SH. Immunology’s foundation: the 100-year anniversary of the Nobel Prize to Paul Ehrlich and Elie Metchnikoff. Nature Immunol. 2008;9:705–712. doi: 10.1038/ni0708-705. [DOI] [PubMed] [Google Scholar]
- 6.Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell–dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 7.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte–stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 8.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 9.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 10.Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc. Natl Acad. Sci. USA. 2003;100:2604–2609. doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousso P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nature Rev. Immunol. 2008;8:675–684. doi: 10.1038/nri2379. [DOI] [PubMed] [Google Scholar]
- 12.Germain RN, et al. Making friends in out-of-the-way places: how cells of the immune system get together and how they conduct their business as revealed by intravital imaging. Immunol. Rev. 2008;221:163–181. doi: 10.1111/j.1600-065X.2008.00591.x. [DOI] [PubMed] [Google Scholar]
- 13.Mempel TR, et al. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity. 2006;25:129–141. doi: 10.1016/j.immuni.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Breart B, Lemaitre F, Celli S, Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J. Clin. Invest. 2008;118:1390–1397. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartholomaus I, et al. Effector T cell interactions with meningeal vascular structures in nascent autoimmune CNS lesions. Nature. 2009;462:94–98. doi: 10.1038/nature08478. [DOI] [PubMed] [Google Scholar]
- 16.Beuneu H, et al. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 2010;33:412–423. doi: 10.1016/j.immuni.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Okada T, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrasco YR, Batista FD. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 2007;27:160–171. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Phan TG, Grigorova I, Okada T, Cyster JG. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nature Immunol. 2007;8:992–1000. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- 20.Qi H, Egen JG, Huang AY, Germain RN. Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science. 2006;312:1672–1676. doi: 10.1126/science.1125703. [DOI] [PubMed] [Google Scholar]
- 21.Junt T, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 22.Germain RN, Miller MJ, Dustin ML, Nussenzweig MC. Dynamic imaging of the immune system: progress, pitfalls and promise. Nature Rev. Immunol. 2006;6:497–507. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- 23.Wilson EH, et al. Behavior of parasite-specific effector CD8+ T cells in the brain and visualization of a kinesis-associated system of reticular fibers. Immunity. 2009;30:300–311. doi: 10.1016/j.immuni.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajenoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azar GA, Lemaitre F, Robey EA, Bousso P. Subcellular dynamics of T cell immunological synapses and kinapses in lymph nodes. Proc. Natl Acad. Sci. USA. 2010;107:3675–3680. doi: 10.1073/pnas.0905901107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J. Exp. Med. 2010;207:2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi M, et al. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Invest. 2010;120:1683–1693. doi: 10.1172/JCI41963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr. Opin. Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Andresen V, et al. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr. Opin. Biotechnol. 2009;20:54–62. doi: 10.1016/j.copbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Herz J, et al. Expanding two-photon intravital microscopy to the infrared by means of optical parametric oscillator. Biophys. J. 2010;98:715–723. doi: 10.1016/j.bpj.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu S, Yan P, Maslov K, Lee JM, Wang LV. Intravital imaging of amyloid plaques in a transgenic mouse model using optical-resolution photoacoustic microscopy. Opt. Lett. 2009;34:3899–3901. doi: 10.1364/OL.34.003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Wang L. Photoacoustic imaging in biomedicine. Rev. Sci. Instrum. 2006;77:041101. [Google Scholar]
- 34.Dustin ML. Hunter to gatherer and back: immunological synapses and kinapses as variations on the theme of amoeboid locomotion. Annu. Rev. Cell Dev. Biol. 2008;24:577–596. doi: 10.1146/annurev.cellbio.24.110707.175226. [DOI] [PubMed] [Google Scholar]
- 35.Kupfer A, Dennert G, Singer SJ. The reorientation of the Golgi apparatus and the microtubule-organizing center in the cytotoxic effector cell is a prerequisite in the lysis of bound target cells. J. Mol. Cell. Immunol. 1985;2:37–49. [PubMed] [Google Scholar]
- 36.Ryser JE, Rungger-Brandle E, Chaponnier C, Gabbiani G, Vassalli P. The area of attachment of cytotoxic T lymphocytes to their target cells shows high motility and polarization of actin, but not myosin. J. Immunol. 1982;128:1159–1162. [PubMed] [Google Scholar]
- 37.Geiger B, Rosen D, Berke G. Spatial relationships of microtubule-organizing centers and the contact area of cytotoxic T lymphocytes and target cells. J. Cell Biol. 1982;95:137–143. doi: 10.1083/jcb.95.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poenie M, Tsien RY, Schmitt-Verhulst AM. Sequential activation and lethal hit measured by [Ca2+]i in individual cytolytic T cells and targets. EMBO J. 1987;6:2223–2232. doi: 10.1002/j.1460-2075.1987.tb02494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donnadieu E, Bismuth G, Trautmann A. Antigen recognition by helper T cells elicits a sequence of distinct changes of their shape and intracellular calcium. Curr. Biol. 1994;4:584–595. doi: 10.1016/s0960-9822(00)00130-5. [DOI] [PubMed] [Google Scholar]
- 40.Negulescu PA, Krasieva TB, Khan A, Kerschbaum HH, Cahalan MD. Polarity of T cell shape, motility, and sensitivity to antigen. Immunity. 1996;4:421–430. doi: 10.1016/s1074-7613(00)80409-4. [DOI] [PubMed] [Google Scholar]
- 41.Brossard C, et al. Multifocal structure of the T cell – dendritic cell synapse. Eur. J. Immunol. 2005;35:1741–1753. doi: 10.1002/eji.200425857. [DOI] [PubMed] [Google Scholar]
- 42.Stinchcombe JC, Griffiths GM. Secretory mechanisms in cell-mediated cytotoxicity. Annu. Rev. Cell Dev. Biol. 2007;23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521. [DOI] [PubMed] [Google Scholar]
- 43.Gomez TS, et al. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;26:177–190. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fleire SJ, et al. B cell ligand discrimination through a spreading and contraction response. Science. 2006;312:738–741. doi: 10.1126/science.1123940. This study uses a combination of multiple light microscopy techniques and scanning electron microscopy to reveal morphological changes in activated B cells.
- 45.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. The seminal first observation of the detailed molecular organization within the immunological synapse of a ‘bull’s eye’ pattern, termed pSMAC and cSMAC, using three-dimensional confocal imaging of cell conjugates.
- 46.Freiberg BA, et al. Staging and resetting T cell activation in SMACs. Nature Immunol. 2002;3:911–917. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- 47.Potter TA, Grebe K, Freiberg B, Kupfer A. Formation of supramolecular activation clusters on fresh ex vivo CD8+ T cells after engagement of the T cell antigen receptor and CD8 by antigen-presenting cells. Proc. Natl Acad. Sci. USA. 2001;98:12624–12629. doi: 10.1073/pnas.221458898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orange JS. Formation and function of the lytic NK-cell immunological synapse. Nature Rev. Immunol. 2008;8:713–725. doi: 10.1038/nri2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carroll-Portillo A, et al. Formation of a mast cell synapse: Fcε RI membrane dynamics upon binding mobile or immobilized ligands on surfaces. J. Immunol. 2010;184:1328–1338. doi: 10.4049/jimmunol.0903071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trautmann A, Valitutti S. The diversity of immunological synapses. Curr. Opin. Immunol. 2003;15:249–254. doi: 10.1016/s0952-7915(03)00040-2. [DOI] [PubMed] [Google Scholar]
- 51.Singleton KL, et al. Spatiotemporal patterning during T cell activation is highly diverse. Sci. Signal. 2009;2:ra15. doi: 10.1126/scisignal.2000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purtic B, Pitcher LA, van Oers NS, Wulfing C. T cell receptor (TCR) clustering in the immunological synapse integrates TCR and costimulatory signaling in selected T cells. Proc. Natl Acad. Sci. USA. 2005;102:2904–2909. doi: 10.1073/pnas.0406867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richie LI, et al. Imaging synapse formation during thymocyte selection: inability of CD3ζ to form a stable central accumulation during negative selection. Immunity. 2002;16:595–606. doi: 10.1016/s1074-7613(02)00299-6. [DOI] [PubMed] [Google Scholar]
- 54.Hailman E, Burack WR, Shaw AS, Dustin ML, Allen PM. Immature CD4+CD8+ thymocytes form a multifocal immunological synapse with sustained tyrosine phosphorylation. Immunity. 2002;16:839–848. doi: 10.1016/s1074-7613(02)00326-6. [DOI] [PubMed] [Google Scholar]
- 55.Dustin ML. Visualization of cell-cell interaction contacts—synapses and kinapses. Adv. Exp. Med. Biol. 2008;640:164–182. doi: 10.1007/978-0-387-09789-3_13. [DOI] [PubMed] [Google Scholar]
- 56.Valitutti S, Dupre L. Plasticity of immunological synapses. Curr. Top. Microbiol. Immunol. 2010;340:209–228. doi: 10.1007/978-3-642-03858-7_11. [DOI] [PubMed] [Google Scholar]
- 57.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. The first use of a glass-supported planar bilayer for real-time imaging of T cell activation with peptide–MHC molecules, revealing the formation and dynamics of TCR microclusters within the immunological synapse.
- 58.Johnson KG, Bromley SK, Dustin ML, Thomas ML. A supramolecular basis for CD45 tyrosine phosphatase regulation in sustained T cell activation. Proc. Natl Acad. Sci. USA. 2000;97:10138–10143. doi: 10.1073/pnas.97.18.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krummel MF, Sjaastad MD, Wulfing C, Davis MM. Differential clustering of CD4 and CD3ζ during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- 60.Lee KH, et al. T cell receptor signaling precedes immunological synapse formation. Science. 2002;295:1539–1542. doi: 10.1126/science.1067710. [DOI] [PubMed] [Google Scholar]
- 61.Cullinan P, Sperling AI, Burkhardt JK. The distal pole complex: a novel membrane domain distal to the immunological synapse. Immunol. Rev. 2002;189:111–122. doi: 10.1034/j.1600-065x.2002.18910.x. [DOI] [PubMed] [Google Scholar]
- 62.Ludford-Menting MJ, et al. A network of PDZ-containing proteins regulates T cell polarity and morphology during migration and immunological synapse formation. Immunity. 2005;22:737–748. doi: 10.1016/j.immuni.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 63.Barr VA, et al. Dynamic movement of the calcium sensor STIM1 and the calcium channel Orai1 in activated T-cells: puncta and distal caps. Mol. Biol. Cell. 2008;19:2802–2817. doi: 10.1091/mbc.E08-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oddos S, et al. High-speed high-resolution imaging of intercellular immune synapses using optical tweezers. Biophys. J. 2008;95:L66–L68. doi: 10.1529/biophysj.108.143198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toomre D, Manstein DJ. Lighting up the cell surface with evanescent wave microscopy. Trends Cell Biol. 2001;11:298–303. doi: 10.1016/s0962-8924(01)02027-x. [DOI] [PubMed] [Google Scholar]
- 66.Barr VA, Bunnell SC. Interference reflection microscopy. Curr. Protoc. Cell Biol. 2009;45:4.23.1–4.23.19. doi: 10.1002/0471143030.cb0423s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bunnell SC, Kapoor V, Trible RP, Zhang W, Samelson LE. Dynamic actin polymerization drives T cell receptor-induced spreading: a role for the signal transduction adaptor LAT. Immunity. 2001;14:315–329. doi: 10.1016/s1074-7613(01)00112-1. [DOI] [PubMed] [Google Scholar]
- 68.Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl Acad. Sci. USA. 2007;104:20296–20301. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nature Immunol. 2009;10:531–539. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quann EJ, Merino E, Furuta T, Huse M. Localized diacylglycerol drives the polarization of the microtubule-organizing center in T cells. Nature Immunol. 2009;10:627–635. doi: 10.1038/ni.1734. [DOI] [PubMed] [Google Scholar]
- 71.Bunnell SC, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. An extensive imaging study showing the proteins that are recruited to microclusters, the dynamics of proteins at microclusters, the sorting of proteins away from TCR microclusters and the dynamics of intracellular calcium levels.
- 72.Barda-Saad M, et al. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nature Immunol. 2005;6:80–89. doi: 10.1038/ni1143. [DOI] [PubMed] [Google Scholar]
- 73.Braiman A, Barda-Saad M, Sommers CL, Samelson LE. Recruitment and activation of PLCγ1 in T cells: a new insight into old domains. EMBO J. 2006;25:774–784. doi: 10.1038/sj.emboj.7600978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huse M, et al. Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity. 2007;27:76–88. doi: 10.1016/j.immuni.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 75.Yokosuka T, et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nature Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 76.Bunnell SC, et al. Persistence of cooperatively stabilized signaling clusters drives T-cell activation. Mol. Cell. Biol. 2006;26:7155–7166. doi: 10.1128/MCB.00507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seminario MC, Bunnell SC. Signal initiation in T-cell receptor microclusters. Immunol. Rev. 2008;221:90–106. doi: 10.1111/j.1600-065X.2008.00593.x. [DOI] [PubMed] [Google Scholar]
- 78.Harwood NE, Batista FD. Early events in B cell activation. Annu. Rev. Immunol. 2010;28:185–210. doi: 10.1146/annurev-immunol-030409-101216. [DOI] [PubMed] [Google Scholar]
- 79.Yokosuka T, et al. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase C θ translocation. Immunity. 2008;29:589–601. doi: 10.1016/j.immuni.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaizuka Y, Douglass AD, Vardhana S, Dustin ML, Vale RD. The coreceptor CD2 uses plasma membrane microdomains to transduce signals in T cells. J. Cell Biol. 2009;185:521–534. doi: 10.1083/jcb.200809136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barr VA, et al. T-cell antigen receptor-induced signaling complexes: internalization via a cholesterol-dependent endocytic pathway. Traffic. 2006;7:1143–1162. doi: 10.1111/j.1600-0854.2006.00464.x. [DOI] [PubMed] [Google Scholar]
- 82.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. In this investigation, imaging of activated T cells on planar bilayers demonstrates the importance of peripheral microclusters in TCR signalling and the role of the cSMAC in its termination.
- 83.Vardhana S, Choudhuri K, Varma R, Dustin ML. Essential role of ubiquitin and TSG101 protein in formation and function of the central supramolecular activation cluster. Immunity. 2010;32:531–540. doi: 10.1016/j.immuni.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purbhoo MA, et al. Dynamics of subsynaptic vesicles and surface microclusters at the immunological synapse. Sci. Signal. 2010;3:ra36. doi: 10.1126/scisignal.2000645. [DOI] [PubMed] [Google Scholar]
- 85.Balagopalan L, et al. c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Mol. Cell. Biol. 2007;27:8622–8636. doi: 10.1128/MCB.00467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balagopalan L, Barr VA, Samelson LE. Endocytic events in TCR signaling: focus on adapters in microclusters. Immunol. Rev. 2009;232:84–98. doi: 10.1111/j.1600-065X.2009.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nguyen K, Sylvain NR, Bunnell SC. T cell costimulation via the integrin VLA-4 inhibits the actin-dependent centralization of signaling microclusters containing the adaptor SLP-76. Immunity. 2008;28:810–821. doi: 10.1016/j.immuni.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 88.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 89.Rodriguez-Fernandez JL, Riol-Blanco L, Delgado-Martin C. What is the function of the dendritic cell side of the immunological synapse? Sci. Signal. 2010;3:re2. doi: 10.1126/scisignal.3105re2. [DOI] [PubMed] [Google Scholar]
- 90.Trautmann A. Microclusters initiate and sustain T cell signaling. Nature Immunol. 2005;6:1213–1214. doi: 10.1038/ni1205-1213. [DOI] [PubMed] [Google Scholar]
- 91.Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nature Immunol. 2005;6:1168–1176. doi: 10.1038/ni1262. An important application of FRET to resolve some of the earliest steps of BCR activation on ligand binding, including clustering and conformational changes.
- 92.Tolar P, Hanna J, Krueger PD, Pierce SK. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity. 2009;30:44–55. doi: 10.1016/j.immuni.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3! cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. The combination of live-cell FRET microscopy and NMR in this study reveals conformational changes in the TCR–CD3ε chain complex on receptor activation.
- 94.Gascoigne NR, et al. Visualizing intermolecular interactions in T cells. Curr. Top. Microbiol. Immunol. 2009;334:31–46. doi: 10.1007/978-3-540-93864-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huppa JB, et al. TCR–peptide–MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. A conceptually novel study that quantifies on-rates and off-rates of the TCR with peptide–MHC complexes at the immunological synapse using single-molecule FRET, and shows surprisingly fast dynamics between these molecules.
- 96.Hashimoto-Tane A, et al. T-cell receptor microclusters critical for T-cell activation are formed independently of lipid raft clustering. Mol. Cell. Biol. 2010;30:3421–3429. doi: 10.1128/MCB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barda-Saad M, et al. Cooperative interactions at the SLP-76 complex are critical for actin polymerization. EMBO J. 2010;29:2315–2328. doi: 10.1038/emboj.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ananthanarayanan B, Ni Q, Zhang J. Molecular sensors based on fluorescence resonance energy transfer to visualize cellular dynamics. Methods Cell Biol. 2008;89:37–57. doi: 10.1016/S0091-679X(08)00602-X. [DOI] [PubMed] [Google Scholar]
- 99.Paster W, et al. Genetically encoded Forster resonance energy transfer sensors for the conformation of the Src family kinase Lck. J. Immunol. 2009;182:2160–2167. doi: 10.4049/jimmunol.0802639. [DOI] [PubMed] [Google Scholar]
- 100.Randriamampita C, et al. A novel ZAP-70 dependent FRET based biosensor reveals kinase activity at both the immunological synapse and the antisynapse. PLoS ONE. 2008;3:e1521. doi: 10.1371/journal.pone.0001521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Treanor B, et al. Microclusters of inhibitory killer immunoglobulin-like receptor signaling at natural killer cell immunological synapses. J. Cell Biol. 2006;174:153–161. doi: 10.1083/jcb.200601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Janes PW, Ley SC, Magee AI, Kabouridis PS. The role of lipid rafts in T cell antigen receptor (TCR) signalling. Semin. Immunol. 2000;12:23–34. doi: 10.1006/smim.2000.0204. [DOI] [PubMed] [Google Scholar]
- 103.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 104.Shaw AS. Lipid rafts: now you see them, now you don’t. Nature Immunol. 2006;7:1139–1142. doi: 10.1038/ni1405. [DOI] [PubMed] [Google Scholar]
- 105.Kenworthy AK. Have we become overly reliant on lipid rafts? Talking point on the involvement of lipid rafts in T-cell activation. EMBO Rep. 2008;9:531–535. doi: 10.1038/embor.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gaus K, Zech T, Harder T. Visualizing membrane microdomains by Laurdan 2-photon microscopy. Mol. Membr. Biol. 2006;23:41–48. doi: 10.1080/09687860500466857. [DOI] [PubMed] [Google Scholar]
- 107.Rentero C, et al. Functional implications of plasma membrane condensation for T cell activation. PLoS ONE. 2008;3:e2262. doi: 10.1371/journal.pone.0002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Owen DM, et al. High plasma membrane lipid order imaged at the immunological synapse periphery in live T cells. Mol. Membr. Biol. 2010;27:178–189. doi: 10.3109/09687688.2010.495353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilson BS, Pfeiffer JR, Surviladze Z, Gaudet EA, Oliver JM. High resolution mapping of mast cell membranes reveals primary and secondary domains of FcεRI and LAT. J. Cell Biol. 2001;154:645–658. doi: 10.1083/jcb.200104049. An important mapping of the distribution of receptors and downstream signalling molecules at the plasma membrane of mast cells using transmission electron microscopy, revealing their organization into segregated signalling domains on receptor activation.
- 110.Wilson BS, Pfeiffer JR, Oliver JM. Observing Fc!RI signaling from the inside of the mast cell membrane. J. Cell Biol. 2000;149:1131–1142. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl Acad. Sci. USA. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sohn HW, Tolar P, Pierce SK. Membrane heterogeneities in the formation of B cell receptor–Lyn kinase microclusters and the immune synapse. J. Cell Biol. 2008;182:367–379. doi: 10.1083/jcb.200802007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Glebov OO, Nichols BJ. Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nature Cell Biol. 2004;6:238–243. doi: 10.1038/ncb1103. [DOI] [PubMed] [Google Scholar]
- 114.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tanimura N, et al. Dynamic changes in the mobility of LAT in aggregated lipid rafts upon T cell activation. J. Cell Biol. 2003;160:125–135. doi: 10.1083/jcb.200207096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tolentino TP, et al. Measuring diffusion and binding kinetics by contact area FRAP. Biophys. J. 2008;95:920–930. doi: 10.1529/biophysj.107.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andrews NL, et al. Small, mobile Fc!RI receptor aggregates are signaling competent. Immunity. 2009;31:469–479. doi: 10.1016/j.immuni.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 119.Lillemeier BF, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nature Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. The introduction of the super-resolution imaging technique PALM to study the distribution of TCR and LAT in live cells, confirming previous indications from electron microscopy on their distribution at the plasma membrane in small sub-diffraction domains that coalesce on cell activation.
- 120.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 121.Ragan T, et al. High-resolution whole organ imaging using two-photon tissue cytometry. J. Biomed. Opt. 2007;12:014015. doi: 10.1117/1.2435626. [DOI] [PubMed] [Google Scholar]
- 122.Dustin ML. Supported bilayers at the vanguard of immune cell activation studies. J. Struct. Biol. 2009;168:152–160. doi: 10.1016/j.jsb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahmed F, Friend S, George TC, Barteneva N, Lieberman J. Numbers matter: quantitative and dynamic analysis of the formation of an immunological synapse using imaging flow cytometry. J. Immunol. Methods. 2009;347:79–86. doi: 10.1016/j.jim.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.George TC, et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J. Immunol. Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 125.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 126.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nature Rev. Mol. Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 127.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 128.Muik M, et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 129.Calloway N, Vig M, Kinet JP, Holowka D, Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol. Biol. Cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.