Abstract
The human immunodeficiency virus type I (HIV) Tat protein, a potent activator of HIV gene expression, is essential for integrated viral genome expression and represents a potential antiviral target. Tat binds the 5′ terminal region of HIV mRNA’s stem-bulge-loop structure, the Trans-activation Responsive (TAR) element to activate transcription. We find that didehydro-Cortistatin A (dCA), an analogue of a natural steroidal alkaloid from a marine sponge inhibits Tat-mediated trans-activation of the integrated provirus by binding specifically to the TAR-binding domain of Tat. Working at subnanomolar concentrations, dCA reduces Tat mediated transcriptional initiation/elongation from the viral promoter to inhibit HIV-1 and HIV-2 replication in acutely and chronically infected cells. Importantly, dCA abrogates spontaneous viral particle release from CD4+T cells from virally suppressed subjects on highly active antiretroviral therapy (HAART). Thus, dCA defines a unique class of anti-HIV drugs that may inhibit viral production from stable reservoirs and reduce residual viremia during HAART.
Keywords: HIV-1, HIV-2, Tat, inhibition, compound, anti-retroviral drugs, Tat activated transcription, HAART, HIV persistence
Introduction
Although treatment with antiretroviral drugs (ARVs) has extended the quality and expectancy of life for people infected with HIV, it has been unsuccessful in curing HIV infection. ARVs fall into the following major classes: fusion inhibitors (FIs), nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), nucleotide reverse transcriptase inhibitors (NtRTIs), protease inhibitors (PIs) and integrase inhibitors (INIs). Highly active antiretroviral therapy (HAART) is based on triple or quadruple combinations of ARVs (Gulick et al., 1997; Hammer et al., 1997); however, while reducing HIV to very low levels, this treatment fails to eliminate the infection completely (Chun et al., 1997; Finzi et al., 1997; Wong et al., 1997). Ultrasensitive assays revealed that HIV persists in latently and productively infected CD4+T cells in the peripheral blood of individuals receiving HAART who have maintained undetectable plasma viremia for prolonged periods of time (Chun et al., 2005; Palmer et al., 2008). Residual viremia is not affected by the addition of an integrase or a fusion inhibitor to a stable HAART regimen (Dinoso et al., 2009; Gandhi et al., 2010; Yukl et al., 2010), suggesting that it originates from long-lived stable reservoirs that contain an integrated provirus that continuously produces viral particles despite HAART. Viral production from these cellular reservoirs, resulting from continuous transcription of an integrated viral genome, is not affected by current ARVs. As such, other classes of ARVs are needed to inhibit this process.
Tat is a 14 kDa protein that is synthesized from an mRNA joined from 2 coding exons. The first exon encodes amino acids 1–72 and (in most strains of HIV-1) the second exon encodes amino acids 73–101. An N-terminal 86-amino acid form of Tat produced in a few laboratory-passaged virus strains, is frequently used to study Tat. This potent activator of HIV gene expression is essential for the synthesis of full-length transcripts of the integrated viral genome by RNA polymerase II (RNAPII) (Okamoto and Wong-Staal, 1986; Selby et al., 1989; Sodroski et al., 1985). Tat mediates association between the host positive transcription elongation factor (pTEFb) complex and the trans-activation-responsive element (TAR) of the nascent viral RNA to promote transcriptional elongation from the viral promoter (Fujinaga et al., 1998; Laspia et al., 1989; Mancebo et al., 1997). Given Tat’s crucial requirement for viral gene expression, it has been the target for the development of several compounds (for review (Baba, 2006; De Clercq, 1995; Richter and Palu, 2006)); however, these compounds have low efficacy in vivo and none have reached the clinic.
Cortistatin A (CA) is a recently discovered natural steroidal alkaloid isolated from the marine sponge Corticium simplex (Aoki et al., 2006), and it has been reported to display anti-proliferative properties towards human umbilical vein endothelial cells (HUVECs) with an average half-maximal inhibitory concentration (IC50) of 0.35 μM (Aoki et al., 2006; Aoki et al., 2007). The scarce natural supply prompted the chemical synthesis of didehydro-Cortistatin A (dCA), the equipotent analogue of CA, starting from the inexpensive and abundant steroid prednisone and requiring only 13 steps for its synthesis of gram quantities of material in a cost-effective manner (Shi et al., 2011; Shi et al., 2009) (Figure 1A).
Figure 1. Structure and activity of dCA on HIV-1 expression.
A) Structure of CA and of its analog dCA (B) Activity of dCA on acute replication of HIV-1 at 3 different MOIs. Quantitative CPRG assay. (C) Effect of pre-treating cells with dCA on acute HIV-1 replication. HeLa-CD4 cells were either treated or not with increasing concentrations of dCA. HIV-1 pNL4-3 was added 24 h later to the cells of both experiment sets (MOI≫10) in the presence of testing compound or DMSO control. CPRG assay performed 48 h later. (D) dCA does not block HIV-1 proviral integration into host cells. HeLa-CD4 cells were infected with pNL4-3 (MOI ≫1) in the presence of dCA. Total DNA was extracted 24 h later and integrated provirus was determined by qPCR. (E) Analysis of viral mRNA expression. Total RNA was extracted 3 days after acute infection with pNL4-3 (MOI≫1) in the presence of dCA. First-strand cDNA was quantified by qPCR using primers directed to the env and LTR regions. Results were normalized as copies of viral mRNA per copy of GAPDH mRNA. The arbitrary value of 1 was assigned to the amount of viral mRNA generated in the absence of dCA. RNA samples not reverse transcribed were used as negative control. Error bars represent standard deviations. (F) Viral mRNA expression levels upon dCA treatment of chronically infected cells. HeLa-CD4 cells chronically infected with pNL4-3 were treated with dCA for 10 or 60 days, total RNA was extracted, reverse transcribed and viral cDNA was quantified as in (E). (G) p24 antigen quantification. Viral supernatants recovered from cells described in (B) grown for 60 days, were assayed for their p24 antigen content using a sandwich ELISA kit. Error bars represent standard deviations. (H) Cell viability of HeLa-CD4 cells chronically infected with pNL43 and long term treated with dCA. MTT assay on HeLa-CD4 cells incubated with increasing concentrations of dCA for 12 months. Results are representative of 3 independent experiments. (I) Viral RNA levels upon treatment of CEM-SS cells with dCA and known antiviral drugs. CEM-SS cells chronically infected with pNL4-3 were treated with the indicated compounds for 7 days. Quantification of viral RNA was performed as in (E). Error bars represent standard deviations. Results are representative of 4 independent experiments. See also Figure S1.
Here we report that dCA potently and selectively inhibits Tat-mediated trans-activation of the integrated HIV provirus. dCA binds specifically to the TAR-binding domain of Tat and as a consequence, reduces cell-associated HIV-1 viral RNA and capsid p24 antigen production in acutely and chronically infected cultured and primary cells at an half-maximal effective concentration (EC50) as low as 0.7 pM. Moreover, in vitro dCA abrogates low-level virus replication from primary cells isolated from patients undergoing HAART treatment. In total, these results define dCA as a potential anti-HIV drug that could be used to decrease residual viremia during HAART.
RESULTS
dCA Inhibits HIV transcriptional activity
We previously reported that eukaryotic initiation factor 3 subunit f (eIF3f) mediates restriction of HIV-1 RNA 3′ end-processing, through the involvement of a set of factors that includes eIF3f, the SR protein 9G8, and cyclin-dependent kinase 11 (CDK11) (Valente et al., 2009a; Valente et al., 2009b). These data suggested that a CDK11 inhibitor might possess anti-HIV activity. Given that CA was reported to bind with high affinity CDK11 (Cee et al., 2009), we examined the ability of its analogue dCA to decrease HIV production by interfering with CDK11 activity. While we did not confirm an effect of dCA on CDK11 activity, we discovered a potent activity as an inhibitor of HIV-1 transcription (Figure S1A, B).
HIV-1 susceptibility to dCA was assayed using a reporter cell line that stably expresses the β-galactosidase (LacZ) gene; LacZ expression is driven by the 5′ long terminal repeat (LTR) of HIV-1 and responds to Tat expressed by an incoming virus. HeLa-CD4-LacZ cells were infected with HIV-1 at different multiplicities of infection (MOIs) in the presence of increasing concentrations of dCA, and β-gal activity was determined (Figure 1B). Inhibition of transcription was dose-dependent, with an EC50 as low as 2.6 nM at the highest, and 0.7 pM at the lowest MOI; the lower MOI is more representative of biological amounts of virus found in infected subjects. Pre-treatment of cells with dCA for 24 h prior to infection resulted in a 7-fold reduction in the EC50 (Figure 1C), suggesting that dCA potency depends on the time of addition or target concentration. Following acute infection, maximal inhibition levelled off at 75–85%, possibly due to the inability of dCA to block Tat-independent HIV transcription. Transcription of the HIV-1 provirus is regulated by both viral and cellular transcription factors. Before Tat is produced, low-level basal transcription from the viral promoter is initiated by cellular factors, such as the nuclear factor-kappa B (NF-κB) (Nabel and Baltimore, 1987), Sp1 (Jones et al., 1986), TATA-binding protein (Olsen and Rosen, 1992) and RNAPII. A desirable Tat inhibitor should block Tat-mediated activation of the viral promoter without affecting its basal transcription, which would result in cellular toxicity, given the shared transcription factors of the HIV promoter and cellular genes. The effective concentrations of dCA did not compromise HeLa-CD4 cell viability, as a half-maximal cytotoxic concentration (CC50) of 20 μM was observed (Figure S1C, D).
To assess whether the viral block mediated by dCA occurs before or after integration of proviral DNA into the host cell chromosome, HeLa-CD4 cells were infected with HIV-1, treated with different concentrations of dCA and total cellular DNA was extracted for quantification of proviral DNA by real time quantitative PCR (qPCR). Such an early time point rules out de novo infections. The presence of dCA did not change integrated HIV DNA copy number as compared with DMSO-treated cells (Figure 1D). These results are consistent with a viral block by dCA at a step following integration of the provirus into the host chromosome. Viral mRNA expression in cells treated with increasing concentrations of dCA was then measured by reverse transcription (RT) qPCR. A drastic dose-dependent reduction in the total amount of viral RNA in infected cells was detected (Figure 1E), suggesting that dCA inhibits transcription from the integrated viral promoter. This conclusion was further supported by the fact that treatment of cells that were chronically infected, and therefore continuously shedding the virus (without incurring new infections due to down-regulation of CD4 from the cell surface), reduced virus production to undetectable levels (Figure 1F). Treatment of chronically HIV-1 infected HeLa-CD4 cells with dCA for 10 or 60 days resulted in a drastic reduction of viral cellular RNA levels with an EC50 as low as 0.1 nM and an EC90 of less than 10 nM (Figure 1F). Moreover, we observed a continuous reduction in viral RNA levels in the cell with increased treatment times with dCA, an expected result since Tat inhibition decreases Tat production. Results were similar when viral release from cells treated for 60 days were measured by p24 enzyme-linked immunosorbent assay (ELISA) (Figure 1G). Prolonged treatment of cells with dCA did not alter cell viability (Figure 1H). Furthermore dCA, but not other ARVs, reduced viral RNA levels in the lymphocytic cell line CEM-SS chronically infected with pNL4-3 (Figure 1I), demonstrating that the effect of dCA is not only cell-type independent but also that they extend to the reduction of viral expression from cells already infected, a result that none of the currently used ARVs is able to achieve.
dCA is a Tat inhibitor
To determine whether dCA directly impacts Tat-mediated transcriptional activation from the viral promoter, recombinant Tat protein was added in the presence or absence of dCA to HeLa-CD4 cells stably expressing a construct containing the HIV-1 5′LTR promoter driving luciferase expression (LTR-Luc). Transcriptional repression was observed in a dCA dose-dependent manner (Figure 2A). Similar results were obtained when a Tat-encoding plasmid was transfected into these cells (Figure 2B). These results suggest that dCA blocks Tat transcriptional activation of the HIV-1 promoter. Reporter transcription from heat-inactivated recombinant Tat protein (Figure 2A), which was used as negative control, reflects Tat-independent activation of the HIV promoter, and accounts for approximately 20% of maximal activation. This result supports the notion that the observed maximum inhibition plateau at 75–85% during acute infection is due to Tat-independent activity of the promoter. In line with these results, dCA did not alter basal transcription from the LTR promoter in the absence of Tat, nor when Phorbol 12-Myristate 13-Acetate (PMA) (Siekevitz et al., 1987) was used to activate promoter transcription via NF-κB (Figure S2A). Furthermore, dCA showed no effect on TNF-α activation of an NF-κB reporter construct (Duh et al., 1989) (Figure S2B), showing that inhibition by dCA is independent of NF-κB. Transcription from CXCR4, herpes simplex thymidine kinase (TK), phosphoglycerate kinase (PGK) or CD4 promoters was also not affected by dCA (Figures S2C and S2D).
Figure 2. dCA binds to Tat and inhibits Tat transactivation of the HIV-1 LTR.
(A) dCA prevents transactivation of the HIV-1 promoter by recombinant Tat. HeLa-CD4-LTR-Luc cells were treated with 0.1 μM recombinant Tat protein with increasing concentrations of dCA. Chloroquine 100 μM was added to increase Tat uptake and was added to all points except untreated wells. Luciferase activity was measured 24 h later using the Luciferase Assay System (Promega). Luciferase activity per protein concentration of each sample is shown as relative light units (RLU). HI: Heat Inactivated. (B) HeLa-LTR-Luc cells were transfected with 2 μg of a construct expressing Tat-flag driven by the thymidine kinase (TK) promoter. Twenty-four hours later cells were split and treated or not with dCA at the indicated concentrations. RLU determined 40 h later as in (A). DMSO point set as 100% activation. (C) Schematic diagram of the HIV-1 Tat protein. Depicted above is the amino acid sequence of the wild-type basic domain or a mutated form deficient in binding to the TAR loop. (D) Structure of biotinylated dCA ( Bio-dCA) (E) dCA binds to TAR but not to TAR non-binding mutant of Tat. HEK293T cells were transfected with flag-tagged Tat 101 a.a. (Tat-F-101-wt), a shorter Tat version with 86 a.a. (Ta(86)-F-wt), Tat 86 a.a. mutated in the basic domain (Tat(86)-F-BRM), flag-tagged CDK11 (CDK11-F), flag-tagged 9G8 (9G8-F) and empty vector control. Protein extracts recovered 40 h later were incubated with DynabeadsMyOne Streptavidin T1 coated with either Biotin or Bio-dCA. Pulled-down proteins were revealed by western bloting with the indicated antibodies. (F) dCA interacts with purified recombinant Tat-wt protein. Recombinant Tat-wt protein was incubated with Bio-dCA streptavidin-coated beads in the presence or absence of an excess of non-biotinylated dCA (represented as molar equivalent [eq.] of Bio-dCA) and Raltegravir used as a negative control competitor. Recombinant GST-9G8 and Tat-BRM were used as negative protein-binding control. Pulled-down proteins were revealed by western blot with anti-Tat and anti-GST antibodies. See also Figure S2.
Tat has several functional domains (Kuppuswamy et al., 1989) (Figure 2C). Domains II and III are essential for transactivation and domain IV mediates TAR RNA binding and nuclear localization. To determine whether Tat binds dCA we synthesized a biotinylated form of the compound (Bio-dCA) (Figure 2D). Despite a tenfold higher EC50 than dCA, higher concentrations of Bio-dCA showed the same efficacy and did not compromise the viability of the cells (Figures S2E–F). Bio-dCA coupled to streptavidin-coated magnetic beads retained flag-tagged Tat (86 a.a.) (Tat(86)-F-wt) or Tat (101 a.a.) (Tat(101)-F-wt) transiently expressed in cells, but not a basic region Tat mutant (Tat(86)-F-BRM) that no longer binds TAR and was therefore transactivation-incompetent (Figures 2E). Bio-dCA did not interact with the RNA-binding protein 9G8, ABCE1 protein (used as negative controls), nor with CDK11, which had been reported to interact with its analogue CA in vitro (Cee et al., 2009). In line with the lack of interaction between dCA and CDK11, dCA was unable to block in vitro CDK11 kinase activity, and failed to alter the cellular expression profile of CDK11 (Figures S1A–B).
To confirm that Bio-dCA interacts directly with Tat we performed pull-down experiments with recombinant purified protein (Figure 2F). Recombinant Tat(86)-wt protein bound directly to Bio-dCA and was competed by dCA but not by Raltegravir, demonstrating the specificity of the interaction. As expected from the results from transfected cells, Bio-dCA did not associate with purified recombinant 9G8-GST, or with the recombinant Tat mutated at the basic region.
While it is well known that Tat accumulates in the nucleolus via its basic region (Hauber et al., 1987; Siomi et al., 1990), Tat function in this compartment is still largely unknown. The nucleolus, a highly organized, non-membrane-bound-sub-compartment, is involved in transcription and maturation of rRNA and ribosome biogenesis as well as in apoptosis and cell cycle control (Gerbi et al., 2003). Some studies suggest that the nucleolus plays a role in HIV-1 infection. First, lymphocytes isolated from infected patients show abnormal nucleolar structures (Galati et al., 2003), and second, nucleolar localization of TAR impairs virus replication (Michienzi et al., 2000; Michienzi et al., 2002).
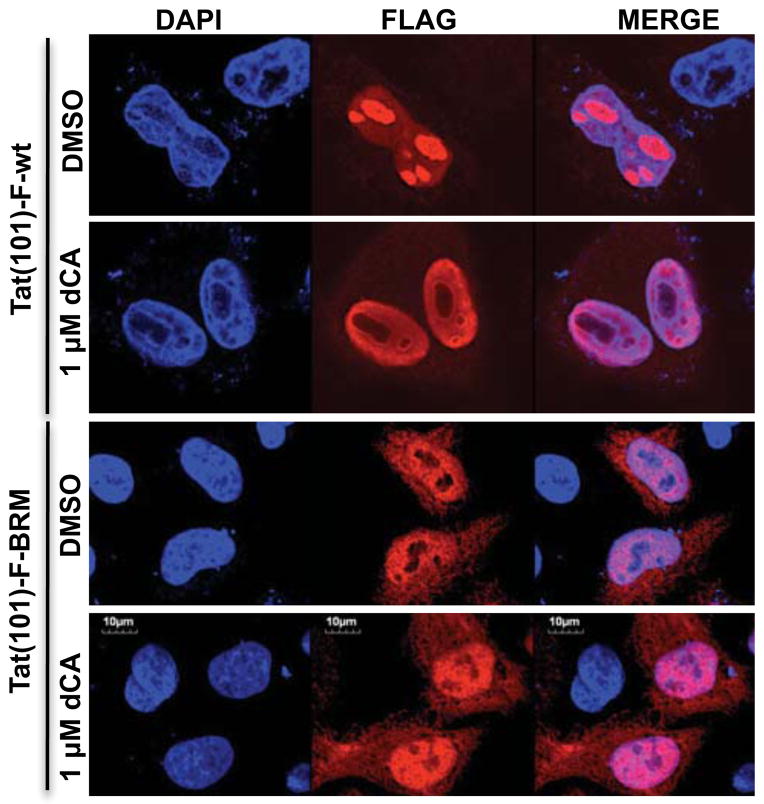
Given that dCA interacts with Tat via its basic domain, which is also the nucleolar localization signal (NoLS), we wondered whether dCA impacts Tat localization. To address this possibility, we transfected HeLa-CD4 cells with a plasmid expressing Tat(101)-F-wt and assessed Tat localization in the presence or absence of dCA by fluorescence microscopy (Figure 3). dCA caused a redistribution of Tat to the periphery of the nucleolus, forming a distinctive ring-like structure (Figures 3 and S3). As expected, dCA caused the same nucleolar exclusion of Tat(86)-F-wt, while cell viability, assessed by tubulin staining, was not perturbed (Figure S3A). Furthermore, the redistribution of Tat was observed in a dose-dependent manner (Figure S3B). The Tat basic mutant was analysed in parallel, and this protein was completely excluded from the nucleolus both in the presence or absence of dCA. The effect of dCA on wild-type Tat appears to mimic the phenotype caused by the basic domain mutation (Figures 3, S3). The Tat-Δ2–26 mutant lacks the transactivation domain but retains the basic domain and shows a predominantly nucleolar localization. As expected, in the presence of dCA, this mutant was excluded from the nucleolus, consistent with the presence of the basic domain (Figures S3C–D). The localization of fibrillarin, a component of small nucleolar ribonucleoproteins (snoRNPs) (Aris and Blobel, 1991; Hiscox, 2002) or of cyclin T1, a Tat-binding protein (Wei et al., 1998), was not altered by dCA (Figures S3B–C). The biotinylated form of dCA had the same effect on Tat localization (Figure S3E). As Tat localizes to the nucleolus via the basic region, these results support the biochemical data showing a direct interaction of dCA with Tat via the TAR binding domain.
Figure 3. dCA alters Tat-Flag cellular localization.
Confocal microscopy analysis of the sub-cellular localization of transfected Tat(86)-F-wt and Tat(86)-F-BRM and where indicated treated (24 h) or not with dCA. Flag epitope-tagged Tat was recognized with anti-flag and AlexaFluor 568 anti-IgG. Transfections were performed in HeLa-CD4 cells. Magnification 300X. See also Figure S3.
dCA blocks transcriptional ini1tiation/elongation
pTEFb is composed of cyclin T1 and cyclin-dependent kinase 9 (CDK9) and is recruited by Tat to the HIV TAR region ((Peng et al., 1998; Wei et al., 1998). pTEFb is used at many promoters, including HIV-1, to phosphorylate serine 2 residues present in the RNAPII C-terminal domain (CTD), converting a non-phosphorylated to a hyperphosphorylated RNAPII form that engages in productive elongation (reviewed in (Peterlin and Price, 2006; Sims et al., 2004)). The effects of dCA on transcription initiation and elongation from the 5′LTR by RNAPII were analysed by RT-qPCR and chromatin immunoprecipitation (ChIP).
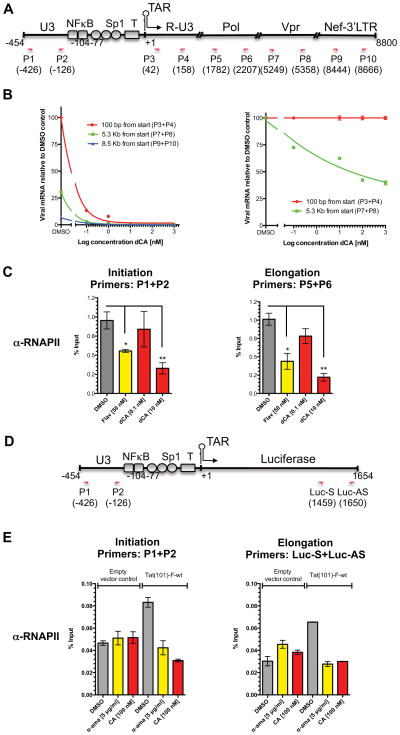
For these studies, the amounts of viral RNA present at different distances from the promoter were measured using sets of primer pairs to quantify transcripts made up to 100 bp, 5.3 kb or 8.5 kb from the transcription start site (Figure 4A). We observed that even in the absence of drug, elongation from the HIV-1 viral promoter is not very efficient. In the presence of DMSO alone only 30% of 5.3 kb long transcripts, and 6% of 8.5 kb long transcripts are produced (Figure 4B-left). The addition of dCA further decreased the production of longer viral RNA species in a dose-dependent manner (Figure 4B-right); supporting the notion that dCA inhibits elongation from the viral promoter.
Figure 4. RNAPII elongation from viral promoter is inhibited by dCA.
(A) Schematic representation of the HIV genome and localization of primers. (B) HIV-1 elongation from HIV-1 promoter measured by qPCR. Left panel: Total RNA was recovered from chronically infected HeLa cells treated with increasing amounts of dCA for 60 days and viral cDNA was measured by relative RT-qPCR using the indicated primers located either at 100 bp, 5.3 kb or 8.5 kb downstream from the transcript start site. All messages normalized to GAPDH mRNA. The amounts of viral mRNA generated in the absence of compound, measured at 100 bp from start site, arbitrarily set at 100%. Right panel: Plotting of data obtained in left panel, setting each data point obtained with the 100 bp primer set at 100% and comparing to results obtained with 5.3 kb primer set. Error bars represent standard deviations of the RT-qPCR assay. Results are representative of 3 independent experiments. (C) ChIP assay of the HIV promoter. HeLa-CD4 cells chronically infected with pNL4-3 were treated with dCA for 4 days and flavopiridol (Flav) for 6–8 h followed by RNAPII ChIP. DNA was measured by qPCR using the indicated set of primers. Results represented as percentages of input. Error bars, ± S.E.M. from 3 independent experiments. *P < 0.05; **P < 0.01 (unpaired t-test). Background was an average of 0.1 ± 0.02 and 0.02 ± 0.01 (S.E.M.) for initiation and elongation respectively. (D) Schematic representation of the LTR-Luciferase transgene and localization of primers. (E) ChIP assay of the LTR-Luciferase. HeLa-CD4-LTR-Luc cells were transfected with pGL4.74-Tat(101)-F-wt or empty vector control and treated with dCA (100 nM) or α-amanitin (α-ama) (5 μg/ml) for 48 h followed by protein-DNA crosslinking and ChIP as in (A) the indicated set of primers. The ChIP background was an average of 0.008± 0.0006 (S.E.M.). Error bars from qPCR (n=3), ± S.E.M. This is representative of 2 independent experiments. See also Figure S4.
Using ChIP with an anti-RNAPII antibody and qPCR, we measured the effect of dCA on RNAPII occupancy of the 5′LTR promoter or viral ORF with the indicated primers in HeLa-CD4 cells chronically infected with HIV-1 pNL4-3 (Figure 4C). Association of RNAPII with the HIV promoter and ORF was significantly decreased in the presence of dCA, in a dose-dependent manner, while the occupancy of RNAPII on the GAPDH promoter and ORF was not affected (Figures 4C and S4A). Flavopiridol (Flav), a CDK9 inhibitor, was used as a positive control and DMSO served as negative control. These results suggest that dCA might block not only elongation but also initiation of transcription in chronically infected cells. As transcription is reduced and less Tat protein is made, the LTR promoter tends to shut off with time, leading to reduced initiation. Furthermore, dCA might reduce Tat mediated recruitment of chromatin-remodelling proteins, such as the histone acetyltransferases (HATs) p300/CBP, to the promoter region (Benkirane et al., 1998; Easley et al., 2010; Marzio et al., 1998). Together these results show dCA’s ability to inhibit RNAPII transcription from the HIV-1 provirus.
Similar results were obtained when ChIP of RNAPII was performed in HeLa-CD4 cells stably expressing the reporter LTR-Luciferase, transfected with either empty vector control or a plasmid encoding Tat(86)-wt and treated or not with dCA or α-amanitin (an RNAPII inhibitor) (Figures 4D–E). RNAPII association with the HIV promoter was minimal in the absence of Tat and was not affected by either α-amanitin or dCA. When Tat was present, an approximate 2-fold enrichment of RNAPII was observed both at the promoter and at the Luciferase ORF. This observed enrichment, in the presence of Tat(86)-wt, was inhibited in by α-amanitin and by dCA. These results confirm that Tat mediated transcription from the HIV promoter is inhibited by dCA. In contrast to α-amanitin used as a positive control for inhibition, dCA did not alter the occupancy of RNAPII at the GAPDH promoter and ORF (Figure S4B).
Potency and scope of dCA inhibition
The potency of dCA inhibition was compared with that of 9 compounds from 4 major ARVs (Figures 5A–D). In a 2-day acute infection assay with HeLa-CD4-LTR-LacZ cells, dCA exhibited a 1.5–3 log lower EC50 than all other ARVs. Although dCA only reduces infectivity by 75–80% (versus 95–100% for the NNRTIs and INIs) this value is comparable to the NRTIs and is better than the PIs (~30%). PIs show low efficacy in this short 48 h assay as they act only upon spreading from the initial infection. Given that dCA blocks a post-integration event, this result is unsurpassing and it compares favourably with late-acting compounds such as PIs. The results were similar when viral production was determined by RT-qPCR of viral RNA in HeLa-CD4 cells infected and treated for 4 days with 1 compound from each class of inhibitors (Figure 5E). Again, dCA consistently showed lower EC50 values (< 0.1 nM) with maximum inhibition around 80% while other ARVs showed complete inhibition, albeit with 100 fold or higher EC50.
Figure 5. Effect of dCA on acute replication of HIV-1 compared with known retroviral inhibitors.
(A–D) HeLa-CD4 cells were infected with HIV-1 pNL4-3 in the presence of the indicated compounds or DMSO. Forty hours post infection a CPRG assay was performed. Same dCA curve plotted in graphs (A–D). Error bars represent standard deviation. (E) Analysis of viral mRNA expression upon treatment with dCA and retroviral inhibitors. Total RNA extracted 4 days after acute infection with pNL4-3 (MOI≫1) in the presence of drugs. Viral cDNA was quantified by qPCR using primers recognizing the env and LTR regions. Results normalized to copies of GAPDH mRNA, with value of 1 assigned to the DMSO control. Error bars represent standard deviation. (F) Activity of dCA on acute HIV-2 replication. HeLa-CD4 cells infected with HIV-2 ROD/A in the presence of the indicated concentrations of dCA. Antigen p27 in the supernatant measured 5 days post-infection. (G) Activity of dCA on chronic HIV-2 replication. HeLa-CD4 cells chronically infected with ROD/A were treated with the indicated concentrations of dCA and antigen p27 measured 7 days post-treatment. (H–I) Activity of dCA on primary cells. Freshly isolated uninfected human PBMCs were stimulated with PHA for 2 days, washed, and grown from then on in IL-2 alone. Cells were infected with pNL4-3 in the presence of increasing concentrations of dCA, saquinavir and raltegravir. Antigen p24 was measured 5 days post infection. (J) No viral rebound upon termination of dCA treatment. Left: HeLa-CD4-LTR-LacZ cells chronically infected with pNL4-3 were treated with dCA for 60 days and treated with dCA for another 10 days (dCA) or stopped dCA treatment for 10 days (dCA Stop). The viral mRNA copies were quantified by qPCR and normalized with GAPDH mRNA. Viral mRNA output from untreated cells was assigned as 1. RNA extracts were used in the qPCR as negative control for the presence of genomic DNA contamination, 0.1% of the amplification of the cDNA samples is due to genomic DNA. The error represents a covariance of less than 5%. Right: Same as left, however treatment was stopped for 27 days before harvest of the cells.
We tested the susceptibility of HIV-2 to dCA inhibition. dCA inhibited acute infection of HeLa-CD4 cells by HIV-2 (ROD/A) with an EC50 of ≈ 5nM (Figure 5F), as well as chronic infection with an EC50 of ≈ 1.7 nM (Figure 5G), demonstrating the broad potential of dCA. Furthermore, dCA excluded equally well HIV-2 Tat protein from the nucleolus (Figures S3C–D). The slightly lower efficiency of dCA in blocking HIV-2 replication as compared to HIV-1 might be explained by the fact that unlike the HIV-1 TAR element, which contains a single stem-loop, the HIV-2 TAR element consists of 2 characteristic stem-loop structures, both of which participate in optimal Tat response.
We measured the ability of dCA to inhibit HIV-1 replication in freshly isolated uninfected human peripheral blood mononuclear cells (PBMCs) stimulated with mitogen phytohaemagglutinin (PHA) and interleukin 2 (IL-2). PBMCs were infected with pNL4-3 and treated with raltegravir, saquinavir or dCA, and viral replication was measured by p24 ELISA. dCA inhibited replication with an EC50 of < 1 nM (Figures 5H and 5I) in primary cells similar to that obtained with raltegravir and saquinavir; however, a maximum inhibition plateau of 86% was observed for dCA. dCA treatment of primary cells did not affect cell viability, as the CC50 value was 100 μM, which is higher than what was observed for Hela-CD4 cells (Figures S5). HIV-1 transcription is intimately linked to T-cell activation due to shared motifs between the HIV-LTR and the regulatory regions in induced genes, namely NF-κB (Alcami et al., 1995). A large variety of T cell stimuli activate NF-κB (Bours et al., 1992), including PHA, IL-2 and tumor necrosis factor alpha (TNF-α) (Arima et al., 1992). In the absence of Tat-independent promoter activity, such as the activation mediated by NF-κB, dCA inhibition of HIV replication would be expected to be higher.
To determine whether virus production by cells treated with dCA rebounds after withdrawal of the drug, we treated HeLa-CD4 cells chronically infected with pNL4-3 for 2 months with dCA and measured viral RNA levels by RT-qPCR at several time points after terminating treatment. No virus rebound was observed even 27 days after the treatment was stopped (Figure 5J), contrary to what is normally observed with ARVs. Overall, these results suggest that dCA promotes rapid and permanent silencing of the HIV promoter, which may drastically limit the emergence of dCA-resistant viruses.
dCA inhibits HIV replication in primary cells from uninfected and infected subjects at different disease stages
The potency of dCA was further tested on spontaneous viral output (p24 production) from primary CD4+T cells isolated from 8 viremic HIV-infected patients (Figure 6A). Cells were grown in IL-2 to increase viability (Figure S6A). The inhibition of viral replication mediated by dCA alone was approximately 25%. ARVs alone, which inhibit all new infections, inhibited replication by approximately 40%, while dCA and ARVs combined inhibited approximately 60%. ARVs or dCA did not affect viability or growth of these primary cells (Figures S6B–C). dCA thus provides a significant additive effect when used with other ARVs to supress replication in primary CD4+T cells from HIV-viremic subjects.
Figure 6. dCA activity on CD4+T cells from viremic and aviremic HIV-infected subjects.
(A) dCA effect on CD4+T cells isolated from viremic subjects. (A) Viral production from CD4+T cells isolated from 8 viremic subjects and grown in IL-2 was measured in the presence or absence of ARVs (RAL+AZT+EFV) supplemented or not with 100 nM dCA (circle) or 1μM dCA (square). Viral production in the supernatant measured by ELISA p24 and normalized to the negative control (Mock). Statistical significance was assessed by a paired t-test. (B) dCA effect on CD4+T cells isolated from aviremic subjects. CD4+T cells isolated from PBMCs from 4 subjects who had been treated with HAART for at least 3 years were cultured for 6 days without IL-2 in the presence of ARVs to block de novo infection. dCA (100 nM) effect on the spontaneous release of HIV particles was assessed by measuring viral RNA in culture supernatants by ultrasensitive RT-qPCR. See also figure S6.
We also investigated whether dCA could impact residual viremia from virally suppressed subjects (plasma viral load less than 50 copies/mL) receiving HAART. We isolated CD4+T cells from PBMCs of 4 subjects treated for at least 3 years who spontaneously released viral particles in vitro and grew them in the absence of IL-2. The non-activated state of these cells confers them viability in vitro in the absence of IL-2 for the duration of the experiment (Figures S6D–E). Using an ultrasensitive RT-qPCR assay, in the presence of ARVs, we observed a reduction of viral production of 99.7% at day 6 (Figure 6B). Importantly, dCA did not affect the viability of the cells at the concentrations used. Altogether, our results suggest that dCA is a highly potent inhibitor of the residual viral production from CD4+T cells of virally suppressed subjects. These results also support the prediction that higher HIV-1 inhibition by dCA occurs in the absence of promoter activation by IL-2 (mediated by NF-κB), when replication is mostly dependent on Tat activity.
dCA pharmacokinetics
To evaluate the in vitro and in vivo stability of dCA an analytical LC-MS/MS method was developed (Table 1). In vitro studies compared murine and human hepatic microsomes (150 donor mixed male/female pool). Sunitinib, an FDA-approved kinase inhibitor with favourable human pharmacokinetics, was included as a positive control. dCA was resistant to hepatic oxidative metabolism in both human and mouse (Table 1A). Based on the encouraging microsomal data, follow-up mouse experiments were conducted to evaluate the ability to dose dCA via oral gavage (PO) and intraperitoneal injection (IP). dCA was easily formulated (1 mg/mL in water) due to its high aqueous solubility. C57Bl6 mice were dosed at 10 mg/kg and drug levels were quantitated in plasma at 1, 6, and 24 h by LC-MS/MS (Table 1B). The results impressively demonstrated that dCA could be given either IP or orally. Drug levels at 1 h after dosing were greater than 1000-fold the EC50 value found in the cell-based assays. dCA concentration decreased at 6 and 24 h, but even 24 h post-dose, plasma drug levels for all mice were above 30 nM (50-fold above the EC50). Most importantly, mice were still healthy after 24 h dCA treatment.
Table 1.
(A) Hepatic microsomal stability evaluated by incubating 1 μM of dCA, with 0.2–1 mg/mL hepatic microsomes. The reaction was initiated by adding NADPH (1 mM). Aliquots were removed at 0, 5, 10, 20, 40, and 60 minutes and samples were analyzed by LC-MS/MS. Data are log transformed and represented as half-lives. (B) Pharmacokinetics of dCA assessed in 3 C57Bl-6 mice. Drug levels were measured by LC-MS/MS. A sample formulation 1 mg/ml dCA in water of 10 mg/kg was dosed intraperitonealy (IP) via the tail vein and orally by gavage (PO) with blood drawn at 1, 6, and 24 h.
| A | ||
|---|---|---|
| Species (T 1/2 in minutes) | ||
| Compound ID | Human | Mouse |
| dCA | 60 | 181 |
| Sunitinib | 77 | 28 |
| B | |||
|---|---|---|---|
| Plasma Concentration | |||
| Time (hr) | Mouse | PO (μM) | IP(μM) |
| 1 | 1 | 0.95 | 2.19 |
| 2 | 0.78 | 1.96 | |
| 3 | 0.84 | 3.19 | |
| Avg. | 0.85 | 2.45 | |
| 6 | 1 | 0.34 | 0.52 |
| 2 | 0.53 | 0.45 | |
| 3 | 0.71 | 0.9 | |
| Avg. | 0.52 | 0.63 | |
| 24 | 1 | 0.03 | 0.04 |
| 2 | 0.05 | 0.03 | |
| 3 | 0.06 | 0.06 | |
| Avg. | 0.05 | 0.04 | |
DISCUSSION
From the findings described herein, dCA is the most potent anti-Tat inhibitor described to date. It binds selectively to the basic domain of HIV Tat, a region also responsible for the Tat-TAR interaction. Importantly, dCA has a drug-like structure, is highly soluble in water, and displays good bioavailability in mice. dCA inhibits both HIV-1 and HIV-2 replication in tissue culture-adapted cells or in primary cells when used at single digit nanomolar concentrations, with no associated toxicity at the cellular level. dCA is very potent in the sense of having an effect at a very low concentration. However, the efficacy of dCA is limited by Tat-independent transcription. For these reasons, it does not achieve the same multilog reductions produced by antiretroviral drugs. It is now clear the slope of the dose response curve is a major determinant of antiviral activity (Shen et al., 2008), and it is the high slope that gives protease inhibitors the ability to inhibit replication by as much as 10 logs. The dose response curve for dCA is unusual and flattens out, resulting in partial inhibition in acute infections. However, even though dCA alone fails to totally inhibit acute HIV infections, due to residual Tat-independent promoter activity, this feature is desirable as it limits off target effects from shared transcription factors binding cellular and viral promoters, such as NF-κB. HIV-1 lacking Tat undergoes some basal transcription; however, does not sustain a spreading infection (Verhoef et al., 1997). Nonetheless, when chronically infected cells were grown in 100 nM dCA for longer periods of time, 99% of viral replication was inhibited.
dCA reduces both transcriptional initiation and elongation from the viral promoter, which is consistent with inhibiting the Tat-mediated conversion of hypophosphorylated RNAPII to the hyperphosphorylated, processive form. Furthermore, termination of dCA treatment does not result in immediate virus rebound because the HIV promoter is transcriptionally silenced in the absence of Tat activity, a feature that is extremely valuable as it may reduce the emergence of resistant HIV-1 strains.
Tat accumulates in the nucleolus via the basic region but whether its function in this compartment is relevant for pathogenesis is still debated. dCA excludes Tat from the nucleolus, most likely because its association with the Tat basic domain inhibits Tat-RNA interactions that cause nucleolar accumulation. Whether dCA-mediated nucleolar Tat exclusion translates into any significant phenotypic outcome in HIV pathogenesis, other than its effect on transcriptional activity, remains to be addressed.
In an effort to understand the molecular basis of the only reported anti-proliferative and anti-migratory activity of CA against HUVECs (Aoki et al., 2007), a high throughput kinome-binding assay was performed by Cee et al. (Cee et al., 2009). CA was reported to bind to CDK11, CDK8 and ROCK I and II kinases in vitro. We were unable to confirm dCA binding to CDK11 and dCA did not inhibit the kinase activity of CDK11 in vitro nor did it bind to biotinylated dCA. Moreover, inhibition of CDK11 activity would be expected to be toxic, as CDK11 knockdown severely impairs cellular viability (Petretti et al., 2006), and at inhibitory concentrations we observed no toxicity. How CA inhibits HUVECs proliferation is still unclear. Unpublished observations (personal communication to Dr Phil Baran) suggest that CA does not inhibit CDK8 activity, nor do our own data confirm inhibition of CDK11. Thus, of the reported kinases to be affected by CA, only ROCK I or II remain as being possibly inhibited by CA resulting in blockage of HUVECs proliferation. However, these displayed the lowest affinity for CA compared to CDK11. Given that dCA inhibits neither CDK8 nor CDK11, the reported binding apparently seen in the in vitro kinome assay might occur outside the kinase active site, and not have any physiologic relevance.
Low levels of viral production persist in HIV-infected subjects taking HAART (Chun et al., 2011) and are a major obstacle for complete eradication of the infection (Hatano et al., 2009). dCA treatment was extremely successful at reducing viral production by a drastic 99.7% from primary CD4+T cells isolated from aviremic patients who had been under HAART treatment for a long period of time. Furthermore, by acting additively with other ARVs dCA further reduced by 20% viral replication from CD4+T cells isolated from viremic patients. Distinct from any currently available ARVs that prevent new rounds of infection, dCA inhibits HIV production from integrated proviral DNA, which by its mode of action may drastically reduce the low levels of persistent viremia observed in treated subjects (Palmer et al., 2008). With a therapeutic index of over 8000, dCA defines a unique class of HIV anti-viral drugs endowed with the ability to decrease residual viremia during HAART, and should be considered as a promising drug to be included in therapeutic eradication strategies.
METHODS
Mitochondrial metabolic activity (MTT) Assay
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay was performed in the presence of increasing concentrations of dCA according to the manufacturer’s protocol (ATCC).
Evaluation of toxicity by flow cytometry
PBMCs from HIV negative subjects were incubated with increasing doses of dCA (0.1 nM to 1 μM). Viability was measured after 24 and 72 h of culture. PBMCs were stained and analyses were performed on a LSRII (BD Bioscience) flow cytometer as described (Trautmann et al., 2006). Antibodies used are listed in supplementary data.
Tat transactivation assay: CPRG
Detailed quantitative chlorophenol red-β-D-galactopyranoside (CPRG)-based (Boehringer Mannheim) assay provided in supplementary information.
HIV production in CD4+T cells from viremic and virally suppressed subjects
All patients provided written informed consent. CD4+T cells were isolated from PBMCs of HIV-infected subjects by negative magnetic selection (StemCell) and cultured for 9 days in the presence of IL-2 (30 UI/mL), ARVs (AZT (180 nM), EFV (100 nM), RALT (200 nM)) and exposed to dCA 100 nM or 1 μM. Viral production was measured in the supernatant by a sensitive in-house ELISA specific for viral capsid protein p24 (Bounou et al., 2002).
CD4+T cells from virally suppressed subjects were cultured for 9 days in the presence of ARVs and exposed to dCA 100 nM. Viral production was measured by quantification of viral particle-associated RNA by ultra-sensitive two-step real-time reverse transcription quantitative PCR (RT-qPCR) as detailed in supplementary information.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed as previously described (Jablonski et al., 2010); detailed protocol in supplementary information.
Supplementary Material
Highlights.
Didehydro-Cortistatin A (dCA) inhibits Tat-mediated HIV trans-activation
dCA binds Tat’s TAR-binding domain and prevents transcriptional elongation
No viral rebound in chronically infected cells upon termination of dCA treatment
dCA suppresses residual viremia in CD4+T cells from virally suppressed subjects
Acknowledgments
This work was supported by NIH K22AI077353, R01AI097012 and the Landenberger Foundation. We are grateful to the AIDS reagent program. We thank Drs. A.D. Strosberg, T. Tellinghuisen, S.P. Goff, C. Weissmann, J.V. Garcia-Martinez and P. Thompson for helpful discussions and critical reading of the manuscript.
Footnotes
Author contributions G.M, W.B. M.A.C, N.N. and S.V. performed experiments, interpreted data and discussed results. M.C performed pharmacokinetic studies. P.B and J.S. synthesized dCA and Bio-dCA. R.F. and N.C. tested dCA in viremic and aviremic PBMCs. S.V. conceived the study and wrote the manuscript. All authors edited the manuscript.
Additional Information
Supplementary information is available online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB, et al. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. Embo J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S, Watanabe Y, Sanagawa M, Setiawan A, Kotoku N, Kobayashi M. Cortistatins A, B, C, and D, anti-angiogenic steroidal alkaloids, from the marine sponge Corticium simplex. J Am Chem Soc. 2006;128:3148–3149. doi: 10.1021/ja057404h. [DOI] [PubMed] [Google Scholar]
- Aoki S, Watanabe Y, Tanabe D, Arai M, Suna H, Miyamoto K, Tsujibo H, Tsujikawa K, Yamamoto H, Kobayashi M. Structure-activity relationship and biological property of cortistatins, anti-angiogenic spongean steroidal alkaloids. Bioorg Med Chem. 2007;15:6758–6762. doi: 10.1016/j.bmc.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Arima N, Kuziel WA, Grdina TA, Greene WC. IL-2-induced signal transduction involves the activation of nuclear NF-kappa B expression. J Immunol. 1992;149:83–91. [PubMed] [Google Scholar]
- Aris JP, Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc Natl Acad Sci U S A. 1991;88:931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M. Recent status of HIV-1 gene expression inhibitors. Antiviral Res. 2006;71:301–306. doi: 10.1016/j.antiviral.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- Bounou S, Leclerc JE, Tremblay MJ. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J Virol. 2002;76:1004–1014. doi: 10.1128/JVI.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours V, Franzoso G, Brown K, Park S, Azarenko V, Tomita-Yamaguchi M, Kelly K, Siebenlist U. Lymphocyte activation and the family of NF-kappa B transcription factor complexes. Curr Top Microbiol Immunol. 1992;182:411–420. doi: 10.1007/978-3-642-77633-5_52. [DOI] [PubMed] [Google Scholar]
- Cee VJ, Chen DY, Lee MR, Nicolaou KC. Cortistatin A is a high-affinity ligand of protein kinases ROCK, CDK8, and CDK11. Angew Chem Int Ed Engl. 2009;48:8952–8957. doi: 10.1002/anie.200904778. [DOI] [PubMed] [Google Scholar]
- Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, Fauci AS. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2011;204:135–138. doi: 10.1093/infdis/jir208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, O’Shea MA, Hallahan CW, Daucher M, Ward DJ, et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. 2005;115:3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Antiviral therapy for human immunodeficiency virus infections. Clinical microbiology reviews. 1995;8:200–239. doi: 10.1128/cmr.8.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O’Shea A, Callender M, Spivak A, Brennan T, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 2009;106:9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci U S A. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley R, Van Duyne R, Coley W, Guendel I, Dadgar S, Kehn-Hall K, Kashanchi F. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta. 2010;1799:275–285. doi: 10.1016/j.bbagrm.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy [see comments] Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Fujinaga K, Cujec TP, Peng J, Garriga J, Price DH, Grana X, Peterlin BM. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati D, Paiardini M, Cervasi B, Albrecht H, Bocchino M, Costantini A, Montroni M, Magnani M, Piedimonte G, Silvestri G. Specific changes in the posttranslational regulation of nucleolin in lymphocytes from patients infected with human immunodeficiency virus. J Infect Dis. 2003;188:1483–1491. doi: 10.1086/379249. [DOI] [PubMed] [Google Scholar]
- Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, Kallungal B, Palmer S, Medvik K, Lederman MM, et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin AV, Lange TS. The nucleolus: a site of ribonucleoprotein maturation. Curr Opin Cell Biol. 2003;15:318–325. doi: 10.1016/s0955-0674(03)00049-8. [DOI] [PubMed] [Google Scholar]
- Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ, Jr, Feinberg JE, Balfour HH, Jr, Deyton LR, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- Hatano H, Delwart EL, Norris PJ, Lee TH, Dunn-Williams J, Hunt PW, Hoh R, Stramer SL, Linnen JM, McCune JM, et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009;83:329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J, Perkins A, Heimer EP, Cullen BR. Trans-activation of human immunodeficiency virus gene expression is mediated by nuclear events. Proc Natl Acad Sci U S A. 1987;84:6364–6368. doi: 10.1073/pnas.84.18.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA. The nucleolus--a gateway to viral infection? Arch Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski JA, Amelio AL, Giacca M, Caputi M. The transcriptional transactivator Tat selectively regulates viral splicing. Nucleic Acids Res. 2010;38:1249–1260. doi: 10.1093/nar/gkp1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Kadonaga JT, Luciw PA, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989;17:3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A, Cagnon L, Bahner I, Rossi JJ. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc Natl Acad Sci U S A. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A, Li S, Zaia JA, Rossi JJ. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc Natl Acad Sci U S A. 2002;99:14047–14052. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Wong-Staal F. Demonstration of virus-specific transcriptional activator(s) in cells infected with HTLV-III by an in vitro cell-free system. Cell. 1986;47:29–35. doi: 10.1016/0092-8674(86)90363-6. [DOI] [PubMed] [Google Scholar]
- Olsen HS, Rosen CA. Contribution of the TATA motif to Tat-mediated transcriptional activation of human immunodeficiency virus gene expression. J Virol. 1992;66:5594–5597. doi: 10.1128/jvi.66.9.5594-5597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Petretti C, Savoian M, Montembault E, Glover DM, Prigent C, Giet R. The PITSLRE/CDK11p58 protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO reports. 2006;7:418–424. doi: 10.1038/sj.embor.7400639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter SN, Palu G. Inhibitors of HIV-1 Tat-mediated transactivation. Current medicinal chemistry. 2006;13:1305–1315. doi: 10.2174/092986706776872989. [DOI] [PubMed] [Google Scholar]
- Selby MJ, Bain ES, Luciw PA, Peterlin BM. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989;3:547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med. 2008;14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Manolikakes G, Yeh CH, Guerrero CA, Shenvi RA, Shigehisa H, Baran PS. Scalable synthesis of cortistatin a and related structures. J Am Chem Soc. 2011;133:8014–8027. doi: 10.1021/ja202103e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Shigehisa H, Guerrero CA, Shenvi RA, Li CC, Baran PS. Stereodivergent synthesis of 17-alpha and 17-beta-alpharyl steroids: application and biological evaluation of D-ring cortistatin analogues. Angew Chem Int Ed Engl. 2009;48:4328–4331. doi: 10.1002/anie.200901116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz M, Josephs SF, Dukovich M, Peffer N, Wong-Staal F, Greene WC. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science. 1987;238:1575–1578. doi: 10.1126/science.2825351. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Siomi H, Shida H, Maki M, Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J, Rosen C, Wong-Staal F, Salahuddin SZ, Popovic M, Arya S, Gallo RC, Haseltine WA. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science. 1985;227:171–173. doi: 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- Valente ST, Gilmartin GM, Mott C, Falkard B, Goff SP. Inhibition of HIV-1 replication by eIF3f. Proc Natl Acad Sci U S A. 2009a;106:4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente ST, Gilmartin GM, Venkatarama K, Arriagada G, Goff SP. HIV-1 mRNA 3′ end processing is distinctively regulated by eIF3f, CDK11, and splice factor 9G8. Mol Cell. 2009b;36:279–289. doi: 10.1016/j.molcel.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Yukl SA, Shergill AK, McQuaid K, Gianella S, Lampiris H, Hare CB, Pandori M, Sinclair E, Gunthard HF, Fischer M, et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS. 2010;24:2451–2460. doi: 10.1097/QAD.0b013e32833ef7bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.