Abstract
Using high resolution magnetic resonance imaging data, we examined the interrelationships between eight cerebral lobar volumetric measures via both explor atory and confirmatory factor analyses in a large sample (N = 484) of pediatric twins and singletons. These analyses suggest the presence of strong genetic correlations between cerebral structures, particularly between regions of like tissue type or in spatial proximity. Structural modeling estimated that most of the variance in all structures is associated with highly correlated lobar latent factors, with differences in genetic covariance and heritability driven by a common genetic factor that influenced gray and white matter differently. Reanalysis including total brain volume as a covariate dramatically reduced the total residual variance and disproportionately influenced the additive genetic variance in all regions of interest.
Keywords: Twin, Cerebrum, MRI, Genetics, Pediatrics
Introduction
The human telencephalon, or cerebrum, is approximately 950 grams at adulthood and represents over 80% of total brain weight (Jenkins and Truex 1963). As a region of dramatic evolutionary expansion in humans, the cerebrum is of particular interest in attempts to understand the biology underlying complex psychological and psychiatric phenomena (Carroll 2003; Hill and Walsh 2005). The human cerebrum is both the defining neuroanatomic structure of our species as well as one of the most important determinants of our identity as individuals. Yet despite the importance of the cerebrum to behavior genetics, neurogenetics, and evolutionary genetics, our understanding of both the etiology of individual differences in cerebral patterning and the genetics underlying human brain evolution remain quite limited (Rakic 2009).
By isolating genetically mediated variance in brain structures, twin and family studies using magnetic resonance imaging (MRI) provide valuable information on the relative importance of genetics on brain anatomy. Prior twin studies have consistently shown that population variability in total brain volumes is almost entirely due to genetic variation (Panizzon et al. 2009; Schmitt et al. 2007b). For example, Baaré et al. estimated very high heritabilities for total brain volume (0.90), total gray matter (0.82) and total white matter (0.88) using high resolution MRI scans from 54 monozygotic (MZ) twin pairs, 58 dizygotic (DZ) twin pairs, and 34 siblings (Baaré et al. 2001). In a pediatric sample of 34 MZ and 32 DZ twin pairs with reading disability, Pennington et al. estimated total cerebral volume heritability at 97% (Pennington et al. 2000). More recently, Peper et al. reported a total brain volume heritability estimate of 0.94 in a sample of 45 MZ and 62 DZ nine year old twin pairs (Peper et al. 2009).
Despite the well-established importance of genetic factors on global brain volumes, the role of genetics on cerebral substructures is far less well-understood. To date, there have been only three twin studies that have examined the genetic contributions to individual cerebral lobar volumes (Carmelli et al. 2002; Geschwind et al. 2002; Wallace et al. 2006). In an older adult sample from the National Heart, Lung, and Blood Institute (NHLBI), Geschwind et al. examined lobar brain volumes in 72 MZ and 67 DZ twins and found that the frontal and temporal lobe volumes had relatively high heritability (0.69–0.77), parietal lobe moderate heritability (0.54–0.57), and occipital lobe the lowest heritability (0.37–0.38) of the cerebral lobes (Geschwind et al. 2002). Carmeli et al. reported similar heritability estimates, also based on data from the NHLBI sample (Carmelli et al. 2002). Using the NIH pediatric twin sample, Wallace et al. estimated that the heritability of the frontal, temporal, and parietal lobes ranged from 0.88 to 0.92 in 90 MZ twin pairs, 38 DZ pairs, and 158 singletons (Wallace et al. 2006).
To date, only a handful of studies have attempted to examine the genetic relationships between brain volumes in a multivariate framework (Baaré et al. 2001; Posthuma et al. 2000; Schmitt et al. 2007a; Wright et al. 2002), and none has focused on cerebral volumes in a typical sample. In other words, despite the evidence that most large brain volumes are highly heritable, there is very little evidence about whether genetic effects within the cerebrum are the consequence of independent genetic factors or caused by shared genetic effects influencing the cerebrum in its entirety. In this article, we attempt to address the question of whether genetically mediated individual differences in cerebral lobar volumes exhibit regional specificity within the cerebrum, or rather are dominated by global or tissue-specific effects.
Methods
These analyses utilize neuroimaging data from the Pediatric Twin Study at the Child Psychiatry Branch of the National Institute of Mental Health (NIMH). The subjects in this study were recruited by means of local and national advertisements for participation in an ongoing longitudinal pediatric imaging study. Advertisements specified that the MRI study sought twins between the ages of 5 and 18, with no learning disabilities, neurological problems or behavioral disorders. The screening process involved phone interviews, behavioral questionnaires mailed to parents and teachers, an in-person clinical interview, family history assessment, as well as a physical and neurological exam. Exclusion criteria included having a lifetime history of physical, neurological, or psychiatric abnormalities, learning disabilities, or psychiatric illness oneself, or in either one-first-degree relative or more than 20% of second-degree relatives. Approximately one in four families responding to the advertisements met inclusion criteria. Twins were included in the analysis only if quantifiable MRI scans free from motion or other artifact were obtained on both twins at the same age. Written assent from the child and written consent from a parent were obtained for each participant. The study protocol was approved by the institutional review board of the National Institute of Mental Health.
Zygosity was determined by DNA analysis of buccal cheek swabs using 9–21 unlinked short tandem repeat loci for a minimum certainty of 99%, by BRT Laboratories, Inc. (Baltimore, MD). The resultant sample consisted of 127 same-sex MZ twin pairs (mean age = 11.6, SD = 3.3; age range = 5.6–18.7; 74 [58%] male, 53 female), 36 same-sex DZ twin pairs (mean age = 11.0, SD = 3.7; age range = 5.5–18.2; 18 [60%] male, 12 female), and 158 singletons (mean age = 11.3, SD = 3.5; age range = 5.2–18.7; 94 [59%] male, 64 female).
Image acquisition
All subjects were scanned on the same GE 1.5 Tesla Signa MRI scanner. A three-dimensional spoiled gradient recalled echo in the steady state (3D SPGR) imaging protocol was used for all subjects (axial slice thickness = 1.5 mm, time to echo = 5 ms, repetition time = 24 ms, flip angle = 45 degrees, acquisition matrix = 192 × 256, number of excitations = 1, and field of view = 24 cm).
Image processing
The native MRI scans were first registered into standardized stereotaxic space using a linear transformation (Collins et al. 1994) and corrected for non-uniformity artifacts (Sled et al. 1998). The registered and corrected volumes were segmented into gray matter, white matter, cerebro-spinal fluid, and background using a neural net classifier (Zijdenbos et al. 2002). The tissue classification information was combined with a probabilistic atlas to provide region of interest measures (Collins et al. 1994). The output measures of this process have shown high agreement with conventional hand tracing measures. The measures used in the present study were cerebral lobar volumes (frontal, parietal, temporal, and occipital lobes), for gray and white tissue compartments separately.
Statistical analyses
Preliminary inspection of normal quantile plots of the observed variables indicated that they met the normality assumptions of structural equation modeling with likelihood based optimization. Using R (Ihaka and Gentleman 1996; R Development Core Team 2005) we calculated correlations between all volumes for MZ and DZ groups separately.
Multivariate analyses were performed in Mx (Neale et al. 2002). We started by constructing a preliminary 8-variable triple Cholesky decomposition, the multivariate analog of the classical ACE model, in order to calculate relatively unbiased genetic and environmental correlations between the observed variables as well as heritability estimates (Neale and Cardon 1992). We also calculated eigenvalues on the decomposed covariance matrices to obtain an approximate number of factors required to explain most of the observed variance. We then performed an exploratory factor analysis (EFA) using independent pathway models (Kendler et al. 1987; Mcardle and Goldsmith 1990). Scree plots of eigenvalues from the Cholesky decomposition suggested that three common factors, at most, would be required to explain most of the covariance for each variance component. Thus, our full model included three factors for each of three variance components (additive genetic, shared environmental, and unique environmental variance), as well as structure-specific factors for each variance component. This model is subsequently referred to as the 333-ACE IPM, indicating 3 additive genetic (A), 3 shared environmental (C), and 3 unique environmental (E) common factors, respectively.
As a complementary approach to IFA, we also performed a confirmatory analysis (CFA) as the nature of the cerebral measures implied a straightforward factor structure. Each of the eight variables measured was hypothesized to be influenced by factors related to tissue type (e.g. gray versus white matter) or spatial location (e.g. lobe). These relationships are visualized in Fig. 1a, in which each observed variable is influenced by two latent factors based on its unique combination of region and tissue. Coincidentally, the factor structure of this model is identical to the multitrait-multimethod model (MTMM) originally proposed by Campbell and Fiske for the assessment of construct validity (Campbell and Fiske 1959). In MTMMs, the variance in each observed measure is assumed to be a combination of latent contributions from one of two or more trait-specific factors (e.g. anxiety, depression) and one of two or more method-specific factors (e.g. test battery). Our parameterization was similar to the correlated traits, correlated methods version of MTMM devised by Jöreskog for CFA using structural equation modeling (Jöreskog 1971) and is virtually identical in specification to prior models used in the behavioral sciences (Coromina and Coenders 2006). Given our twin sample, the MTMM model was expanded in order to decompose observed intercerebral covariances into genetic and nongenetic sources (Fig. 1b). Thus, intercerebral relationships due to genetic (A), shared environmental (C), and unique environmental (E) influences could be estimated separately.
Fig. 1.
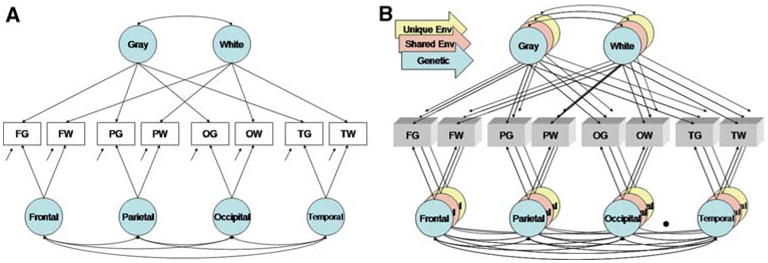
Confirmatory factor models. Panel A is a sample path diagram of the MTMM model factor pattern. Two classes of latent variables are defined, those pertaining to tissue or spatial location. Though not shown, each path corresponds to a unique, freely estimated parameter. The first letter of the observed variable name corresponds to spatial location (F, frontal; P, parietal; O, occipital; T temporal) and the second to tissue (G, gray; W, white). Given data from twins and family members, the variance can be decomposed into genetic and nongenetic sources (Panel B). For simplicity, the model from only one twin is shown
The MTMM has been extensively used in the social sciences, though the principles of the model can be easily generalized (Bechger and Maris 2004). The widespread use of MTMM over several decades has revealed many unfortunate limitations of the approach, such as identification problems for small numbers of factors, easy misspecification, unidentified submodels due to inadequate local information, and errors in estimation. Some of the problems are specific to the use of ordinal data, however (Tomás et al. 2000). Additive MTMM models, for example, have been shown to fit well for continuous, normally distributed variables even if the fit is poor for ordinal data (Corten et al. 2002). Nevertheless, because of the inherent problems associated with the MTMM factor structure, we elected to pursue a cautious approach in our own data. Since prior research has demonstrated at most a trivial role of the shared environment on absolute cerebral volumes (Wallace et al. 2006), we attempted to remove all factors related to the shared environment. But because of the risk of over-fitting, we did not attempt more subtle simplifications of the model (e.g. removing tissue-specific factors).
We employed maximum likelihood for numeric optimization (Edwards 1972). In these analyses, best-fit models were chosen using AIC (Akaike 1987), though we also performed likelihood ratio chi-squared tests to compare model fits without rewarding parsimony. Since preliminary simple linear regressions demonstrated a significant effect of age, race, and sex, we used residuals from multiple regressions including age sex and race as explanatory variables for all models.
Covarying for global effects
Univariate ACE modeling was used to decompose the phenotypic variance of total brain volume (TBV). We then repeated our statistical analyses, but included TBV as a regressor on mean ROI volumes.
Results
Within individuals, correlations between cerebral structures were high (Table 1). The highest correlations, approximately 0.80–0.95, were between frontal gray matter and both temporal and parietal gray, as well as between frontal white and parietal and temporal white matter. The lowest correlations, approximately 0.30–0.60, were between occipital structures and structures of different tissue type in different lobes. In general, variables that differed only by one qualitative trait were more correlated than if they differed by both tissue type and spatial location. For example, frontal gray matter was highly correlated with frontal white, as well as to temporal and parietal gray, but to a lesser extent to parietal and temporal white. The most prominent exception to this observation was a high correlation between temporal gray and parietal white matter.
Table 1. Within and cross twin correlations between cerebral volumes for MZ (below diagonal) and DZ (above diagonal) groups.
| FG1 | FW1 | OG1 | OW1 | PG1 | PW1 | TG1 | TW1 | FG2 | FW2 | OG2 | OW2 | PG2 | PW2 | TG2 | TW2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FG1 | 1.00 | 0.75 | 0.54 | 0.60 | 0.80 | 0.67 | 0.87 | 0.68 | 0.46 | 0.32 | 0.20 | 0.22 | 0.37 | 0.23 | 0.36 | 0.24 |
| FW1 | 0.67 | 1.00 | 0.48 | 0.77 | 0.59 | 0.94 | 0.79 | 0.92 | 0.29 | 0.49 | 0.10 | 0.43 | 0.19 | 0.48 | 0.34 | 0.53 |
| OG1 | 0.64 | 0.58 | 1.00 | 0.69 | 0.59 | 0.52 | 0.61 | 0.52 | 0.23 | 0.13 | 0.44 | 0.31 | 0.25 | 0.14 | 0.28 | 0.25 |
| OW1 | 0.39 | 0.73 | 0.70 | 1.00 | 0.54 | 0.84 | 0.71 | 0.82 | 0.20 | 0.32 | 0.33 | 0.51 | 0.19 | 0.45 | 0.26 | 0.44 |
| PG1 | 0.84 | 0.61 | 0.73 | 0.51 | 1.00 | 0.62 | 0.70 | 0.53 | 0.23 | 0.19 | 0.14 | 0.21 | 0.28 | 0.19 | 0.14 | 0.11 |
| PW1 | 0.44 | 0.86 | 0.60 | 0.83 | 0.55 | 1.00 | 0.71 | 0.90 | 0.13 | 0.40 | 0.03 | 0.40 | 0.07 | 0.48 | 0.19 | 0.47 |
| TG1 | 0.85 | 0.67 | 0.71 | 0.51 | 0.74 | 0.53 | 1.00 | 0.83 | 0.38 | 0.39 | 0.37 | 0.46 | 0.36 | 0.38 | 0.45 | 0.48 |
| TW1 | 0.48 | 0.83 | 0.52 | 0.77 | 0.46 | 0.84 | 0.64 | 1.00 | 0.30 | 0.50 | 0.27 | 0.57 | 0.26 | 0.56 | 0.42 | 0.65 |
| FG2 | 0.82 | 0.62 | 0.62 | 0.50 | 0.74 | 0.49 | 0.70 | 0.53 | 1.00 | 0.58 | 0.51 | 0.22 | 0.89 | 0.42 | 0.88 | 0.43 |
| FW2 | 0.62 | 0.90 | 0.58 | 0.69 | 0.55 | 0.80 | 0.68 | 0.81 | 0.63 | 1.00 | 0.29 | 0.59 | 0.49 | 0.84 | 0.68 | 0.77 |
| OG2 | 0.53 | 0.57 | 0.71 | 0.65 | 0.59 | 0.57 | 0.58 | 0.60 | 0.63 | 0.55 | 1.00 | 0.60 | 0.66 | 0.39 | 0.63 | 0.39 |
| OW2 | 0.39 | 0.66 | 0.59 | 0.82 | 0.43 | 0.72 | 0.51 | 0.74 | 0.42 | 0.70 | 0.73 | 1.00 | 0.32 | 0.71 | 0.41 | 0.72 |
| PG2 | 0.66 | 0.57 | 0.60 | 0.58 | 0.80 | 0.54 | 0.56 | 0.52 | 0.87 | 0.57 | 0.70 | 0.49 | 1.00 | 0.46 | 0.82 | 0.39 |
| PW2 | 0.43 | 0.81 | 0.56 | 0.76 | 0.50 | 0.90 | 0.55 | 0.83 | 0.50 | 0.87 | 0.61 | 0.80 | 0.57 | 1.00 | 0.60 | 0.79 |
| TG2 | 0.71 | 0.66 | 0.63 | 0.60 | 0.66 | 0.57 | 0.83 | 0.70 | 0.86 | 0.68 | 0.69 | 0.53 | 0.76 | 0.61 | 1.00 | 0.68 |
| TW2 | 0.47 | 0.74 | 0.52 | 0.68 | 0.42 | 0.74 | 0.67 | 0.91 | 0.50 | 0.82 | 0.60 | 0.76 | 0.47 | 0.85 | 0.70 | 1.00 |
Cross twin correlations are shown in shaded boxes; cross-twin, within-trait correlations are in boldface
Similar to our prior univariate studies on a slightly smaller sample, the within-region, cross twin correlations were substantially higher in MZ twins relative to DZ twins for cerebral volumes. The differences between groups were strongest for white matter structures, though these descriptive statistics suggest that both gray and white matter are highly heritable. Heritability estimates from the Cholesky decomposition (Table 2) also suggested generally high heritability values for cerebral ROIs. Variance attributable to the shared environment was small for all cerebral regions, ranging from 0.02 to 0.24.
Table 2. Variance components estimates for eight cerebral lobar volumes.
| No global covariate | TBV-adjusted | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| a2 | c2 | e2 | a2 | c2 | e2 | |
| FG | 0.55 (0.31–0.75) | 0.21 (0.03–0.44) | 0.23 (0.17–0.32) | 0.17 (0.00–0.49) | 0.45 (0.15–0.62) | 0.38 (0.29–0.51) |
| FW | 0.75 (0.51–0.89) | 0.10 (0.00–0.35) | 0.14 (0.10–0.19) | 0.55 (0.24–0.74) | 0.15 (0.00–0.44) | 0.30 (0.22–0.42) |
| OG | 0.47 (0.20–0.69) | 0.22 (0.03–0.49) | 0.30 (0.22–0.41) | 0.27 (0.05–0.55) | 0.29 (0.04–0.52) | 0.44 (0.33–0.58) |
| OW | 0.57 (0.19–0.79) | 0.17 (0.00–0.53) | 0.26 (0.19–0.35) | 0.37 (0.02–0.60) | 0.16 (0.00–0.50) | 0.46 (0.35–0.61) |
| PG | 0.76 (0.48–0.84) | 0.02 (0.00–0.30) | 0.21 (0.15–0.31) | 0.72 (0.37–0.83) | 0.05 (0.00–0.38) | 0.23 (0.17–0.34) |
| PW | 0.71 (0.47–0.88) | 0.15 (0.00–0.34) | 0.14 (0.10–0.19) | 0.27 (0.06–0.60) | 0.48 (0.16–0.69) | 0.25 (0.18–0.34) |
| TG | 0.65 (0.43–0.80) | 0.13 (0.01–0.34) | 0.21 (0.15–0.30) | 0.43 (0.12–0.65) | 0.22 (0.17–0.52) | 0.35 (0.26–0.48) |
| TW | 0.61 (0.31–0.88) | 0.24 (0.00–0.54) | 0.15 (0.11–0.20) | 0.33 (0.01–0.71) | 0.35 (0.00–0.67) | 0.32 (0.23–0.45) |
95% confidence intervals are given in parentheses. Values represent the proportional variance in brain volumes attributable to genetic (a2), shared environmental (c2) and individual specific (e2) factors
Genetic and environmental correlations
Cross-twin, cross-structure correlations tended to be substantially higher in the MZ group, suggesting that most of the covariance between variables is genetically mediated. Genetic and unique environmental correlations generated with Cholesky decomposition are given in Table 3; the covariances attributable to the shared environment were substantially smaller than the other two variance components, and the corresponding correlation matrix therefore is not shown. Maximum likelihood estimates for the genetic correlations were quite high, ranging from 0.56 to 0.94. As with the within-individual phenotypic correlations, structures of like tissue type or location tended to be slightly more correlated via genetic factors than those without these similarities. In contrast, unique environmental correlations were, in general, lower and more variable, ranging from −0.22 to 0.86. By far the strongest environmental correlations were between gray matter structures (.0.80), excepting the occipital lobe. Unique environmental correlations between regions of unlike tissue composition tended to be small or even negative.
Table 3. Genetic and unique environmental correlations between cerebral substructures, with 95% confidence intervals in parenthesis.
| FG | FW | OG | OW | PG | PW | TG | TW | |
|---|---|---|---|---|---|---|---|---|
| FG | 1 | .20 (0 .39) | .26 (.07 .43) | −.28 (−.44 −.09) | .82 (.74 .88) | .11 (−.09 .31 | .86 (.80 .91) | −.08 (−.28 .12) |
| FW | .75 (.57 .94) | 1 | −.05 (−.24 .16) | .32 (.14 .49) | .12 (−.09 .32) | .66 (.54 .76) | .04 (−.17 .24) | .52 (.36 .65) |
| OG | .83 (.57 .98) | .69 (.43 .93) | 1 | .41 (.24 .56) | .47 (.30 .62) | .27 (.08 .47) | .39 (.21 .55) | .03 (−.17 .23) |
| OW | .76 (.44 1) | .74 (.56 .96) | .86 (.63 .98) | 1 | −.11 (−.31 .09) | .54 (.38 .67) | −.22 (−.40 −.01) | .43 (.26 .58) |
| PG | .91 (.79 .98) | .58 (.39 .79) | .86 (.68 .97) | .66 (.41 .95) | 1 | .29 (.08 .47) | .81 (.73 .88) | .10 (−.30 .12) |
| PW | .88 (.64 .99) | .91 (.84 .98) | .89 (.68 .99) | .83 (.69 .98) | .74 (.57 .93) | 1 | .11 (−.09 .32) | .63 (.48 .74) |
| TG | .94 (.85 .99) | .75 (.57 .93) | .63 (.40 .85) | .64 (.40 .98) | .79 (.67 .89) | .79 (.60 .94) | 1 | −.13 (−.33 .08) |
| TW | .82 (.49 1) | .92 (.80 1) | .61 (.25 .90) | .72 (.45 .97) | .56 (.29 .88) | .84 (.70 .96) | .81 (.55 1) | 1 |
The diagonal divides genetic (below) from unique environmental (above) correlations. Cross-tissue, within-lobe correlations are in boldface
Exploratory factor analysis
The full 333-ACE IPM fit the data significantly worse than the Cholesky decomposition ( , p-value = .0153) but was preferred by fit statistics favoring parsimony (AIC = −4.65, Δ BIC = −40.80). Attempts to simplify this model resulted in the elimination of one shared environmental common factor and the shared environmental structure-specific factors, with little deterioration in model fit compared to the full IPM (compared to 333-ACE: , p-value = .99, AIC = −24.90). Maximum likelihood parameter estimates from this best-fit model (a 323:AE IPM) are shown in Table 4.
Table 4. Standardized, varimax rotated factor loadings for the best fit exploratory model (3-2-3-AE).
| ROI | Shared factors | Specific factors | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| A1 | A2 | A3 | C1 | C2 | E1 | E2 | E3 | As | Es | |
| FG | 0.57 | 0.37 | 0.28 | 0.43 | −0.15 | 0.47 | 0.01 | −0.04 | 0.00 | 0.13 |
| FW | 0.24 | 0.78 | 0.37 | 0.02 | −0.06 | 0.06 | 0.30 | −0.05 | 0.24 | 0.21 |
| OG | 0.28 | 0.18 | 0.57 | 0.39 | 0.32 | 0.18 | 0.00 | 0.47 | 0.00 | 0.22 |
| OW | 0.22 | 0.46 | 0.48 | 0.08 | 0.28 | −0.15 | 0.26 | 0.31 | 0.38 | 0.31 |
| PG | 0.64 | 0.15 | 0.50 | 0.16 | −0.01 | 0.41 | 0.03 | 0.10 | 0.26 | 0.19 |
| PW | 0.32 | 0.59 | 0.59 | −0.22 | 0.04 | 0.04 | 0.32 | 0.10 | 0.08 | 0.15 |
| TG | 0.58 | 0.39 | 0.21 | 0.34 | 0.08 | 0.44 | −0.03 | 0.04 | 0.34 | 0.18 |
| TW | 0.35 | 0.79 | 0.14 | -0.05 | 0.29 | −0.06 | 0.27 | 0.04 | 0.00 | 0.27 |
As and Es represent ROI-specific factor loadings for additive genetic and unique environmental variance, respectively
For all structures, the largest factor loadings were on genetic factors. The first common genetic factor loaded at least moderately on all structures, but gray structures in particular. The second genetic factor clearly loaded preferentially on white matter structures, while the third had high loadings primarily on the posterior cerebrum (occipital and parietal lobes), irrespective of tissue type. The shared environmental factor loadings were substantially smaller than the genetic loadings. The first factor primarily loaded on frontal, occipital, and temporal gray, with the second on the occipital lobe and temporal white matter. The three unique environmental common factors were clearly identifiable as a gray matter, white matter, and an occipital lobe factor, respectively.
Confirmatory factor analysis
The full ACE MTMM model did not fit the data significantly worse than the more parameterized Cholesky decomposition (Cholesky decomposition: −2LL 20882.58, 180 parameters; MTMM Full model −2LL 20893.70, 165 parameters, , p-value = 0.74, AIC = −18.88). Similarly, the removal of shared environmental factors had little effect on the fit of the model (compared to Full MTMM Model: , p-value = 0.80, AIC = −37.81) and had a substantially better fit than the best-fit exploratory model. As the correlation between the gray and white matter factors was near unity for both genetic and environmental sources of variance, these could be fixed to one without a significant effect on model fit or parameter estimates (compared to Full model: −2LL 20919.36, , p-value = .8192, AIC = −40.34). Fixing these values does, however, simplify presentation of the complex model since the two tissue factors can be represented as a single, common factor. Though many correlations between the lobar factors were high, these could not be fixed to unity without substantial deterioration in model fit for both genetic ( , p-value ,.0001, AIC = 10.47) and environmental ( , p-value, .0006, AIC = −2.634) variance components.
The standardized maximum likelihood estimates from this model are presented in Fig. 2. The vast majority of the variance in all cerebral structures was attributable to genetic factors common to lobes. These factors, in turn, were highly correlated. The subtle differences in pheno-typic correlations between gray and white matter could be explained by the second genetic factor which had modest loadings, but with negative loadings on white matter structures. These negative correlations produced negative partial covariances between ROIs of differing tissue type. Though loadings were substantially lower, a very similar pattern emerged for environmental variance.
Fig. 2.
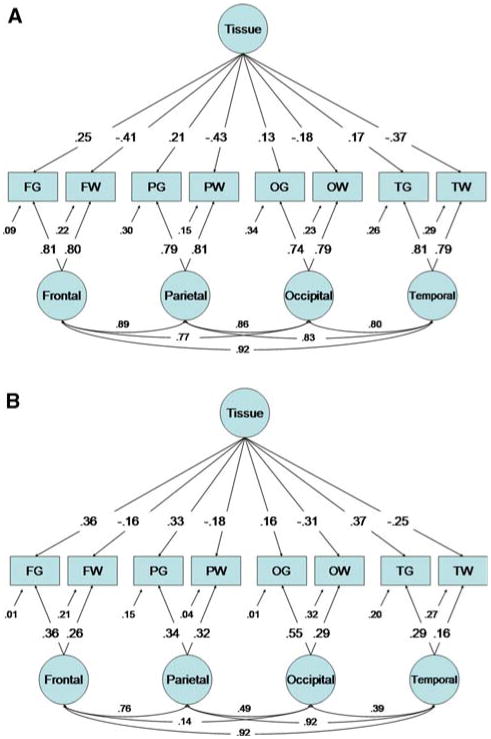
Maximum likelihood parameter estimates for the best-fit MTMM model. For simplicity, genetic (panel A) and unique environmental (panel B) factor loadings are shown separately
Models including global covariates
Univariate analysis of TBV demonstrated high heritability (a2 = 0.89 [.65–0.92], c2 = 0.00 [0.00–0.23], e2 = 0.11 [0.08–0.16]). The Cholesky decomposition allowing for linear effects oftotal brain volume on mean cerebral volumes fit the data substantially better (−2LL = 19,102.57) than the model without this regressor (−2LL = 20,882.58). After adding the covariate, the total residual variance decreased precipitously for all regions (Fig. 3). Without a global covariate, additive genetic was the largest variance component for all ROIs; the use of a global covariate had a disproportionately greater influence on the genetic variance compared to the environmental variance components (Table 2). Thus, after controlling for global effects, additive genetic factors became relatively less important in explaining the residual variance. This was expected because the covariate itself was both strongly heritable and highly correlated with lobar volumes.
Fig. 3.
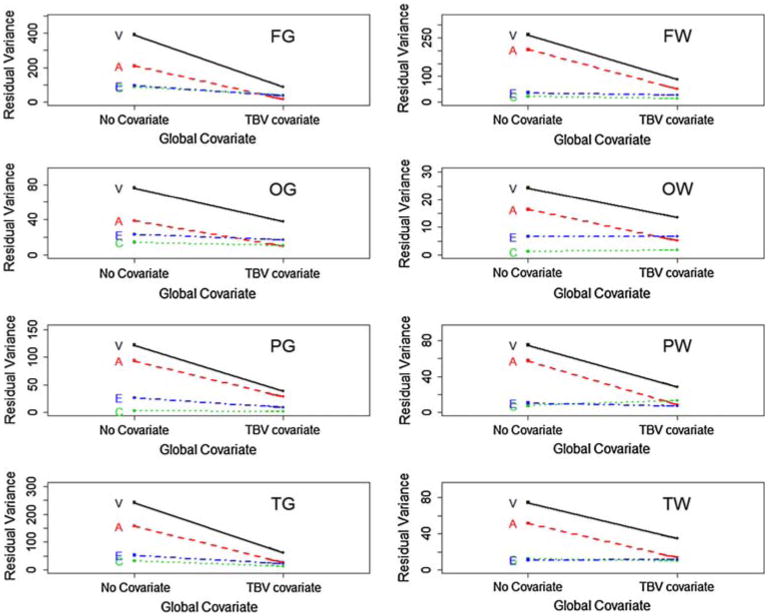
Residual variance for variance components with and without a global covariate, organized by cerebral region of interest. Variance components (A, C, and E) are labeled and shown as broken lines. Total variance (V) is shown as a solid line
Table 5 provides correlations for individual variance components from this model. Unlike the strong positive genetic correlations observed for absolute volumes, genetic correlations between TBV-covaried structures varied widely and were often negative. For all variance components, negative correlations tendedtobebetween gray and white matter structures. In effect, controlling for TBV removed a sizable, positive partial correlation between all structures.
Table 5. Genetic, shared environmental, and unique environmental correlations after use of a global covariate on mean volumes for all structures.
| FG | FW | OG | OW | PG | PW | TG | TW | |
|---|---|---|---|---|---|---|---|---|
| Genetic | ||||||||
| FG | 1.00 | |||||||
| FW | −0.94 | 1.00 | ||||||
| OG | 0.21 | −0.33 | 1.00 | |||||
| OW | −0.36 | 0.22 | 0.34 | 1.00 | ||||
| PG | 0.80 | −0.63 | 0.46 | −0.21 | 1.00 | |||
| PW | −0.67 | 0.63 | 0.26 | 0.34 | −0.22 | 1.00 | ||
| TG | 0.52 | −0.41 | −0.62 | −0.56 | 0.19 | −0.56 | 1.00 | |
| TW | −0.66 | 0.67 | −0.56 | 0.26 | −0.65 | 0.21 | −0.25 | 1.00 |
| Shared environmental | ||||||||
| FG | 1.00 | |||||||
| FW | −0.12 | 1.00 | ||||||
| OG | −0.15 | −0.96 | 1.00 | |||||
| OW | −0.91 | −0.19 | 0.40 | 1.00 | ||||
| PG | −0.32 | 0.11 | −0.07 | 0.41 | 1.00 | |||
| PW | −0.69 | 0.62 | −0.47 | 0.60 | 0.38 | 1.00 | ||
| TG | 0.34 | −0.89 | 0.81 | −0.12 | −0.53 | −0.76 | 1.00 | |
| TW | −0.70 | 0.33 | −0.13 | 0.51 | −0.36 | 0.63 | −0.22 | 1.00 |
| Unique environmental | ||||||||
| FG | 1.00 | |||||||
| FW | −0.45 | 1.00 | ||||||
| OG | −0.30 | −0.41 | 1.00 | |||||
| OW | −0.76 | 0.30 | 0.37 | 1.00 | ||||
| PG | 0.51 | −0.66 | 0.12 | −0.51 | 1.00 | |||
| PW | −0.72 | 0.54 | −0.02 | 0.56 | −0.43 | 1.00 | ||
| TG | 0.66 | −0.67 | −0.01 | −0.59 | 0.52 | −0.63 | 1.00 | |
| TW | −0.58 | 0.49 | −0.13 | 0.44 | −0.64 | 0.62 | −0.58 | 1.00 |
Within-lobe correlations are shown in boldface
Attempts to model the relationships between TBV-covaried structures were only marginally successful. The 333-IPM fit worse than Cholesky decomposition based on χ2 but not by AIC ( , p-value = .0048, AIC = −0.47). As with the initial analyses on absolute volumes, the 323-AE IPM was found to be the most parsimonious model after a stepwise removal of latent factors (compared to Cholesky: , p-value = 0.13, AIC = −25.55; compared to 333-IPM: , p-value = 0.99, AIC = −25.09).
Table 6 reports the parameter estimates from this model, which were substantially more difficult to interpret than those on absolute volumes. Genetic effects were no longer predominant. Additionally, strong negative factor loadings were a prominent feature of the TBV-covaried factor models. The common factors still demonstrated evidence of the gray/white dichotomy, but these effects were somewhat obscured by loadings that did not fit the pattern. For example, the most prominent loadings for the third genetic factor were a positive loading on occipital gray matter, and a negative loading on temporal gray; these structures were uncorrelated via other common genetic factors.
Table 6. Varimax-rotated, standardized maximum likelihood parameter estimates of best-fit exploratory model after covarying for total cerebral volume. 1.
| ROI | A1 | A2 | A3 | C1 | C2 | E1 | E2 | E3 | As | Es |
|---|---|---|---|---|---|---|---|---|---|---|
| FG | −0.28 | 0.27 | −0.08 | 0.64 | 0.00 | 0.52 | 0.20 | −0.18 | 0.19 | 0.24 |
| FW | 0.62 | −0.37 | 0.01 | −0.07 | 0.40 | −0.31 | 0.28 | 0.26 | 0.00 | 0.26 |
| OG | 0.00 | 0.29 | 0.43 | −0.09 | −0.52 | −0.04 | −0.57 | −0.06 | 0.00 | 0.34 |
| OW | 0.04 | −0.09 | 0.35 | −0.39 | −0.05 | −0.45 | −0.29 | 0.26 | 0.47 | 0.38 |
| PG | −0.20 | 0.76 | −0.04 | −0.04 | 0.03 | 0.16 | −0.04 | −0.46 | 0.38 | 0.00 |
| PW | 0.34 | -0.02 | 0.11 | −0.53 | 0.38 | −0.42 | 0.02 | 0.09 | 0.44 | 0.27 |
| TG | −0.27 | 0.01 | −0.58 | 0.18 | −0.46 | 0.44 | −0.04 | 0.17 | 0.00 | 0.35 |
| TW | 0.09 | −0.51 | 0.03 | −0.40 | 0.16 | −0.33 | 0.07 | 0.28 | 0.44 | 0.39 |
Confirmatory analyses were somewhat more successful. The Full MTMM did not fit significantly worse than Cholesky decomposition (−2LL = 19,119.070, , p-value = .42, AIC = −16.50). Parameter estimates from the best-fit MTMM submodel (−2LL 19149.989; relative to Cholesky , p-value = .79, AIC = −64.581; relative to full MTMM , p-value = .85, AIC = −49.01) are shown in Fig. 4. The most striking differences compared to the MTMM without a global covariate were (1) the substantial decrease in variance explained by genetic lobar factors, (2) a reversal in the correlations between these factors from strongly positive to negative, and (3) a relative increase in the importance of the path coefficients of the common genetic factor with a similar pattern of positive and negative values.
Fig. 4.
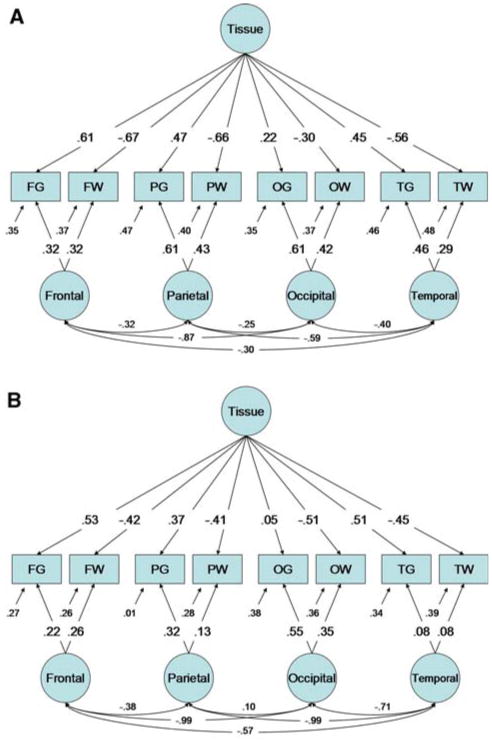
Maximum likelihood parameter estimates for the best-fit MTMM model after covarying for total brain volume. For clarity, genetic (panel A) and unique environmental (panel B) factor loadings are shown separately
Discussion
A multivariate analysis of cerebral volumes suggests that genetic factors play the dominant role in the generation of interlobar covariance. These findings are similar to a prior volumetric study on more ontogenetically diverse regions of interest, which found a single genetic factor explaining most of the observed covariance between cerebrum, cerebellum, caudate, and corpus callosum (Schmitt et al. 2007a). Because of the availability of segmentation data in the cerebrum, however, in the analyses reported here we were able to detect subtle differences in the relationships between gray and white matter. Genetic correlation matrices, EFA, and CFA all suggest that genetically mediated covariance is greater between brain regions of similar tissue type. Since segmentation data from the rest of the brain are not available, it is unclear whether these relationships are unique to the cerebrum, or rather are part of more widespread genetically mediated tissue patterning. Though a few studies also have reported cerebral lobar heritability (Carmelli et al. 2002; Geschwind et al. 2002; Wallace et al. 2006), the present analyses are unfortunately the only extant information on intralobar volumetric relationships. However, the prior observation by Barré et al. of a 0.68 genetic correlation between total brain gray and white matter is, broadly speaking, consistent with the more detailed analyses reported here (Baaré et al. 2001).
Despite overwhelming evidence that brain volumes are highly heritable, the specific genetic contributors to typical human brain variation are unknown at present. However, numerous genes have been linked to neurological diseases with abnormalities in brain size and morphology, including several forms of microcephaly and lissencephaly (Gaitanis and Walsh 2004; Guerrini and Marini 2006). There is increasing evidence that several of these genes experienced rapid evolutionary change in humans relative to other primates (Preuss et al. 2004). Given their profound influence on brain volumes, the genes implicated in microcephaly are of particular relevance to the present study. Several genes that cause microcephaly, including microcephalin (MCPH1), ASPM, CENPJ, and NDE1 have well-documented roles in cell division and cell growth (Chae and Walsh 2007). As neocortical expansion throughout evolution has been hypothesized to have occurred via increases in the number of progenitor cell divisions (Rakic 1995), intra-species variation in genes such as these may have driven the evolution of the relatively large human brain.
Further, recent studies of evolutionarily novel polymorphisms have suggested that the human brain continues to evolve. For example, a novel variant in MCPH1 that appeared approximately 37,000 years ago has spread throughout the human population at a rate that suggests it was positively selected (Evans et al. 2005). Similar evidence exists for a polymorphism of ASPM that originated only 6,000 years ago (Mekel-Bobrov et al. 2005). Behavioral genetic data supports the hypothesis that the human brain is a work in progress, as the extraordinarily high metabolic cost of brain tissue should place a selective pressure towards smaller brains and ultimate fixation of genetic polymorphisms affecting brain volumes. In contrast, imaging data has established that genetic variance not only persists but by far the predominant determinant of brain size. Although decades of neuropsychological research has established that brain volumes are generally poor predictors of cognitive abilities in human populations, the persistence of genetic variation within the cortex suggests that through much of human history cortical size produced a selective advantage that was greater than its metabolic cost.
Although of great interest to scientists and laypersons alike, the genetics of brain evolution remains an enigmatic field that intersects a diverse cadre of other scholarly fields including anthropology, the neurosciences, molecular and population genetics, philosophy, and medicine. A central question in brain evolution is whether neuroanatomic regions tend to evolve independently of one another based on their behavioral specializations, or rather evolve together as part of a more global evolutionary process. Finlay and Darlington established that TBV is correlated .96% with regional substructure volumes (with the exception of the olfactory bulb) when they examined data from 131 mammalian species (Darlington et al. 1999; Finlay and Darlington 1995). Such strong correlations have been hypothesized to reflect a generalized adaption to specific selective pressures; although it is more energetically expensive to expand the computational resources of the entire brain when only specific functions are needed, the molecular adjustments required are far fewer than those required to completely repattern gross neural architecture. The multivariate analyses reported here are generally concordant with this hypothesis, as these data suggest that most of the variance in lobar brain volumes is caused by genetic factors that are shared between regions. This finding is expected if a significant fraction of the genes that cause inter-species variation in neuroanatomic volumes also cause individual differences in human brain volumes. Further molecular genetic studies are required, however, to confirm this hypothesis.
As the cerebrum is the largest structure in the brain, it was not surprising that its eight major subcomponents were highly influenced by total brain volume. It is interesting, however, that the use of the TBV disproportionately decreased the additive genetic variance component with respect to the environmental components, reducing the heritability of all regions of interest substantially. This effect suggests strong genetic correlations between TBV and the cerebral ROIs in this study. In other words, a strong—single genetic factor or several highly correlated factors drives covariation between the observed variables. When the effects of this factor are largely removed by adding the TBV covariate, the positive partial correlations it generates also disappear, producing more complex interrelationships within the residual variance. The MTMM factor structure still fit well after the covariate was added, but the importance of the lobar factors relative to a common ‘‘tissue’’ factor was reversed and the interlobar correlations flipped to negative values. Thus, both the exploratory and hypothesis-driven models suggest strong global genetic effects mask more subtle and competing genetic and nongenetic factors in the pediatric brain.
Limitations
Although provocative, the results of the present study must be considered in the light of several limitations inherent in the methods. First, our sample was limited to healthy children and findings may be different in other populations. In particular, our stringent selection criteria, which excluded children with psychiatric or neurological risk factors, likely eliminated a substantial fraction of the true population variance in the measured endophenotypes. As no complete population based pediatric twin imaging sample currently exists, it is difficult to estimate the effect of these selection factors. Secondly, the analyses reported here are complex, involving sophisticated computational algorithms for the generation of MRI images, segmentation and parcellation of these images into brain volumes, and optimization of behavioral genetic models. Although these techniques give us the ability to ask more sophisticated questions about the genetics of brain structure than many other methods, they also rely on far more assumptions. Third, our study focused on brain volumes alone. A recent study of Vietnam era twins has suggested that the genetic factors exert their effects on brain volumes through at least two largely independent pathways, one that influences cortical thickness and one surface area (Panizzon et al. 2009). From the present data, it is unclear whether genetic factors influencing thickness, surface area, or both, are responsible for the observed genetic covariation between cerebral lobes. It is noteworthy that our group has identified a large common genetic factor influencing 54 gyral-level measures of cortical thickness (Schmitt et al. 2008) previously, but it is likely that surface area also is patterned at least in part via global genetic factors. Fourth, although one of the largest imaging studies currently available, our sample size is modest by the standards of population genetics.
Finally, although highly heritable, it is important to remember that the role of environment can not be over-stressed in the process of brain development. For example, environmental factors have well-documented effects on synaptogenesis and pruning, gray matter volumes, and downstream cognitive measures such as memory and intelligence. Our data suggests that environmental factors play a relatively lesser role in the variance in volumes within the population studied (American children), but it is possible that samples with a broader range of environmental exposures (international or older populations) may exhibit a more pronounced effect of the environment on neuroanatomic endophenotypes.
Acknowledgments
This research was supported by the intramural research program of the NIH as well as NIH grants MH-65322, MH-20030, and DA-18673.
Contributor Information
J. Eric Schmitt, Email: eric.schmitt@stanfordalumni.org, schmittje@vcu.edu, Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, 800 East Leigh Street, suite 100, PO Box 980003, Richmond, VA 23298-0003, USA.
Gregory L. Wallace, Pediatric Imaging Unit, Child Psychiatry Branch, National Institute of Mental Health, Bethesda, MD, USA
Rhoshel K. Lenroot, Pediatric Imaging Unit, Child Psychiatry Branch, National Institute of Mental Health, Bethesda, MD, USA
Sarah E. Ordaz, Department of Psychology, University of Pittsburgh, Pittsburgh, PA, USA
Dede Greenstein, Pediatric Imaging Unit, Child Psychiatry Branch, National Institute of Mental Health, Bethesda, MD, USA.
Liv Clasen, Pediatric Imaging Unit, Child Psychiatry Branch, National Institute of Mental Health, Bethesda, MD, USA.
Kenneth S. Kendler, Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, 800 East Leigh Street, suite 100, PO Box 980003, Richmond, VA 23298-0003, USA
Michael C. Neale, Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, 800 East Leigh Street, suite 100, PO Box 980003, Richmond, VA 23298-0003, USA
Jay N. Giedd, Pediatric Imaging Unit, Child Psychiatry Branch, National Institute of Mental Health, Bethesda, MD, USA
References
- Akaike H. Factor analysis and AIC. Psychometrika. 1987;52:317–332. [Google Scholar]
- Baaré WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, De Geus EJ, Schnack HG, Van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Bechger TM, Maris G. Structural equation modeling of multiple facet data: extending models for multitrait-multimethod data. Psicologica. 2004;25:253–274. [Google Scholar]
- Campbell DT, Fiske DW. Convergent and discriminant validation by the multitrait-multimethod matrix. Psychol Bull. 1959;56:81–105. [PubMed] [Google Scholar]
- Carmelli D, Swan GE, DeCarli C, Reed T. Quantitative genetic modeling of regional brain volumes and cognitive performance in older male twins. Biol Psychol. 2002;61:139–155. doi: 10.1016/s0301-0511(02)00056-x. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- Chae TH, Walsh CA. Genes that control the size of the cerebral cortex. In: Bock G, Goode J, editors. Cortical development: genes and genetic abnormalities. Wiley, Chichester; 2007. pp. 79–90. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in Standardized Talairach Space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Coromina L, Coenders G. Reliability and validity of egocen-tered network data collected via web—a meta-analysis of multilevel multitrait, multimethod studies. Soc Netw. 2006;28:209–231. [Google Scholar]
- Corten IW, Saris WE, Coenders G, van der Veld W, Aalberts CE, Kornelis C. Fit of different models for multitrait-multimethod experiments. Struct Equ Model. 2002;9:213–232. [Google Scholar]
- Darlington RB, Dunlop SA, Finlay BL. Neural development in metatherian and eutherian mammals: variation and constraint. J Comp Neurol. 1999;411:359–368. [PubMed] [Google Scholar]
- Edwards AWF. Likelihood. Cambridge; London: 1972. [Google Scholar]
- Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, Tishkoff SA, Hudson RR, Lahn BT. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Gaitanis JN, Walsh CA. Genetics of disorders of cortical development. Neuroimaging Clin N Am. 2004;14:219–229. doi: 10.1016/j.nic.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci USA. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini R, Marini C. Genetic malformations of cortical development. Exp Brain Res. 2006;173:322–333. doi: 10.1007/s00221-006-0501-z. [DOI] [PubMed] [Google Scholar]
- Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Statist. 1996:299–314. [Google Scholar]
- Jenkins TW, Truex RC. Dissection of the human brain as a method for its fractionation by weight. Anat Rec. 1963;147:359–366. doi: 10.1002/ar.1091470307. [DOI] [PubMed] [Google Scholar]
- Jöreskog KG. Statistical analysis of sets of congeneric tests. Psychometrika. 1971;52:101. [Google Scholar]
- Kendler KS, Heath AC, Martin NG, Eaves LJ. Symptoms of anxiety and symptoms of depression—same genes, different environments. Arch Gen Psychiatry. 1987;44:451–457. doi: 10.1001/archpsyc.1987.01800170073010. [DOI] [PubMed] [Google Scholar]
- Mcardle JJ, Goldsmith HH. Alternative common factor models for multivariate biometric analyses. Behav Genet. 1990;20:569–608. doi: 10.1007/BF01065873. [DOI] [PubMed] [Google Scholar]
- Mekel-Bobrov N, Gilbert SL, Evans PD, Vallender EJ, Anderson JR, Hudson RR, Tishkoff SA, Lahn BT. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Kluwer, Dordrecht: 1992. [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: statistical modeling. Department of Psychiatry, Medical College of Virginia; Virginia Commonwealth University, Richmond, VA: 2002. [Google Scholar]
- Panizzon M, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, Xian H, Tsuang M, Fischl B, Seidman LJ, Dale A, Kremen WS. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC. A twin MRI study of size variations in human brain. J Cogn Neurosci. 2000;12:223–232. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Peper J, Schnack H, Brouwer R, van Baal C, van Leeuwen M, Collins L, Evans A, Boomsma D, Kahn R, Poll HH. Heritability of brain structure at the onset of puberty: an MRI study in 9-year old twin-pairs. Human Brain Map. 2009;30:2184–2196. doi: 10.1002/hbm.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJ, Neale MC, Hulshoff Pol HE, Baare WEC, Kahn RS, Boomsma D. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 2000;30:311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Caceres M, Oldham MC, Geschwind DH. Human brain evolution: insights from microarrays. Nat Rev Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: 2005. [Google Scholar]
- Rakic P. A small step for the cell, a giant leap for mankind—a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Wallace GL, Rosenthal MA, Molloy EA, Ordaz S, Lenroot R, Clasen LS, Blumenthal J, Kendler KS, Neale MC, Giedd JN. A multivariate analysis of neuroanatomic relationships in a genetically informative pediatric sample. Neuroimage. 2007a;35:70–82. doi: 10.1016/j.neuroimage.2006.04.232. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Eyler LT, Giedd JN, Kremen WS, Kendler KS, Neale MC. Review of twin and family studies on neuroanatomic phenotypes and typical neurodevelopment. Twin Res Hum Genet. 2007b;10:683–694. doi: 10.1375/twin.10.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Lenroot RK, Wallace GL, Ordaz S, Taylor KN, Kabani N, Greenstein D, Lerch JP, Kendler KS, Neale MC, Giedd JN. Identification of genetically mediated cortical networks: a multivariate study of pediatric twins and siblings. Cereb Cortex. 2008;18:1737–1747. doi: 10.1093/cercor/bhm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Tomás JM, Hontangas PM, Oliver A. Linear confirmatory factor models to evaluate multitrait-multimethod matrices: the effects of number of indicators and correlation among methods. Multivar Behav Res. 2000;35:469–499. doi: 10.1207/S15327906MBR3504_03. [DOI] [PubMed] [Google Scholar]
- Wallace GL, Schmitt JE, Lenroot R, Viding E, Ordaz S, Rosenthal MA, Molloy EA, Clasen LS, Kendler KS, Neale MC, Giedd JN. A pediatric twin study of brain morphometry. J Child Psychol Psychiatry. 2006;47:987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- Zijdenbos AP, Forghani R, Evans AC. Automatic ‘‘pipeline’’ analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans Med Imaging. 2002;21:1280–1291. doi: 10.1109/TMI.2002.806283. [DOI] [PubMed] [Google Scholar]


