Abstract
Porphyromonas gingivalis, a major periodontal pathogen, must acquire nutrients from host derived substrates, overcome oxidative stress and subvert the immune system. These activities can be coordinated via the gingipains which represent the most significant virulence factor produced by this organism. In the context of our contribution to this field, we will review the current understanding of gingipain biogenesis, glycosylation, and regulation, as well as discuss their role in oxidative stress resistance and apoptosis. We can postulate a model, in which gingipains may be part of the mechanism for P. gingivalis virulence.
Keywords: Porphyromonas gingivalis, gingipains, apoptosis, caspase-independent apoptosis, oxidative stress, VimA, anoikis, N-cadherin, VE-cadherin, integrin β1, VimA, VimE, VimF, DNA repair, glycosylation, virulence, host cell survival, HRgpA, RgpB, Kgp, Review
2. INTRODUCTION
P. gingivalis, a black-pigmented, Gram-negative anaerobe, is an important etiological agent of periodontal disease and is also linked to cardiovascular disease and other systemic diseases [reviewed in (43,103)]. The inflammatory nature of periodontal disease implies that innate host defense mechanisms play a vital role in limiting bacterial growth. During active infection including tissue invasion, toxic reactive oxygen metabolites (e.g. superoxides, hydrogen peroxide and hydroxyl radicals) are mostly generated by polymorphonuclear leukocytes and macrophages (27,29). In order to survive in the inflammatory environment of the periodontal pocket, the bacterium must not only obtain nutrients for growth, but also overcome oxidative stress and subvert the immune defense system. Coordinated regulation of these activities would be beneficial to the bacterium. While there is documented evidence of the response of P. gingivalis to environmental stimulus (56,117,126), one possible mechanism for coordination may occur via the gingipains produced by this bacterium. The presence of P. gingivalis in the periodontal pocket and the high levels of gingipain activity detected in gingival crevicular fluid could implicate a role for gingipains in the destruction of the highly vascular periodontal tissue. Protease-associated degradation of cell-cell adhesion proteins on epithelial and endothelial cells resulting in cell death will compromise tissue integrity and facilitate spreading of the bacterium. Furthermore, degradation of the immune response components will facilitate the survival of the organism. Together, these activities may also satisfy the nutritional requirements of the organism. Gingipain-dependent heme accumulation on the bacterium cell surface may also act as an “oxidative sink”, thus, leading to protection against oxidative stress (191,193). We will discuss the role of the gingipains in the survival/pathogenicity of P. gingivalis and the unique vim (virulence modulating) locus that is involved in regulation of the gingipains. We will highlight some of our observations that are consistent with the hypothesis that the gingipains are central to an effective survival strategy for the organism.
3. P. GINGIVALIS VIRULENCE FACTORS - GINGIPAINS
While several virulence factors [(fimbriae (adhesins), capsule (antiphagocytosis), lipopolysaccharide (bone resorption), proteases (specific and generalized tissue destruction) and a variety of toxic by-products (e.g., ammonia)] have been implicated in the pathogenicity of P. gingivalis, the high proteolytic abilities of this organism are considered to play the most significant role in virulence [reviewed in (85,142)]. Proteolytic enzymes that have been secreted by this organism include endopeptidases, aminopeptidases, carboxypeptidase, oligopeptidase, and diand tri-peptidyl peptidases [reviewed in (161)]. The major proteases are endopeptidases called gingipains, (P. gingivalis +clostripain) and are both extracellular and cell-associated. They consist of arginine-specific proteases [Arg-gingipain, (Rgp)] and lysine-specific protease [Lysgingipain, (Kgp)]. The Rgp is encoded by two genes rgpA and rgpB and Kgp is encoded by a single gene kgp (142). The gingipains have multiple functions [reviewed in (50,80,142,161)]. In addition to being essential for growth, because of the asaccharolytic nature of this organism, they play a role in complement and immunoglobulin degradation, inactivation of cytokines and their receptors, platelet aggregation, attenuation of neutrophil antibacterial activities, and increasing vascular permeability, as well as, prevention of blood clotting. We and others have also demonstrated the ability of the gingipains to disrupt cell-cell and cell-matrix adhesion and induce apoptosis in several cell types (34,99,129,182,183). An involvement in hemoglobin binding and adsorption and heme accumulation further confirms their multiple contributions to bacterial survival, including oxidative stress resistance (3,107,114,152,191).
3.1. Gingipain biogenesis/activation
The translated product of the rgpA gene is composed of a pre-pro-fragment, followed by a proteinase domain, and a large hemagglutinin/adhesion (HA) domain at the C-terminus (40,162). The kgp gene product shares a similar size and structure to the RgpA gingipain. While it displays a 20% identity in the catalytic domain compared to RgpA, the amino acid sequence of the C-terminal HA domains are identical (157,158). As demonstrated in Kgp, the C-terminal HA domains of RgpA are not only essential for full expression of gingipain activity, but also for proper processing of the multiprotein complex assembly on the P. gingivalis outer membrane (199). In comparison to rgpA, the rgpB gene is missing almost the entire section coding for the HA domain, with the exception of a small C-terminal segment (132,141). Collectively, these observations indicate that the mature gingipains are derived from extensive processing from the nascent polypeptide precursors. Studies comparing the gene structure with the N-terminal sequences of different isoforms of the purified gingipain have demonstrated specific post-translational cleavage at Arg-Xaa and Lys-Xaa bonds (20). There is emerging evidence that this post-translational process is achieved by an autoproteolytic mechanism (131). Further, both gingipains R and K can be mutually involved in the efficient maturation of each other in addition to a carboxypeptidase (CPG70) that can play a role in the C-terminus processing of RgpA and Kgp HA domains (219).
As a common theme, the expression of extracellular proteolytic activities is highly regulated in both prokaryotic and eukaryotic systems [reviewed in (39,221)]. This regulation can occur at multiple levels including processing of an inactive secreted precursor to its active form and/or the post-translational glycosylation of the proteins (60,210). The multiple layers of regulation are vital to ensure that expression is tightly controlled in the appropriate temporal and spatial patterns. Currently, information is known only on the activation pathway of pro-RgpB (131). The recombinant, full length zymogen undergoes three sequential autoproteolytic, intermolecular processing steps that involve processing at Arg129, followed by Arg229, then the C-terminal extensions, to generate full activity. Each step in the processing requires the previous step, and enzyme activity is enhanced in a stepwise manner (131). If gingipain activation occurs by an autoproteolytic mechanism, questions are raised on how this process is regulated and the involvement of specific bacterial host factors. In fact, in previous reports, we have demonstrated the secretion of the inactive proenzyme gingipain species in vimA-, vimE- and vimF-defective mutants (150,213,214,215). In both the vimE and vimF isogenic mutants, activation of the gingipain proenzyme species could not be achieved (213,214). Taken together, these observations suggest a role for the Vim proteins in the regulation of gingipain activation; however, their mechanisms are unclear and are the subject of ongoing investigation (see further discussion below in section 3.3.).
3.2. Gingipain glycosylation
Glycosylation is one of the important ways by which protein maturation and other cellular processes are regulated (71,201,217). There is emerging evidence that this process may also be important in gingipain biogenesis in P. gingivalis (60,213,214,215). For glycosylation, different glycosyl transferases catalyze the transfer of different carbohydrate moieties from active donors to specific acceptors (including lipids, proteins and nucleic acids) (26). In proteins, for example, this can have significant effects on the intrinsic properties that can influence their stability, resistance to proteolysis, and tertiary structure which can affect the rate of processes that involve conformational changes (112,134,200). The level of glycosylation can also modulate the interaction of the glycoconjugate with other molecules (14,189). Given the significant impact of carbohydrate modifications on protein structure/function, glycosylation of bacterial proteins is of major relevance to the molecular basis of pathogenesis. There is a growing list of bacterial virulence factors that are glycoproteins (60,201). Curtis and colleagues were the first to demonstrate that the gingipains are post-translationally modified with carbohydrate additions that are cross-reactive with monoclonal antibodies to P. gingivalis lipopolysaccharide (LPS) (41,60). Our recent publications confirm and extend these results (213,214,215). Most of the LPS-like glycan moieties appear to occur at the C-terminus of the polypeptide chain and seem to serve to anchor the gingipain molecule into the outer membrane (146,178,188). There is also diversity and variable levels of carbohydrate modifications demonstrated in the various isoforms of the catalytic domains of Rgp (41,60). This may partly account for the heterogeneity in the RgpA and RgpB isoforms.
There is currently no information on the enzymology of gingipain glycosylation. However, the gene products of wpbB and several genes in the porR locus (31,60,188) have been shown to participate in modification of the gingipains. These mostly conserved groups of enzymes in Gram-negative bacteria are known to be involved in the biosynthesis of the O-antigen side chains of the LPS component of the outer membrane. Isogenic mutants defective in these genes result in the loss of pigmentation, glycan synthesis and glycosylation, and reduced gingipain activity, which is mostly soluble. In our studies, we have identified other novel genes (vimA, vimE and vimF) that also appear to participate in modification of the gingipains (213,214,215). While the vimA-defective mutant displayed a similar phenotype to the porR mutant, the gingipains in the vimE and vimF mutants, although inactive, were cell-associated. Further, there was no observed late onset of gingipain activity in vimE and vimF mutants suggesting that other modifications beyond those that facilitate membrane anchorage are needed for activation. VimF is a putative glycosyl transferase; however, its specific role in glycosylation leading to gingipain activation is unclear. Ongoing studies are testing the hypothesis that regions of the gingipain require specific glycosylation to facilitate activation/maturation. Consistent with this hypothesis is the recent observation that truncation of the last two residues (valyl-lysine) from the C terminus of RgpB is sufficient to create an inactive version of the protein that lacks the post-translational glycosylation seen in the wild type, and the protein remains trapped behind the outer membrane. Alanine scanning of the last five residues also revealed the importance of the C-terminal motif in mediating correct post-translational modification of the protein (146). RgpB is required for the normal post-translational glycosylation of Arg-gingipains derived from rgpA (165). It is unclear how the vim genes are involved in this process.
3.3. Regulation of gingipain biogenesis and virulence in P. gingivalis - Role of the bcp-recA-vimA region
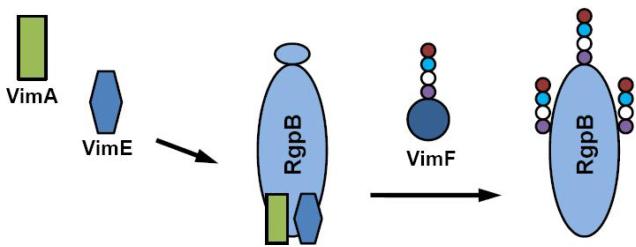
Although several virulence regulators have been described in P. gingivalis [reviewed in (56,81)], there is an emerging view that modulation of virulence in P. gingivalis may be coordinated via an ability to modulate proteolytic activity. For example, there is evidence that the gingipains can modulate heme uptake/utilization which is vital for its survival [(151) and reviewed in (152)]. Fimbrial expression is also regulated by the gingipains (229). Other studies in our laboratory have also shed light on the relationship of virulence and the ability of P. gingivalis to manage the oxidative stress typical of the inflammatory microenvironment of the periodontal pocket. A coordinate relationship between gingipain activity, oxidative stress resistance and virulence is further confirmed by the gene cluster at the bcp-recA-vimA locus. We have demonstrated that the recA gene plays the expected role in DNA repair (55) (see section 4. below). The bcp and vimA (virulence modulating) genes are part of the same recA transcriptional unit (1) and upstream of the vimE and vimF genes (213,214). While P. gingivalis FLL301, the bcp-defective mutant, showed in vitro sensitivity to hydrogen peroxide, its virulence potential in a mouse model was unaltered when compared to the wild-type (95). In contrast, the vimA- vimE- and vimF-defective mutants were non-black-pigmented and showed decreased gingipain activity (1,213,214). The gingipain proenzyme species were observed in these mutants providing some of the first evidence for post-translational regulation of protease activity in P. gingivalis. It is also consistent with other observations which suggest that glycosylation plays an important role in post-translational regulation of gingipain activity (60,188). In in vivo experiments using a mouse model, the vimA-defective mutant (P. gingivalis FLL92) was dramatically reduced in virulence when compared with wild-type W83 strain (1). We have also demonstrated that VimA interacts with the gingipains and other proteins known to be associated with sugar metabolism and protease maturation and is not cleaved by purified RgpB (216). In our bioinformatic studies, VimA appears to be a unique protein that lacks DNA binding motifs or known enzyme domains. It may be a putative membrane protein and the C-terminus may also have a putative protein binding domain (216). These data suggest that VimA may be part of a protein complex that is involved in gingipain biogenesis/glycosylation (Figure 1).
Figure 1.
Putative model for VimA-dependent glycosylation of the gingipains. Glycosylation of gingipains are altered by inactivation of the vimA gene. It is likely that VimA could function as an accessory protein that will facilitate the interaction of other proteins (e.g. VimF) that is vital for glycosylation/activation. Possible defects of specific glycosyl transferases may prevent the gingipains from folding correctly resulting in lack of activation.
Collectively, our observations indeed support the hypothesis that regulation of virulence in P. gingivalis may be coordinated via an ability to modulate proteolytic activity, although we cannot rule out any other direct or indirect effect of the vim genes on the expression of other virulence factors. It is interesting that the recA and bcp genes are part of the same transcriptional unit with the vim genes that are involved in modulating proteolytic activity. This kind of association may be significant since a response to oxidative stress will involve binding of oxygen and its toxic derivatives to iron accumulated on the surface of the cell via the gingipains (193). The bound heme can be involved in the catalytic destruction of the toxic oxygen derivative species (191). Since proteolytic activity in P. gingivalis is associated with heme accumulation (152) via the HA2 hemagglutinin domains of Arg- and Lysgingipains (192), it might be considered an important strategy for the organism to coordinate its oxidative stress and proteolytic activities. Under conditions of oxidative stress, there is an increased proportion of cell-bound gingipain (RgpA and Kgp) activity (45). This importance is further underscored by the observation that the recA gene is expressed during infection of the murine host (118). Further, expression of the recA gene is regulated by temperature, iron, and calcium (117) which are components known to coordinately regulate the expression of other bacterial virulence genes [reviewed in (56,126)].
4. THE SIGNIFICANCE OF OXIDATIVE STRESS IN THE SURVIVAL OF P. GINGIVALIS
In the inflammatory microenvironment of the mouth, reactive oxygen species (ROS) which are mostly produced by polymorphonuclear leukocytes and macrophages (30), constitute an important component (133). An increase in ROS or depletion of antioxidant molecules and/or enzymes results in oxidative stress. Thus, oxidative stress occurs when there is an imbalance between oxidant exposure and antioxidant protection. In addition, the occasional exposure of P. gingivalis to air can give rise to the metabolic conversion of atmospheric oxygen to ROS inside bacterial cells. ROS are highly reactive and can damage proteins, lipids, RNA and DNA (27,133). The generation and accumulation of O2*, H2O2 and hydroxyl radicals (16,30,133) that target DNA, cellular membranes, metalloproteases and transcription factors (69,86,87,123,195) can be lethal to the organism. In bacteria, DNA damage appears to be the most significant consequence of oxidative stress [reviewed in (133)]. While oxidant-induced DNA damage generates over 20 different oxidatively altered bases (44), 8-oxoG is the major product of DNA oxidation (179). Unlike some other modified DNA bases, 8-oxoG does not block replication. Instead, it can Watson-Crick base pair with cytosine, as well as, Hoogsteen base pair with adenine (68,205). The polymerases can efficiently incorporate both cytosine and adenine across from 8-oxoG. Mispairing with adenine often leads to GC → TA transversion mutations that can be dangerous for the cell (68). Because the average G + C content of the genome of P. gingivalis is 49% [(145), (www.oralgen.lanl.gov)], a mechanism(s) to prevent or repair lesions resulting from guanine oxidation is imperative. This is further underscored by the observations that the salivary levels of 8-oxoG; P. gingivalis and T. forsythia in the periodontitis patients were significantly higher than those in healthy subjects (173). In addition, 8-oxoG was significantly correlated with the presence of P. gingivalis (173). Thus, it is crucial that P. gingivalis utilize an arsenal of mechanisms to either prevent or fix oxidative damage resulting from ROS in order to survive in this hostile environment. P. gingivalis has developed various strategies some of which involve the gingipains in protecting itself from oxidative stress. These strategies include antioxidant enzymes, the hemin layer, DNA repair enzymes, the bcp-recA-vimA locus, htrA, feoB2, and other hypothetical genes of unknown function.
4.1. Oxidative stress resistance: antioxidant enzymes and hemin accumulation
In the first report to describe oxidative DNA damage in an anaerobe, Prevotella melaninogenica is shown to be highly sensitive to O2 or H2O2 exposure (203). Under these conditions, the elevated 8-oxoG detected in that strain correlated with its decreased survival. This suggests that oxidative DNA damage is an important cause of oxygen intolerance in P. melaninogenica. In our studies, we have also identified the presence of 8-oxoG in the chromosomal DNA of P. gingivalis W83 and the non-pigmented isogenic mutant FLL92 after exposure to H2O2 (97).
Two cellular systems function in bacteria to protect against oxidative stress (12,119,171). In one system, antioxidant enzymes such as superoxide dismutase (SOD), catalase, peroxidase, and oxidase attempt to neutralize molecular oxygen and ROS (2,4,51,177) before they can damage cellular components. The other system, which will be discussed later, involves endonucleases which are involved in the repair of nucleic acids that have already been damaged (21,105,119), as seen in Escherichia coli. Although P. gingivalis is oxygen tolerant and is missing catalase activity, it has been shown to express cytosolic SOD activity (35). This SOD activity, however, is protective only for atmospheric oxygen but ineffective against H2O2 or exogenously generated ROS (121,143). Recently, rubrerythrin (a non-heme iron protein which functions as a cytoplasmic peroxidase), encoded by the rbr gene, and DNA binding proteins, encoded by the dps gene, were shown to provide oxidative stress protection against H2O2 (198,209). Our own studies (96) have investigated the ability of Ahp (alkyl hydroperoxide reductase), a peroxide-scavenging enzyme, to protect against oxidative damage. This enzyme consists of two components, AhpC with peroxidase activity and the flavoprotein AhpF. Our findings suggest that the ahpC gene may play an important role in peroxide resistance in P. gingivalis in vitro, but does not contribute to its virulence in vivo. These results were partly confirmed recently by Diaz, et al (47) who demonstrated that P. gingivalis seems to lack a protective NADH oxidase but AhpF-C could contribute to its moderate tolerance to reactive oxygen species by metabolizing H2O2.
Protection against oxidative damage may also utilize another unique mechanism in P. gingivalis. Cell surface heme acquisition has been postulated to be a defense mechanism against ROS in P. gingivalis (191,193). The storage of the heme on the cell surface which gives the organism its characteristic black pigmentation, can form μ-oxo dimers in the presence of ROS and can give rise to the catalytic degradation of H2O2 (191). An important component of the heme acquisition system in P. gingivalis involves the gingipains (65,115,148). Recent reports suggest that gingipains Kgp and RgpA are the major proteases involved in hemin acquisition, binding and accumulation in P. gingivalis (24,42,115,163,186,192). This raises an important question as to whether this organism would be more susceptible to oxidative stress in the absence or reduction of this “oxidative sink” layer. Because the gingipains are downregulated at elevated temperature, typical of the inflammatory microenvironment of the periodontal pocket (104,159), or when P. gingivalis contacts host cells (156), this may have implications for its capacity to adapt to conditions of oxidative stress. In our studies, we sought to determine if this organism was more susceptible to oxidative stress in the absence or reduction of this hemin layer. Thus, we examined a non-pigmented vimA-defective isogenic mutant of P. gingivalis (FLL92) which has reduced cell surface associated gingipain activity. As expected, the 8-oxoG and its repair activity were elevated in the non-pigmented isogenic mutant (97), which could further support a protective role of the pigmented layer and, consequently, gingipain involvement in oxidative stress resistance.
4.2. DNA repair mechanisms in oxidative stress
Although vital for survival, there is currently a gap in our understanding of mechanism(s) used by P. gingivalis or other anaerobes to repair oxidative DNA damage. It is unclear if the gingipains are coordinately regulated with this process given its association with the formation of the “oxidative sink” layer (191,193). In other bacteria DNA damage can be repaired by several mechanisms including base excision repair (BER) and nucleotide excision repair (NER) (21,106,139,174).
In general, most oxidative DNA damage, whether it is base or sugar damage or the formation of abasic sites, is repaired by BER (106). DNA lesions generated as a result of ROS damage are usually not bulky DNA lesions and are excellent substrates for the process of BER (69). BER involves MutM, MutY, and MutT. MutM removes 8-oxoG paired with cytosine. If replication occurs without the removal of 8-oxoG, MutY, a glycosylase, removes adenine mispaired with 8-oxoG. Thus, MutM and MutY work together to prevent GC → TA transversions associated with 8-oxoG. MutT hydrolyzes 8-oxodGTP to 8-oxodGMP, thus depleting the nucleotide pool of 8-oxodGTP. DNA N-glycosylases are the effectors of BER and are more specific in lesion recognition and repair than the process of NER (106).
NER is a universal DNA repair mechanism found in all kingdoms of life (15,206). It is different from all the other forms of DNA repair in its ability to act on a wide variety of substrates (15). NER was one of the first repair mechanisms discovered in bacteria (180) and is mediated by the products of the following genes: uvrABC, uvrD, polA and lig (15,172). NER recognizes distortions in DNA caused by bulky adducts that also alter the chemistry of the DNA. Damaged DNA is recognized, cleaved 3’ and 5’ to the DNA lesion, and repaired in a sequential multistep process that requires ATP (15,172).
In a previous report, we have determined the repair of 8-oxoG-induced DNA damage in P. gingivalis (97). Bacterial extracts from the P. gingivalis isogenic strains grown in the presence or absence of H2O2 were used in glycosylase assays with a 5’-end labeled [γ-32P]-ATP 8-oxodG:C containing oligonucleotide (24mer) (97). If 8-oxoG is removed by a BER mechanism, a cleavage product corresponding to a 12mer would be observed because the 8-oxodG:C was placed in the middle of the 24mer. The Formamidopyrimidine-DNA glycosylase (Fpg) enzyme generated the expected 12mer fragment in oligonucleotide containing 8-oxoG; however, a fragment of a similar size was missing in P. gingivalis strains W83 and FLL92. Instead, a cleavage product of approximately 17 bases was observed. In addition, when compared to other anaerobic periodontal pathogens, the removal of 8-oxoG was unique to P. gingivalis. Collectively, these data indicate that the repair mechanism of 8-oxoG is different in P. gingivalis when compared to E. coli and suggests a mechanism that needs to be defined. In contrast to MutY and MutT, there is no detectable evidence of the E. coli mutM homologue in the P. gingivalis genome [(145), (http://www.oralgen.lanl.gov/)]. Preliminary studies in the laboratory have demonstrated that as in E. coli, MutY appears to play a similar role in protecting against oxidative stress in P. gingivalis (168).
The P. gingivalis genome contains genes which encode for the UvrA, UvrB, and UvrC proteins (145) (http://www.oralgen.lanl.gov/). UvrB is the central component of bacterial NER. It is directly involved in distinguishing damaged from undamaged DNA and guides the DNA from recognition to repair synthesis [reviewed in (206)]. To further evaluate if NER may play a role in this repair activity given the size of the cleavage product (97), the uvrB gene in P. gingivalis was inactivated (78). In contrast to the wild-type P. gingivalis W83, the uvrB-deficient mutant FLL144 was significantly more sensitive to UV irradiation. However, the enzymatic removal of 8-oxoG was unaffected by the inactivation of the uvrB gene. Collectively, these results suggest that the uvrB gene in P. gingivalis may not be involved in the removal of 8-oxoG and that another yet unidentified mechanism may be employed in its repair.
4.3. Other mechanisms: role of the bcp-recA-vimA locus, htrA, and hypothetical genes of unknown function in oxidative stress resistance
In P. gingivalis, oxidative stress was found to modulate the expression of several proteins including HtpG, GroEL, DnaK, AhpC, FeoB2, TPR domain protein, and trigger factor (8,46,77,82,101,149,184). Since several of these proteins are also involved in heat shock, it implies that the organism uses a multifunctional approach that may involve the gingipains to maintain cell viability under adverse environmental conditions.
Since the recA gene product is a key protein in DNA repair, we hypothesized that this DNA repair ability may play a role in the survival and virulence of P. gingivalis. We constructed a recA-deficient mutant by allelic exchange mutagenesis and showed that in contrast to the wild-type strain, this mutant was more sensitive to UV irradiation (55). Thus, this gene may play the expected role in DNA damage repair (55). We have also identified other genes, bcp and vimA-vimE-vimF that are part of the same recA transcriptional unit and can be differentially expressed (1,213,214). The bacterioferritin comigratory protein (Bcp) is a part of the thiol-specific antioxidant (TSA)/alkyl hydroperoxidase family and functions in a similar way to the AhpC in detoxifying H2O2 (92,96). Preliminary evidence has shown that P. gingivalis FLL301, a bcp-defective mutant, showed increased in vitro sensitivity to H2O2 (95). The vimA-defective mutant also showed increased sensitivity to H2O2 compared to the wild-type strain; however, the level of resistance is inducible (125).
In protein-protein interaction studies, the VimA protein was shown to interact with gingipains and other proteins including high temperature requirement A (HtrA) (216). In several organisms, including bacteria, yeast, plant, and humans, HtrA is considered an important factor that is involved in protein folding and maturation as well as in the degradation of proteins that are misfolded (160). It functions as a molecular chaperone at low temperatures and has proteolytic activity at elevated temperatures (102). Inactivation of the htrA gene has been shown to affect the sensitivity of many organisms to thermal and oxidative stress (116,155). In P. gingivalis, HtrA is also involved in resistance to thermal and oxidative stress (169). P. gingivalis FLL203, an htrA-defective mutant, had increased sensitivity to H2O2 and decreased Rgp activity. HtrA was also shown to physically interact with the gingipains RgpA, RgpB and Kgp (169). This further underscores the importance of the gingipains and their regulator vim genes in oxidative stress resistance in P. gingivalis. Thus, we can envision a scenario in P. gingivalis where the HtrA protein may be important for regulation of gingipain activity in the inflammatory microenvironment of the periodontal pocket. A specific mechanism for this interaction is unclear and should be further investigated.
Transcriptome analysis of P. gingivalis in the presence of hydrogen peroxide has further extended our understanding of the complex response of this organism to oxidative stress (8,46,184). Diaz, et al (46) in their DNA microarray analysis of P. gingivalis isogenic mutants, have demonstrated that common OxyR-regulated genes such as dps and ahpFC were not positively regulated in response to H2O2, although, their expression was dependent on the presence of a functional OxyR protein. As confirmed elsewhere (147), they concluded that OxyR does not act as a sensor of H2O2 in P. gingivalis. In preliminary experiments, we have shown that the duration of oxidative stress can modulate different sets of genes (125). As expected, an up-regulation of DNA repair/modification genes and several hypothetical genes which have not been previously characterized were mostly seen at a shorter H2O2 exposure time. However, during prolonged exposure to H2O2, several genes some of which are known to be involved in protein repair and many previously uncharacterized were up-regulated. Characterization of the hypothetical genes should lead to a more comprehensive understanding of mechanisms of oxidative stress resistance in P. gingivalis. It is of particular interest to identify transcriptional regulators that activate transcription of oxidative-stress-related genes in response to H2O2. This is being further being investigated in the laboratory. The mechanisms that P. gingivalis employs to combat the effects of ROS are just beginning to be elucidated. It will be interesting to determine the extent of gingipain involvement in these processes.
5. GINGIPAIN-INDUCED EFFECTS ON HOST CELL SURVIVAL
5.1. Gingipain-induced degradation of cell adhesion molecules
5.1.1. Gingipains directly cleave Cell Adhesion Molecules (CAMs)
CAMs mediate cell-cell interaction, cell-matrix interactions, and cell signals for growth, differentiation, and apoptosis [reviewed in (164)]. There are four major families of CAMs: cadherins, integrins, immunoglobulin gene superfamily members, and selectins. Proteolytic cleavage of these proteins can modulate their signaling functions (130,226). Thus, gingipain-induced degradation of CAMs could be implicated in periodontal disease pathogenesis. There is evidence that degradation of proteins of cell-cell junctional complexes by P. gingivalis may facilitate the organism's invasion and spread to deeper structures via a paracellular pathway (98).
We and others have shown that gingipains cleave or modulate the expression of CAMs in various cell types (34,79,83,99,100,182,183,202,220,222). There is specific affinity and cleavage efficiency of the gingipains for CAMs. HRgpA was found to be 12 times more effective than RgpA in cleaving integrin α5β1 on human gingival fibroblasts, while αvβ3 was unaffected (176). This study also established that the adhesion domain of HRgpA facilitates binding to integrin β1 (176). However, in human gingival fibroblasts Rgp was implicated in the cleavage of integrins α2, β1, and β3, but not α5 and αv (10). Yun et al. (236) showed that in human umbilical vein endothelial cells (HUVEC), RgpA and Kgp degrade CD99, a CAM that has been implicated in multifactorial cellular events producing prominent contraction and progressive intercellular gap formation in the endothelial monolayer (232). These investigators also demonstrated that endothelial leukocyte adhesion molecule-1 (ELAM-1 also known as E-selectin), VCAM-1, and ICAM-1 were not degraded by the gingipains (232). These effects may contribute to the successful colonization of P. gingivalis by limiting the host inflammatory response (232).
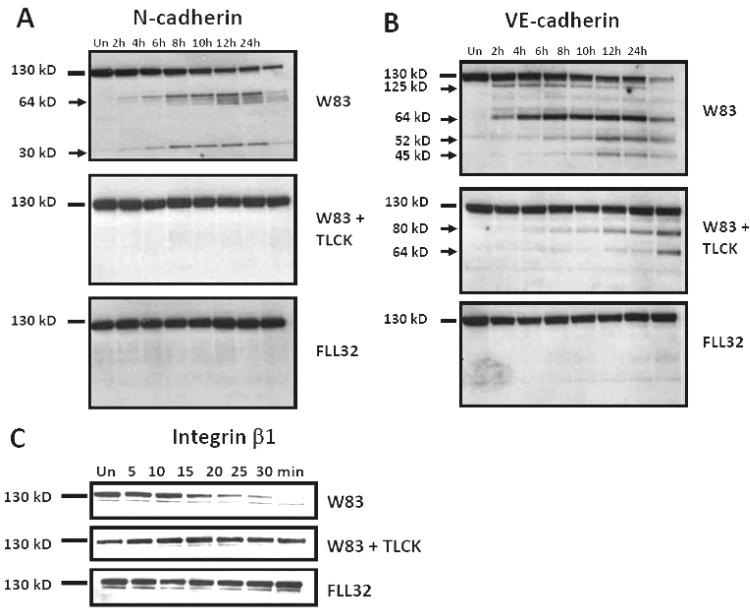
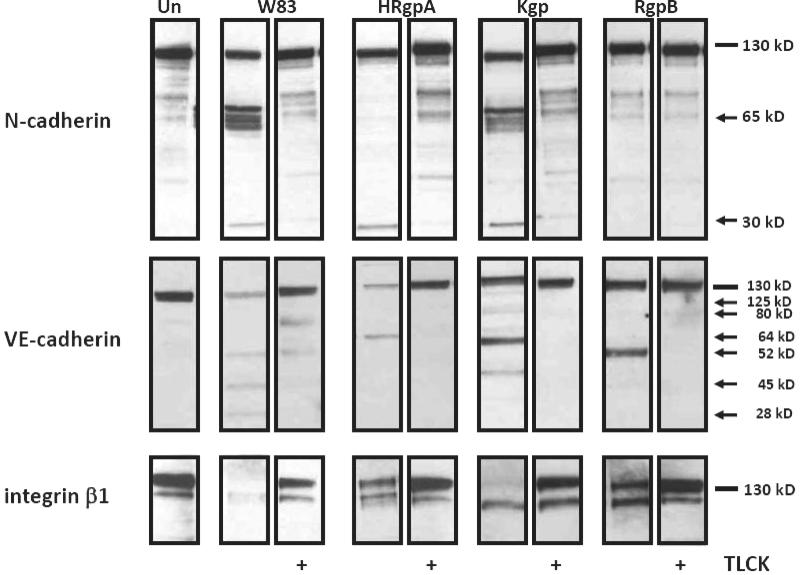
We have also demonstrated that treatment with gingipain-active W83 extracts of both coronary artery (BCAEC) and microvascular (HMVEC) endothelial cells resulted in detachment and BCAEC displayed cleaved N-cadherin, VE-cadherin, and degraded integrin β1 (182,183). BCAEC integrin β1 was cleaved rapidly after exposure to gingipain-active W83 extracts with complete degradation by approximately 30 min (Figure 2C). This degradation occurred when BCAEC were still fully adherent. VE-cadherin cleavage was evident at 2 h and progressed to almost total disappearance of the full length protein by 24 h (Figure 2B). N-cadherin cleavage occurred somewhat later than VE-cadherin cleavage (Figure 2A). These cleavages were confirmed in vitro by direct exposure of immunoprecipitated N- and VE-cadherin to gingipains (182). Using selective gingipain inhibitors, we demonstrated that Kgp activity was responsible for degradation of integrin β1 while both Kgp and Rgp activity were responsible for cleavage of N-cadherin and VE-cadherin (183). In experiments were BCAEC were exposed to purified gingipains, Kgp was found to be the most efficient gingipain at cleaving integrin β1 while N-cadherin was cleaved by both HRgpA and Kgp but not by RgpB (Figure 3). VE-cadherin was cleaved by HRgpA, Kgp, and RgpB, (Figure 3). BCAEC treated with purified Kgp reattached and remained viable during the time course but still exhibited cleaved/degraded CAMs, most notably integrin β1 (Figure 3). This indicated that re-attachment was not because of the loss of Kgp activity.
Figure 2.
Cleavage of N-cadherin, VE-cadherin, and integrin β1 by gingipain-active extracts from strain W83. 10μg of protein from BCAEC treated, in the presence of 5 mM L-cysteine, for the indicated times with W83 extracts (70 Units Rgp activity/ml media and 5.3 units Kgp activity/ml media), TLCK-treated W83 and FLL32 extracts (0.73 Units of Rgp activity/ml media and 0.49 Units of Kgp activity/ml media) were separated by SDS-PAGE and immunoblotted with a monoclonal antibody to (A) N-cadherin or (B) a polyclonal antibody to VE-cadherin. (C) BCAEC treated with 200μg/ml W83 extract (230 Units of Rgp activity/ml media and 19.4 Units of Kgp activity/ml media), FLL32 extract (0.73 Units of Rgp activity/ml media and 0.49 Units of Kgp activity/ml media), and W83 extract pretreated with 10 mM TLCK were harvested, after treatment for the indicated times, and 20 μg was separated by SDS-PAGE. Membranes were reacted with a monoclonal antibody to integrin β1. Arrows indicate cleavage products. Un, untreated. Reproduced with permission from ASM) (182).
Figure 3.
Synergism of purified gingipains in induction of CAM cleavage. 15 μg of total protein from BCAEC treated for 6 h (N-cadherin), 24h (VE-cadherin), or 12h (integrin β1), in the presence of 5 mM L-cysteine, with purified HRgpA (8 μg /ml), Kgp (3 μg /ml), or RgpB (5.2 μg /ml) (all equivalent to 113 Units of Rgp activity/ml media or 12.4 Units of Kgp activity/ml media) were separated by SDS-PAGE and immunoblotted with antibodies to N-cadherin, VE-cadherin, and integrin β1. Arrows indicate cleavage products, whereas lines indicate intact proteins. Un, untreated; +, present. Reproduced with permission from ASM (183).
Taken together, these results showed that the gingipains work together to detach cells and cleave specific CAMs (183). Although it cannot be ruled out that these observations may be cell type dependent, they implicate a role for gingipains in the pathogenesis of periodontitis and possibly modulation of host cell survival through detachment-induced apoptosis.
5.1.2. CAM degradation may implicate anoikis in gingipain-induced apoptosis
Anoikis is a type of caspase-dependent apoptosis that can be triggered by cellular detachment that is associated with loss of integrin survival signaling or inappropriate cell-matrix contacts (57,233). Anoikis has been described in numerous cell types, including epithelial cells (57), endothelial cells (127,166), keratinocytes, thyroid cells, fibroblasts, and osteoblasts (233).
Numerous kinase and phosphatase signaling molecules have been implicated as major regulators of anoikis [reviewed in (58)]. It has been reported that gingipains cleave focal adhesion kinase (FAK) and p130Cas (79), which suggests the possibility that gingipains can induce anoikis in BCAEC. FAK acts as a bridge protein linking growth factor receptor and integrin stimulated signaling pathways and is potentially able to modulate anoikis because it is connected to signaling pathways involving the cytoskeleton and transcriptional activity in the nucleus (136). It has been shown that gingipains cleave FAK in immortalized human oral keratinocytes, which could be inhibited by peptide caspase inhibitors (79), suggesting Kgp involvement in this cleavage (183). The cleavage of FAK has several consequences related to apoptosis induction. For example, loss of FAK signaling triggers Bid and Bax translocation (167). In addition, it was also reported that FAK interacts directly with receptor-interacting protein (RIP), which is a Fas receptor binding protein that induces apoptosis, and that the caspase cleavage fragment of FAK inhibits FAK survival signaling (167). This raises the possibility that a gingipain-induced FAK cleavage fragment may also be self inhibitory. Furthermore, the cleavage of FAK may disrupt its ability to be phosphorylated and thereby activating p53 and cell death (84). Further studies demonstrating that FAK is cleaved in BCAEC, determining whether Kgp is responsible for FAK cleavage, and exploring the inability of Kgp to induce apoptosis at the levels used in our models system need to be performed.
Viable BCAEC treated with purified Kgp displayed no full length integrin β1, whereas detached apoptotic BCAEC treated with HRgpA or RgpB still had some full length protein (Figure 3). This would suggest that signaling pathways triggered by the loss of functional integrin β1 do not play a role in gingipain-induced apoptosis. P. gingivalis activates the PI3-K/Akt pathway to increase host cell survival in response to apoptotic triggers (230), raising the possibility that in BCAEC treated with purified Kgp these same pathways are activated to prevent initiation of apoptosis in response to cell detachment. In addition, the cytosolic domain of integrin requires membrane localization to promote apoptosis, and it is possible that cleavage of the β integrin cytosolic domain observed in extracellular matrix (ECM)-deprived cells may even act to delay apoptosis by preventing integrin mediated death (196).
In intestinal epithelial cells, anoikis involved the activation of the initiator caspases-2 and -9 within minutes of detachment followed by the activation of caspases-7, -3, and -6, with caspase-8 activation occurring downstream of caspase-9 and -2 activation (70). Caspase-1 and -10 remained inactive (70). Caspase-8 activation occurred independently of downstream caspases or caspase-9 (70). Yet, the caspase-8 inhibitor z-IETD was also able to block anoikis, suggesting that at least two caspase pathways were activated for induction and execution of anoikis (70). By contrast, in gingival fibroblasts infected with P. gingivalis, caspases-3, -6, -7, -9, but not -8, -10, or -12, were activated which strongly points to a mitochondrial mediated apoptosis pathway (211). Further analysis of the hierarchy of caspase activation in BCAEC treated with gingipains will clarify the role that anoikis plays in gingipain-induced apoptosis.
5.2. P. gingivalis-induced apoptosis
5.2.1. Apoptosis can occur with and without caspase involvement
5.2.1.1. Caspase-dependent P. gingivalis-induced apoptosis
Because of the close proximity between oral pathogens and host cells in the gingiva, the interplay of P. gingivalis and the various cell types present in the gingival crevice has been under intense scrutiny. It is becoming increasingly clear that host cell responses to pathogenic bacteria can involve either elevated apoptosis or suppression of cell death (144). The body of evidence reveals that P. gingivalis-induced apoptosis is cell type-specific.
Murine infection studies using live whole cells of P. gingivalis found that apoptosis is induced in fibroblasts and that it is modulated by TNF-α (67). Moreover, immortalized human keratinocytes infected with P. gingivalis rapidly exhibit significant levels of apoptosis (79). In order to ascertain the active constituent(s) in P. gingivalis that induces apoptosis, many have not used whole P. gingivalis, but rather different fractions of the bacterium or its culture fluid. Heat-stable molecules secreted in the culture medium of P. gingivalis induced apoptosis in peripheral blood mononuclear cells (63). In addition, studies in T-cells demonstrated that activation of caspase-3 by P. gingivalis culture fluid was not inhibited by protease inhibitors (76). Myocardial cells treated with P. gingivalis culture supernatant became apoptotic and had increased levels of Bad, calcineurin, a calmodulin-dependent serine/threonine phosphatase that is a mediator of cardiomyocyte hypertrophy and apoptosis, and the calcineurin downstream effector nuclear factor of activated T cells-3 (NFAT-3) (110). These results suggested that calcineurin dephosphorylates NFAT-3 and Bad, causing myocyte hypertrophy and triggering apoptosis (110). In addition, in this same system P. gingivalis culture supernatant induced DNA fragmentation and activation of caspases-3, -8, and -9. These effects could be blocked by inhibitors of p38 and ERK, but enhanced by a JNK inhibitor (111), suggesting that P. gingivalis signals through p38 and ERK to initiate apoptosis and that the JNK pathway may counteract this signaling.
One of the metabolites in P. gingivalis culture supernatant is butyric acid which induced calmodulin-sensitive apoptosis in two different B lymphoma cell lines (108). Butyric acid induced caspase-8 and -9 dependent apoptosis in T-cells that is independent of Fas (109). Another feature of the culture supernatant is that it can contain P. gingivalis cell membrane, which has been shown to induce apoptosis in T cells (64). P. gingivalis LPS has been demonstrated to induce apoptosis in the thymus, spleen, and lymph nodes in a mouse model (90). Culture supernatant can also include vesicles that contain both cell membrane and gingipains. In HeLa cells (human epithelial-like cells) treated with microspheres coated with P. gingivalis vesicles, a portion of the cells detached and displayed apoptotic morphology within 6 h, but detachment and cell death were not demonstrated with longer treatment or with gingipain-deficient mutants (88).
The relationship between gingipains and apoptosis has also been intensely studied for more than a decade. The first reports investigated the cytotoxicity of gingipains on different epithelial cell lines and found that culture supernatants with gingipain activity caused the cells to round up and detach from the monolayer concomitant with apoptotic morphology in some cell lines (181). Cultured human gingival fibroblasts also rounded up, detached, and lost cell viability when exposed to outer membrane vesicles that possessed gingipain activity (137). However, low levels of cytotoxicity were induced in peripheral blood mononuclear cells by the gingipains (197).
Studies with gingival fibroblasts and epithelial cells treated with culture supernatants in the presence and absence of leupeptin, which inhibits Rgp activity, revealed that both Rgp and Kgp activities are responsible for cell detachment (93). More specific assays for apoptotic cells established that culture supernatants with gingipain activity were responsible for human gingival fibroblast detachment and apoptosis (223). Our group has also shown that gingipain active extracts induce epithelial cell detachment and apoptosis (34). Using extracts of wild type P. gingivalis, in the presence and absence of Rgp and/or Kgp inhibitors, along with Rgp and/or Kgp mutants, Baba and colleagues reported that both gingipains, especially Rgp, were responsible for cell detachment and loss of viability in human gingival fibroblasts (10) and endothelial cells (11). Our recent work extended these results by using purified gingipains to further elucidate the individual roles of the different gingipains in BCAEC apoptosis (182,183). Despite the ability of HRgpA, Kgp, and RgpB to cleave CAMs to varying degrees, only HRgpA and RgpB were able to induce apoptosis in BCAEC, with RgpB-induced apoptosis occurring slower (182,183). Unexpectedly, BCAEC treated with Kgp, after initially detaching and displaying cellular blebs, were able to re-adhere to the culture surface with no apparent loss of cell viability (182). In addition, detached cells produced by Kgp appear to still adhere to one another, whereas cells detached in the presence of HRgpA or RgpB subsequently died by apoptosis. The slower detachment and apoptosis induced by RgpB is not unexpected and may be explained by the absence of an adhesin domain. The adhesin domain of HRgpA, and most likely Kgp as well, provides a means of attachment to cells (32,33) through integrin β1 (176) that RgpB does not have. Moreover, HRgpA has been shown to be more effective than RgpA (cat) in decreasing the expression of integrin β1 (176) because the adhesion domain enhances the binding of HRgpA to the cell.
N-cadherin signaling exerts an anti-apoptotic function by complexing with pro-caspase-8 which interferes with the recruitment of procaspase-8 to the DISC and initiation of apoptosis (74). Therefore, it is tempting to speculate that cleavage of N-cadherin, and the possible loss of its function, may be integral to the initiation of gingipain-induced apoptosis. In support of this, our results demonstrated that N-cadherin cleavage by HRgpA produced a single cleavage product of 30 kD (Figure 3). However, cleavage by Kgp generated fragments of approximately 65 kD and 30 kD and RgpB did not cleave N-cadherin within the time frame tested (Figure 3). These results suggest that cleavage of N-cadherin is not vital to the apoptotic trigger by gingipains since BCAEC treated with RgpB had no cleaved N-cadherin, but were apoptotic, and BCAEC treated with Kgp had cleaved N-cadherin, but were viable. As stated earlier, in this system Kgp was functional throughout the time course since it was able to cleave integrin β1 at all time points through 24 h. A potential role for N-cadherin in gingipain induced BCAEC apoptosis cannot be completely ruled out solely on the basis of its apparent lack of cleavage in apoptotic cells induced by treatment with RgpB. At 24 h there appeared to be decreased levels of N-cadherin cleavage by Kgp, as evidenced by the disappearance of some of the cleavage products in the 65kD group and the 30kD cleavage product (our data not shown). This suggests that BCAEC are in some way able to protect N-cadherin from cleavage by Kgp activity at later time points in order to counteract the apoptotic signaling and/or stimulate survival signaling. A similar phenomenon was observed in the gingipain-mediated cleavage of TNF-α in fibroblasts, in which HRgpA was shown to have the most activity toward TNF-α, but Kgp-induced cleavage of TNF-α was greatly increased if TNF-α re-expression was prevented, suggesting that the loss of TNF-α through Kgp cleavage could be compensated for by de novo synthesis (128), allowing cells to offset the Kgp-induced effects. Moreover, we have observed that a high concentration of Kgp can cause BCAEC detachment and apoptosis (our unpublished observations) suggesting that there is a threshold level of gingipain activity below which a cell can remain viable, but at gingipain activity above this level the cell cannot overcome gingipain-induced effects and becomes apoptotic. This threshold phenomenon has been demonstrated in epithelial cells treated with HRgpA (175).
Because of the differing abilities of the gingipains to cleave N-cadherin, VE-cadherin and integrin β1, induce cell detachment, and apoptosis, it can be assumed that HRgpA, RgpB, and Kgp have other substrate specificities, which could explain the dramatic divergence in cell survival and death produced by these gingipains. It is conceivable that there may be differences in the ability of the specific gingipains to cleave one or more CAMs that may be pivotal for gingipain-induced apoptosis in BCAEC. Further investigation to define their substrate specificity and the apoptotic pathway components modulated by the purified gingipains will help pinpoint the nature of the Rgp-induced apoptotic trigger.
5.2.1.2. Caspase-independent P. gingivalis –induced apoptosis
The involvement of non-caspase proteases in some cases of apoptosis is suggested by observations that inhibition of caspases generally causes delays, but does not fully block cell death resulting from most apoptotic stimuli (94). While the ability of proteases to implement apoptotic signaling in the absence of caspase activity is not uncommon, our research work is the first to present initial evidence that HRgpA and RgpB can induce a caspase-3 and -7 independent apoptosis (183). The non-caspase proteases most closely associated with apoptosis are cathepsins, calpains, and granzymes (94). Cathepsins are translocated from lysosomes into the cytoplasm during apoptosis, where they intersect with and augment other apoptosis signaling mechanisms (94). Since HRgpA has been shown to rapidly traverse the plasma membrane (175), there is the possibility that the gingipains could interact with apoptotic signaling pathways in a manner similar to the cathepsins. Furthermore, HRgpA enters the nucleus (175) which may provide even more possibilities for HRgpA and possibly Kgp, since catalytic activity was not required for protease translocation, to modulate host cell death and survival pathways.
Calpains are known to cleave many important proteins that are involved in cellular architecture which may be particularly important during apoptosis [reviewed in (94)]. Because of the ability of gingipains to cleave FAK, a substrate of both caspases (54) and calpains (94), paxillin, β-catenin, γ-catenin, p120, integrin β4, Src, and p130Cas (79) and as our work demonstrates other CAMs, such as N-cadherin, VE-cadherin, integrin β1 (182,183), gingipains may have a similar role in apoptosis as calpains. Further similarities reside in the ability of calpains and gingipains to cleave pro-caspases, although the calpain-induced cleavage neither activates nor inactivates caspase-3 and -9 (94), whereas gingipain cleavage does activate procaspase-3 (211). Calpain and caspase-mediated apoptotic pathways can be inter-related (94) which raises a similar possibility for gingipains in light of our discovery of the caspase-independence of gingipain-induced apoptosis (183).
Because of the ability of granzyme A to cleave after lysine and arginine residues (91,124) and its ability to induce caspase-independent cell death (194), it is tempting to make a case that gingipains may behave similar to granzyme A. While granzyme A is the most abundant protease found in the granules of CTL cells it induces apoptosis after prolonged exposure making its role in apoptosis induction far more subtle than that of granzyme B (94). However, treatment of cells with granzyme A and perforin, a CTL granule protein that permeabilizes cell membranes, induces a caspase-independent, rapid accumulation of DNA strand breaks with subsequent nuclear condensation without the cleavage of poly(ADP-ribose) polymerase (PARP) and lamin B (94). However, there appears to be cell-type specific variance in some of the Granzyme A-induced apoptotic features (234). Granzyme A-induced cell death is marked by rapid induction of other apoptotic features, including loss of membrane integrity with blebbing, loss of mitochondrial membrane potential and production of ROS, chromatin condensation, and nuclear fragmentation (18,52,53). Mitochondrial cytochrome c release, oligonucleosomal DNA fragmentation, caspase activation, and Bcl-2 over-expression, all hallmarks of caspase-dependent apoptosis, are absent from granzyme A-induced apoptosis (52). Independent of caspases and Bcl-2, granzyme A cleaves lamins A, B, and C disrupting the nuclear lamina, a critical step in the progression of apoptosis (234). After gaining access to the nucleus (52,53), perhaps through the cleavage of lamins (234), granzyme A rapidly degrades histone H1 and the core histones disturbing the chromatin and facilitating exogenous DNase access to the DNA (235). In comparison, granzyme B/perforin-mediated apoptosis usually involves caspase activation, which granzyme B can initiate by the processing of pro-caspase-3, -7, and -9 to the active forms, and produces cleaved PARP and lamin B (94). However, granzyme A can also induce apoptotic features in a caspase-independent manner (94). A recent report showed that the cleavage of Bid by granzyme B occurs at a residue distinct from the caspase cleavage site. This initiates apoptosis mediated by cytochrome c, SMAC release upstream of caspase activation, and loss of mitochondrial transmembrane potential
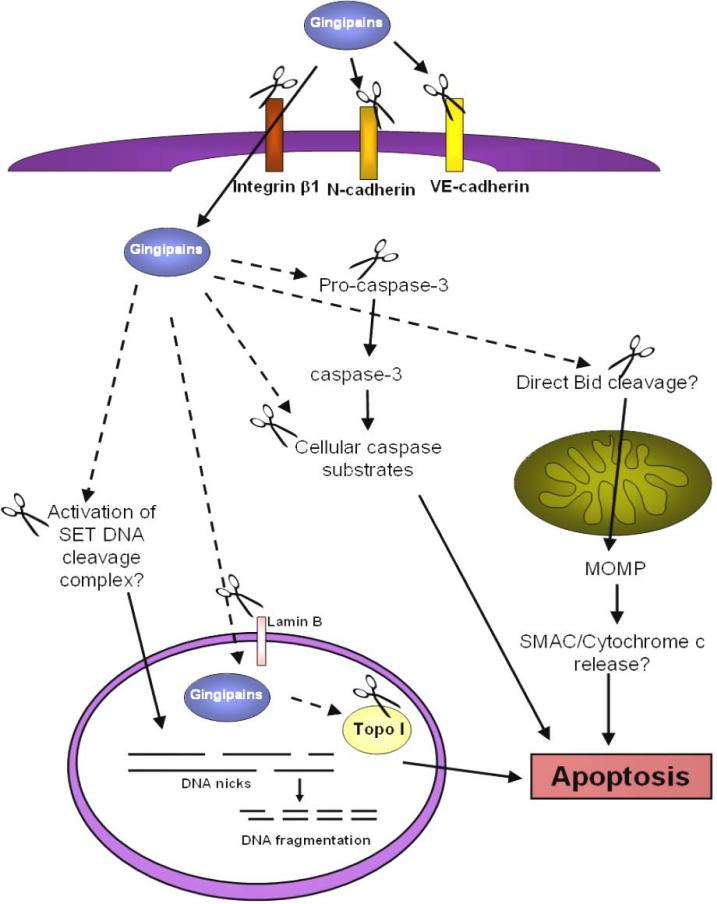
There are several commonalities between granzyme A and gingipains such as cleavage after lysine and arginine residues, translocation to the nucleus, and induction of caspase-independent cell death. However, the apoptotic features induced by gingipains needs to be further characterized. For example, it is unknown whether ssDNA nicks are produced during caspase-independent gingipain-induced apoptosis in BCAEC or if only oligosomal DNA fragmentation is produced in both caspase-dependent and -independent gingipain-induced apoptosis. The morphology of the nucleus and chromatin during both caspase-dependent and -independent gingipain-induced apoptosis in BCAEC needs to be carefully analyzed for more detailed classification of the type of cell death induced. In addition, the effects of gingipains on the mitochondria of BCAEC are completely unknown. It would also be interesting to ascertain what classical apoptotic signaling molecules are activated during both types of apoptosis triggered by gingipains in BCAEC. For example, granzyme A-induced apoptosis is not characterized by Bcl-2 upregulation, since it is caspase-independent (52). However, Bcl-2 can block caspase-independent apoptosis by preventing the release of cytochrome c in some systems (48) and can also prevent granzyme B-mediated apoptosis (208). It is tempting to speculate that Rgp activity could induce caspase-independent apoptosis via proteolytic activation of Bid at Arg65, similar to cathepsins (208), with subsequent cytochrome c release and caspase-9 activation. A recent report demonstrated that Clostridium difficile Toxin-A-induced apoptosis is mediated by the cleavage of Bid prior to the activation of caspase-8, -9, -6, and -3 (28), so a bacterial mediated activation of Bid in the absence of active caspases is not improbable. We have summarized in Figure 4 a potential model of a granzyme-like mechanism for gingipain-induced caspase-independent apoptosis in BCAEC.
Figure 4.
Potential granzyme-like model of gingipain-induced apoptosis. Through adhesion and cleavage of CAMs, most likely integrin β1 (176), gingipains gain access to the host cell cytoplasm. In the cytoplasm gingipains may cleave pro-caspase-3 to the active caspase-3 (211) (as indicated by the scissors) that can cleave other cellular substrates, which may be cleaved by gingipains as well, to produce apoptotic morphology. It remains to be seen if gingipain activity could cleave Bid (208) (as indicated by the dashed arrow) producing MOMP, cytochrome c release, and apoptosis. Similar to granzyme A (19), gingipain activity could activate the SET complex that once inside the nucleus produces DNA nicks and fragmentation and apoptosis. Finally, it is conceivable that the possible degradation of lamin B by gingipain activity provides a means for gingipains to enter the nucleus and interact with nuclear proteins, such as Topo I, or allows escape of nuclear proteins to the cytoplasm where gingipains could cause their cleavage. Modified from (225) and (52).
Gingipain activity may be responsible for caspase-like activity in cells (23) and it appears that gingipains may be able to cleave some of the same cellular substrates as caspases, such as FAK (79), N-cadherin (182,183), Topoisomerase I (Topo I) (182), and Lamin B (our unpublished observations). These results make it tempting to speculate that the gingipains can aid and possibly initiate apoptosis. A recent report described two independent apoptotic mechanisms simultaneously induced by a single stimulus. In the first pathway, there was breakdown of focal adhesion components leading to anoikis, while the second was a direct effect through receptor-mediated signal transduction pathways initiated by a cell adhesion domain (204). Conceivably, this scenario could be occurring during gingipain-induced apoptosis in BCAEC. Gingipains cleave many proteins involved in focal adhesion which can lead to anoikis. Secondly, the cleavage of CAMs by gingipains, such as N-cadherin which is involved in pro-caspase-8 recruitment to the DISC (74), may induce apoptosis concurrently. Additionally, Gingipains can signal a third pathway of cell death that is caspase-independent (183). This pathway is not caused by cell detachment since the broad caspase inhibitor z-VADFMK has been shown to block anoikis (170), and is not mediated by the JNK pathway since active caspases are needed for JNK to respond to the loss of cell-matrix contact (59).
Our studies demonstrated that gingipain-active W83 extracts, HRgpA, and RgpB induce apoptosis with a more pronounced blebbing in the absence of active caspases (182). It will be interesting to determine if death-associated protein kinase (DAP kinase) as well as DAP kinase-related kinase (DRP kinase) are involved in this type of cell death. They are members of a family of calcium/calmodulin-regulated serine/threonine death kinases (72) that have been shown to be involved in membrane blebbing and function independent of caspase activity (238). Other proteins that may be activated by gingipains in the presence of z-VAD-FMK are p21-activated kinase 2 or Rho-activated serine/threonine kinase, which also produce cellular blebbing (113).
Other features of caspase-independent cell death involve loss of mitochondrial membrane potential, cytochrome c and apoptosis-inducing factor (AIF) release from the mitochondria, zeiosis, phosphatidylserine translocation, chromatin cleavage to 50 kD pieces, and chromatin condensation and margination (228). While the nature of caspase-independent gingipain-induced apoptosis in BCAEC needs to be further characterized, there are several proteins that maybe involved in this pathway that merit initial studies. For example, AIF was identified in human coronary artery endothelial cells outside the nucleus in the basal state and translocated to the nucleus upon induction of apoptosis in a caspase-independent manner (236) Moreover, treatment of BCAEC with gingipains may affect mitochondrial function (17), leading to the release AIF and/or endoG, both of which induce apoptotic morphology (89). Another possible protein involved in gingipain-induced caspase-independent signaling pathway could be Bax since it induces cytochrome c release that is not inhibited by z-VAD-FMK (9). P. gingivalis also modulates Bcl-2 levels (144) and Bcl-2 has been shown to block caspase-independent apoptosis via antioxidant effects (22). It will be interesting to compare the modulation of Bcl-2 in gingipain-induced caspase-dependent and -independent apoptosis.
Besides apoptosis-related proteins AIF, Bax and Bcl-2, gingipains may modulate other proteins to activate caspase-independent signaling. Interestingly, Topo I, a substrate for both caspases and cathepsins, was cleaved to a 45 kD fragment in the presence of active gingipains (182,183). A fragment of this size usually appears in caspase-independent, necrotic cell death and is generated by cathepsins (154), however, we did not observe necrosis during gingipain-induced cell death in BCAEC. This suggests that gingipains might be able to directly cleave Topo I. This pattern of Topo I cleavage was observed in BCAEC cells exposed to purified gingipains or gingipain-active W83 extracts in the presence and absence of selective inhibitors of Rgp and Kgp (183), suggesting that both Rgp and Kgp can cleave Topo I to varying degrees. However, Topo I cleavage may not be relevant to gingipain-induced apoptosis since endothelial cells treated with Kgp that are attached and viable also display this 45 kD cleavage fragment.
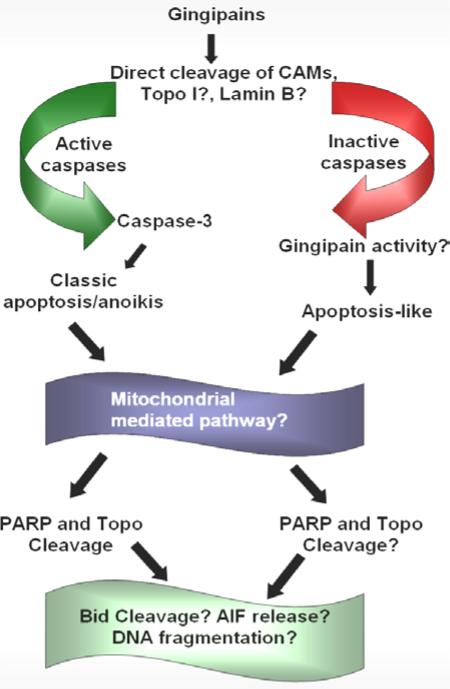
Lamins are major structural proteins of the nuclear envelope (37) that are cleaved by caspases to induce chromatin condensation (207) and possibly disrupt lamin-lamin interactions, as well as, interactions of lamins with other nuclear components (37). Cleavage of lamins also results in nuclear shrinkage (113). HRgpA localizes to the nucleus (175) and may be able to cleave lamin B and subsequently gain access to the nucleus to cleave Topo I and possibly modulate other nuclear proteins, producing the nuclear apoptotic morphology. Kgp may function similarly to and work with HRgpA, since its nuclear targeting ability was not dependent on proteolytic activity (175). Figure 5 is a comparison of potential caspase-independent apoptosis pathway members induced by gingipain activity.
Figure 5.
Comparison of potential caspase-dependent and -independent apoptosis pathway members induced by gingipain activity. Treatment of endothelial cells with gingipains causes the cleavage of CAMs and possibly Topo I and lamin B. In the presence of active caspases, this produces classically defined apoptosis and/or anoikis that is most likely mediated by the mitochondrial pathway of apoptosis (211). Apoptosis triggers the cleavage of PARP and Topo I. Other proteins that have yet to have their role defined in gingipain-induced apoptosis are Bid and AIF. In addition, it is unknown at this time whether DNA cleavage is a significant feature of gingipain-induced caspase-dependent apoptosis. Alternatively, the cleavage of CAMs and possibly Topo I and lamin B in the absence of active caspases, may provide the opportunity for gingipain-activity to initiate apoptosis that may also be mitochondrial in nature. The roles of PARP and Topo I cleavage in caspase-independent apoptosis, as well as, Bid, AIF, and DNA fragmentation need to be clarified.
5.3. P. gingivalis-induced resistance to apoptosis
Despite several reports of the ability of P. gingivalis whole cells, culture supernatants, membranes, and gingipains to induce apoptosis, there is also data indicating that P. gingivalis inhibits apoptosis in some cell types. Furthermore, there is evidence that P. gingivalis infection may initiate divergent survival and death pathways (36). P. gingivalis fimbriae inhibited growth factor deprivation-induced apoptosis via ERK-dependent expression of p21 in a human monocytic cell line (153). In addition, it has been demonstrated that P. gingivalis LPS can prevent apoptosis of neutrophils derived from a human promyelocytic cell line, which may prolong an acute inflammatory response resulting in an increased potential for tissue destruction (140). Experimental results of Nakhjiri, et al., suggested that the first response to P. gingivalis infection in primary gingival epithelial cells was apoptosis; however, over time P. gingivalis initiated anti-apoptotic signaling via increased levels of Bcl-2 and decreased levels of Bax that canceled the cells’ death response and was even able to promote cell survival in the presence of the apoptotic inducer camptothecin (144). Furthermore, inhibition of the PI3-K/Akt pathway abolished the P. gingivalis-induced protection from apoptosis (230). We believe that these studies demonstrated that apoptosis was induced first since a reversible phosphatidylserine exposure triggered by P. gingivalis invasion could be inhibited by z-VAD-FMK (230), suggesting that caspases are activated but the anti-apoptotic signaling overrides the apoptotic signaling. Yilmaz, et al proposed that P. gingivalis inhibited mitochondrial-dependent apoptosis in gingival epithelial cells in order to maintain its intracellular lifestyle long enough to allow the successful spread of the bacteria to adjacent cells (231) and deeper host tissues (5,230). In contrast, another report found that heat-killed P. gingivalis induced caspase-3 and -8 activation and apoptosis in gingival epithelial cells through FasL-mediated apoptosis that was mediated by nuclear factor-κB (NF-κB) and Toll-like receptor-2 (TLR-2) (25). Brozovic, et al surmised that NF-κB activation was likely to be both anti-apoptotic and pro-apoptotic, depending on the stimulus, such as variances in bacterial concentrations, and the specific cell type involved (25).
Evidence is emerging that P. gingivalis can modulate several different monocytic cell survival mechanisms in order to evade host defenses such as, signal transducer and activator of transcription 6 (STAT6), which permits nitric oxide synthesis in macrophages; interferon-dependent positive-acting transcription factor (STAT1), which is essential in caspase-independent cell death of activated macrophages; double stranded RNA (dsRNA) -activated protein kinase (PRKR), which may be important in the anti-apoptotic signaling by P. gingivalis; calreticulin precursor (CALR), which binds complement C1q modulating the host immune response, and gene associated with retinoid-interferon induced mortality (GRIM19) (237). GRIM19 appears to be a nuclear protein that is expressed in response to interferon and retinoic acid, induces cell death (7), and is involved in pathogen recognition and invasion (13). Moreover, in human immortalized gingival keratinocytes P. gingivalis modulated 25 apoptosis-associated genes linked to p53 and the mitochondrial pathway of apoptosis (75). A recent study defined the anti-apoptotic pathways mediated by P. gingivalis in response to camptothecin treatment. It was demonstrated that P. gingivalis transiently activates the JAK1/Stat3 pathways with Akt pathway modulation also being involved. These pathways up-regulate Survivin, at the mRNA level, and ultimately block activation of caspase-3 (122). Considering the many diverse signaling pathways engaged in apoptosis, it is not surprising that some bacteria inhibit or delay apoptosis while others induce it (230).
Interestingly, P. gingivalis can recognize different host cell types and is capable of targeting specific and distinct signaling pathways (6). For example, P. gingivalis has been shown to be uniquely capable of selectively activating one MAPK pathway and down-regulating another which may cause responses of epithelial cells to be species and even strain specific (224). In addition, the effects that are produced in a particular host cell may be specific to the infecting bacterial strain. Out of 15 clinical isolates and a reference American Type Culture Collection (ATCC) strain 33277 of P. gingivalis only one strain was cytotoxic to KB epithelial cells (49). Interestingly, the strains used in this study had a higher lysine-specific cysteine protease activity than the reference strain and there was no decrease in arginine-specific activity in strains that did not possess an rgpB gene (49). These results suggest that not only may the effects triggered be dependent on the host cell, but may also be related to the strain of P. gingivalis used, especially since our group has shown that gingipain-active extracts from strain W83 induced apoptosis in KB cells (34). P. gingivalis has also exhibited the ability to modulate the NF-κB pathway differently during the course of infection. In gingival fibroblasts early NF-κB activation occurred with a later decrease in activity via a PI3-K mediated pathway that appeared not to be mediated by the gingipains (34,211). It may be possible that the anti-apoptotic effects of P. gingivalis infection in some cell types signal initially through NF-κB similar to Rickettsia rickettsii, whose primary target is vascular endothelial cells (227).
The recent findings that fimbriae from P. gingivalis modulated both pro-apoptotic and anti-apoptotic genes in human aortic endothelial cells (36) and in gingival fibroblasts (211) confirmed that P. gingivalis could induce either apoptosis or resistance to apoptosis. It appears that the pathway that prevails in a given cell type depends on the balance of cell death and survival modulators that are present. These results raise some questions. In cells in which P. gingivalis can induce apoptosis, is this because of direct pro-apoptotic signaling by P. gingivalis or because of a specific survival signal that is absent in those cells and is essential to prevent P. gingivalis-induced apoptosis? For instance, a recent report demonstrated that the same apoptotic stimulus induced an intrinsic apoptosis that was caspase-dependent in one cell type and caspase-independent in a different cell type (89), implying cellular differences and not apoptotic stimulus differences. In cells that survive despite P. gingivalis infection, is this because P. gingivalis is overriding the host cell's apoptotic reflex or because the cell is ‘ambivalent’ to the presence of the invading bacterium? It has been proposed that cells respond to changes that bacteria cause by activating the apoptotic mechanism, although they are not acting primarily to cause apoptosis (218).
There is overwhelming evidence that host cell responses to pathogenic bacteria can involve either apoptosis induction or suppression of cell death (144) and that host cells appear to be able to distinguish between infecting organisms and their components (237). Recently, it has been proposed that the possibly finite number of pathways that the pathogen may have evolved to modulate could be characteristic of the organism's genus (75). This suggests that the outcome of infection may be more determined by the presence or absence of the host cells’ repertoire of defensive molecules than by components of the pathogen's virulence arsenal. A review of the literature indicates that P. gingivalis induces apoptosis in only some cell types. The molecules involved in triggering apoptosis by P. gingivalis are only beginning to be elucidated. It will be interesting to determine if the same molecules are involved in both gingipain-induced apoptosis and gingipain-induced resistance to apoptosis and to compare the morphological characteristics and pathways of caspase-dependent and -independent apoptosis mediated by the gingipains.
5.4. The purpose of pathogen-induced effects on host cell survival
Numerous microbes are known to affect host cell survival; some trigger apoptosis while others provide increased cell survival in host cells [reviewed in (61,62,66,138,227)]. Overall, there appears to be two general mechanisms for Gram-negative pathogen-induced cell death. In the first, effector proteins delivered by a Type III secretion system activate caspase-1 producing both features of apoptosis and necrosis. In the second mechanism, activation of TLR-4 by LPS triggers apoptosis (73).
Apoptosis induced in any host cell type by bacteria acting external to the cell is likely to be a pathogenic mechanism for promoting invasiveness (38). It would be beneficial to the bacteria to induce apoptosis of the host cell in order to evade the immune system by eliminating key defense cells like lymphocytes, monocytes, and neutrophils (227); however, cell death may also be a protective mechanism signaled by the host cell to induce inflammation through caspase-1 activation (61,62), clear the pathogen, and limit intracellular replication (38). For example, apoptosis of lung epithelial cells upon infection with Pseudomonas aeruginosa constitutes a pivotal part of host defense against P. aeruginosa infections. However, this apoptosis crucially contributes to the host defense against the bacteria (66). It is also possible that apoptosis contributes to the spread of intracellular bacteria, as appears to be the case for Salmonella-induced apoptosis of macrophages (62,135) through successive rounds of intracellular growth, apoptosis, and phagocytosis of infected apoptotic bodies, perpetuating the infection and ensuring cell to cell spread (73). In addition, apoptosis may play a role in bacterial trafficking into a niche permissive for intracellular replication (62), as in the case of Shigella flexneri, which kills macrophages by apoptosis and thereby gains access to Peyer patches, permitting the bacteria to spread and to infect enterocytes from the basolateral side (66). A recent report speculated that P. gingivalis invaded HeLa cells and did not cause cell death to acquire intracellular persistence, but if the invasion is rather quick there was probably not enough time for complete P. gingivalis-induced anti-apoptotic signaling to initiate before pro-apoptotic signals induced some apoptotic morphology (88). However, it has also been suggested that an initial delay of apoptosis would allow P. gingivalis extra time to replicate intracellularly to a higher yield while providing protection from the host immune system before inducing apoptosis for bacterial spread to other cells (211). Anoikis could be triggered to prevent the lateral spread of bacteria and aid in the removal of the infected host cell (185). Collectively, these findings suggest that there is no universal function for pathogen regulated cell death (138).
Host cells infected by some pathogens may actually have increased cell survival that aids in pathogenesis. Inhibiting host cell apoptosis would provide a safe haven for an intracellular pathogen (227) and help to maintain the metabolic activity of the infected cell (62). For example, inhibiting host cell death facilitates the intracellular development of Toxoplasma gondii, which increases parasitemia and may lead to an enhanced inflammatory response to the parasite resulting in host mortality (120). Experimental evidence suggests that T. gondii inhibits apoptosis of host cells by different mechanisms, both direct inhibition of infected cells and indirect mechanisms, to protect both infected and uninfected host cells (120). Direct mechanisms interfere with caspase cascade activation, increase levels of anti-apoptotic Bcl-2 family members, increase inhibitor of apoptosis proteins (IAPs), and decrease levels of PARP that may inhibit apoptosis in a caspase-independent fashion (120). Remarkably, a single viable parasite was sufficient to block apoptosis of the host cell and this inhibition was reversed when the parasite was killed (120). Some pathogens signal both cell death and cell survival. For example, Mycobacterium tuberculosis induces apoptosis in macrophages via a TNF-α and caspase-1 dependent pathway, but also protects cells from apoptosis via NF-κB and neutralization of TNF-α pro-apoptotic activity (62). Chronic infection with Helicobacter pylori is associated with gastric cancer suggesting that this bacterium would induce apoptosis resistance in gastric cells. However, it induces apoptosis in the majority of gastric cells, but a small percentage of cells are resistant to H. pylori-induced apoptosis, as well as, other apoptotic stimuli (187). The bacterial-related factors responsible for this effect are unclear. Furthermore, Chlamydia induces resistance to apoptosis during the initial stages of infection and then later induces apoptosis in host cells (227). This indicates that intracellular bacteria can fine tune the balance between anti- and pro-apoptotic activities even at discrete stages of infection (62), with the ultimate goal to facilitate the spread of infection.
6. SUMMARY
In order for P. gingivalis to survive and grow in the host, it must be able to obtain nutrients and overcome lethal factors, such as oxidative stress which may result from the host's immune response. The gingipains of P. gingivalis are vital for the survival of the bacterium. They degrade host tissue to provide a source of nutrients and through degradation of cell adhesion molecules, induce apoptosis and access deeper tissue layers evading some aspects of the host immune system. Gingipains are involved in the accumulation of heme on the bacterial surface creating a shield from ROS, as well as, interacting with other gene products that are involved in oxidative stress resistance.
In summary, based on our observations, we can postulate a tentative model, which may be part of the mechanism for P. gingivalis virulence. Central to this model are the gingipains and the regulatory role of the vim gene products. 1) The accumulation of the earliest colonizers on the tooth surface initiates an inflammatory reaction known as gingivitis. 2) As the ecological succession continues, conditions in the subgingival area will facilitate the colonization of the anaerobes, in particular, P. gingivalis. Gingipains and the expression of the genes in the recA region in this organism are an important strategy in an environment with increasing inflammation and neutrophil infiltration. These gene products are required to facilitate survival of P. gingivalis in this oxidative stress environment. 3) The recA gene product is involved in repair of DNA damage while the products of the vim genes facilitate the appropriate production and cell-associated localization of the gingipains. The cell-associated localization of the gingipains may be important because protection from oxidative damage may also be facilitated by surface accumulation of hemin which will form μ-oxo dimers in the presence of oxygen and its toxic reactive derivatives (190,193). P. gingivalis gingipains are known to be associated with heme accumulation [reviewed in (152)]. 4) Survival in the oxidative environment requires that P. gingivalis also utilize a combination of other mechanisms for growth and proliferation. Other mechanisms utilized by the bacterium such as Hydrogen peroxide reductase and Bcp can guard against oxidative damage which would curtail the growth of P. gingivalis. Gingipain-associated degradation of cell-cell adhesion proteins (cadherins and integrin) on epithelial and endothelial cells resulting in cell death will compromise tissue integrity. Increased tissue permeability will facilitate access of P. gingivalis to the vascular system, giving rise to other systemic effects (e.g. cardiovascular disease) potentially providing a more hospitable niche. Degradation of host tissues also provides nutrients for the organism. Since the mouth reflects the health of the whole body, as suggested by the Surgeon General (212), preventing gingipain activity in the oral cavity and, thus, inhibiting the key mediator of survival for P. gingivalis may be a means to greatly improve the overall health status.
ACKNOWLEDGMENTS
This work was supported by Loma Linda University, Schools of Medicine and Dentistry and by Public Health Service grants DE13664 from the National Institute of Dental and Craniofacial Research (to H.M.F).
REFERENCES
- 1.Abaibou H, Chen Z, Olango GJ, Liu Y, Edwards J, Fletcher HM. vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect Immun. 2001;69(1):325–335. doi: 10.1128/IAI.69.1.325-335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano A, Ishimoto T, Tamagawa H, Shizukuishi S. Role of superoxide dismutase in resistance of Porphyromonas gingivalis to killing by polymorphonuclear leukocytes. Infect Immun. 1992;60(2):712–714. doi: 10.1128/iai.60.2.712-714.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Kuboniwa M, Kataoka K, Tazaki K, Inoshita E, Nagata H, Tamagawa H, Shizukuishi S. Binding of hemoglobin by Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;134(1):63–67. doi: 10.1111/j.1574-6968.1995.tb07915.x. [DOI] [PubMed] [Google Scholar]
- 4.Amano A, Tamagawa H, Shizukuishi S, Tsunemitsu A. Superoxide dismutase, catalase and peroxidases in oral anaerobic bacteria. J Osaka Univ Dent Sch. 1986;26:187–192. [PubMed] [Google Scholar]
- 5.Andrian E, Grenier D, Rouabhia M. In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infect Immun. 2004;72(8):4689–4698. doi: 10.1128/IAI.72.8.4689-4698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrian E, Grenier D, Rouabhia M. Porphyromonas gingivalis-Epithelial Cell Interactions in Periodontitis. J Dent Res. 2006;85(5):392–403. doi: 10.1177/154405910608500502. [DOI] [PubMed] [Google Scholar]
- 7.Angell JE, Lindner DJ, Shapiro PS, Hofmann ER, Kalvakolanu DV. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J Biol Chem. 2000;275(43):33416–33426. doi: 10.1074/jbc.M003929200. [DOI] [PubMed] [Google Scholar]
- 8.Araki M, Hiratsuka K, Kiyama-Kishikawa M, Abiko Y. Monitoring of dnaK gene expression in Porphyromonas gingivalis by oxygen stress using DNA microarray. J Oral Sci. 2004;46(2):93–100. doi: 10.2334/josnusd.46.93. [DOI] [PubMed] [Google Scholar]
- 9.Arnoult D, Karbowski M, Youle RJ. Caspase inhibition prevents the mitochondrial release of apoptosis-inducing factor. Cell Death Differ. 2003;10(7):845–849. doi: 10.1038/sj.cdd.4401240. [DOI] [PubMed] [Google Scholar]
- 10.Baba A, Abe N, Kadowaki T, Nakanishi H, Ohishi M, Asao T, Yamamoto K. Arg-gingipain is responsible for the degradation of cell adhesion molecules of human gingival fibroblasts and their death induced by Porphyromonas gingivalis. Biol Chem. 2001;382(5):817–824. doi: 10.1515/BC.2001.099. [DOI] [PubMed] [Google Scholar]
- 11.Baba A, Kadowaki T, Asao T, Yamamoto K. Roles for Arg- and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol Chem. 2002;383(7-8):1223–1230. doi: 10.1515/BC.2002.135. [DOI] [PubMed] [Google Scholar]
- 12.Baillon ML, van Vliet AH, Ketley JM, Constantinidou C, Penn CW. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J Bacteriol. 1999;181(16):4798–4804. doi: 10.1128/jb.181.16.4798-4804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnich N, Hisamatsu T, Aguirre JE, Xavier R, Reinecker HC, Podolsky DK. GRIM-19 interacts with nucleotide oligomerization domain 2 and serves as downstream effector of anti-bacterial function in intestinal epithelial cells. J Biol Chem. 2005;280(19):19021–19026. doi: 10.1074/jbc.M413776200. [DOI] [PubMed] [Google Scholar]
- 14.Bate C, Williams A. Role of glycosylphosphatidylinositols in the activation of phospholipase A2 and the neurotoxicity of prions. J Gen Virol. 2004;85(Pt 12):3797–3804. doi: 10.1099/vir.0.80366-0. [DOI] [PubMed] [Google Scholar]
- 15.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241(2):193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 16.Beaman L, Beaman BL. The role of oxygen and its derivatives in microbial pathogenesis and host defense. Annu Rev Microbiol. 1984;38:27–48. doi: 10.1146/annurev.mi.38.100184.000331. [DOI] [PubMed] [Google Scholar]
- 17.Belton CM, Goodwin PC, Fatherazi S, Schubert MM, Lamont RJ, Izutsu KT. Calcium oscillations in gingival epithelial cells infected with Porphyromonas gingivalis. Microbes Infect. 2004;6(5):440–447. doi: 10.1016/j.micinf.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Beresford PJ, Xia Z, Greenberg AH, Lieberman J. Granzyme A loading induces rapid cytolysis and a novel form of DNA damage independently of caspase activation. Immunity. 1999;10(5):585–594. doi: 10.1016/s1074-7613(00)80058-8. [DOI] [PubMed] [Google Scholar]
- 19.Beresford PJ, Zhang D, Oh DY, Fan Z, Greer EL, Russo ML, Jaju M, Lieberman J. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276(46):43285–43293. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- 20.Bhogal PS, Slakeski N, Reynolds EC. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143(Pt 7):2485–2495. doi: 10.1099/00221287-143-7-2485. [DOI] [PubMed] [Google Scholar]
- 21.Boiteux S, Radicella JP. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie. 1999;81(1-2):59–67. doi: 10.1016/s0300-9084(99)80039-x. [DOI] [PubMed] [Google Scholar]
- 22.Borner C, Monney L. Apoptosis without caspases: an inefficient molecular guillotine? Cell Death Differ. 1999;6(6):497–507. doi: 10.1038/sj.cdd.4400525. [DOI] [PubMed] [Google Scholar]
- 23.Boyce M, Degterev A, Yuan J. Caspases: an ancient cellular sword of Damocles. Cell Death Differ. 2004;11(1):29–37. doi: 10.1038/sj.cdd.4401339. [DOI] [PubMed] [Google Scholar]
- 24.Brochu V, Grenier D, Nakayama K, Mayrand D. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol Immunol. 2001;16(2):79–87. doi: 10.1034/j.1399-302x.2001.016002079.x. [DOI] [PubMed] [Google Scholar]
- 25.Brozovic S, Sahoo R, Barve S, Shiba H, Uriarte S, Blumberg RS, Kinane DF. Porphyromonas gingivalis enhances FasL expression via up-regulation of NFkappaB-mediated gene transcription and induces apoptotic cell death in human gingival epithelial cells. Microbiology. 2006;152(Pt 3):797–806. doi: 10.1099/mic.0.28472-0. [DOI] [PubMed] [Google Scholar]
- 26.Campbell JA, Davies GJ, Bulone, V V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1998;329(Pt 3):719. doi: 10.1042/bj3290719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canakci CF, Cicek Y, Canakci V. Reactive oxygen species and human inflammatory periodontal diseases. Biochemistry (Mosc ) 2005;70(6):619–628. doi: 10.1007/s10541-005-0161-9. [DOI] [PubMed] [Google Scholar]
- 28.Carneiro BA, Fujii J, Brito GA, Alcantara C, Oria RB, Lima AA, Obrig T, Guerrant RL. Caspase and bid involvement in Clostridium difficile toxin A-induced apoptosis and modulation of toxin A effects by glutamine and alanyl-glutamine in vivo and in vitro. Infect Immun. 2006;74(1):81–87. doi: 10.1128/IAI.74.1.81-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapple IL, Matthews JB. The role of reactive oxygen and antioxidant species in periodontal tissue destruction. Periodontol 2000. 2007;43:160–232. doi: 10.1111/j.1600-0757.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 30.Chapple ILC. Role of free radicals and antioxidants in the pathogenesis of the inflammatory periodontal diseases. J Clin Pathol Clin Mol Pathol. 1996;49(5):M247–M255. doi: 10.1136/mp.49.5.m247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T, Dong H, Yong R, Duncan MJ. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb Pathog. 2000;28(4):235–247. doi: 10.1006/mpat.1999.0338. [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Duncan MJ. Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells. Microb Pathog. 2004;36(4):205–209. doi: 10.1016/j.micpath.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Chen T, Nakayama K, Belliveau L, Duncan MJ. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect Immun. 2001;69(5):3048–3056. doi: 10.1128/IAI.69.5.3048-3056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Casiano CA, Fletcher HM. Protease-active extracellular protein preparations from Porphyromonas gingivalis W83 induce N-cadherin proteolysis, loss of cell adhesion and apoptosis in human epithelial cells. J Periodontol. 2001;72:641–650. doi: 10.1902/jop.2001.72.5.641. [DOI] [PubMed] [Google Scholar]
- 35.Choi JI, Takahashi N, Kato T, Kuramitsu HK. Isolation, expression, and nucleotide sequence of the sod gene from Porphyromonas gingivalis. Infect Immun. 1991;59(4):1564–1566. doi: 10.1128/iai.59.4.1564-1566.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou HH, Yumoto H, Davey M, Takahashi Y, Miyamoto T, Gibson FC, III, Genco CA. Porphyromonas gingivalis fimbria-dependent activation of inflammatory genes in human aortic endothelial cells. Infect Immun. 2005;73(9):5367–5378. doi: 10.1128/IAI.73.9.5367-5378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326(Pt 1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colino J, Snapper CM. Two distinct mechanisms for induction of dendritic cell apoptosis in response to intact Streptococcus pneumoniae. J Immunol. 2003;171(5):2354–2365. doi: 10.4049/jimmunol.171.5.2354. [DOI] [PubMed] [Google Scholar]
- 39.Corcoran ML, Kleiner DE, Jr., Stetler-Stevenson WG. Regulation of matrix metalloproteinases during extracellular matrix turnover. Adv Exp Med Biol. 1995;385:151–159. doi: 10.1007/978-1-4899-1585-6_18. [DOI] [PubMed] [Google Scholar]
- 40.Curtis MA, Kuramitsu HK, Lantz M, Macrina FL, Nakayama K, Potempa J, Reynolds EC, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodont Res. 1999;34(8):464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 41.Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, Paramonov N, Hounsell EF. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect Immun. 1999;67(8):3816–3823. doi: 10.1128/iai.67.8.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dashper SG, Cross KJ, Slakeski N, Lissel P, Aulakh P, Moore C, Reynolds EC. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19(1):50–56. doi: 10.1046/j.0902-0055.2003.00113.x. [DOI] [PubMed] [Google Scholar]
- 43.Demmer RT, Desvarieux M. Periodontal infections and cardiovascular disease: the heart of the matter. J Am Dent Assoc. 2006;137(Suppl):14S–20S. doi: 10.14219/jada.archive.2006.0402. [DOI] [PubMed] [Google Scholar]
- 44.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 45.Diaz PI, Rogers AH. The effect of oxygen on the growth and physiology of Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19(2):88–94. doi: 10.1046/j.0902-0055.2003.00121.x. [DOI] [PubMed] [Google Scholar]
- 46.Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol. 2006;188(7):2454–2462. doi: 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz PI, Zilm PS, Wasinger V, Corthals GL, Rogers AH. Studies on NADH oxidase and alkyl hydroperoxide reductase produced by Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19(3):137–143. doi: 10.1111/j.0902-0055.2004.00120.x. [DOI] [PubMed] [Google Scholar]
- 48.Egger L, Schneider J, Rheme C, Tapernoux M, Hacki J, Borner C. Serine proteases mediate apoptosis-like cell death and phagocytosis under caspase-inhibiting conditions. Cell Death Differ. 2003;10(10):1188–1203. doi: 10.1038/sj.cdd.4401288. [DOI] [PubMed] [Google Scholar]
- 49.Eick S, Rodel J, Einax JW, Pfister W. Interaction of Porphyromonas gingivalis with KB cells: comparison of different clinical isolates. Oral Microbiol Immunol. 2002;17(4):201–208. doi: 10.1034/j.1399-302x.2002.170401.x. [DOI] [PubMed] [Google Scholar]
- 50.Eley BM, Cox SW. Proteolytic and hydrolytic enzymes from putative periodontal pathogens: characterization, molecular genetics, effects on host defenses and tissues and detection in gingival crevice fluid. Periodontol 2000. 2003;31:105–124. doi: 10.1034/j.1600-0757.2003.03107.x. [DOI] [PubMed] [Google Scholar]
- 51.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol. 1999;65(10):4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112(5):659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 53.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4(2):145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 54.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10(1):76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fletcher HM, Morgan RM, Macrina FL. Nucleotide sequence of Porphyromonas gingivalis W83 recA homolog and construction of a recA-deficient mutant. Infect Immun. 1997;65(11):4592–4597. doi: 10.1128/iai.65.11.4592-4597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forng RY, Champagne C, Simpson W, Genco CA. Environmental cues and gene expression in Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Dis. 2000;6(6):351–365. doi: 10.1111/j.1601-0825.2000.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 57.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124(4):619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13(5):555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 59.Frisch SM, Vuori K, Kelaita D, Sicks S. A role for Jun-N-terminal kinase in anoikis; suppression by bcl-2 and crmA. J Cell Biol. 1996;135(5):1377–1382. doi: 10.1083/jcb.135.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallagher A, Aduse-Opoku J, Rangarajan M, Slaney JM, Curtis MA. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr Protein Pept Sci. 2003;4(6):427–441. doi: 10.2174/1389203033486974. [DOI] [PubMed] [Google Scholar]
- 61.Gao L, Abu KY. Hijacking of apoptotic pathwaysby bacterial pathogens. Microbes Infect. 2000;2(14):1705–1719. doi: 10.1016/s1286-4579(00)01326-5. [DOI] [PubMed] [Google Scholar]
- 62.Gao LY, Kwaik YA. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 2000;8(7):306–313. doi: 10.1016/s0966-842x(00)01784-4. [DOI] [PubMed] [Google Scholar]
- 63.Geatch DR, Harris JI, Heasman PA, Taylor JJ. In vitro studies of lymphocyte apoptosis induced by the periodontal pathogen Porphyromonas gingivalis. J Periodont Res. 1999;34(2):70–78. doi: 10.1111/j.1600-0765.1999.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 64.Gemmell E, Prajaneh S, Grieco DA, Taylor JJ, Seymour GJ. Apoptosis in Porphyromonas gingivalis-specific T-cell lines. Oral Microbiol Immunol. 1999;14(6):331–338. doi: 10.1034/j.1399-302x.1999.140601.x. [DOI] [PubMed] [Google Scholar]
- 65.Genco CA, Dixon DW. Emerging strategies in microbial haem capture. Mol Microbiol. 2001;39(1):1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 66.Grassme H, Jendrossek V, Gulbins E. Molecular mechanisms of bacteria induced apoptosis. Apoptosis. 2001;6(6):441–445. doi: 10.1023/a:1012485506972. [DOI] [PubMed] [Google Scholar]
- 67.Graves DT, Oskoui M, Volejnikova S, Naguib G, Cai S, Desta T, Kakouras A, Jiang Y. Tumor necrosis factor modulates fibroblast apoptosis, PMN recruitment, and osteoclast formation in response to P. gingivalis infection. J Dent Res. 2001;80(10):1875–1879. doi: 10.1177/00220345010800100301. [DOI] [PubMed] [Google Scholar]
- 68.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9(7):246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 69.Gros L, Saparbaev MK, Laval J. Enzymology of the repair of free radicals-induced DNA damage. Oncogene. 2002;21(58):8905–8925. doi: 10.1038/sj.onc.1206005. [DOI] [PubMed] [Google Scholar]
- 70.Grossmann J. Molecular mechanisms of “detachment-induced apoptosis--Anoikis”. Apoptosis. 2002;7(3):247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- 71.Gu J, Taniguchi N. Regulation of integrin functions by N-glycans. Glycoconj J. 2004;21(1-2):9–15. doi: 10.1023/B:GLYC.0000043741.47559.30. [DOI] [PubMed] [Google Scholar]
- 72.Guimaraes CA, Linden R. Programmed cell deaths. Apoptosis and alternative deathstyles. Eur J Biochem. 2004;271(9):1638–1650. doi: 10.1111/j.1432-1033.2004.04084.x. [DOI] [PubMed] [Google Scholar]
- 73.Guiney DG. The role of host cell death in Salmonella infections. Curr Top Microbiol Immunol. 2005;289:131–150. doi: 10.1007/3-540-27320-4_6. [DOI] [PubMed] [Google Scholar]
- 74.Gwak GY, Yoon JH, Yu SJ, Park SC, Jang JJ, Lee KB, Lee SH, Lee SM, Shin CS, Suh KS, Lee HS. Anti-apoptotic N-cadherin signaling and its prognostic implication in human hepatocellular carcinomas. Oncol Rep. 2006;15(5):1117–1123. [PubMed] [Google Scholar]
- 75.Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, Narasimhan G, Baker HV, Lamont RJ. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005;7(6):811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 76.Harris JI, Russell RRB, Curtis MA, Aduse-Opoku J, Taylor JJ. Molecular mediators of Porphyromonas gingivalis-induced T-cell apoptosis. Oral Microbiol Immunol. 2002;17(4):224–230. doi: 10.1034/j.1399-302x.2002.170404.x. [DOI] [PubMed] [Google Scholar]
- 77.He J, Miyazaki H, Anaya C, Yu F, Yeudall WA, Lewis JP. Role of Porphyromonas gingivalis FeoB2 in metal uptake and oxidative stress protection. Infect Immun. 2006;74(7):4214–4223. doi: 10.1128/IAI.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henry L, Johnson NA, Fletcher HM. The role of uvrB in UV sensitvity and oxidative stress in Porphyromonas gingivalis W83. Journal of Dental Research 86 (Spec Iss A) Abstract# 1819 IADR 85th General Session, New Orleans, LA. 2007;86(Special Issue A) [Google Scholar]
- 79.Hintermann E, Haake SK, Christen U, Sharabi A, Quaranta V. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect Immun. 2002;70(10):5846–5856. doi: 10.1128/IAI.70.10.5846-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 81.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 82.Hosogi Y, Duncan MJ. Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect Immun. 2005;73(4):2327–2335. doi: 10.1128/IAI.73.4.2327-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang GT, Kim D, Lee JK, Kuramitsu HK, Haake SK. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect Immun. 2001;69(3):1364–1372. doi: 10.1128/IAI.69.3.1364-1372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ilic D, Almeida EA, Schlaepfer DD, Dazin P, Aizawa S, Damsky CH. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J Cell Biol. 1998;143(2):547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74(1):111–118. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- 86.Imlay JA. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol. 2002;46:111–153. doi: 10.1016/s0065-2911(02)46003-1. [DOI] [PubMed] [Google Scholar]
- 87.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240(4857):1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 88.Inaba H, Kawai S, Kato T, Nakagawa I, Amano A. Association between epithelial cell death and invasion by microspheres conjugated to Porphyromonas gingivalis vesicles with different types of fimbriae. Infect Immun. 2006;74(1):734–739. doi: 10.1128/IAI.74.1.734-739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inayat-Hussain SH, Ross D. Intrinsic pathway of hydroquinone induced apoptosis occurs via both caspase-dependent and caspase-independent mechanisms. Chem Res Toxicol. 2005;18(3):420–427. doi: 10.1021/tx049762o. [DOI] [PubMed] [Google Scholar]
- 90.Isogai E, Isogal H, Kimura K, Fujii N, Takagi S, Hirose K, Hayashi M. In vivo induction of apoptosis and immune responses in mice by administration of lipopolysaccharide from Porphyromonas gingivalis. Infect Immun. 1996;64(4):1461–1466. doi: 10.1128/iai.64.4.1461-1466.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jaattela M. Programmed cell death: many ways for cells to die decently. Ann Med. 2002;34(6):480–488. doi: 10.1080/078538902321012423. [DOI] [PubMed] [Google Scholar]
- 92.Jeong W, Cha MK, Kim IH. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol- specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275(4):2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- 93.Johansson A, Kalfas S. Characterization of the proteinase-dependent cytotoxicity of Porphyromonas gingivalis. Eur J Oral Sci. 1998;106(4):863–871. doi: 10.1046/j.0909-8836.1998.eos106405.x. [DOI] [PubMed] [Google Scholar]
- 94.Johnson DE. Noncaspase proteases in apoptosis. Leukemia. 2000;14(9):1695–1703. doi: 10.1038/sj.leu.2401879. [DOI] [PubMed] [Google Scholar]
- 95.Johnson NA, Fletcher HM. The bcp gene in the bcp-recA-vimA operon is important to oxidative stress resistance in Porphyromonas gingivalis W83. American Society for Microbiology 104th General Meeting Abstract # I-023 New Orleans, LA. 2004 doi: 10.1111/j.2041-1014.2010.00596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson NA, Liu Y, Fletcher HM. Alkyl hydroperoxide peroxidase subunit C (ahpC) protects against organic peroxides but does not affect the virulence of Porphyromonas gingivalis W83. Oral Microbiol Immunol. 2004;19(4):233–239. doi: 10.1111/j.1399-302X.2004.00145.x. [DOI] [PubMed] [Google Scholar]
- 97.Johnson NA, McKenzie R, McLean L, Sowers LC, Fletcher HM. 8-Oxo-7,8-dihydroguanine is removed by a nucleotide excision repair-like mechanism in Porphyromonas gingivalis W83. J Bacteriol. 2004;186(22):7697–7703. doi: 10.1128/JB.186.22.7697-7703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katz J, Sambandam V, Wu JH, Michalek SM, Balkovetz DF. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect Immun. 2000;68(3):1441–1449. doi: 10.1128/iai.68.3.1441-1449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Katz J, Yang QB, Zhang P, Potempa J, Travis J, Michalek SM, Balkovetz DF. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infection and Immunity. 2002;70(5):2512–2518. doi: 10.1128/IAI.70.5.2512-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khlgatian M, Nassar H, Chou HH, Gibson FC, III, Genco CA. Fimbria-dependent activation of cell adhesion molecule expression in Porphyromonas gingivalis-infected endothelial cells. Infect Immun. 2002;70(1):257–267. doi: 10.1128/IAI.70.1.257-267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kikuchi Y, Ohara N, Sato K, Yoshimura M, Yukitake H, Sakai E, Shoji M, Naito M, Nakayama K. Novel stationary-phase-upregulated protein of Porphyromonas gingivalis influences production of superoxide dismutase, thiol peroxidase and thioredoxin. Microbiology. 2005;151(Pt 3):841–853. doi: 10.1099/mic.0.27589-0. [DOI] [PubMed] [Google Scholar]
- 102.Kim DY, Kim KK. Crystallization and preliminary X-ray studies of the protease domain of the heat-shock protein HtrA from Thermotoga maritima. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 1):170–172. doi: 10.1107/s0907444901018248. [DOI] [PubMed] [Google Scholar]
- 103.Kim J, Amar S. Periodontal disease and systemic conditions: a bidirectional relationship. Odontology. 2006;94(1):10–21. doi: 10.1007/s10266-006-0060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Konkel ME, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbe Infect. 2000;2(2):157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 105.Konola JT, Sargent KE, Gow JB. Efficient repair of hydrogen peroxide-induced DNA damage by Escherichia coli requires SOS induction of RecA and RuvA proteins. Mutat Res. 2000;459(3):187–194. doi: 10.1016/s0921-8777(99)00073-7. [DOI] [PubMed] [Google Scholar]
- 106.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys-Gingipain). Biochem Biophys Res Commun. 1998;249(1):38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 108.Kurita-Ochiai T, Ochiai K, Fukushima K. Volatile fatty acid, metabolic by-product of periodontopathic bacteria, induces apoptosis in WEHI 231 and RAJI B lymphoma cells and splenic B cells. Infect Immun. 1998;66(6):2587–2594. doi: 10.1128/iai.66.6.2587-2594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kurita-Ochiai T, Ochiai K, Fukushima K. Butyric acid-induced T-cell apoptosis is mediated by caspase-8 and -9 activation in a Fas-independent manner. Clin Diagn Lab Immunol. 2001;8(2):325–332. doi: 10.1128/CDLI.8.2.325-332.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee SD, Kuo WW, Lin DY, Chen TH, Kuo WH, Hsu HH, Chen JZ, Liu JY, Yeh YL, Huang CY. Role of calcineurin in Porphyromonas gingivalis-induced myocardial cell hypertrophy and apoptosis. J Biomed Sci. 2005:1–10. doi: 10.1007/s11373-005-9048-4. [DOI] [PubMed] [Google Scholar]
- 111.Lee SD, Wu CC, Kuo WW, Lin JA, Hwang JM, Lu MC, Chen LM, Hsu HH, Wang CK, Chang SH, Huang CY. Porphyromonas gingivalis-related cardiac cell apoptosis was majorly co-activated by p38 and extracellular signal-regulated kinase pathways. J Periodontal Res. 2006;41(1):39–46. doi: 10.1111/j.1600-0765.2005.00842.x. [DOI] [PubMed] [Google Scholar]
- 112.Lee SG, Pancholi V, Fischetti VA. Characterization of a unique glycosylated anchor endopeptidase that cleaves the LPXTG sequence motif of cell surface proteins of Gram-positive bacteria. J Biol Chem. 2002;277(49):46912–46922. doi: 10.1074/jbc.M208660200. [DOI] [PubMed] [Google Scholar]
- 113.Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol. 2001;2(8):589–598. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
- 114.Lewis JP, Dawson J, Jones K, Hannis J, Muddiman D, Birkedal-Hansen H, Macrina FL. Hemoglobinase activity of the lysine protease of Porphyromonas gingivalis. J Dent Res. 1999;78:506. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181(16):4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172(4):1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, Fletcher HM. Environmental regulation of recA gene expression in Porphyromonas gingivalis. Oral Microbiol Immunol. 2001;16(3):136–143. doi: 10.1034/j.1399-302x.2001.016003136.x. [DOI] [PubMed] [Google Scholar]
- 118.Liu Y, Fletcher HM. The recA gene in Porphyromonas gingivalis is expressed during infection of the murine host. Oral Microbiol Immunol. 2001;16(4):218–223. doi: 10.1034/j.1399-302x.2001.160404.x. [DOI] [PubMed] [Google Scholar]
- 119.Lu AL, Li X, Gu Y, Wright PM, Chang DY. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem Biophys. 2001;35(2):141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 120.Luder CG, Gross U. Apoptosis and its modulation during infection with Toxoplasma gondii: molecular mechanisms and role in pathogenesis. Curr Top Microbiol Immunol. 2005;289:219–237. doi: 10.1007/3-540-27320-4_10. [DOI] [PubMed] [Google Scholar]
- 121.Lynch MC, Kuramitsu HK. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect Immun. 1999;67(7):3367–3375. doi: 10.1128/iai.67.7.3367-3375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz O, Lamont RJ. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007 doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marnett LJ. Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 2002;181-182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]
- 124.Mathiasen IS, Jaattela M. Triggering caspase-independent cell death to combat cancer. Trends Mol Med. 2002;8(5):212–220. doi: 10.1016/s1471-4914(02)02328-6. [DOI] [PubMed] [Google Scholar]
- 125.McKenzie R, Fletcher HM. Studies in oxidative stress resistance in Porphyromonas gingivalis. Journal of Dental Research 86 (Spec Iss A) Abstract# 2353 IADR 85th General Session New Orleans, LA. 2007 [Google Scholar]
- 126.Mekalanos JJ. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meredith JE, Jr., Fazeli B, Schwartz MA. The extracellular matrix as a cell survival factor. Mol Biol Cell. 1993;4(9):953–961. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mezyk-Kopec R, Bzowska M, Potempa J, Bzowska M, Jura N, Sroka A, Black RA, Bereta J. Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis. Infect Immun. 2005;73(3):1506–1514. doi: 10.1128/IAI.73.3.1506-1514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Michalek SM, Katz J, Childers NK, Martin M, Balkovetz DF. Microbial/host interactions: mechanisms involved in host responses to microbial antigens. Immunol Res. 2002;26(1-3):223–234. doi: 10.1385/IR:26:1-3:223. [DOI] [PubMed] [Google Scholar]
- 130.Michel JB. Anoikis in the Cardiovascular System. Known and Unknown Extracellular Mediators. Arterioscler Thromb Vasc Biol. 2003 doi: 10.1161/01.ATV.0000099882.52647.E4. [DOI] [PubMed] [Google Scholar]
- 131.Mikolajczyk J, Boatright KM, Stennicke HR, Nazif T, Potempa J, Bogyo M, Salvesen GS. Sequential autolytic processing activates the zymogen of Arg-gingipain. J Biol Chem. 2003;278(12):10458–10464. doi: 10.1074/jbc.M210564200. [DOI] [PubMed] [Google Scholar]
- 132.Mikolajczyk-Pawlinska J, Kordula T, Pavloff N, Pemberton PA, Chen WC, Travis J, Potempa J. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol Chem Hoppe Seyler. 1998;379(2):205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- 133.Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10(1):1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moens S, Vanderleyden J. Glycoproteins in prokaryotes. Arch Microbiol. 1997;168(3):169–175. doi: 10.1007/s002030050484. [DOI] [PubMed] [Google Scholar]
- 135.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2(9):747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 136.Monteiro HP, Silva EF, Stern A. Nitric oxide: a potential inducer of adhesion-related apoptosis--anoikis. Nitric Oxide. 2004;10(1):1–10. doi: 10.1016/j.niox.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 137.Morioka M, Hinode D, Nagata A, Hayashi H, Ichimiya S, Ueda M, Kido R, Nakamura R. Cytotoxicity of Porphyromonas gingivalis toward cultured human gingival fibroblasts. Oral Microbiol Immunol. 1993;8(4):203–207. doi: 10.1111/j.1399-302x.1993.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 138.Moss JE, Aliprantis AO, Zychlinsky A. The regulation of apoptosis by microbial pathogens. Int Rev Cytol. 1999;187:203–259. doi: 10.1016/s0074-7696(08)62419-5. [DOI] [PubMed] [Google Scholar]
- 139.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997;17(2):760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Murray DA, Wilton JM. Lipopolysaccharide from the periodontal pathogen Porphyromonas gingivalis prevents apoptosis of HL60-derived neutrophils in vitro. Infect Immun. 2003;71(12):7232–7235. doi: 10.1128/IAI.71.12.7232-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nakayama K. Domain-specific rearrangement between the two Arg-gingipain- encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol Immunol. 1997;41(3):185–196. doi: 10.1111/j.1348-0421.1997.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 142.Nakayama K. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Curr Protein Pept Sci. 2003;4(6):389–395. doi: 10.2174/1389203033486983. [DOI] [PubMed] [Google Scholar]
- 143.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178(10):2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, Watanabe K, El Sabaeny A, Park K, Lamont RJ. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol Lett. 2001;200(2):145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 145.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185(18):5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria? J Bacteriol. 2007;189(3):833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ohara N, Kikuchi Y, Shoji M, Naito M, Nakayama K. Superoxide dismutase-encoding gene of the obligate anaerobe Porphyromonas gingivalis is regulated by the redox-sensing transcription activator OxyR. Microbiology. 2006;152(Pt 4):955–966. doi: 10.1099/mic.0.28537-0. [DOI] [PubMed] [Google Scholar]
- 148.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273(33):21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 149.Okano S, Shibata Y, Shiroza T, Abiko Y. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics. 2006;6(1):251–258. doi: 10.1002/pmic.200401338. [DOI] [PubMed] [Google Scholar]
- 150.Olango GJ, Roy F, Sheets SM, Young MK, Fletcher HM. Gingipain RgpB is excreted as a proenzyme in the vimA-defective mutant Porphyromonas gingivalis FLL92. Infect Immun. 2003;71(7):3740–3747. doi: 10.1128/IAI.71.7.3740-3747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Olczak T, Dixon DW, Genco CA. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 2001;183(19):5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Olczak T, Simpson W, Liu X, Genco CA. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 2005;29(1):119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 153.Ozaki K, Hanazawa S. Porphyromonas gingivalis fimbriae inhibit caspase-3-mediated apoptosis of monocytic THP-1 cells under growth factor deprivation via extracellular signal-regulated kinase-dependent expression of p21 Cip/WAF1. Infect Immun. 2001;69(8):4944–4950. doi: 10.1128/IAI.69.8.4944-4950.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pacheco FJ, Servin J, Dang D, Kim J, Molinaro C, Daniels T, Brown-Bryan TA, Imoto-Egami M, Casiano CA. Involvement of lysosomal cathepsins in the cleavage of DNA topoisomerase I during necrotic cell death. Arthritis Rheum. 2005;52(7):2133–2145. doi: 10.1002/art.21147. [DOI] [PubMed] [Google Scholar]
- 155.Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol. 1997;26(2):209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 156.Park Y, Lamont RJ. Contact-dependent protein secretion in Porphyromonas gingivalis. Infection and Immunity. 1998;66(10):4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pavloff N, Pemberton PA, Potempa J, Chen WCA, Pike RN, Prochazka V, Kiefer MC, Travis J, Barr PJ. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain - A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272(3):1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 158.Pavloff N, Potempa J, Pike RN, Prochazka V, Kiefer MC, Travis J, Barr PJ. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 159.Percival RS, Marsh PD, Devine DA, Rangarajan M, Aduse-Opoku J, Shepherd P, Curtis MA. Effect of temperature on growth, hemagglutination, and protease activity of Porphyromonas gingivalis. Infect Immun. 1999;67(4):1917–1921. doi: 10.1128/iai.67.4.1917-1921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Poquet I, Saint V, Seznec E, Simoes N, Bolotin A, Gruss A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol. 2000;35(5):1042–1051. doi: 10.1046/j.1365-2958.2000.01757.x. [DOI] [PubMed] [Google Scholar]
- 161.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;12:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 162.Potempa J, Pavloff N, Travis J. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 1995;3(11):430–434. doi: 10.1016/s0966-842x(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 163.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4(6):397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 164.Price DT, Loscalzo J. Cellular adhesion molecules and atherogenesis. Am J Med. 1999;107(1):85–97. doi: 10.1016/s0002-9343(99)00153-9. [DOI] [PubMed] [Google Scholar]
- 165.Rangarajan M, Hashim A, duse-Opoku J, Paramonov N, Hounsell EF, Curtis MA. Expression of Arg-Gingipain RgpB is required for correct glycosylation and stability of monomeric Arg-gingipain RgpA from Porphyromonas gingivalis W50. Infect Immun. 2005;73(8):4864–4878. doi: 10.1128/IAI.73.8.4864-4878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Re F, Zanetti A, Sironi M, Polentarutti N, Lanfrancone L, Dejana E, Colotta F. Inhibition of anchorage-dependent cell spreading triggers apoptosis in cultured human endothelial cells. J Cell Biol. 1994;127(2):537–546. doi: 10.1083/jcb.127.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Reddig PJ, Juliano RL. Clinging to life: cell to matrix adhesion and cell survival. Cancer Metastasis Rev. 2005;24(3):425–439. doi: 10.1007/s10555-005-5134-3. [DOI] [PubMed] [Google Scholar]
- 168.Robles-Price A, Reid K, Fletcher HM. The role of MutY in Porphyromonas gingivalis W83. Journal of Dental Research 86 (Spec Iss A) Abstract# 2354 IADR 85th General Session New Orleans, LA. 2007;86 [Google Scholar]
- 169.Roy F, Vanterpool E, Fletcher HM. HtrA in Porphyromonas gingivalis can regulate growth and gingipain activity under stressful environmental conditions. Microbiology. 2006;152(Pt 11):3391–3398. doi: 10.1099/mic.0.29147-0. [DOI] [PubMed] [Google Scholar]
- 170.Rytomaa M, Martins LM, Downward J. Involvement of FADD and caspase-8 signalling in detachment-induced apoptosis. Curr Biol. 1999;9(18):1043–1046. doi: 10.1016/s0960-9822(99)80454-0. [DOI] [PubMed] [Google Scholar]
- 171.Salyers AA. Bacterial pathogenesis: a molecular approach. 1994. pp. 1–418.
- 172.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 173.Sawamoto Y, Sugano N, Tanaka H, Ito K. Detection of periodontopathic bacteria and an oxidative stress marker in saliva from periodontitis patients. Oral Microbiol Immunol. 2005;20(4):216–220. doi: 10.1111/j.1399-302X.2005.00215.x. [DOI] [PubMed] [Google Scholar]
- 174.Scott AD, Neishabury M, Jones DH, Reed SH, Boiteux S, Waters R. Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast. 1999;15(3):205–218. doi: 10.1002/(SICI)1097-0061(199902)15:3<205::AID-YEA361>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 175.Scragg MA, Alsam A, Rangarajan M, Slaney JM, Shepherd P, Williams DM, Curtis MA. Nuclear targeting of Porphyromonas gingivalis W50 protease in epithelial cells. Infect Immun. 2002;70(10):5740–5750. doi: 10.1128/IAI.70.10.5740-5750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Scragg MA, Cannon SJ, Rangarajan M, Williams DM, Curtis MA. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect Immun. 1999;67(4):1837–1843. doi: 10.1128/iai.67.4.1837-1843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183(24):7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol. 2006;188(17):6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sekiguchi M, Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21(58):8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- 180.SETLOW RB, CARRIER WL. The disappearance of thymine dimers from DNA: An error correcting mechanism. Proc Natl Acad Sci U S A. 1964;51:226–231. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shah HN, Gharbia SE, O'Toole CM. Assessment of the relative cytotoxicity of Porphyromonas gingivalis cells, products, and components on human epithelial cell lines. J Periodontol. 1992;63(1):44–51. doi: 10.1902/jop.1992.63.1.44. [DOI] [PubMed] [Google Scholar]
- 182.Sheets SM, Potempa J, Travis J, Casiano CA, Fletcher HM. Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect Immun. 2005;73(3):1543–1552. doi: 10.1128/IAI.73.3.1543-1552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sheets SM, Potempa J, Travis J, Fletcher HM, Casiano CA. Gingipains from Porphyromonas gingivalis W83 synergistically disrupt endothelial cell adhesion and can induces caspase-independent apoptosis. Infect Immun. 2006 doi: 10.1128/IAI.01140-05. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shelburne CE, Gleason RM, Coulter WA, Lantz MS, Lopatin DE. Differential display analysis of Porphyromonas gingivalis gene activation response to heat and oxidative stress. Oral Microbiol Immunol. 2005;20(4):233–238. doi: 10.1111/j.1399-302X.2005.00219.x. [DOI] [PubMed] [Google Scholar]
- 185.Sherman MP, Petrak K. Lactoferrin-enhanced anoikis: a defense against neonatal necrotizing enterocolitis. Med Hypotheses. 2005;65(3):478–482. doi: 10.1016/j.mehy.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 186.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin finding, and hemagglutination of Porphyromonas gingivalis - Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274(25):17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 187.Shirin H, Sordillo EM, Kolevska TK, Hibshoosh H, Kawabata Y, Oh SH, Kuebler JF, Delohery T, Weghorst CM, Weinstein IB, Moss SF. Chronic Helicobacter pylori infection induces an apoptosis-resistant phenotype associated with decreased expression of p27 (kip1). Infect Immun. 2000;68(9):5321–5328. doi: 10.1128/iai.68.9.5321-5328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology. 2002;148(Pt 4):1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- 189.Silveira JR, Kalafatis M, Tracy PB. Carbohydrate moieties on the procofactor factor V, but not the derived cofactor factor Va, regulate its inactivation by activated protein C. Biochemistry. 2002;41(5):1672–1680. doi: 10.1021/bi011304g. [DOI] [PubMed] [Google Scholar]
- 190.Smalley JW, Birss AJ. Iron protoporphyrin IX albumin complexing increases the capacity and avidity of its binding to the periodontopathogen Porphyromonas gingivalis. Microb Pathog. 1999;26(3):131–137. doi: 10.1006/mpat.1998.0259. [DOI] [PubMed] [Google Scholar]
- 191.Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the m-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183(1):159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 192.Smalley JW, Birss AJ, Szmigielski B, Potempa J. The HA2 haemagglutinin domain of the lysine-specific gingipain (Kgp) of Porphyromonas gingivalis promotes micro-oxo bishaem formation from monomeric iron (III) protoporphyrin IX. Microbiology. 2006;152(Pt 6):1839–1845. doi: 10.1099/mic.0.28835-0. [DOI] [PubMed] [Google Scholar]
- 193.Smalley JW, Silver J, Marsh PJ, Birss AJ. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the m-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331(3):681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sreedhar AS, Csermely P. Heat shock proteins in the regulation of apoptosis: new strategies in tumor therapy: a comprehensive review. Pharmacol Ther. 2004;101(3):227–257. doi: 10.1016/j.pharmthera.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 195.Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2(2):188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 196.Stupack DG. Integrins as a distinct subtype of dependence receptors. Cell Death Differ. 2005;12(8):1021–1030. doi: 10.1038/sj.cdd.4401658. [DOI] [PubMed] [Google Scholar]
- 197.Sugawara S, Nemoto E, Tada H, Miyake K, Imamura T, Takada H. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. J Immunol. 2000;165(1):411–418. doi: 10.4049/jimmunol.165.1.411. [DOI] [PubMed] [Google Scholar]
- 198.Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM., Jr. Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol. 2002;44(2):479–488. doi: 10.1046/j.1365-2958.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- 199.Sztukowska M, Sroka A, Bugno M, Banbula A, Takahashi Y, Pike RN, Genco CA, Travis J, Potempa J. The C-terminal domains of the gingipain K polyprotein are necessary for assembly of the active enzyme and expression of associated activities. Mol Microbiol. 2004;54(5):1393–1408. doi: 10.1111/j.1365-2958.2004.04357.x. [DOI] [PubMed] [Google Scholar]
- 200.Szymanski CM, Burr DH, Guerry P. Campylobacter protein glycosylation affects host cell interactions. Infect Immun. 2002;70(4):2242–2244. doi: 10.1128/IAI.70.4.2242-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3(3):225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 202.Tada H, Sugawara S, Nemoto E, Imamura T, Potempa J, Travis J, Shimauchi H, Takada H. Proteolysis of icam-1 on human oral epithelial cells by gingipains. J Dent Res. 2003;82(10):796–801. doi: 10.1177/154405910308201007. [DOI] [PubMed] [Google Scholar]
- 203.Takeuchi T, Kato N, Watanabe K, Morimoto K. Mechanism of oxidative DNA damage induction in a strict anaerobe, Prevotella melaninogenica. FEMS Microbiol Lett. 2000;192(1):133–138. doi: 10.1111/j.1574-6968.2000.tb09371.x. [DOI] [PubMed] [Google Scholar]
- 204.Tanjoni I, Weinlich R, Della-Casa MS, Clissa PB, Saldanha-Gama RF, de Freitas MS, Barja-Fidalgo C, Amarante-Mendes GP, Moura-da-Silva AM. Jararhagin, a snake venom metalloproteinase, induces a specialized form of apoptosis (anoikis) selective to endothelial cells. Apoptosis. 2005;10(4):851–861. doi: 10.1007/s10495-005-2945-1. [DOI] [PubMed] [Google Scholar]
- 205.Tchou J, Grollman AP. Repair of DNA containing the oxidatively-damaged base, 8-oxoguanine. Mutat Res. 1993;299(3-4):277–287. doi: 10.1016/0165-1218(93)90104-l. [DOI] [PubMed] [Google Scholar]
- 206.Theis K, Skorvaga M, Machius M, Nakagawa N, Van Houten B, Kisker C. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat Res. 2000;460(3-4):277–300. doi: 10.1016/s0921-8777(00)00032-x. [DOI] [PubMed] [Google Scholar]
- 207.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281(5381):1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 208.Turk B, Stoka V, Rozman-Pungercar J, Cirman T, Droga-Mazovec G, Oresic K, Turk V. Apoptotic pathways: involvement of lysosomal proteases. Biol Chem. 2002;383(7-8):1035–1044. doi: 10.1515/BC.2002.112. [DOI] [PubMed] [Google Scholar]
- 209.Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect Immun. 2003;71(3):1170–1178. doi: 10.1128/IAI.71.3.1170-1178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Upreti RK, Kumar M, Shankar V. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics. 2003;3(4):363–379. doi: 10.1002/pmic.200390052. [DOI] [PubMed] [Google Scholar]
- 211.Urnowey S, Ansai T, Bitko V, Nakayama K, Takehara T, Barik S. Temporal activation of anti- and pro-apoptotic factors in human gingival fibroblasts infected with the periodontal pathogen, Porphyromonas gingivalis: potential role of bacterial proteases in host signalling. BMC Microbiol. 2006;6:26. doi: 10.1186/1471-2180-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 212.US Department of Health and Human Services Oral Health in America:A Report of the Surgeon General. 2000.
- 213.Vanterpool E, Roy F, Fletcher HM. The vimE gene downstream of vimA is independently expressed and is involved in modulating proteolytic activity in Porphyromonas gingivalis W83. Infect Immun. 2004;72(10):5555–5564. doi: 10.1128/IAI.72.10.5555-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vanterpool E, Roy F, Fletcher HM. Inactivation of vimF, a Putative Glycosyltransferase Gene Downstream of vimE, Alters Glycosylation and Activation of the Gingipains in Porphyromonas gingivalis W83. Infect Immun. 2005;73(7):3971–3982. doi: 10.1128/IAI.73.7.3971-3982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vanterpool E, Roy F, Sandberg L, Fletcher HM. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect Immun. 2005;73(3):1357–1366. doi: 10.1128/IAI.73.3.1357-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Vanterpool E, Roy F, Zhan W, Sheets SM, Sangberg L, Fletcher HM. VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology. 2006;152(Pt 11):3383–3389. doi: 10.1099/mic.0.29146-0. [DOI] [PubMed] [Google Scholar]
- 217.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3(2):97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vaux DL, Strasser A. The molecular biology of apoptosis. Proc Natl Acad Sci U S A. 1996;93(6):2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Veith PD, Chen YY, Reynolds EC. Porphyromonas gingivalis RgpA and Kgp proteinases and adhesins are C terminally processed by the carboxypeptidase CPG70. Infect Immun. 2004;72(6):3655–3657. doi: 10.1128/IAI.72.6.3655-3657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Walter C, Zahlten J, Schmeck B, Schaudinn C, Hippenstiel S, Frisch E, Hocke AC, Pischon N, Kuramitsu HK, Bernimoulin JP, Suttorp N, Krull M. Porphyromonas gingivalis strain-dependent activation of human endothelial cells. Infect Immun. 2004;72(10):5910–5918. doi: 10.1128/IAI.72.10.5910-5918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wandersman C. Secretion, processing and activation of bacterial extracellular proteases. Mol Microbiol. 1989;3(12):1825–1831. doi: 10.1111/j.1365-2958.1989.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 222.Wang PL, Shinohara M, Murakawa N, Endo M, Sakata S, Okamura M, Ohura K. Effect of cysteine protease of Porphyromonas gingivalis on adhesion molecules in gingival epithelial cells. Jpn J Pharmacol. 1999;80(1):75–79. doi: 10.1254/jjp.80.75. [DOI] [PubMed] [Google Scholar]
- 223.Wang PL, Shirasu S, Shinohara M, Daito M, Oido M, Kowashi Y, Ohura K. Induction of apoptosis in human gingival fibroblasts by a Porphyromonas gingivalis protease preparation. Arch Oral Biol. 1999;44(4):337–342. doi: 10.1016/s0003-9969(99)00002-3. [DOI] [PubMed] [Google Scholar]
- 224.Watanabe K, Yilmaz O, Nakhjiri SF, Belton CM, Lamont RJ. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect Immun. 2001;69(11):6731–6737. doi: 10.1128/IAI.69.11.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Waterhouse NJ, Sedelies KA, Sutton VR, Pinkoski MJ, Thia KY, Johnstone R, Bird PI, Green DR, Trapani JA. Functional dissociation of DeltaPsim and cytochrome c release defines the contribution of mitochondria upstream of caspase activation during granzyme B-induced apoptosis. Cell Death Differ. 2006;13(4):607–618. doi: 10.1038/sj.cdd.4401772. [DOI] [PubMed] [Google Scholar]
- 226.Wei L, Yang Y, Zhang X, Yu Q. Cleavage of p130Cas in anoikis. J Cell Biochem. 2004;91(2):325–335. doi: 10.1002/jcb.10760. [DOI] [PubMed] [Google Scholar]
- 227.Weinrauch Y, Zychlinsky A. The induction of apoptosis by bacterial pathogens. Annu Rev Microbiol. 1999;53:155–187. doi: 10.1146/annurev.micro.53.1.155. [DOI] [PubMed] [Google Scholar]
- 228.Wilson CA, Browning JL. Death of HT29 adenocarcinoma cells induced by TNF family receptor activation is caspase-independent and displays features of both apoptosis and necrosis. Cell Death Differ. 2002;9(12):1321–1333. doi: 10.1038/sj.cdd.4401107. [DOI] [PubMed] [Google Scholar]
- 229.Xie H, Chung WO, Park Y, Lamont RJ. Regulation of the Porphyromonas gingivalis fimA (fimbrillin) gene. Infection and Immunity. 2000;68(12):6574–6579. doi: 10.1128/iai.68.12.6574-6579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72(7):3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74(1):703–710. doi: 10.1128/IAI.74.1.703-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yun PL, Decarlo AA, Hunter N. Gingipains of Porphyromonas gingivalis modulate leukocyte adhesion molecule expression induced in human endothelial cells by ligation of CD99. Infect Immun. 2006;74(3):1661–1672. doi: 10.1128/IAI.74.3.1661-1672.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhan M, Zhao H, Han ZC. Signalling mechanisms of anoikis. Histol Histopathol. 2004;19(3):973–983. doi: 10.14670/HH-19.973. [DOI] [PubMed] [Google Scholar]
- 234.Zhang D, Beresford PJ, Greenberg AH, Lieberman J. Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc Natl Acad Sci U S A. 2001;98(10):5746–5751. doi: 10.1073/pnas.101329598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhang D, Pasternack MS, Beresford PJ, Wagner L, Greenberg AH, Lieberman J. Induction of rapid histone degradation by the cytotoxic T lymphocyte protease Granzyme A. J Biol Chem. 2001;276(5):3683–3690. doi: 10.1074/jbc.M005390200. [DOI] [PubMed] [Google Scholar]
- 236.Zhang W, Shokeen M, Li D, Mehta JL. Identification of apoptosis-inducing factor in human coronary artery endothelial cells. Biochem Biophys Res Commun. 2003;301(1):147–151. doi: 10.1016/s0006-291x(02)02981-9. [DOI] [PubMed] [Google Scholar]
- 237.Zhou Q, Amar S. Identification of proteins differentially expressed in human monocytes exposed to Porphyromonas gingivalis and its purified components by high-throughput immunoblotting. Infect Immun. 2006;74(2):1204–1214. doi: 10.1128/IAI.74.2.1204-1214.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ziegler U, Groscurth P. Morphological features of cell death. News Physiol Sci. 2004;19:124–128. doi: 10.1152/nips.01519.2004. [DOI] [PubMed] [Google Scholar]