Abstract
We consider a Bayesian analysis using WinBUGS to estimate the distribution of usual intake for episodically consumed foods and energy (calories). The model uses measures of nutrition and energy intakes via a food frequency questionnaire (FFQ) along with repeated 24 hour recalls and adjusting covariates. In order to estimate the usual intake of the food, we phrase usual intake in terms of person-specific random effects, along with day-to-day variability in food and energy consumption. Three levels are incorporated in the model. The first level incorporates information about whether an individual in fact reported consumption of a particular food item. The second level incorporates the amount of intake from those individuals who reported consumption of the food, and the third level incorporates the energy intake. Estimates of posterior means of parameters and distributions of usual intakes are obtained by using Markov chain Monte Carlo calculations. This R function reports to users point estimates and credible intervals for parameters in the model, samples from their posterior distribution, samples from the distribution of usual intake and usual energy intake, trace plots of parameters and summary statistics of usual intake, usual energy intake and energy adjusted usual intake.
Keywords: excess zero models, MCMC, nonlinear mixed models, R, R2WinBUGS, zero-inflation
1. Introduction
There are many statistical challenges when modeling food intakes reported on two or more 24 hours recalls. Some of the challenges involve the presence of measurement error because estimating the distribution of usual intake of nutrients and foods in the population involves monitoring and measuring such intakes over time and their associated recall biases. In addition, many consumers report episodically consumed foods, foods that are not typically consumed every day. For example, fish may be reported in a particular day with a positive intake, but on a different day fish intake may equal zero. Consequently, it is difficult to estimate nutrition intake with recall surveys when one of these recalls incorporate excess zero measurements. Data of this nature is often modeled with measurement error models with zero inflated data.
Recently, in nutritional surveillance (Tooze et al. 2006) and nutritional epidemiology (Kipnis et al. 2009) two-part methods had been developed for analyzing episodically consumed foods. In the first part of the model, the probability of an episodically consumed food is estimated using logistic regression with a person-specific random effect. Then, in the second part of the model the amount of that episodically consumed food per day is modeled using linear regression on a transformed scale with also a person-specific effect. These two parts are linked allowing that the two person-specific effects are correlated as well as by allowing common covariates in both parts of the model. This method is known as the “NCI method” (http://riskfactor.cancer.gov/diet/usualintakes/).
An extension into a three-part method that also incorporates the estimation of the amount of energy intake consumed per day is described in detail by Kipnis et al. (2010). These authors estimated this three-part method using nonlinear mixed effects models with likelihoods computed by adaptive Gaussian quadrature in SAS software. However, computationally it was found to have serious convergence issues within the context of nutritional epidemiology and nutritional surveillance as indicated by Kipnis et al. (2010) and Zhang et al. (2010).
The goal of this article is to present a function for the implementation of this nonlinear mixed effects model. The function Intake_epis_food() allows readers to input their data in R (R Development Core Team 2009) to generate and run the script of the three part model in WinBUGS (Spiegelhalter et al. 1999) and to run and obtain output simulations to R using the package R2WinBUGS (Sturtz et al. 2005). While the most important functions of the package R2WinBUGS are illustrated, we do not provide comprehensive documentation here; instead the reader is referred to the manual and online documentation with that package available from the Comprehensive R Archive Network at http://CRAN.R-project.org/package=plink. In the next sections, the function and corresponding algorithms are explained and an example is provided.
2. Method and parameter inference
The computational details of the Bayesian approach to fit the three-part model for an episodically consumed food and energy model (Kipnis et al. 2010) through Markov Chain Monte Carlo techniques are provided by Zhang et al. (2010) with a brief summary given here. Our model takes into consideration measures of nutrition and energy intakes via a food frequency questionnaire (FFQ) along with two repeated 24 hour recalls (24hr) and adjustment covariates. These two repeated 24hr provide information on the amounts of food and energy consumed by each individual. Consequently, an indicator variable of whether the food was consumed can be generated from the reported amount of food consumed. In addition, because two or more 24hr require distinguishing between and within person random error (Eckert et al. 1997), a type of classical measurement error is needed. Generally nutritional data are skewed, which may require transformations to reach normality and standardization. For person i = 1, …, n, and for the k = 1, 2 repeats of the 24hr, the data are Ỹik = (Yi1k, …, Yi3k)T, where
Yi1k = Indicator of whether the food is consumed.
Yi2k = Amount of the food consumed as reported by the 24hr, which equals zero if the food is not consumed.
Yi3k = Amount of energy consumed as reported by the 24hr.
We will use a Box-Cox transformation to account for the skewness in the data. The Box-Cox transformation with transformation parameter λ, is h(y, λ) = (yλ −1)/λ if λ ≠ 0, and h(y, λ) = log(y) if λ = 0. We will allow for the user to specify different transformation parameters λF and λE for food and energy intake, respectively.
After Box-Cox transformation, we further standardize and center these transformed variables to have a mean of zero and variance of one. This is useful for making the prior distribution specifications given below to be sensible and allows rapid convergence of the posterior samples. Specifically, let μλF and σλF be the mean and standard deviation of the transformed non-zero food data h(Yi2k, λF), and let μλE and σλE be the mean and standard deviation of the transformed energy data h(Yi3k, λE). Then, our analysis is performed using the following transformations:
| (1) |
| (2) |
| (3) |
There are also covariates such as age category, ethnic status and, in many cases the results of reported intakes from a food frequency questionnaire. In principle, the covariates can differ based on the three types of data, so we denote them as Vi1, Vi2, Vi3, which are vector-valued. To improve linearity and homocedasticity of the model, we follow the recommendations of (i) implementing Box-Cox transformations on all or some covariates like intakes from a food frequency questionnaire (Kipnis et al. 2009), (ii) centering and (iii) scaling on all covariates. Without loss of generality, let λc represent a vector containing the Box-Cox transformation parameter used for covariates. Let μλc and σλc be the vector means and standard deviations of the transformed covariates. Then, these transformed, center and scaled covariates are denoted by:
| (4) |
| (5) |
| (6) |
Let (Ui1, Ui2, Ui3) = Normal(0, Σu) be random effects associated with consumption, amount (if the food is consumed), and energy. Similarly, for k = 1, 2, (εi1k, εi2k, εi3k) = Normal(0, Σε) accounts for day-to-day variation. The model for whether there is consumption can be stated as:
| (7) |
where Φ(·) is the standard normal distribution function and (Wi1k, Wi2k, Wi3k) represent their corresponding latent variables as follows. A food being consumed at visit k is equivalent to
| (8) |
The food when consumed and energy intake are modeled as
| (9) |
| (10) |
The prior distribution of parameters β1, β2, β3 is assumed to be multivariate Normal with vector mean 0 and variance covariance Σu. An inverse Wishart denoted as IW(Ωu, mu) prior was specified for Σu. We set Ωu to have an exchangeable correlation structure with starting values with diagonal entries all equal to 1 and correlations 0.5. We have two necessary restrictions on Σε: (i) εi1k and εi2k are independent; and (ii) var(εi1k) = 1, so that β1 is identifiable, along with the distribution of Ui1. Furthermore, we constrained this covariance matrix using a polar coordinate representation with γ ∈ (−1, 1) and θ ∈ (−π, π). With these considerations Σε can be written as:
| (11) |
where p1 = γcos(θ) and p2 = γsin (θ). The recommended prior distributions for s22 and s33 are Uniform(0,3), for γ is Uniform(−1,1) and for θ is Uniform(−π, π) (Zhang et al. 2010). The corresponding inverse of Σu is a Wishart distribution denoted as , and the inverse of Σε can be written as:
| (12) |
where .
2.1. Usual intake analysis
Besides the parameters and the random effects (Ui1, Ui2, Ui3) in this context it is vital to also have the posterior samples of usual intake TFi, usual energy intake TEi, and energy adjusted usual intake 1000TFi/TEi, all on the original scale. We used the best-power method (Dodd et al. 2006) for estimating both usual energy intake and usual intake for person i. We first consider energy:
| (13) |
| (14) |
where
Similarly, a person’s usual intake of the dietary component on the original scale is defined as
| (15) |
When λ = 0, the back-transformation and second derivative are:
Similarly, when λ ≠ 0, the back-transformation and second derivative are:
3. Using the Intake_epis_food() function in R
Users must install the R2WinBUGS (Sturtz et al. 2005) package in R and WinBUGS (Spiegelhalter et al. 1999) before running Intake_epis_food() function. This function assumes that there are no missing observations. Intake_epis_food() loads the libraries R2WinBUGS, stats and fSeries automatically.
The Intake_epis_food() function incorporates this three-part model, with truncated normal random variables for generating the latent Wi1k, and Metropolis-Hastings computations for α1, α2, α3, β1, β2, β3, the elements of Σu, and Σε. This function generates and runs the script of this model in WinBUGS and returns Markov Chain Monte Carlo (MCMC) computation. We emphasize that MCMC computation can either be thought of as a strictly Bayesian computation with ordinary Bayesian inference, or as a means of developing frequentist estimators of the crucial parameters. These MCMC produces estimates which are known to be asymptotically equivalent to maximum likelihood estimators, which are difficult to obtain (Zhang et al. 2010). In Bayesian terms we used the posterior means of the model. Users are mainly interested in estimates of equations (14), (15) as well as the energy-adjusted usual intake 1000TFi/TEi.
The Intake_epis_food() function produces seven output files. The first output file contains parameter estimates (posterior means) of α1, α2, α3, β1, β2, β3 from equations (8), (9) and (10) as well as Σμ, , Σε, and . The second output file contains summary statistics of Bayesian estimates of equations (14), (15) and the energy-adjusted usual intake 1000TFi/TEi. The third output file contains the iterations from the MCMC computations. The fourth output file contains summary statistics of Bayesian estimates for Ui1, Ui2, Ui3. The fifth output file contains trace plots of the intercept and coefficients in equations (8), (9) and (10). The sixth output file contains trace plots of the variance-covariances Σμ, , Σε and . The seventh output file contains density plots of equations (15) and the energy adjusted usual intake 1000TFi/TEi.
The default input for the Intake_epis_food() function in R is:
Intake_epis_food(data.file.name="data.file.name", numcov=4, df=5,lambdaf=0.32,lambdae=0.23,lvar1=999, lvar2=999, lvar3=0.33, lvar4=0, n.iter=11000,n.burnin=1000, n.thin=10, bugs.seed=123456, working.directory="C:/", file.estimates="estimates.csv", file.istats="estimates_intake.csv", file.iterations="iterations.csv", file.uis="uis.csv", file.tracep="trace_plots.pdf", file.tpvarcov="trace_plot_var_cov_matrices.pdf", file.densityp="density_plots.pdf", bugs.directory="c:/Program Files/WinBUGS14/")
with the following arguments:
- data.file.name : name of the file containing the data (default=data.file.name) in the following order:
- the first variable that identifies individuals, e.g. an ID number
- the next two variables are the 24hr variables of food intake, in order
- the next two variables are the 24hr variables of energy intake, in order
- the next variables are the covariates, and cannot have a column of ones. We limited the number of covariates to four, given that each covariate may have a different Box-Cox transformation parameter.
numcov : number of covariates in user’s dataset (default=4)
df : number of the degrees of freedom of mu (default=5)
lambdaf : value of λF to be used for transformation of food intake (default=1 indicating no transformation done)
lambdae : value of λE to be used for transformation of energy intake (default=1 indicating no transformation done)
lvar1 : value of λc to be used for Box-Cox transformation of first covariable (default=999 indicating no transformation done)
lvar2 : value of λc to be used for Box-Cox transformation of second covariable (default=999 indicating no transformation done)
lvar3 : value of λc to be used for Box-Cox transformation of third covariable (default=999 indicating no transformation done)
lvar4 : value of λc to be used for Box-Cox transformation of fourth covariable (default=999 indicating no transformation done)
n.iter : number of iterations of the MCMC including burning iterations (default=11,000)
n.burnin : number of burn-in iterations of the MCMC (default=1,000)
n.thin : thinning rate of MCMC (default=10)
bugs.seed : random seed to be use for MCMC (default=123456)
working.directory : drive and folder location for output files (default=”C:”)
file.estimates : name of the file to save posterior summary statistics including mean, standard deviation as well as percentiles: 2.5th, 25th, 50th, 75th and 97.5th (default = estimates.csv), of α1, α2, α3, β1, β2, β3 from equations (8), (9) and (10) as well as Σμ, , Σε, and .
file.istats : name of the file to save summary statistics of usual food intake, (15), usual energy intake, (14) and usual food intake per 1000 calories (1000TFi/TEi) (default = estimates intake.csv).
file.iterations : name of the file to save iterations (default=iterations.csv). The number of iterations saved corresponds to (n.iter − n.burnin)/(n.thin)
file.uis : name of the file to save summary statistics of Bayesian estimates for Ui1, Ui2, Ui3 including mean, standard deviation as well as percentiles: 2.5th, 25th, 50th, 75th and 97.5th for person i = 1, …, n. The first variable given in the dataset that identifies individuals is included as the first variable in this file for linking purposes to users.
file.tracep : name of the file to save trace plots of parameters (default = trace plots.pdf)
file.tpvarcov : name of the file to save trace plots of variance-covariances Σμ, , Σε and (default = trace plot var cov matrices.pdf)
file.densityp : name of the file to save density plots of usual food intake and usual food intake per 1000 calories. Each one is generated individually and an overlayed plot is generated as well (default = density plots.pdf)
bugs.directory : drive and folder location of WinBUGS (default=”c:/Program Files /Win-BUGS14/”)
Once the data is input, Intake_epis_food() invokes WinBUGS from R. WinBUGS requires a file describing the model in equations (1)–(5) in Section 2. This model is included as file intake_program.txt and is loaded in R using its bugs() argument. Although, the default priors of the constrained variance-covariance matrix of random errors are geared to the pre-standardization of the data as described in Section 2, the user may change those prior entries within the file intake_program.txt, possibly changing:
| Constrained variance-covariance matrix of random errors | ||
|---|---|---|
| Matrix notation | Default distribution and range | Code in intake_program.txt |
| s22 | Uniform(0,3) | tau.e1 ~ dunif(0,3) |
| s33 | Uniform(0,3) | tau.e2 ~ dunif(0,3) |
| γ | Uniform (−1,1) | a.e1 ~ dunif(−0.99,0.99) |
| θ | Uniform (−1,1) | a.e2 ~ dunif(−3.11,3.11) |
Users may change any of the defaults of Intake_epis_food() function when the function is called to run by replacing those arguments in the last sentence of the file accompanied by this paper. MCMC posterior means of α1, α2, α3, β1, β2, β3, the elements of Σu, Σε, are displayed on the screen of R by Intake_epis_food() and saved automatically in numeral matrices on file names previously listed. See Section 4 for an example.
4. Application of the Intake_epis_food() function
We simulated data using parameter values identified from the calibration sub-study of the National Institutes of Health (NIH) and the American Association of Retired Persons (AARP) Diet and Health Study (Schatzkin et al. 2001). The NIH-AARP study is a cohort composed of people who resided in one of six states: California, Florida, Pennsylvania, New Jersey, North Carolina, and Louisiana or in two metropolitan areas: Atlanta, Georgia and Detroit, Michigan. From 1995 through 1996, 3.5 million questionnaires were mailed to members of the AARP, aged 50–71 years. The questionnaire included a dietary section as well as some lifestyle questions. Over 500,000 people returned the questionnaire after three mailing waves. This, then, is the largest study of diet and health ever conducted in the USA. In 1996–1997, these participants received a Risk-Factor Questionnaire which asked additional questions about lifestyle and behavior, and in 2004–2006 these participants received another follow-up questionnaire (http://dietandhealth.cancer.gov/history.html).
Participants for the calibration study were randomly selected from the 46,970 subjects who had responded to the first wave as of January 1996. Two thousand participants was the targeted sample size to attempt recruitment calls for a baseline and 24 hours follow up. The sample available to us included 920 men. Because the data are not publicly available, we simulated 920 food intakes that are similar in distribution to the men in the NIH-AARP calibration study. We used as four covariates: age, body mass index (BMI), consumption of servings of whole grains from the FFQ, and energy intake from whole grains from the FFQ. Therefore, the degrees of freedom of the inverse Wishart were setup as mu = 5. The latter two covariates were transformed by the cube root and the logarithm, respectively.
We present results from Intake_epis_food() for this simulated data example with a sample size of 920 men, modeling whole grains consumption. We used a burn-in of 1, 000 steps followed by 10, 000 MCMC iterations; our Intake_epis_food() function took approximately 6 hours and 16 minutes (Pentium computer (R) D CPU 3.5GHz and 1.99GB of RAM). We used only every 10th value of the chain. The first output file contains the average over 10,000 MCMC of posterior mean, posterior standard deviation, posteriors: 2.5th percentile, 25th percentile, 50th percentile, 75th percentile and 97.5th percentile. The intercepts of the model are α1, α2, α3 corresponding to each level of the three-part model. Also, estimates of regression parameters for each covariate appear for each level of the model. This is the notation used for the output:
alpha_1 : represents the intercept in equation (8).
alpha_2 : represents the intercept in equation (9).
alpha_3 : represents the intercept in equation (10).
beta_1_j : represents the estimated coefficient of the jth covariate in equation (8), j=1,‥,numcov.
beta_2_j : represents the estimated coefficient of the jth covariate in equation (9), j=1,‥,numcov.
beta_3_j : represents the estimated coefficient of the jth covariate in equation (10), j=1,‥,numcov.
tau_j_p : represents the (j,p) entry estimate of , j=p=1,2,3.
sigma2_j_p : represents the (j,p) entry estimate of Σu, j=p=1,2,3.
a.e1 : represents the estimate of γ
a.e2 : represents the estimate of θ
sigmaemat_j_p : represents the (j,p) entry estimate of Σε, j=p=1,2,3, with Σε11=1 and Σε12=Σε21=0.
tauemat_j_p : represents the the (j,p) entry estimate of , j=p=1,2,3.
We used λF = 0.32 as the Box-Cox transformation parameter of food intake λE = 0.23 as the Box-Cox transformation of energy intake. We setup our working directory in R with the command setwd(’C:’) and using Intake_epis_food() function:
Intake_epis_food(data.file.name="Sim_AARP_wg_Men.csv", numcov=4, df=5,lambdaf=0.32,lambdae=0.23,lvar1=999, lvar2=999, lvar3=0.33, lvar4=0, n.iter=11000,n.burnin=1000, n.thin=10, bugs.seed=123456, working.directory="C:/", file.estimates="estimates.csv", file.istats="estimates_intake.csv", file.iterations="iterations.csv", file.uis="uis.csv", file.tracep="trace_plots.pdf", file.tpvarcov="trace_plot_var_cov_matrices.pdf", file.densityp="density_plots.pdf", bugs.directory="c:/Program Files/WinBUGS14/")
After calculations, estimates of α1, α2, α3, β1, β2, β3 from equations (8), (9) and (10), Σμ, , Σε, are shown as well as summary statistics using a Bayesian approach for three variables are provided immediately in the console screen (a) food usual intake, (b) usual food intake per 1000 calories, and (c) energy usual intake. The following shows the results for whole grains.
Inference for Bugs model at ' Intake_program.txt ', fit using WinBUGS,
1 chains, each with 11000 iterations (first 1000 discarded)
n.thin= 10 n.sims= 1000 iterations saved
mean sd 2.5% 25% 50% 75% 97.5%
alpha_1 0.89 0.06 0.79 0.85 0.89 0.92 1.00
alpha_2 −0.04 0.05 −0.13 −0.07 −0.04 −0.01 0.05
alpha_3 0.00 0.04 −0.07 −0.03 0.00 0.02 0.07
beta_1_1 0.16 0.05 0.07 0.13 0.16 0.19 0.25
beta_1_2 −0.08 0.05 −0.18 −0.12 −0.08 −0.05 0.00
beta_1_3 0.57 0.05 0.47 0.54 0.57 0.61 0.67
beta_1_4 −0.29 0.05 −0.38 −0.32 −0.29 −0.26 −0.19
beta_2_1 −0.07 0.04 −0.15 −0.09 −0.06 −0.04 0.01
beta_2_2 −0.08 0.04 −0.16 −0.11 −0.08 −0.06 −0.01
beta_2_3 0.42 0.04 0.34 0.40 0.42 0.45 0.50
beta_2_4 −0.11 0.04 −0.18 −0.13 −0.11 −0.08 −0.03
beta_3_1 −0.07 0.04 −0.15 −0.10 −0.07 −0.05 0.00
beta_3_2 −0.06 0.04 −0.14 −0.09 −0.06 −0.04 0.00
beta_3_3 0.00 0.04 −0.07 −0.03 0.00 0.03 0.08
beta_3_4 0.34 0.04 0.27 0.32 0.34 0.37 0.42
tau_1_1 2.05 0.57 1.26 1.66 1.94 2.31 3.50
tau_1_2 −0.33 0.60 −1.64 −0.66 −0.31 0.04 0.82
tau_1_3 −0.11 0.19 −0.47 −0.23 −0.11 0.01 0.29
tau_2_1 −0.33 0.60 −1.64 −0.66 −0.31 0.04 0.82
tau_2_2 3.29 0.79 2.21 2.77 3.13 3.62 5.31
tau_2_3 −0.57 0.28 −1.21 −0.73 −0.54 −0.38 −0.09
tau_3_1 −0.11 0.19 −0.47 −0.23 −0.11 0.01 0.29
tau_3_2 −0.57 0.28 −1.21 −0.73 −0.54 −0.38 −0.09
tau_3_3 1.53 0.16 1.26 1.42 1.51 1.63 1.89
sigma2_1_1 0.56 0.13 0.33 0.47 0.55 0.64 0.83
sigma2_1_2 0.06 0.09 −0.12 0.00 0.06 0.12 0.22
sigma2_1_3 0.06 0.06 −0.06 0.02 0.06 0.10 0.18
sigma2_2_1 0.06 0.09 −0.12 0.00 0.06 0.12 0.22
sigma2_2_2 0.37 0.07 0.23 0.32 0.36 0.41 0.51
sigma2_2_3 0.13 0.05 0.03 0.10 0.13 0.16 0.23
sigma2_3_1 0.06 0.06 −0.06 0.02 0.06 0.10 0.18
sigma2_3_2 0.13 0.05 0.03 0.10 0.13 0.16 0.23
sigma2_3_3 0.72 0.07 0.60 0.68 0.72 0.77 0.86
a.e1 0.17 0.05 0.09 0.14 0.17 0.20 0.26
a.e2 0.81 0.29 0.28 0.62 0.80 0.99 1.37
sigmaemat_1_3 0.12 0.05 0.03 0.09 0.12 0.16 0.23
sigmaemat_2_2 1.26 0.08 1.11 1.21 1.26 1.31 1.42
sigmaemat_2_3 0.14 0.05 0.04 0.11 0.14 0.18 0.24
sigmaemat_3_1 0.12 0.05 0.03 0.09 0.12 0.16 0.23
sigmaemat_3_2 0.14 0.05 0.04 0.11 0.14 0.18 0.24
sigmaemat_3_3 1.16 0.06 1.06 1.12 1.16 1.19 1.27
tauemat_1_1 1.02 0.01 1.00 1.01 1.01 1.02 1.05
tauemat_1_2 0.01 0.01 0.00 0.01 0.01 0.02 0.03
tauemat_1_3 −0.11 0.05 −0.21 −0.14 −0.11 −0.08 −0.03
tauemat_2_1 0.01 0.01 0.00 0.01 0.01 0.02 0.03
tauemat_2_2 0.81 0.05 0.72 0.78 0.81 0.84 0.91
tauemat_2_3 −0.10 0.04 −0.17 −0.13 −0.10 −0.08 −0.03
tauemat_3_1 −0.11 0.05 −0.21 −0.14 −0.11 −0.08 −0.03
tauemat_3_2 −0.10 0.04 −0.17 −0.13 −0.10 −0.08 −0.03
tauemat_3_3 0.89 0.04 0.81 0.87 0.89 0.92 0.98
Usual food intake Usual food intake per 1000 calories Energy usual intake
Mean 0.9359701 0.4072328 2298.3852
S.d. 0.6947905 0.2849965 446.3253
5th 0.1887330 0.0823138 1636.7102
10th 0.2632069 0.1156591 1755.9317
25th 0.4359173 0.1992969 1982.9681
50th 0.7421419 0.3408097 2267.2364
75th 1.2117088 0.5350990 2568.3280
90th 1.9042402 0.7920862 2891.3676
95th 2.3828280 0.9484864 3088.9744
This is the only output presented in the console screen. These results are saved in file.estimates and file.istats respectively. Five additional output files are stored as mentioned before, but examples of these files are not presented here. Instead, we present the posterior density of the mean of usual whole grains intake plot and the posterior density of the mean of usual whole grains intake per 1000 calories in figure 1 (file.densityp). We present trace plots of the intercept and coefficients in equations (8), (9) and (10) in figure 2 (file.tracep). We present trace plots of entry estimates saved in file.tpvarcov of variance-covariance matrices (i) for Σμ in figure 3, (ii) for in figure 4, (iii) for Σε in figure 5 and (iv) for in figure 6.
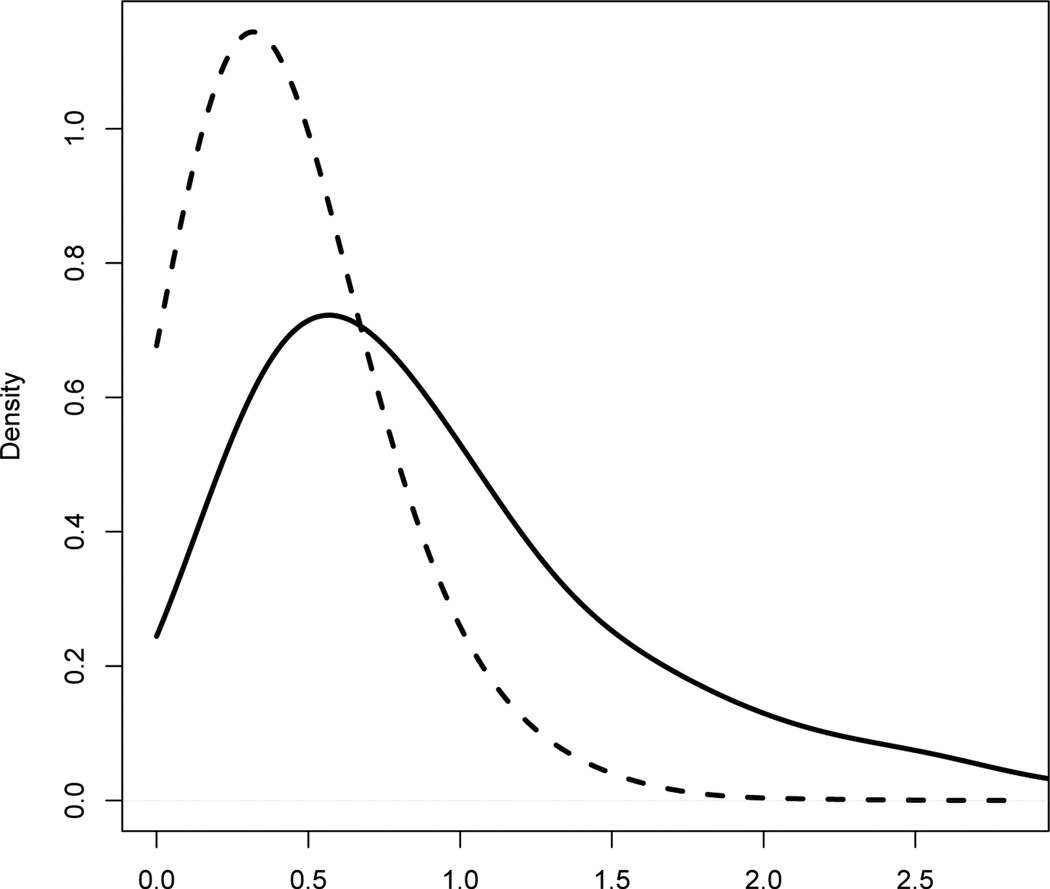
Figure 1.
Posterior density of the mean for whole grains. The solid line is the density estimate for usual intake from 1000 MCMC. The dashed line is the density estimate for usual intake per 1000 calories.
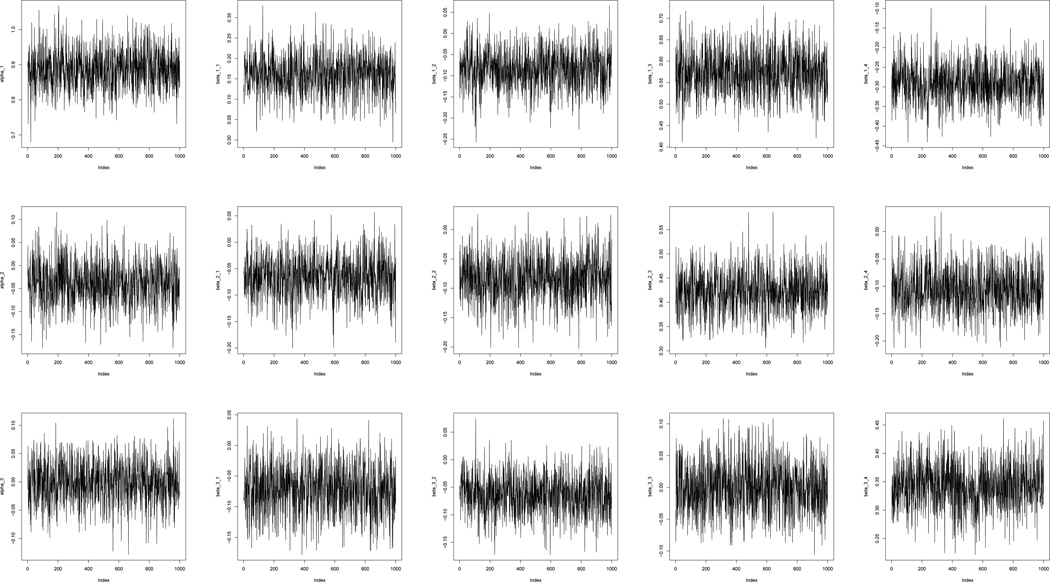
Figure 2.
Trace plot from 1000 MCMC of intercepts and estimated coefficients. First row shows trace plots of parameters for the intercept, age, body mass index, consumption of whole grains and energy intake from whole grains in equation (8). Second row shows trace plots of parameters for the intercept, age, body mass index, consumption of whole grains and energy intake from whole grains in equation (9). Third row shows trace plots of parameters for the intercept, age, body mass index, consumption of whole grains and energy intake from whole grains in equation (10).
Figure 3.
Trace plot from 1000 MCMC for each entry estimate of variance-covariance matrix Σμ j=p=1,2,3.
Figure 4.
Trace plot from 1000 MCMC for each entry estimate of variance-covariance matrix j=p=1,2,3.
Figure 5.
Trace plot from 1000 MCMC for each entry estimate of variance-covariance matrix Σε j=p=1,2,3. The following parameters are neither estimated nor plotted because they are fixed: Σε11=1 and Σε12=Σε21=0.
Figure 6.
Trace plot for each entry estimate of variance-covariance matrix j=p=1,2,3.
5. Discussion
This paper was motivated by the AARP calibration sub-study in nutritional epidemiology. Our main aim when implementing this function was to help users to estimate this recent nonlinear mixed three-part model of measurement error for an episodically food consumed in a Bayesian manner.
Acknowledgments
This research was supported by a grant from the National Cancer Institute (R37-CA-057030).
Contributor Information
Adriana Pérez, Division of Biostatistics and Michael & Susan Dell Center for Healthy Living, The University of Texas Health Science Center at Houston, School of Public Health, 1616 Guadalupe Street, Suite 6.340, Austin, TX 78705, Telephone: +1-(512)-391-2524, Fax: +1-(512)-482-6185, adriana.perez@uth.tmc.edu.
Saijuan Zhang, Department of Statistics, Blocker Building, Room, Texas A&M University, 3143 TAMU, College Station TX 77843-3143, Telephone: +1-979-845-3141, Fax: +1-979-845-3144, sjzhang@stat.tamu.edu.
Victor Kipnis, Division of Cancer Prevention, National Cancer Institute, 6130 Executive Boulevard, Bethesda, Maryland 20892-7354, Telephone: +1-301-496-7464, Fax: +1-301-496-7463, vk3b@nih.gov.
Douglas Midthune, Division of Cancer Prevention, National Cancer Institute, 6130 Executive Boulevard, Bethesda, Maryland 20892-7354, Telephone: +1-301-496-7464, Fax: +1-301-496-7463, dm76q@nih.gov.
Laurence S. Freedman, Gertner Institute for Epidemiology and Health Policy Research, Biostatistics Unit, Sheba Medical Center, Tel Hashomer 52161, Israel, Telephone: +972-3-530-5390, Fax: +972-3-534-9607, lsf@actcom.co.il
Raymond J. Carroll, Department of Statistics, Blocker Building, Room, Texas A&M University, 3143 TAMU, College Station TX 77843-3143, Telephone: +1-979-845-3141, Fax: +1-979-845-3144, carroll@stat.tamu.edu
References
- Dodd KW, Guenther PM, Freedman LS, Subar AF, Kipnis V, Midthune D, Tooze JA, Krebs-Smith SM. Statistical Methods for Estimating Usual Intake of Nutrients and Foods: a Review of the Theory. Journal of the American Dietetic Association. 2006;106(10):1640–1650. doi: 10.1016/j.jada.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Eckert RS, Carroll RJ, Wang N. Transformations to Additivity in Measurement Error Models. Biometrics. 1997;53:262–272. [PubMed] [Google Scholar]
- Kipnis V, Freedman LS, Carroll RJ, Midthune D. A measurement error model for episodically consumed foods and energy. 2010 doi: 10.18637/jss.v046.c03. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipnis V, Midthune D, Buckman DW, Dodd KW, Guenther PM, Krebs-Smith SM, Subar AF, Tooze JA, Carroll RJ, Freedman LS. Modeling Data with Excess Zeros and Measurement Error: Application to Evaluating Relationships between Episodically Consumed Foods and Health Outcomes. Biometrics. 2009;65(4):1003–1010. doi: 10.1111/j.1541-0420.2009.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2009 ISBN 3-900051-07-0, URL http://www.R-project.org.
- Schatzkin A, Subar AF, Thompson FE, Harlan LC, Tangrea J, Hollenbeck AR, Hurwitz PE, Coyle L, Schussler N, Michaud DS, Freedman LS, Brown CC, Midthune D, Kipnis V. Design and serendipity in establishing a large cohort with wide dietary intake distributions: the National Institutes of Health-AARP Diet and Health Study. American Journal of Epidemiology. 2001;154:1159–1125. doi: 10.1093/aje/154.12.1119. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Thomas A, Best NG. WinBUGS Version 1.2 User Manual. Cambridge, UK: MRC Biostatistics Unit; 1999. URL http://www.mrc-bsu.cam.ac.uk/bugs/. [Google Scholar]
- Sturtz S, Ligges U, Gelman A. R2WinBUGS: A Package for Running WinBUGS from R. Journal of Statistical Software. 2005;12(3):1–16. URL http://www.jstatsoft.org. [Google Scholar]
- Tooze JA, Midthune D, Dodd KW, Freedman LS, Krebs-Smith SM, Subar AF, Guenther PM, Carroll RJ, Kipnis V. A New Statistical Method for Estimating the Usual Intake of Episodically Consumed Foods With Application to Their Distribution. Journal of the American Dietetic Association. 2006;106(10):1575–1587. doi: 10.1016/j.jada.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Midthune D, Pérez A, Buckman DW, Kipnis V, Freedman LS, Dodd KW, Krebs-Smith SM, Carroll RJ. A Bivariate Nonlinear Measurement Error Model for Episodically Consumed Dietary Components. 2010 doi: 10.2202/1557-4679.1267. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]