Abstract
HIV-epitope-specific T cell responses are often comprised of clonotypic expansions with distinct functional properties. In HIV+ individuals, we measured PD-1 and IL-7Rα expression, MHC-I tetramer binding, cytokine production, and proliferation profiles of dominant and sub-dominant T cell receptor clonotypes to evaluate the relationship between the composition of the HIV-specific T cell repertoire and clonotypic phenotype and function. Dominant clonotypes are characterized by higher PD-1 expression and lower C127 expression compared to sub-dominant clonotypes and TCR avidity positively correlates with PD-1 expression. At low peptide concentrations, dominant clonotypes fail to survive in culture. In response to stimulation with peptides representing variant epitopes, sub-dominant clonotypes produce higher relative levels of cytokines and display greater capacity for cross-recognition compared to dominant clonotypes. These data indicate that dominant clonotypes within HIV-specific T cell responses display a phenotype consistent with ongoing exposure to cognate viral epitopes and suggest that cross-reactive, sub-dominant clonotypes may retain greater capacity to suppress replication of viral variants as well as to survive in the absence of strong antigenic signaling.
Introduction
Evidence indicates that CD8+ T cell responses are a critical component of the natural immune responses to HIV (1–3). Epitope-specific CD8+ T cell responses appear to be impaired as a result of unique conditions present in HIV infection, namely constant antigen exposure (4) and overwhelming immune activation leading to exhaustion and eventual deletion of HIV-specific T cell responses (5). Our understanding of the mechanisms that underlie impaired T cell responses and their contributions to viral control remains incomplete.
Reversible T cell exhaustion has been associated with the expression of high levels of Programmed Death-1 receptor (PD-1), especially on epitope-specific CD8+ T cells (6). PD-1 is a surface-expressed transmembrane signaling protein with extracellular homology to CD28 superfamily molecules and is upregulated on activated lymphocytes (7). The role of PD-1 in the development of functional T cell memory and resolution of acute infections is increasingly well defined using model systems such as lymphocytic choriomeningitis virus (LCMV) and in human infections such as human hepatitis B (6, 8). In the setting of chronic viral infection, however, the immunomodulatory role of PD-1 signaling becomes more complex as the necessity to limit immunopathology can also dampen effective T cell responses that might contribute to viral clearance (9, 10).
In HIV infection, PD-1 expression on T cell populations correlates positively with viral load (11) and likely contributes to increased sensitivity to apoptosis (12, 13). PD-1 signaling blockade has been shown to restore some T cell function in LCMV infection as well as in vitro with T cells from HIV+ individuals (6, 14). A reduction in the expression of cytokine receptor molecules such as IL-7Rα (CD127) on epitope-specific T cells may also play an important role in the natural control of HIV (15, 16). Reduced T cell capacity to respond to homeostatic cytokines such as IL-7 represents a point of dysregulation in the maintenance of functional, long-lived antigen-specific memory (17, 18).
Both quantitative and qualitative features of T cell responses are likely important for control of chronic viremia. The frequency of T cells that produce cytokine or proliferate in response to activation by cognate antigen is an important measure of the magnitude of the immune response(19–21), but qualitative aspects of CD8+ T cell responses such as the composition of the HIV-specific T cell receptor repertoire have been shown to be important in chronic viral infections such as hepatitis C virus infection (22) and HIV-1 infection (23). Activation or antigen exposure profiles of T cell subsets (15, 24), differentiation (25), or clonotypic antigen sensitivity (26) continue to provide important insight into potential mechanisms governing the generation and maintenance of optimal T cell responses to chronic viral infections. Our previous work suggests that individual T cell clonotypes within HIV-epitope-specific responses are capable of responding independently to changes in viral load (23) and recognizing circulating viral variants (27).
The relationship between the composition of the clonotypic T cell receptor repertoire and clonotypic phenotype or function has not been clearly defined in model systems or natural infections. We found that dominant clonotypes express relatively higher levels of PD-1 and relatively lower levels of CD127 in comparison to corresponding sub-dominant clonotypes. PD-1 expression correlated strongly with the ability of clonotypes to bind MHC-I tetramers, and while dominant and sub-dominant clonotypes were able to respond to stimulation with HIV peptide epitopes matching circulating sequence, sub-dominant clonotypes were more cross-reactive in response to common variant peptide epitopes. Additionally, dominant clonotypes displayed an impaired ability to survive in culture at low levels of antigen stimulation. These data provide insight into the relationships between the structural composition of HIV-specific CD8+ T cell responses, the relative antigen exposure of clonotypes within the epitope-specific TCR repertoire, and the functional capacity of these clonotypes in ongoing HIV infection.
Materials and Methods
Individual Cohort and HLA-typing
This cohort was organized within the Vanderbilt-Meharry CFAR and was comprised of anti-retroviral therapy naïve patients recruited through the Comprehensive Care Center (Nashville, TN). All individuals were typed for HLA Class I by DCI Tissue Typing Laboratory (Nashville, TN). This study was approved by the Institutional Review Board at Vanderbilt University, and all participating individuals provided written informed consent.
Flow cytometric evaluation of lymphocyte surface molecules
Gating strategy shown in Supplemental Figure 1. Lymphocyte subsets were evaluated using fresh and cryopreserved peripheral blood mononuclear cells and a combination of monoclonal antibodies. CD3-AlexaFluor-700 (BD), CD4-PE-Texas Red (Caltag), CD8-Pacific Orange (Caltag), CD14-PerCP (BD), CD19-PerCP (BD), CD56-PE-Cy5 (BD), Viaprobe (BD), CD127-biotin (eBioScience), Streptavidin-APCCy7 (BD), PD-1-pure (Mouse IgG1, clone EH12:2H7, BioLegend), goat-anti-mouse IgG-Pacific Blue (Molecular Probes), anti-TRBV-PE/FITC (Beckman-Coulter) and MHC-I tetramers-PE/APC. MHC-I tetramers: HLA-B*08-EI8 (EIYKRWII), HLA-B*08-FL8 (FLKEKGGL), HLA-B*15-GY9 (GLNKIVRMY), HLA-B*15-TY11 (TQGYFPDWQNY), HLA-B*27-KK10 (KRWIILGLNK) – synthesized by the NIH Tetramer Core Facility, Atlanta, GA. HLA-B*57-KF11 (KAFSPEVIPMF), HLA-B*57-IW9 (ISPRTLNAW), and HLA-B*57-QW9 (QASQEVKNW) – synthesized by Beckman-Coulter.
Cells were labeled with MHC-I tetramers at 21°C for 10 minutes. Anti-PD-1 antibody was added to the suspension and incubated for a further 20 minutes. Cells were washed and labeled in separate steps with intervening washes with pacific blue conjugated goat anti-mouse antibody, normal goat Ig blocking antibody, anti-CD127-biotin, streptavidin APC-Cy7, and the remaining directly conjugated surface antibodies listed above.
Identification of dominant and sub-dominant clonotypes and TRBV populations
The phenotype of T cell clonotypes was determined by a combination of labeling with tetramer, anti-TRBV antibodies, and antibodies to cell surface markers. Single TCR clonotypes identified by sequencing, and which comprised more than 50% of the epitope-specific population were considered dominant. In TCR repertoires where no clonotype comprised more than 50% of the total, the largest population was considered dominant, and the remaining populations were considered sub-dominant. Monoclonal antibodies are not available to label TRBV7, so in the five cases where the dominant TRBV7 clonotype was not directly labeled, TCR beta chain sequence data informed the identification of sub-dominant populations that were directly labeled. In these cases the unlabeled fraction of tetramer+ cells represented the dominant clonotypes. We determined TRBV repertoires for 11 epitopes in this study by using TRBV antibody panels (IOTest Beta Mark, TCR V-beta repertoire kit, Beckman Coulter). Dominant TRBV populations were definitively labeled within these responses, and sub-dominant populations were defined as tetramer+/TRBV−.
cDNA synthesis and TCR sequencing
Epitope-specific T cells were labeled with appropriate MHC-I tetramers and sorted by FACS to >95% purity on a FACSAria cell sorter (BD). RNA was extracted from sorted cells and anchored RT-PCR was performed with from total RNA as previously described (28). PCR product was cloned into E.coli and sequenced on an ABI 3130xl automated sequencer (PE Applied Biosystems, Norwalk, CT). After editing and alignment using Sequencher (Gene Codes Corp., Ann Arbor, MI), TRBV/TRBJ usage was determined using the human TCR gene database (http://imgt.cines.fr/). T cell receptor variable region classification system of the ImMunoGeneTics database (IMGT) is used throughout this manuscript.
Sequencing of autologous virus
Population viral sequence was obtained using viral RNA isolated from plasma (Qiagen) and reverse transcribed in one step (Qiagen) using HIV-Gag and HIV-Nef specific primers. DNA was amplified by PCR with the following primers: 5gag5–28 5’-GCG AGA GCG TCA GTA TTA AGC G-3’, 3gag1668–1693 5’-TCT GAG GGA AGC TAA AGG ATA CAG TT-3’, 3gag1398-1420 5’-AAA ATT AGC CTG TCT CTC CCC AT-3’, 5nef1-19 5’-ATG GGT GGC AAG TGG TCA A-3’, 3nef691-708 5’-TGC TAG GCG GCT GTC AAA-3’. Resulting PCR fragments were gel purified (Qiagen) and sequenced bi-directionally on an ABI 3130xl automated sequencer using the same primers. Sequencher (Gene Codes) was used to edit and align sequences and identification was made using the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/).
Intracellular Cytokine Staining
Intracellular cytokine staining assays were performed using 10ug/ml of indicated peptide, anti-CD28 and anti-CD49d MAbs (1 ug/mL each; BD) and GolgiPlug at 1ug/mL (BD). Cells were stimulated for 6 hours and labeled with surface and intracellular antibodies. Surface staining panel: CD3 (AlexaFluor-700, BD), CD4 (PE-Texas Red, Caltag), CD8 (Pacific Blue, BD), CD14/CD19/CD56 (PerCP, BD), Fixable Live-Dead Aqua (Invitrogen). Intracellular cytokine production: IFN-γ (PE-Cy7, BD) and TNF-α (APC, BD). Positive (staphylococcus enterotoxin B) and negative (unstimulated/media) controls were included for each individual. Reported cytokine production was subtracted from negative control values. Epitope variant panels: B*08-FL8 [Consensus-FLKEKGGL, Variant 1-FLrEKGGL, Variant 2-FLKdKGGL], B*08-EI8 [Consensus-EIYKRWII, Variant 1-dIYKRWII, Variant 2-EIYKRWIv], B*27-KK10 [Consensus-KRWIILGLNK, Variant 1-KRWIImGLNK, Variant 2-KRWIvLGLNK], B*57-QW9 [Consensus-QASQEVKNW, Variant 1-QAtQdVKNW, Variant 2-QAtQEVKNW] (peptide synthesis – Genemed, CA).
Tetramer binding analysis
PBMC were washed in FACS buffer, resuspended, aliquoted, and labeled for 30 minutes at RT with tetramer (APC-conjugated) at the following dilutions from manufactured stock – 1:25, 1:50, 1:100, 1:200, 1:400 final concentrations (~16uM to 4uM). With 5 minutes remaining for tetramer incubation, Live/Dead Fixable Aqua Dead Cell stain (Invitrogen) was added to each aliquot of PBMC. At 30 minutes, labeled cells were immediately washed with PBS and resuspended. Cells were fixed with 2% paraformaldehyde and washed in PBS. Fixed PBMC were first labeled with anti-TRBV-FITC conjugated antibodies and subsequently with antibodies to surface markers CD3, CD4, CD8, and CD14/19/56 (fluorescent antibodies and manufacturers as detailed above) for 30 minutes at room temperature. Surface antibodies were fixed to cells a final time and analyzed immediately.
In vitro culture and proliferation
PBMC were labeled with CFSE and cultured for 4 days in the presence or absence of peptide epitopes at the indicated concentrations. Cell culture media was supplemented with 1U/ml IL-2. Epitope-specific and clonotypic proliferation was assessed by costaining live cells with tetramer and anti-TRBV antibodies and measuring CFSE dilution.
Statistical analysis
Comparisons between whole CD4+, CD8+, and epitope-specific T cell populations were performed using Mann-Whitney tests. All paired comparisons were made using Wilcoxon matched pairs test. Fisher’s exact test for proportions was used to determine significance between PD-1 and CD127 expression on dominant and sub-dominant populations. Spearman rank correlation was used to test for the relationship between PD-1 expression and avidity for tetramer. All statistics were calculated using GraphPad Prism, v5.01.
Flow cytometry
All samples were sorted and data acquired on a FACSAria (BD) cell sorter. Data was analyzed using FACSDiva (BD) software. Plots shown using log10 fluorescence; histograms are log10 fluorescence vs. count.
Results
Epitope-specific T cell populations express high levels of PD-1
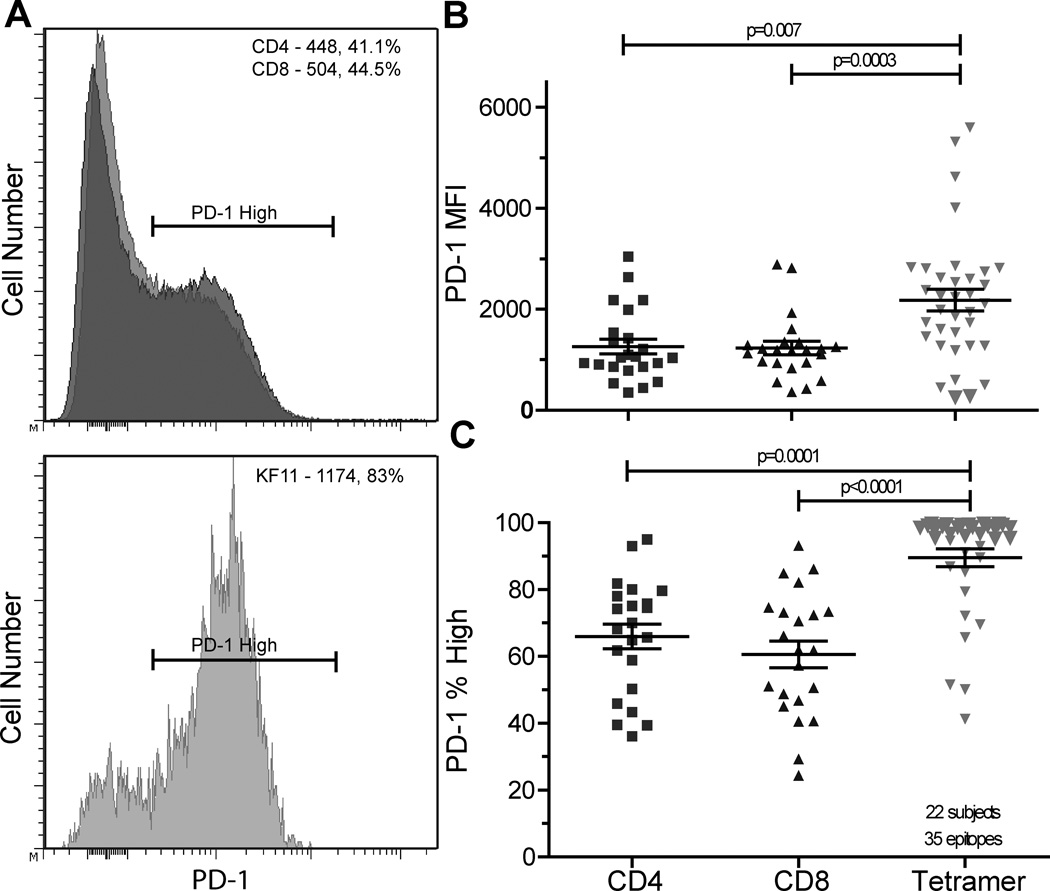
We evaluated the degree of PD-1 expression on total CD4+, CD8+, and HIV-specific CD8+ T cell populations in 22 chronic HIV+ patients off anti-retroviral therapy (Figure 1). These individuals had varying levels of disease progression (Table I, median VL=2474 copies/ml, range=<50-382,000; median CD4=688, range=132-1374). PD-1 expression (Mean fluorescence intensity, MFI) was measured on CD4+, CD8+, and 35 CD8+, HIV-epitope-specific T cell populations identified by MHC class I tetramers (Table I, mean 1.6 epitopes/individual, range 1–5 epitopes/individual). As has been observed by other groups(11, 12), we found PD-1 expression to be higher on HIV-specific CD8+ T cell populations when compared to total CD4+ (p=0.007, mean 2.4 fold higher) and CD8+ (p=0.0003, mean 1.9 fold higher) T cell populations (Figure 1A and 1B).
Figure 1. PD-1 is highly expressed in a bi-modal pattern on epitope-specific T cells in HIV+ individuals.
Histograms showing PD-1 expression on T cell populations in a single individual, PD-1 MFI and percentage PD-1high values are provided in the corner of each histogram, CD8+ (top panel, light grey), CD4+ (top panel, dark grey), and HIV-specific, tetramer+ (bottom panel), A. PD-1 MFI is higher on tetramer+ cells compared to CD4+ T cells (p=0.007) and CD8+ T cells (p=0.0003), B. Percentage of tetramer+ PD-1high cells is higher than the percentage CD4+ PD-1high T cells (p=0.0001) and CD8+ PD-1high T cells (p<0.0001), C. N=35 epitope-specific responses in 22 HIV+ individuals.
Table I.
PD-1 Study Cohort Demographic Data
| Patient ID |
# epitopes studied |
CD4 Count |
CD8 Count |
Plasma Viral Load (HIV copies/ml) |
Sex | Race# | Age (years) |
Duration of Infection (years) |
HLA-A1, -A2, -B1, -B2 |
|---|---|---|---|---|---|---|---|---|---|
| 10001** | 2 | 378 | 1539 | 32030 | M | W | 32 | 2 | 2, 2, 14, 15 |
| 10002†** | 3 | 664 | 1199 | 94 | M | W | 43 | 21 | 3, 31, 27, 57 |
| 10004†** | 2 | 203 | 1091 | 50 | M | W | 59 | 21 | 3, 30, 7, 57 |
| 10015** | 1 | 717 | 1462 | 98778 | M | W | 42 | 3 | 1, 3, 8, 52 |
| 10022†** | 2 | 724 | 1254 | 292 | M | W | 36 | 16 | 1, 31, 8, 27 |
| 10027†** | 5 | 775 | 1300 | 1893 | M | W | 66 | 14 | 1, 2, 8, 57 |
| 10035 | 1 | 429 | 663 | 2679 | M | AA | 52 | 21 | 3, 33, 27, 44 |
| 10038 | 1 | 510 | 646 | 1888 | M | W | 48 | 9 | 2, 2, 27, 44 |
| 10040 | 2 | 1161 | 972 | 50 | F | W | 49 | 14 | 1, 31, 44, 57 |
| 10060 | 1 | 688 | 1042 | 722 | M | AA | 30 | 4 | 1, 33, 42, 57 |
| 10069 | 1 | 1032 | 2005 | 2474 | M | AA | 45 | 4 | 1, 30, 53, 57 |
| 10070** | 1 | 782 | 391 | 7115 | F | AA | 28 | 10 | 23, 74, 57, 58 |
| 10071† | 2 | 743 | 414 | 50 | F | AA | 45 | 14 | 1, 66, 8, 57 |
| 10076 | 1 | 700 | 1500 | 21339 | M | AA | 54 | 6 | 2, 30, 35, 57 |
| 10086** | 1 | 132 | 876 | 76427 | M | W | 39 | 17 | 1, 29, 8, 44 |
| 10094 | 1 | 546 | 928 | 14621 | M | W | 44 | 8 | 1, 3, 7, 8 |
| 10105†** | 2 | 289 | 513 | 35050 | F | AA | 54 | 4 | 1, 23, 8, 44 |
| 10138†** | 2 | 231 | NA | 46800 | M | W | 46 | 6 | 24, 29, 15, 44 |
| 10141** | 1 | 542 | NA | 382000 | M | W | 46 | 6 | 1, 2, 15, 37 |
| 20002 | 1 | 612 | 578 | 1556 | M | W | 56 | 2 | 1, 2, 7, 27 |
| 20004 | 1 | 720 | 1094 | 3714 | M | W | 42 | 1 | 3, 32, 18, 27 |
| 20018** | 1 | 1374 | 1035 | 1886 | M | AA | 24 | 2 | 2, 26, 40, 57 |
W - white, AA - African American
subjects followed longitudinally
subjects in CD127 sub-cohort
NA - not available
PD-1 expression on CD4+, CD8+, and HIV-specific CD8+ T cell populations was often bi-modal, and we were able to measure the percentage of PD-1high cells within a given T cell population. Tetramer+, HIV-specific populations have a larger fraction of PD-1high cells than CD4+ or parent CD8+ T cell populations (p=0.0001 and p<0.0001, figure 1A and 1C). Despite overall high levels of PD-1 expression on epitope-specific T cells, we observed PD-1 expression as low as 40% on some epitope-specific populations, which may represent a subset of epitope-specific cells capable of greater function than PD-1high populations.
Dominant TRBV populations within HIV-specific T cell responses are predominantly clonotypic and express higher levels of PD-1 and lower levels of CD127 compared to sub-dominant TRBV populations
We next evaluated TRBV usage and clonotypic composition within HIV-specific PD-1high and PD1low populations. To identify T cell receptor usage within HIV-specific CD8+ T cell populations, we sequenced FACS-isolated HIV-specific CD8+ T cell in combination with direct staining of PBMCs with HIV-epitope-specific MHC-I tetramers and an anti-TRBV monoclonal antibody panel as previously described (25, 27). Twenty-one of 35 HIV-specific CD8+ T cell responses were sequenced to determine TRBV, CDR3, and corresponding TRBJ regions (Table II) with subsequent repertoire confirmation using monoclonal anti-TRBV antibodies. Within each epitope-specific TCR repertoire, we identified a single, dominant CDR3 clonotype, although sometimes this dominant clonotype was found with other clonotypes within a single TRBV family (Table II). For example, subject 10002 recognizes the HLA B*5701-restricted epitope IW9. Although we identified 8 clonotypes responding to this epitope, one TRBV27-TRJ27 clonotype comprises 64% of the sequences. Two other clonotypes also use TRBV27, but combined, they only contribute to 6% of the total sequences. In this case, staining with anti-TRBV27 antibody was used to identify the dominant T cell clonotype for phenotypic analysis. We noted highly significant concordance between these two methods used to identify clonotypes within the TCR repertoire (Supplemental figure 2A, r=0.86, p<0.0001).
Table II.
TRBV CDR3 Sequences of Epitope-specific Populations
| PID | Epitope | TRBV | CDR3 | TRBJ | Sequence % freq | TRBV labeling | TRBV% | # |
|---|---|---|---|---|---|---|---|---|
| 10001 | TY11 | TRBV10-3 | CAISERAIRGTSGLTDTQYF | TRBJ2-3 | 56 | Vb12 | 64 | 50/67 |
| TRBV10-3 | CAISECAIRGTSGLTDTQYF | TRBJ2-3 | 2 | 1/67 | ||||
| TRBV10-3 | CAISERAIRGTSGLTDTQYL | TRBJ2-3 | 2 | 1/67 | ||||
| TRBV4-2 | CASSQAAGGRAFF | TRBJ1-1 | 25 | Vb7.1 | 36 | 11/67 | ||
| TRBV4-2 | CASSQAEGGQNHF | TRBJ2-7 | 9 | 1/67 | ||||
| TRBV4-2 | CASSLAPGGIAFF | TRBJ1-1 | 5 | 3/67 | ||||
| TRBV4-2 | CASSQAAGGRAIF | TRBJ1-1 | 2 | 1/67 | ||||
| 10001 | GY9 | TRBV11-2 | CASSLDSGFLEQYF | TRBJ2-7 | 44 | Vb21.3 | 44 | 18/36 |
| TRBV10-3 | CAISESGGRVDEQYF | TRBJ2-7 | 36 | Vb12 | 21 | 13/36 | ||
| TRBV12-3 | CASSPPSSYNEQFF | TRBJ2-7 | 3 | Vb8 | 11 | 1/36 | ||
| TRBV4-3 | CASSLQGAPEQFF | TRBJ2-1 | 8 | Vb7.2 | 3/36 | |||
| TRBV11-3 | CTSRLDPGFLEQYF | TRBJ2-7 | 3 | Vb21 | 1/36 | |||
| 10002 | KK10 | TRBV7-2 | CASSLYGEYEQYF | TRBJ2-7 | 79 | Ab unavailable | 30/38 | |
| TRBV13 | not identified by seq | Vb23 | 4 | |||||
| TRBV6-6 | CASSQGTTDTQYF | TRBJ2-3 | 21 | 8/38 | ||||
| 10002 | IW9 | TRBV27 | CASRPGQGGYEQY | TRBJ2-7 | 64 | Vb14 | 62 | 29/45 |
| TRBV27 | CASSSSTGQQPQH | TRBJ1-5 | 4 | 2/45 | ||||
| TRBV27 | CASRTQRWETQY | TRBJ2-5 | 2 | 1/45 | ||||
| TRBV7-9 | CASSLAQGWKTQY | TRBJ2-5 | 20 | Ab unavailable | 9/45 | |||
| TRBV7-9 | CASSIQGLRATNEKLF | TRBJ1-4 | 2 | 1/45 | ||||
| TRBV7-8 | CASRSPLGYEQY | TRBJ2-7 | 2 | 1/45 | ||||
| TRBV12-4 | CASSSGTSGSAGYNEQF | TRBJ2-1 | 2 | 1/45 | ||||
| TRBV5-1 | CASSTNNEQF | TRBJ2-1 | 2 | 1/45 | ||||
| 10002 | QW9 | TRBV27 | CASRTQRWETQY | TRBJ2-5 | 100 | Vb14 | 93 | 13/13 |
| TRBV4 | not identified by seq | Vb7.2 | 7 | |||||
| 10004 | QW9 | TRBV3-1 | CASSQGPGERAGFNYEQY | TRBJ2-7 | 56 | Vb9 | 55 | 4/16 |
| TRBV28 | CASSLGYGYT | TRBJ1-2 | 19 | Vb3 | 29 | 3/16 | ||
| TRBV27 | CASSKGRYNEQF | TRBJ2-1 | 25 | Vb14 | 12 | 9/16 | ||
| 10004 | KF11 | TRBV7-9 | CASPHPDRPNYGYT | TRBJ1-2 | 78 | Ab unavailable | 39/50 | |
| TRBV7-9 | CASGGEFYGYT | TRBJ1-2 | 16 | 8/50 | ||||
| TRBV19 | CASSLTYGYT | TRBJ1-2 | 2 | Vb17 | 26 | 1/50 | ||
| TRBV19 | CASSSRTGGYGYT | TRBJ1-2 | 2 | 1/50 | ||||
| TRBV24 | CATSDRMDNEQF | TRBJ2-1 | 2 | 1//50 | ||||
| 10022 | KK10 | TRBV12-4 | CASSIAGGGEDTQY | TRBJ2-3 | 31 | Vb8 | 45 | 26/84 |
| TRBV6-5 | CASRKGQGDWEAF | TRBJ1-1 | 5 | Vb13.1 | 35 | 4/84 | ||
| TRBV20-1 | CSARGWVSNNQETQY | TRBJ2-5 | 57 | Vb2 | 20 | 48/84 | ||
| TRBV20-1 | CSARDPLPEASGGAGTDTQY | TRBJ2-3 | 2 | Vb2 | 1/84 | |||
| TRBV19 | CASTPPGF | TRBJ1-2 | 1 | Vb17 | 2/84 | |||
| TRBV27 | CASSQWTGELF | TRBJ2-2 | 4 | Vb14 | 1/84 | |||
| 10022 | FL8 | TRBV6-2 | CASSFIPGQGTHYSNQPQH | TRBJ1-5 | 78 | Vb13.2 | 75 | 7/9 |
| TRBV10-3 | CAIRPFLGQDDNYGYT | TRBJ1-2 | 11 | Vb12 | 11 | 1/9 | ||
| TRBV28 | CASSLRGTGELF | TRBJ2-2 | 11 | Vb3 | 14 | 1/9 | ||
| 10027 | KF11 | TRBV10-3 | CAIGGHDYGYT | TRBJ1-2 | 45 | Vb12 | 54 | 40/88 |
| TRBV6-5 | CASSSLVNTGELF | TRBJ2-2 | 23 | Vb13.1 | 10 | 20/88 | ||
| TRBV6-5 | CALTGGDYGYT | TRBJ1-2 | 7 | 6/88 | ||||
| TRBV20-1 | CSARGWVSNNRETQY | TRBJ2-5 | 14 | Vb2 | 10 | 12/88 | ||
| TRBV20-1 | CAASTSAVLGKKGSQETQY | TRBJ2-5 | 2 | 2/88 | ||||
| TRBV20-1 | CSAREKGSQETQY | TRBJ2-5 | 8 | 7/88 | ||||
| TRBV28-1 | CASSGPGGEQY | TRBJ2-7 | 1 | 1/88 | ||||
| 10027 | FL8 | TRBV2 | CASSELGARVYEQYF | TRBJ2-7 | 67 | Vb22 | 70 | 37/42 |
| TRBV10-1 | CASSESSREVSYNSPLHF | TRBJ1-6 | 6 | Vb12 | 11 | 3/42 | ||
| TRBV4-2 | CASKEELSNTGELFF | TRBJ2-2 | 23 | 1/42 | ||||
| TRBV29-1 | CSVGDQGGSEQYF | TRBJ2-7 | 4 | 1/42 | ||||
| 10027 | EI8 | TRBV13-6 | CASTGGRGSPLHF | TRBJ1-6 | 100 | Vb13 .1 | 95 | 45/45 |
| TRBV9 | Not identified by sequence | Vb1 | 2 | |||||
| TRBV20 | Not identified by sequence | Vb2 | 1 | |||||
| TRBV28 | Not identified by sequence | Vb3 | 1 | |||||
| 10060 | IW9 | TRBV7-8 | CASSQDRIHTEAF | TRBJ1-1 | 94 | Ab unavailable | 52/54 | |
| TRBV13-2 | CASSLGLDETQYF | TRBJ2-5 | 6 | 5 | 2/54 | |||
| 10070 | KF11 | TRBV7-9 | CASSLGGGYT | TRBJ1-2 | 50 | Ab unavailable | 22/44 | |
| TRBV7-8 | CASEDFKNIQY | TRBJ2-4 | 16 | 7/44 | ||||
| TRBV7-9 | CASSPGQTNYGYT | TRBJ1-2 | 14 | 6/44 | ||||
| TRBV7-9 | CATPGEVLSPNYGYT | TRBJ1-2 | 2 | 1/44 | ||||
| TRBV7-9 | CASSLGGGQNGYT | TRBJ1-2 | 2 | 1/44 | ||||
| TRBV7-6 | CASSSMGGGTDTQY | TRBJ2-2 | 2 | 1/44 | ||||
| TRBV7-9 | CASSLAGGYT | TRBJ2-2 | 2 | 1/44 | ||||
| TRBV11-2 | CASSDGTGVGLGYT | TRBJ1-2 | 5 | Vb21.3 | 12 | 2/44 | ||
| TRBV11-2 | CASSDGQGRLGYT | TRBJ1-2 | 2 | 1/44 | ||||
| TRBV14 | CASSPRDSQETQY | TRBJ2-5 | 2 | Vb16 | 8 | 1/44 | ||
| 10071 | KF11 | TRBV5-1 | CASYNFGQYGYT | TRBJ1-2 | 75 | Vb5 | 67 | 20/25 |
| TRBV7-6 | CASSPMDLLDEQY | TRBJ2-7 | 25 | Ab unavailable | 5/25 | |||
| 10071 | FL8 | TRBV2 | CASSELGATIYEQY | TRBJ2-7 | 40 | Vb22 | 46 | 8/20 |
| TRBV4-1 | CASSQEMNRVVGNEQF | TRBJ2-1 | 25 | Vb7.1 | 26 | 7/20 | ||
| TRBV6-9 | CASTRPGQGTYNEFQ | TRBJ2-1 | 35 | Ab unavailable | 5/20 | |||
| 10076 | KF11 | TRBV2 | CASRGGSGELF | TRBJ2-2 | 63 | Vb22 | 87 | 34/54 |
| TRBJ7-9 | CASSGFRDRVNEQY | TRBJ2-7 | 33 | Ab unavailable | 18/54 | |||
| 10086 | EI8 | TRBV9 | CASSVVGDSRETQYF | TRBJ2-5 | 52 | Vb1 | 80 | 24/46 |
| TRBV9 | CGSSVVGDSRETQYF | TRBJ2-5 | 2 | 1/46 | ||||
| TRBV9 | CASSTLRDSREKLFF | TRBJ1-4 | 4 | 2/46 | ||||
| TRBV9 | CASSTLGDSREKLFF | TRBJ1-4 | 7 | 3/46 | ||||
| TRBV9 | CASSADGSFYEQYF | TRBJ2-7 | 2 | 1/46 | ||||
| TRBV27 | CASSLVGQGARQPQHF | TRBJ1-5 | 2 | Vb14 | 9 | 1/46 | ||
| TRBV27 | CASSLGSAKNIQYF | TRBJ2-4 | 2 | 1/46 | ||||
| TRBV2 | CASSEPPGVRGEAFF | TRBJ1-1 | 9 | 4/46 | ||||
| TRBV4-1 | CAGSQEFLNRRYF | TRBJ2-5 | 7 | 3/46 | ||||
| TRBV5-1 | CASSLSGSGWQETQYF | TRBJ2-5 | 9 | 4/46 | ||||
| TRBV7-2 | CASSLLPDPRSSGGYTF | TRBJ1-2 | 2 | 1/46 | ||||
| TRBV7-8 | CASSLLDGTRDQQYF | TRBJ2-7 | 2 | 1/46 | ||||
| 10138 | GY9 | TRBV5-1 | CASSEAGGTEAFF | TRBJ1-1 | 46 | Vb5.1 | 47 | 21/46 |
| TRBV9 | CASSVEGTILTDTQYF | TRBJ2-3 | 13 | Vb1 | 47 | 6/46 | ||
| TRBV9 | CASSVEGTIHTDTQYF | TRBJ2-3 | 9 | 4/46 | ||||
| TRBV13 | CASSLQQTLGAFF | TRBJ1-1 | 9 | Vb23.1 | 3 | 4/46 | ||
| TRBV13 | CASSPQQTLGAFF | TRBJ1-1 | 9 | 4/46 | ||||
| TRBV20-1 | CSALVEGDEQFF | TRBJ2-1 | 4 | 2/46 | ||||
| TRBV20 | CSAIVGSAYEQYF | TRBJ2-7 | 2 | 1/46 | ||||
| TRBV29 | CSVDGPTGGYTF | TRBJ1-2 | 4 | 2/46 | ||||
| TRBV29 | CASSQGLAGDEQYF | TRBJ1-2 | 2 | 1/46 | ||||
| TRBV29 | CSASLGGRISGANVLTF | TRBJ1-2 | 2 | 1/46 | ||||
| TRBV14 | CASSQDLRGARGYTF | TRBJ1-2 | 2 | 1/46 | ||||
| TRBV14 | CASSQGTGSTDTQYF | TRBJ2-3 | 2 | 1/46 | ||||
| TRBV4-2 | CASSQDSSGRVTGELFF | TRBJ2-2 | 2 | 1/46 | ||||
| 10141 | GY9 | TRBV27 | CASSDNGGDRSPGELFF | TRBJ2-2 | 54 | Vb14.1 | 67 | 21/39 |
| TRBV27 | CASSPSFPPDTQYF | TRBJ2-3 | 10 | 4/39 | ||||
| TRBV27 | CASSPGGGELFF | TRBJ2-2 | 3 | 1/39 | ||||
| TRBV6-6 | CASSSPGGVTEAFF | TRBJ1-1 | 10 | Vb13.6 | 10 | 4/39 | ||
| TRBV6-6 | CASSYSVVEAAAEAFF | TRBJ1-1 | 2 | 1/39 | ||||
| TRBV20-1 | CSARDRADRVLIPDTQYF | TRBJ2-3 | 8 | Vb2.1 | 6 | 3/39 | ||
| TRBV20-1 | CSASPVGGAYEQYF | TRBJ2-7 | 5 | 2/39 | ||||
| TRBV6-5 | CASRLGRLAYEQYF | TRBJ2-7 | 3 | 1/39 | ||||
| TRBV6-5 | CASSTLTGEDSGPQHF | TRBJ1-5 | 3 | 1/39 | ||||
| TRBV3-1 | CASSQGLAGDEQFF | TRBJ2-1 | 3 | 1/39 | ||||
| 20018 | KF11 | TRBV7-9 | CASELSGNTIY | TRBJ1-3 | 70 | Ab unavailable | 35/51 | |
| TRBV7-9 | CASSYLNTIY | TRBJ1-3 | 2 | 1/51 | ||||
| TRBV7-9 | CASEGGNTIY | TRBJ1-3 | 2 | 1/51 | ||||
| TRBV7-9 | CATEASGNTIY | TRBJ1-3 | 12 | 6/51 | ||||
| TRBV7-9 | CASEITRDRRNTIY | TRBJ1-3 | 2 | 1/51 | ||||
| TRBV7-6 | CASSSWTGQDEQF | TRBJ2-1 | 2 | 1/51 | ||||
| TRBV7-9 | CASSGFTGFANEAF | TRBJ2-6 | 2 | 1/51 | ||||
| TRBV28-1 | CATSDLMDNEQF | TRBJ2-1 | 4 | Vb3 | 7 | 2/51 | ||
| TRBV24-1 | CATSDLMDNEQF | TRBJ2-1 | 2 | 1/51 | ||||
| TRBV5-6 | CASILTSGRNEQF | TRBJ2-1 | 2 | Vb5.2 | 3 | 1/51 | ||
Dark shading identifies dominant clonotype
Light shading identifies sub-dominant population(s) used for comparison (unshaded clonotypes were unlabeled)
TRBV populations directly labeled with antibodies are bordered
IMGT nomenclature for TRBV designation is used throughout the table and accompanying text
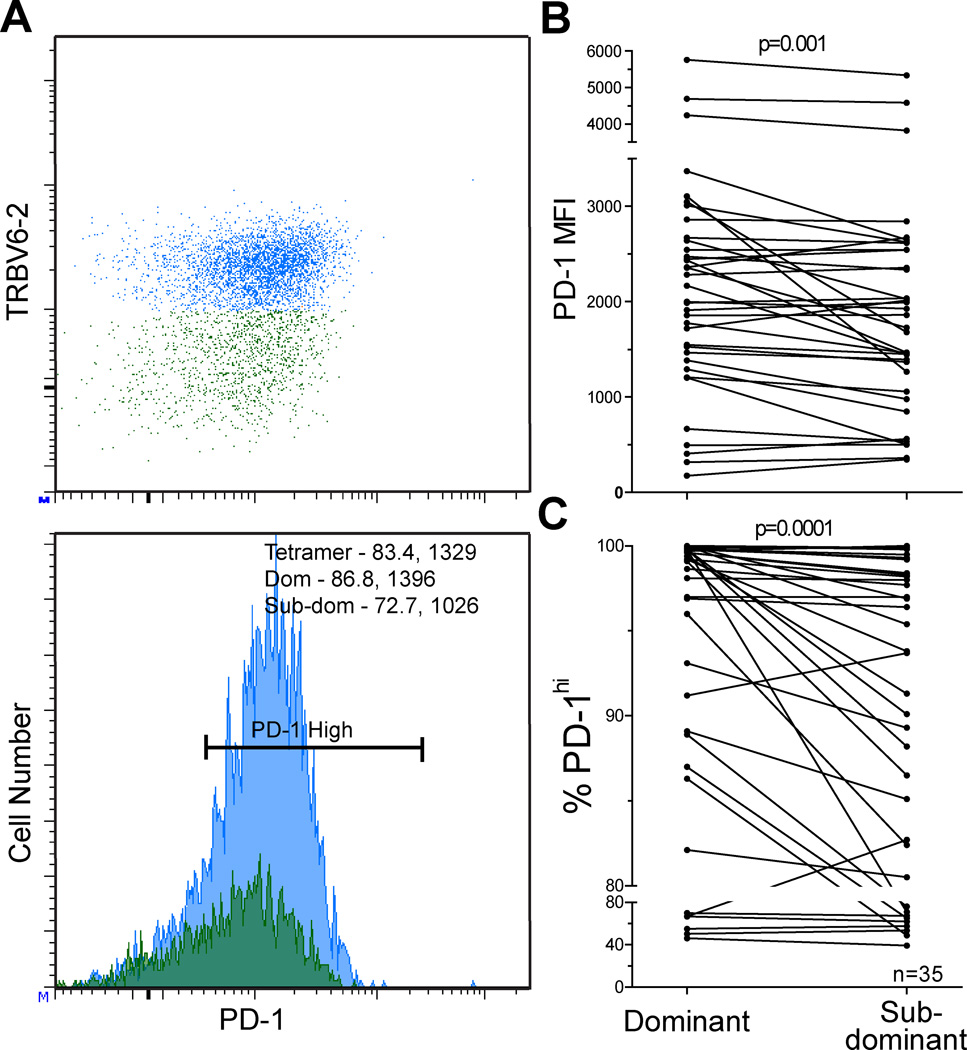
We show representative plots of PD-1 expression on corresponding dominant and sub-dominant TRBV populations in a single HIV-epitope-specific T cell response (Figure 2A). Within HIV-epitope-specific responses, PD-1 expression is higher on dominant TRBV populations compared to sub-dominant TRBV populations when measured by MFI (p=0.001, Figure 2B) or frequency of PD-1high cells (p=0.0001, Figure 2C). We evaluated multiple HIV-epitope-specific populations in 9 of 22 individuals studied (range 2–5 epitopes/individual, Table I and Table III). We did not find a correlation between the degree of dominance within the repertoire and the degree of PD-1 expression on dominant and sub-dominant clonotypes within epitopes, suggesting that the magnitude of expansion within a parent population is not the sole determinant of PD-1 expression.
Figure 2. PD-1 expression is higher on dominant TRBV compared to sub-dominant TRBV populations within epitope-specific responses.
Dot plot and histogram showing PD-1 expression on dominant (blue) and sub-dominant (green) TRBV populations in a single epitope-specific response. PD-1 MFI and percentage PD-1high values are provided in the upper corner histogram for the dominant and sub-dominant TRBV populations, A. PD-1 expression is higher on dominant TRBV compared to subdominant TRBV as measured by MFI, B (p=0.001) and percentage PD-1high, C (p=0.0001). N= 35 epitope-specific populations in 22 HIV+ individuals.
Table III.
TRBV Repertoire Data
| PID | Epitope | % of CD8 | TRBV Repertoirea | Methodb,c |
|---|---|---|---|---|
| 10001 | TY11 | 2.2 | 10-3 (64%), 4-2 (36%) | sort-sequence |
| GY9 | 1.5 | 11-2 (44%), 10-3 (21%), 12 (11%) | sort-sequence | |
| 10002 | KK10 | 5.6 | 13-1 (4%), 7-2 (96%) | sort-sequence |
| IW9 | 2.9 | 27 (62%), 7-9, 12-4, 5-1 (38%) | sort-sequence | |
| QW9 | 1.6 | 27 (92%), 4 (7%) | sort-sequence | |
| 10004 | KF11 | 1.4 | 19 (26%), 7-9, 24 (74%) | sort-sequence |
| QW9 | 1.1 | 3-1 (55%), 28 (29%), 27 (12%) | sort-sequence | |
| 10015 | EI8 | 1.2 | 9 (71%), 2 (10%), 4 (5%) | TRBV |
| 10022 | KK10 | 19 | 12 (45%), 6-5 (40%), 2 (15%) | sort-sequence |
| FL8 | 1.3 | 6-2 (75%), 10-3 (11%), 28 (14%) | sort-sequence | |
| 10027 | KF11 | 0.5 | 10-3 (54%), 6-5 (10%, mult cdr3), 20-1 (10% multi cdr3) | sort-sequence |
| FL8 | 0.8 | 22 (70%), 12 (11%) | sort-sequence | |
| EI8 | 0.4 | 6 (95%), 9 (2%) | sort-sequence | |
| QW9 | 0.8 | 28 (85%) | TRBV | |
| IW9 | 0.7 | 27 (86%), 4 (5% multi-Vb) | TRBV | |
| 10035 | KK10 | 5.5 | 6 (62%), 19 (16%), 20 (15%), 12 (4%), 27 (3%) | TRBV |
| 10038 | KK10 | 1.3 | 6-2 (32%), 3 (24%), 4-2 (11%), 19 (20%) | TRBV |
| 10040 | QW9 | 2 | 6 (95%) | TRBV |
| KF11 | 2 | 9 (94%) | TRBV | |
| 10060 | IW9 | 1.5 | 13-1 (4%), 7-8 (96%) | sort-sequence |
| 10069 | QW9 | 2.6 | 6 (90%) | TRBV |
| 10070 | KF11 | 10 | 11-2 (12%), 27 (8%), 7 (80%) | sort-sequence |
| 10071 | KF11 | 1.8 | 5 (67%), 7 (33%) | sort-sequence |
| FL8 | 3.7 | 2 (46%), 4-1 (26%) | sort-sequence | |
| 10076 | KF11 | 7.4 | 2 (87%), 7 (13%) | sort-sequence |
| 10086 | EI8 | 1.8 | 9 (80%), 27 (9%) | sort-sequence |
| 10094 | FL8 | 3.7 | 27 (77%), 28 (7%) | TRBV |
| 10105 | FL8 | 1.8 | 6-2 (70%) | sort-sequence |
| EI8 | 1.8 | 8-1 (45%) | sort-sequence | |
| 10138 | GY9 | 2.1 | 5-1 (47%), 9 (47%), 13 (3%) | sort-sequence |
| TY11 | 2.2 | 29 (52%), 9 (13%), 5-1 (12%), 13-2 (10%), 12 (8%) | sort-sequence | |
| 10141 | GY9 | 0.8 | 27 (67%), 6-6 (10%), 20 (6%) | sort-sequence |
| 20002 | KK10 | 5.3 | 5-2 (45%) | TRBV |
| 20004 | KK10 | 2.4 | 27 (68%), 4-3 (12%) | TRBV |
| 20018 | KF11 | 11.2 | 28-1 (7%), 5-6 (3%), 7 (90%) | sort-sequence |
TRBV Repertoire - as determined by antibody labeling
sort-sequence - tetramer+, epitope-specific cells were sorted and subjected to TCR sequence analysis before co-staining with anti-TRBV antibodies
TRBV - tetramer+, epitope-specific cells were co-stained with anti-TRBV antibodies
If we limit our phenotypic analysis to those epitopes for which we have sequence confirmation that the dominant TRBV population is monoclonal, the relationships we highlight between clonotypic dominance and PD-1 and CD127 expression remain statistically significant (PD-1 MFI, p=0.0398; CD127 MFI, p=0.0342). Additionally, there are several ways to define clonotypic dominance within epitope-specific TCR repertoires in the absence of a single, highly dominant clonotype; however, even using a more stringent criterion that dominant clonotypes must comprise more than 70% of the TRBV repertoire (19 epitope-specific responses in 14 individuals fit this criteria), comparison between dominant and sub-dominant clonotypes yields significant relationships for MFI and % PD-1high (p=0.03, MFI and p=0.001, % PD-1high, supplemental figure 2B and 2C). These data support our observations that dominant clonotypes express higher levels of PD-1 despite relative differences in dominance within the clonotypic repertoire.
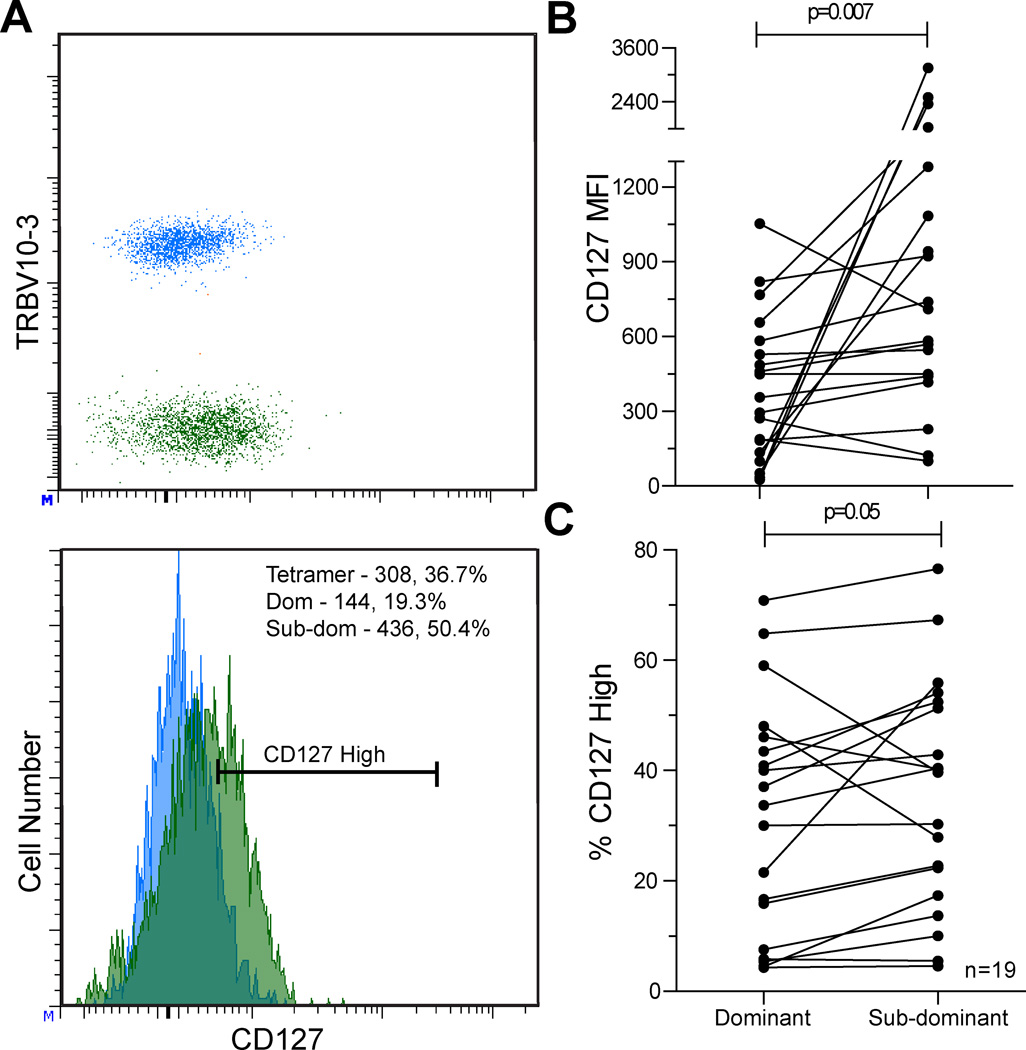
We also evaluated CD127 expression on dominant and sub-dominant TRBV populations in a sub-cohort of 12 individuals, which included analysis of 19 epitope-specific responses (noted in Table I). In contrast to higher PD-1 expression observed on dominant TRBV populations, CD127 expression was lower on dominant TRBV populations as measured by MFI (p=0.007, Figure 3B) and frequency of CD127hi (p=0.05, Figure 3C) compared to corresponding sub-dominant TRBV populations. The PD-1 expression pattern described above on dominant and sub-dominant TRBV populations remains intact in this smaller cohort (p=0.006, PD-1 MFI).
Figure 3. CD127 expression is lower on dominant TRBV compared to sub-dominant TRBV populations within epitope-specific responses.
Dot plot and histogram showing CD127 expression on dominant (blue) and sub-dominant (green) TRBV populations for a single epitope-specific response. MFI and percentage CD127high values are provided in the corner of the histogram for the dominant and sub-dominant TRBV populations, A. CD127 expression is lower on dominant TRBV compared to sub-dominant TRBV as measured by MFI, B (p=0.007) and percentage CD127high, C (p=0.05). Measurements from 19 epitope-specific populations in 12 HIV+ individuals.
Within this sub-cohort of epitopes labeled with PD-1 and CD127, 15 of 19 dominant TRBV populations displayed a PD-1high phenotype and 15 of 19 displayed a CD127low phenotype compared to their corresponding sub-dominant population. However, there was not complete concordance between these populations. The majority of dominant clonotypes (11 of 19) displayed the combination of higher PD-1 expression and lower CD127 expression. In contrast, there were no instances (0 of 19) in which the sub-dominant clonotype had both higher PD-1 expression and lower CD127 expression (p<0.0001). In summary, our data indicate that clonotypic dominance within the epitope-specific TCR repertoire is associated with a PD-1high/CD127low phenotype.
PD-1high/CD127low phenotype on dominant clonotypes in HIV-specific responses is stable over time
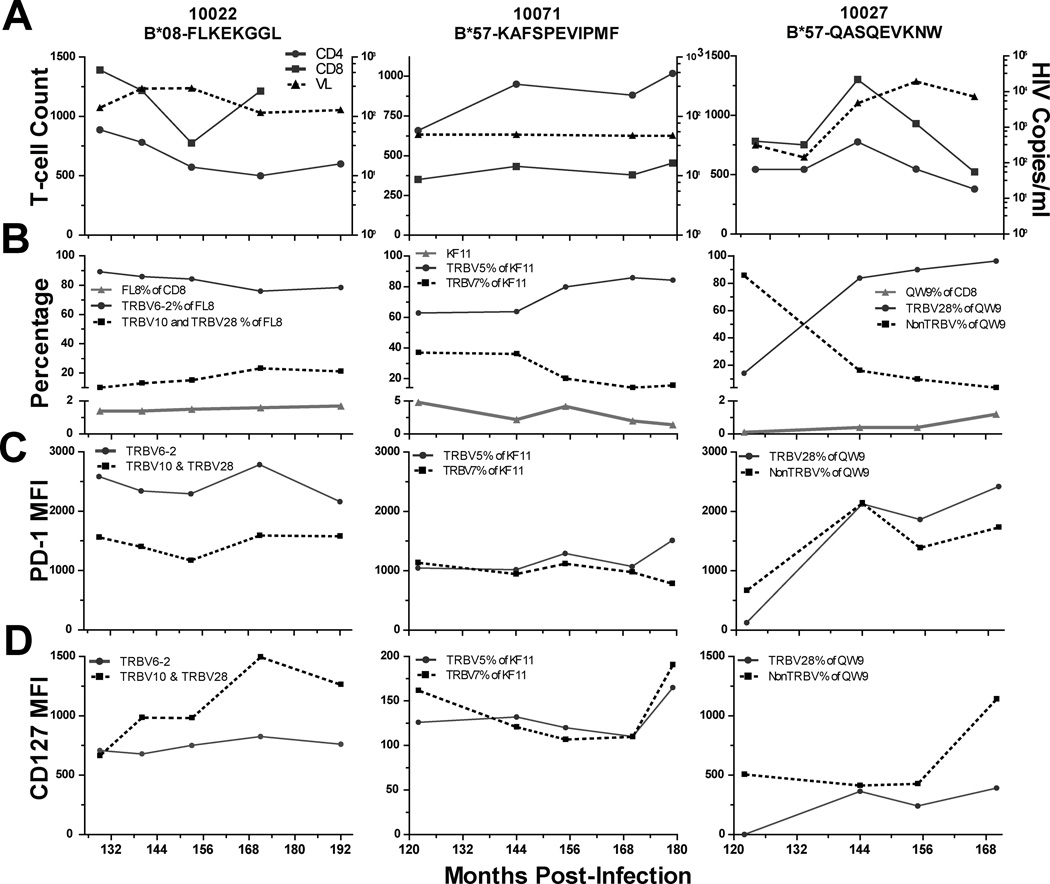
To characterize the stability of PD-1 and CD127 expression on dominant and subdominant TRBV population, we performed a longitudinal analysis of HIV-specific responses from 3 individuals. Figure 4 details longitudinal viral load and CD4+ and CD8+ T cell number (4A), epitope-specific CD8+ T cell frequency and corresponding TRBV repertoire composition (4B), and PD-1 and CD127 expression (4C and 4D) on TRBV populations for the dominant clonotype within the HLA-B*08-FL8 response in 10022, the dominant TRBV population within the HLA-B*57-QW9 response in 10027 (this epitope-specific T cell population was not sequenced), and the dominant clonotype within the HLA-B*57-KF11 response in 10071 for the most recent 6 years of their infections (duration of infection 16, 16, and 15 years, respectively). 10022 and 10071 are long-term controllers with stable viral loads and CD4+ T cell counts and 10027 is a chronically infected individual with progressive disease (increasing viral load and decreasing CD4+ T cell counts).
Figure 4. Longitudinal analysis of epitope-specific TCR repertoire dynamics and clonotypic PD-1 and CD127 expression.
Absolute CD4+ (circles) and CD8+ (squares) T cell counts (left-hand axis) and viral load (dashed line, triangles, RNA copies/ml, right-hand axis), A. Epitope-specific responses as a percentage of total CD8+ parent population (triangles, below y-axis split) and TRBV % of epitope-specific response (above y-axis split, dominant clonotype, solid line, circles and subdominant clonotype, dashed line, squares), B. PD-1 expression, C, and CD127 expression, D, on dominant (solid line, circles) and sub-dominant (dashed line, squares) TRBV populations within epitope-specific responses.
Although expression levels of PD-1 and CD127 on the TRBV clonotypes within these HIV-specific responses are dynamic, the association of higher PD-1 expression and lower CD127 expression with TRBV dominance remains consistent over the 6 years of our analysis. The B*08-FL8-specific TRBV repertoire in 10022 is relatively stable over time. The dominant TRBV2 population in this individual maintains higher PD-1 expression over time whereas the sub-dominant TRBV populations have higher and increasing CD127 levels over the same period. The B*57-KF11-specific TRBV repertoire in 10071 is characterized by an increasingly dominant TRBV5 population and a corresponding increase in PD-1 expression compared to the subdominant TRBV7 population. In 10027, the B*57-QW9-specific TCR repertoire fluctuates early in our observations and as the TRBV28 population becomes dominant, its PD-1 expression levels increase. Over this time period, 10027 experienced declining T cell counts and increasing viral load with an overall increase in PD-1 expression on CD8+ T cells. The dominant circulating viral sequence in 10022 and 10027 was determined for the B*27-FL8 and B*57-QW9 epitopes at a midpoint in this analysis and corresponded to the peptide sequence within the tetramers in each case. 10071 maintained viral loads of <50 copies/ml during this study, and we were unable to generate viral sequences from this individual.
We also evaluated mean PD-1 and CD127 expression levels at early and late timepoints on 10 additional epitope-specific responses and determined a similar and statistically significant expression pattern on dominant and sub-dominant TRBV populations (Supplemental Figure 3). These longitudinal data suggest that dominance within the epitope-specific TRBV repertoire is associated with a more pronounced PD-1hi/CD127lo phenotype over time and may be related to the course of disease.
Tetramer binding characteristics of TRBV populations correlate with PD-1 expression but are not directly related to dominance within the epitope-specific TCR repertoire
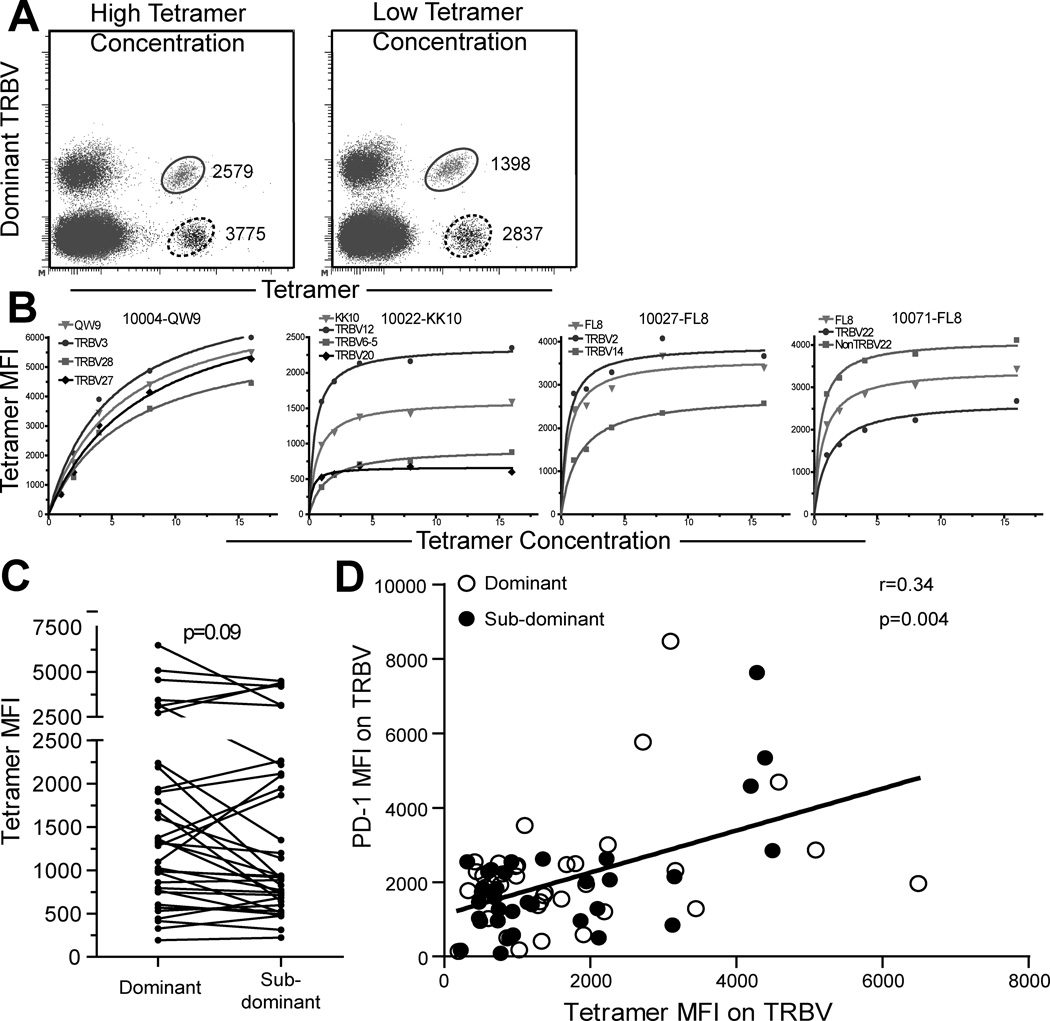
We next investigated whether differences in tetramer binding characteristics were related to dominance within the TRBV repertoire. Our group and others have previously described differential tetramer binding on epitope-specific T cell clonotypes (23, 29) and we observed a similar phenomenon in this study (Figure 5A). Several groups have previously used differential levels of tetramer binding to define T cell receptor avidity (29, 30), and so we measured tetramer binding (MFI) on TRBV populations over a 16-fold range of tetramer concentration and determined tetramer-binding curves for dominant and sub-dominant clonotypes of 9 epitope-specific responses in 4 individuals (Figure 5B and Supplementary Table I). Non-linear regression analysis indicated that TRBV populations with lower half-maximal values have higher maximal binding values in 8 of the 9 epitopes tested. Thus, we used tetramer MFI on labeled TRBV populations as a surrogate measure of TCR avidity for tetramer complexes.
Figure 5. Tetramer binding correlates to PD-1 expression on epitope-specific T cell clonotypes.
MHC-I tetramers were used to label epitope-specific T cell populations at a range of tetramer concentrations. Dot plots from highest and lowest tetramer concentrations show variable tetramer binding on clonotypes, A. Epitope-specific clonotypes were labeled at increasing tetramer concentrations from ~0–16uM. Representative graphs of tetramer binding curves are shown for 10004-QW9, 10022-KK10, 10027-FL8, and 10071-FL8 (subject-epitope) for whole epitope-specific populations (triangles), and TRBV populations (dominant-circles, sub-dominant-squares, sub-sub-dominant-diamonds), B. Comparison of tetramer binding levels (tetramer MFI) on dominant and sub-dominant TRBV populations for 35 epitopes and 22 individuals (p=0.09), C. Spearman correlation of tetramer binding and PD-1 expression on dominant (open circles) and sub-dominant (closed circles) clonotypes, (r=0.34, p=0.004), D.
We compared tetramer binding levels on corresponding dominant and sub-dominant TRBV populations. While there was a trend suggesting that dominant TRBV populations have higher avidity for tetramer than corresponding sub-dominant populations, this pairing was not statistically significant (Figure 5C, p=.09). We found a positive and significant correlation between clonotypic avidity for tetramer and clonotypic PD-1 expression (Figure 5D, r=0.34, p=0.004). These data indicate that while clonotypic avidity for tetramer does not strictly govern dominance within the repertoire, it may influence the degree of PD-1 expression.
Sub-dominant TRBV populations display greater cytokine production capacity and cross-recognition in responses to epitope variant peptides
We assessed the capacity of dominant and sub-dominant TRBV populations to produce cytokines after stimulation with consensus and variant peptides. Two common viral sequence variants for each of 4 HIV-epitopes were tested in 7 individuals. We performed viral sequencing in these individuals and found that circulating viral sequence matched the consensus epitopes used in the tetramer reagents in each individual except for 10094, who harbored a circulating sequence variant at the FL8 epitope which matched the FLKdKGGL variant we used in our functional assay.
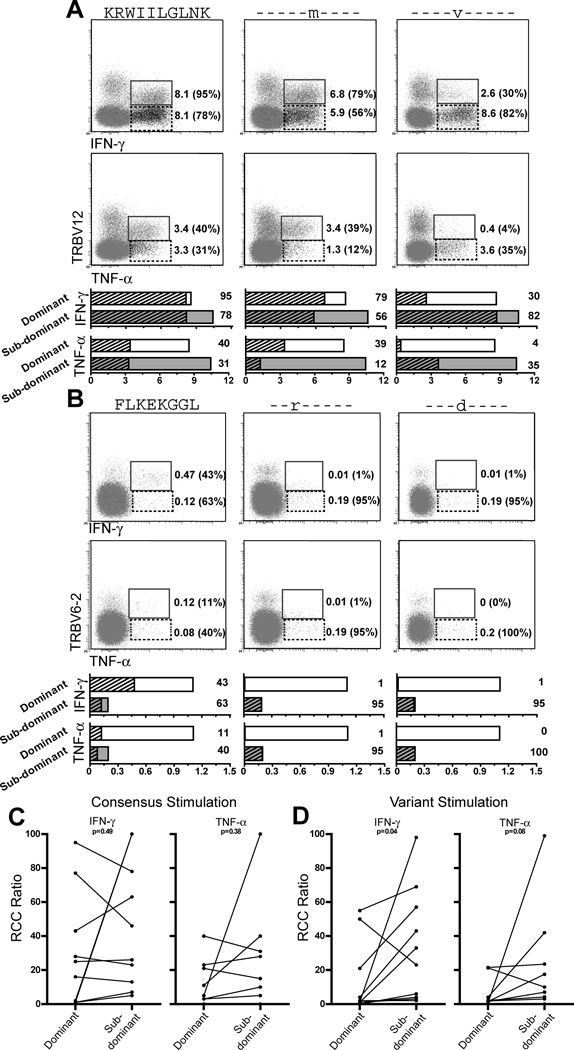
Taking our analysis of the B*27-KK10 response in 10022 as an example, the dominant TRBV12 clonotype comprises 45% of the B*27-KK10 response which is 19% of total CD8+ T cells. The maximal possible cytokine production by the TRBV12 clonotype is therefore 8.6% of total CD8+ T cells. Likewise, maximal cytokine production for the sub-dominant clonotypes (TRBV6-5 and TRBV20-1, together 55% of the KK10-tetramer+ population) is 10.4% of total CD8+ T cells. We determined the relative cytokine capacity (RCC) of dominant and sub-dominant TRBV populations by dividing cytokine production of the TRBV population by the frequency of that TRBV population within the tetramer population. By virtue of being a ratio, the RCC value for each TRBV population illustrates the extent to which it reaches its own maximal cytokine production potential without regard to its absolute percentage within the TCR repertoire.
Representative plots are shown in Figure 6A detailing cytokine production (IFN-γ – upper plots; TNF-α – lower plots) by the dominant TRBV12 clonotype and the sub-dominant clonotypes in response to stimulation with consensus and variant peptides for the HLA-B*27-KK10 epitope. In response to stimulation with consensus peptide, the TRBV12 clonotype reached absolute cytokine production levels of 8.1% (IFN-γ) and 3.4% (TNF-α) of total CD8+ T cells and the sub-dominant clonotypes reached cytokine production levels of 8.1% (IFN-γ) and 3.3% (TNF-α) of total CD8+ T cells. TRBV12 RCC values are 95% (IFN-γ) and 40% (TNF-α). The sub-dominant clonotypes together comprise a larger part of the TCR repertoire than the dominant TRBV12 clonotype, and so despite similar levels of absolute cytokine production, their corresponding RCC values are lower at 78% (IFN-γ) and 31% (TNF-α). The strong cytokine response and high RCC values for the dominant TRBV12 clonotype suggest that these cells recognize consensus peptide more effectively than the sub-dominant clonotypes.
Figure 6. Sub-dominant TRBV populations have high cytokine production potential in response to stimulation with variant peptides.
IFN-γ and TNF-α production was assessed by ICS on dominant and sub-dominant TRBV populations. Dot plots showing dominant (solid box) and sub-dominant (dashed box) clonotypic cytokine production (IFN-γ, upper plots and TNF-α, lower plots) in response to stimulation with consensus and variant peptides (indicated above each column) for the B*27-KK10, A, and B*08-FL8, B, responses in 10022. Within each plot, absolute cytokine production for clonotypic populations as a percentage of total CD8+ T cells is shown to the right of each indicated population as well as relative cytokine capacity (RCC, in parentheses). Graphs representing cytokine production for each response are located below corresponding plots. Bars represent maximal cytokine production for dominant (unfilled) and sub-dominant (grey) clonotypes, absolute cytokine production (% of total CD8+ T cells) is represented by the hatched area within each bar, and RCC for each clonotype and condition is noted to the right of each bar, A and B. Comparison of clonotypic RCC ratios for IFN-γ and TNF-α production in response to stimulations using peptide matching consensus, C (p=0.42 IFN-γ and p=0.38 TNF-α), and variant (p=0.04 IFN-γ and p=0.08 TNF-α), D. Measurements from 10 epitopes in 7 HIV+ individuals.
Stimulation with the KRWIImGLNK variant peptide yielded similar results to those from consensus stimulation. In response to stimulation with the KRWIvLGLNK peptide, the dominant TRBV12 clonotype reached lower levels of absolute cytokine production and had lower RCC ratios for both IFN-γ and TNF-α compared to the sub-dominant clonotypes. The sub-dominant clonotypes preferentially recognized the KRWIvLGLNK peptide, produced their highest levels of absolute cytokine at 8.6% (IFN-γ) and 3.6% (TNF-α) of total CD8+ T cells, and reached their highest RCC ratios of 82% (IFN-γ) and 35% (TNF-α). The B*08-FL8 response in this individual is represented in similar fashion in Figure 6B and yields similar results.
Comparison of RCC values for the clonotypic cytokine responses in a further 8 epitopes from 6 additional individuals (total, 10 epitopes in 7 individuals; Figure 6C–D) reveals that both dominant and sub-dominant TRBV populations are capable of cytokine production to consensus peptides (Figure 6C, p>0.05 for IFN-γ and TNF-α production). In response to stimulation with common variant peptides, sub-dominant TRBV populations have higher RCC ratios for IFN-γ production (Figure 6D, p=0.04) with a trend toward higher sub-dominant RCC ratios for TNF-α production as well (Figure 6D, p=0.08). These results indicate that while dominant and sub-dominant clonotypes are capable of producing cytokines in response to stimulation with consensus and variant peptide epitopes, sub-dominant clonotypes seem to retain greater capacity for cross-recognition and secretion of multiple cytokines in response to the common viral epitope variants we tested.
Dominant TRBV populations display a survival defect in culture
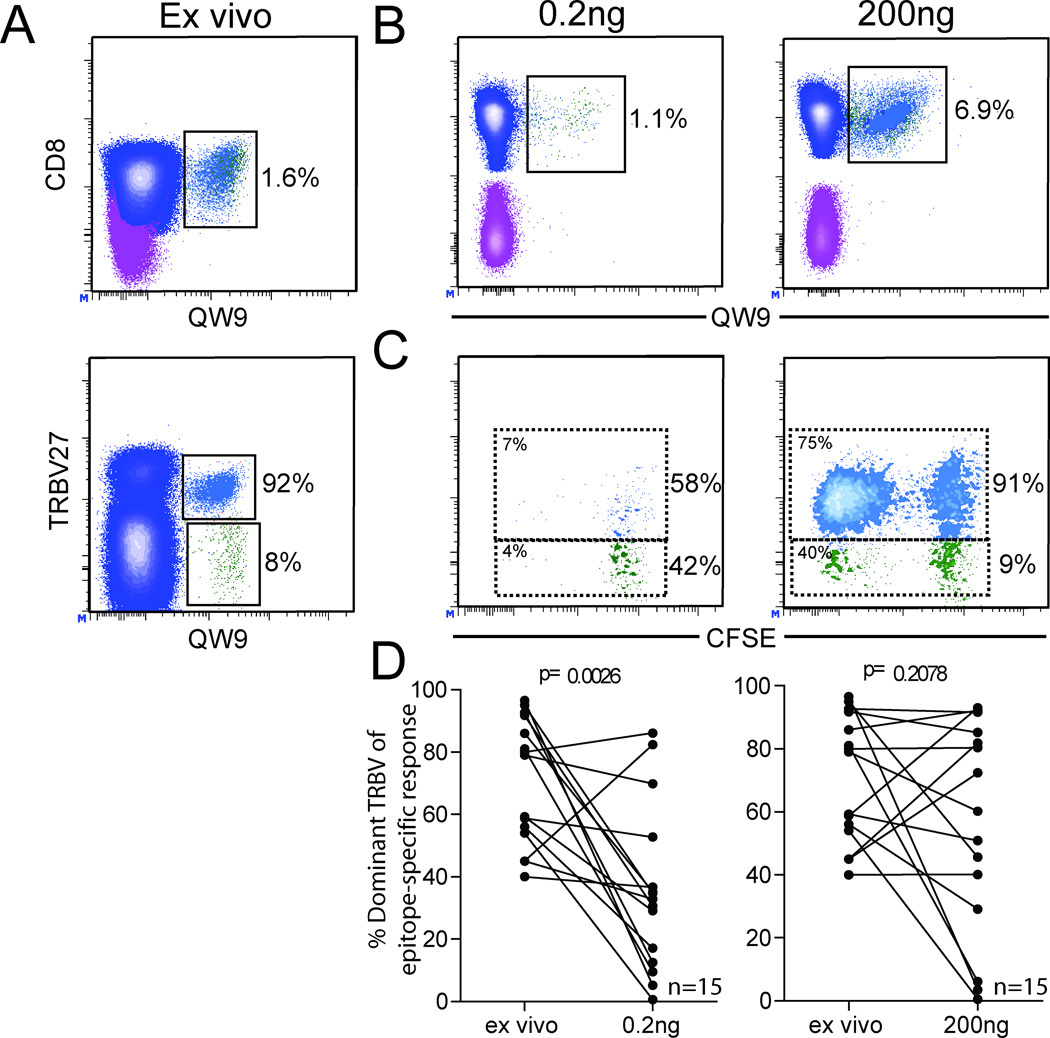
Proliferation upon antigen exposure is an important measure of T cell function and has been associated with improved control of viral replication (21). We labeled T cells with CFSE and cultured them with varying concentrations of peptide for 4 days to assess changes to the epitope-specific TRBV repertoire and capacity for proliferation of dominant and sub-dominant TRBV populations. The ex vivo epitope-specific response and its clonotypic repertoire is shown for the B*57-restricted-QW9 response in 10002 (Figure 7A). Representative plots are shown to illustrate epitope-specific populations (Figure 7B) and CFSE dilution (Figure 7C) for the dominant and sub-dominant clonotypes after 4 days of culture with low (0.2ng/ml) and high (200ng/ml) concentrations of optimal peptide antigen. At the 200 ng/ml peptide concentration, the dominant TRBV27 clonotype made up 91% of the total repertoire at the end of the 4 day stimulation period, reflecting the ex-vivo repertoire. However, at the 200 pg/ml concentration, the TRBV27 clonotype comprised 58% of the repertoire. Therefore while both dominant and sub-dominant TRBV clonotypes proliferate well in response to stimulation with higher concentrations of consensus peptide, the dominant clonotype does not survive as well at lower peptide concentrations and therefore does not maintain the same degree of dominance in vitro. Moreover, as measured by the percentage of CFSElow cells, the dominant TRBV27 clonotype proliferates better than the sub-dominant clonotypes in response to stimulation with consensus peptides, reflecting in vitro what happens naturally in vivo.
Figure 7. Dominant epitope-specific TRBV populations display a survival defect at low peptide concentrations which is alleviated by increasing antigen stimulation.
PBMC were cultured in the presence of peptide antigen at the concentrations indicated above each set of plots and TRBV repertoire composition was assessed by flow cytometry on day 4. Relevant percentages of parent are shown to the right of each population. Representative contour plots showing ex vivo T cell populations: B*57-QW9+ T cells (upper plot) as a percentage of CD8+ T cells; B*57-QW9 epitope-specific population and its constituent dominant (blue) and sub-dominant (green) TRBV populations (lower plot) as a percentage of the epitope-specific population, A. MHC-I tetramer labeling after 4 day culture in the presence of different concentrations of peptide antigen, B. TRBV repertoire composition was determined by antibody labeling for the dominant TRBV as a part of the B*57-QW9+ population. Dominant (blue) and sub-dominant (green) TRBV populations are indicated on each plot and their percentage composition of the B*57-QW9 response is shown at the right of each box. CFSElow percentages are shown for each population in the upper left corner of each box, C. Aggregate data was compiled, and statistical comparisons were made between epitope-specific TRBV repertoire composition ex vivo and after 4 days of proliferation in culture. Dominant populations fail to maintain dominance at low peptide concentrations (p=0.0026), but repertoire composition is not significantly altered at higher concentrations (p=0.2078), D. Wilcoxon signed rank test. Measurements from 15 epitope-specific populations in 7 HIV+ subjects.
Aggregate data from 15 epitopes in 7 subjects indicate that dominant TRBV populations fail to maintain their dominance at low concentrations of peptide (p=0.0026, Figure 7D). Conversely, dominant TRBV populations more effectively maintain their level of dominance at higher concentrations of peptide stimulation (p=0.2078, Figure 7D). In this series of experiments, the addition of antibody to block PD-1/PD-L1 interaction did not significantly alter the relative proliferative capacity of dominant and subdominant TRBV populations over the short duration of this assay (data not shown). These results suggest that while clonotypic constituents may not expand well at low concentrations of stimulation, sub-dominant clonotypic populations are better able to survive culture conditions with low levels of antigen.
Discussion
Several groups have observed enhanced global expression of PD-1 on T cells in HIV+ individuals, with the highest level of PD-1 expression on HIV epitope-specific cells (11, 12, 14, 31). A detailed analysis by Day et. al. found that different epitope-specific responses, even within the same individual, had differing degrees of PD-1 expression (11). This has led to speculation that the degree of PD-1 expression could be linked to the efficacy of viral control for individual epitopes (11, 32). In this study, we evaluated constituent clonotypes within epitope-specific responses and determined that clonal dominance within epitope-specific responses is associated with a PD-1high/CD127low phenotype, that PD-1 expression correlates with clonotypic TCR avidity for tetramer, and that dominant clonotypes display defects in their ability to respond to variant peptide epitopes and survive in the absence of strong antigen signals.
We found that the most dominant clonotype within an epitope-specific response tended to have the highest level of PD-1 expression (p=0.001) and the lowest level of CD127 expression (p=0.007). We did not see a relationship between the overall magnitude of a response (or the degree of clonotypic expansion within a response) and PD-1 expression, suggesting that PD-1 expression may not be directed related to the level of T cell expansion or exhaustion, but could mark T cells which have recently been exposed to their cognate antigen (15, 33, 34). In LCMV infection downregulation of PD-1 and upregulation of CD127 occurs after viral epitope escape (15), suggesting that ongoing antigen exposure is a key factor in pushing T cells toward a PD-1high/CD127low phenotype. Lichterfeld et al described progressive reductions in CD127 expression on high avidity HIV-epitope-specific clonotypes which were eventually deleted (35), and more recent work by Steeck et. al found PD-1 expression on HIV-1 epitope-specific T cells decreased after in-vivo selection for escape mutations (4). While this recent work highlights the relationship between, PD-1, and CD127 expression on epitope specific responses (15, 18), the data we present here is the first to our knowledge which describes differential expression of these markers on individual T cell clonotypes and links dominance to specific differences in clonotypic function.
We have shown that epitope-specific T cell populations are often comprised of a single dominant and various sub-dominant clonotypic populations that can respond variably to changes in viremia (23) and that these clonotypes have differing abilities to recognize epitope variants (27). Our more recent work demonstrates a relationship between TCR use and memory phenotype (28). Thus, our new finding that dominant and sub-dominant T cell clonotypes have phenotypic and functional characteristics linked to antigen sensing is yet another indicator that the fine-specificity of individual T cell clones plays a role in the evolution of epitope-specific immune responses.
The majority of individuals we sequenced had dominant circulating sequences matching HIV Clade-B consensus, with the exception of 10094 (Supplementary Table I and data not shown), and this subject still preferentially recognized the consensus peptide over the circulating variant. Despite their PD-1high/CD127low phenotype, we present evidence that dominant T cell clonotypes able to recognize circulating viral sequences have the capacity to produce multiple cytokines after stimulation with consensus and variant peptide epitopes and that subdominant clonotypes have increased ability to recognize common HIV-1 epitope variants. Improved recognition of viral variants by sub-dominant clonotypes might also be influenced by the diversity of TCR clonotypes within these sub-dominant populations. Each of the epitope-specific responses we assessed is comprised of a single dominant clonotype and at least one and in some cases more than one sub-dominant clonotypes. Effective recognition of variant epitopes may also be a reflection of increased diversity within sub-dominant TRBV populations.
Immune selection pressure mediated by CD8+ T cells can lead to viral mutation and epitope escape from immune recognition (36–38), therefore the frequency of circulating epitope variants and the degree to which individual clonotypes are able to recognize these variants may also play a role in the development and maintenance of the epitope-specific TCR repertoire. A recent study from van Bockel et. al. offers insight into the relationship between clonal evolution within the TCR repertoire in HIV+ individuals and viral epitope variation (39). Their work highlights TCR repertoire remodeling within HLA-B*27-restricted responses to a viral epitope known to consistently undergo immune-mediated mutational escape (25). The authors in this study found that in the presence of epitopes that varied from consensus, dominant T cell clonotypes were maintained over time and expressed higher levels of CD127 compared to subdominant clonotypes. In contrast to van Bockel et. al. we found dominant clonotypes to have lower levels of CD127 compared to subdominant clonotypes. Our study was different in that we evaluated 35 different epitope responses (representing 8 discrete HIV epitopes) in 22 individuals, and in the majority of cases the circulating viral sequence corresponded to the tetramer peptide sequence. These findings are broadly complementary to our own; both data sets indicate that dominant clonotypes are surprisingly persistent in vivo over time and support the notion that broad epitope-specific TCR repertoires may contain clonotypes capable of recognizing and suppressing viral sequence variants.
While we cannot rule out the possibility that some HIV+ individuals in our cohort harbored viral variants not covered by the consensus or variant epitope sequences we selected, the recently reported associations between PD-1 expression and epitope escape (15) highlight the importance of this line of inquiry for future longitudinal in vivo and in vitro studies. The relationship between epitope exposure, recognition, escape, and corresponding epitope-specific T cell phenotype and functional capacity seems to be tightly related, although the effects of persistent exposure to antigen and viral escape on repertoire composition or clonotypic impairment have yet to be determined. In this cross-sectional study, we were unable to assess whether higher avidity clones had been deleted earlier in infection or whether circulating virus had already escaped immune control for all the epitopes studied.
Despite the higher expression of PD-1 on dominant clonotypes, and the relative failure of these dominant clonotypes to survive at low peptide concentrations in vitro, blockade of the PD-1 signaling pathway did not result in significant enhancement of clonotypic proliferation or survival. Studies evaluating the effect of PD-1 blockade on proliferative capacity have typically found modest increases in proliferation (6, 11, 14). The lack of enhanced proliferation we saw may be due to the short duration of our assays, and to the inclusion of relatively healthy subjects with low viral loads. Future studies with combinations of PD-1/PD-L blockade and cytokine combinations may help us determine to what extent dominant clonotypes can be “rescued” in vitro.
Previous reports in mouse influenza models (40) and human EBV/CMV infection (29) indicate that T cell avidity for antigen is positively correlated with dominance in the epitope-specific TRBV repertoire. Our data support the notion that clonotypic TCR avidity is associated with higher expression of PD-1, but suggest that the association between overall TCR avidity and clonal dominance may be weaker in the setting of chronic HIV infection. Prior studies evaluated either acutely resolved or chronic viral infections with limited antigen variability and low levels of ongoing antigen exposure during chronic infection, and those conditions could account for the discrepancies between our study and this previously published work. It remains to be determined if the associations between epitope-specific clonotypic dominance, phenotype, and function which we report in HIV infection also apply to other infections in humans and model systems.
T cell phenotype and function is determined not only by the fundamental interaction between TCR:pMHC but represents a sum of inhibitory and stimulatory signals emanating from surface receptor molecules such as PD-1 and CD127. Recent work from Almeida et al. (26)suggests that a composite measure for T cell function such as ‘antigen sensitivity’ might encompass not only avidity for antigen but a wide range of influential factors such as antigen receptor density, coreceptor-mediated signals, as well as activation status and expression of inhibitory signaling molecules. We suggest that the composition, phenotype, and functional profile of the clonotypic repertoire may be necessarily dynamic in order to respond to a highly variable pathogen such as HIV.
The following model accommodates our observations and experimental results: dominant clonotypes preferentially expand to circulating viral epitopes in vivo. Dominant clonotypes express a surface phenotype consistent with ongoing antigen exposure and activation. Continued exposure to cognate antigen may erode the capacity of dominant clonotypic responses as a result of accumulated PD-1 signal inhibition and a reduction in homeostatic turnover from reduced CD127 expression. Sub-dominant clonotypes expand sub-optimally to circulating viral epitopes in vivo and express a phenotype consistent with reduced exposure to antigen. Sub-dominant populations may recognize non-circulating or low-level variants more effectively than dominant populations and are exposed to relatively lower levels of their preferred cognate antigen resulting in lower overall antigen exposure and concomitant activation. This sparing effect results in the maintenance of a population of cells better able to survive in the absence of strong antigenic signaling. These data also suggest that higher avidity clonotypes develop a relatively PD-1high phenotype compared to lower avidity clonotypes and is consistent with the observation that higher avidity responses are deleted early in infection (35). It remains to be determined whether TCR repertoire composition or clonotypic phenotype in HIV is significantly different in individuals with confirmed viral escape or in the absence of antigen, although data from LCMV infection and HIV infection suggests that this might be the case (4, 15, 39).
A diverse epitope-specific TCR repertoire comprised of clonotypes capable of recognizing and suppressing both circulating and variant epitopes would be a beneficial outcome from either prophylactic vaccine strategies or for strategies seeking to broaden existing immune responses in established HIV infections. Furthermore, manipulation of immunomodulatory surface proteins such as PD-1 or CD127 as a part of vaccination protocols could influence qualitative and quantitative aspects of the epitope-specific immune response including antigen sensitivity or clonotypic repertoire (9). Effective immunological strategies to control chronic infections like HIV may require not only the generation or stimulation of antigen-specific cells but also a coordinated manipulation of inhibitory pathways.
Supplementary Material
Acknowledgements
Flow cytometric cell acquisition or sorting was performed by the Vanderbilt-Meharry Center for AIDS Research (CFAR) Immunopathogenesis Core. TRBV and HIV sequences were generated by the Vanderbilt-Meharry Center for AIDS Research Sequencing Core.
Abbreviations used in this manuscript
- LCMV
Lymphocytic choriomeningitis virus
- PBMC
peripheral blood mononuclear cells
- TRBV
T cell receptor beta variable chain
- MFI
mean fluorescence intensity
- RCC
relative cytokine capacity
Footnotes
This work was supported by the NIH (SAK R01AI39966). Flow cytometric cell acquisition or sorting was performed by the Vanderbilt-Meharry Center for AIDS Research (CFAR) Immunopathogenesis Core, an NIH funded program (P30 AI 54999).
References
- 1.Jin X, Bauer D, Tuttleton S, Lewin S, Gettie A, Blanchard J, Irwin C, Safrit J, Mittler J, Weinberger L. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. Journal of Experimental Medicine. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 4.Streeck H, Brumme Z, Anastario M, Cohen K, Jolin J, Meier A, Brumme C, Rosenberg E, Alter G, Allen T. Antigen load and viral sequence diversification determine the functional profile of HIV-1–specific CD8 T cells. PLoS Med. 2008;5:e100. doi: 10.1371/journal.pmed.0050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douek D, Roederer M, Koup R. Emerging Concepts in the Immunopathogenesis of AIDS*. Annual Review of Medicine. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2005;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 7.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8:239–245. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Jin B, Zhang J, Xu B, Wang H, Shi M, Wherry E, Lau G, Wang F. Dynamic decrease in PD-1 expression correlates with HBV-specific memory CD8 T-cell development in acute self-limited hepatitis B patients. Journal of Hepatology. 2009;50:1163–1173. doi: 10.1016/j.jhep.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 9.Ha S, West E, Araki K, Smith K, Ahmed R. Manipulating both the inhibitory and stimulatory immune system towards the success of therapeutic vaccination against chronic viral infections. Immunological Reviews. 2008;223:317. doi: 10.1111/j.1600-065X.2008.00638.x. [DOI] [PubMed] [Google Scholar]
- 10.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 12.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, Precopio ML, Schacker T, Roederer M, Douek DC, Koup RA. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J.Exp.Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrovas C, Chaon B, Ambrozak D, Price D, Melenhorst J, Hill B, Geldmacher C, Casazza J, Chattopadhyay P, Roederer M. Differential Association of Programmed Death-1 and CD57 with Ex Vivo Survival of CD8+ T Cells in HIV Infection. The Journal of Immunology. 2009;183:1120. doi: 10.4049/jimmunol.0900182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, Routy JP, Haddad EK, Sekaly RP. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat.Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 15.Blattman JN, Wherry EJ, Ha SJ, van der Most RG, Ahmed R. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J.Virol. 2009;83:4386–4394. doi: 10.1128/JVI.02524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radziewicz H, Ibegbu CC, Fernandez ML, Workowski KA, Obideen K, Wehbi M, Hanson HL, Steinberg JP, Masopust D, Wherry EJ, Altman JD, Rouse BT, Freeman GJ, Ahmed R, Grakoui A. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J.Virol. 2007;81:2545–2553. doi: 10.1128/JVI.02021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin H, Blackburn SD, Blattman JN, Wherry EJ. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J.Exp.Med. 2007;204:941–949. doi: 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wherry EJ, Day CL, Draenert R, Miller JD, Kiepiela P, Woodberry T, Brander C, Addo M, Klenerman P, Ahmed R, Walker BD. HIV-specific CD8 T cells express low levels of IL-7Ralpha: implications for HIV-specific T cell memory. Virology. 2006;353:366–373. doi: 10.1016/j.virol.2006.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJ, Rosenberg ES, Altfeld M, Walker BD. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nature medicine. 2006;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 21.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 22.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Olson D, Brady KW, Bartman MT, O'Sullivan KM, Simons BC, Conrad JA, Duncan CB, Lorey S, Siddique A, Draenert R, Addo M, Altfeld M, Rosenberg E, Allen TM, Walker BD, Kalams SA. Fluctuations of functionally distinct CD8+ T-cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood. 2006;107:2373–2383. doi: 10.1182/blood-2005-04-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27–restricted cytotoxic T lymphocyte responses. The Journal of Experimental Medicine. 2001;193:375. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almeida J, Sauce D, Price D, Papagno L, Shin S, Moris A, Larsen M, Pancino G, Douek D, Autran B. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons BC, Vancompernolle SE, Smith RM, Wei J, Barnett L, Lorey SL, Meyer-Olson D, Kalams SA. Despite biased TRBV gene usage against a dominant HLA B57-restricted epitope, TCR diversity can provide recognition of circulating epitope variants. J Immunol. 2008;181:5137–5146. doi: 10.4049/jimmunol.181.7.5137. [DOI] [PubMed] [Google Scholar]
- 28.Meyer-Olson D, Simons B, Conrad J, Smith R, Barnett L, Lorey S, Duncan C, Ramalingam R, Kalams S. Clonal expansion and TCR-independent differentiation shape the HIV-specific CD8+ effector-memory T-cell repertoire in vivo. Blood. 2010;116:396. doi: 10.1182/blood-2009-11-254136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JY, Zhang Z, Wang X, Fu JL, Yao J, Jiao Y, Chen L, Zhang H, Wei J, Jin L, Shi M, Gao GF, Wu H, Wang FS. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood. 2007;109:4671–4678. doi: 10.1182/blood-2006-09-044826. [DOI] [PubMed] [Google Scholar]
- 32.Hansen T, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nature Reviews Immunology. 2009;9:503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 33.Bengsch B, Seigel B, Ruhl M, Timm J, Kuntz M, Blum HE, Pircher H, Thimme R. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2010;6:e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salisch NC, Kaufmann DE, Awad AS, Reeves RK, Tighe DP, Li Y, Piatak M, Lifson JD, Evans DT, Pereyra F. Inhibitory TCR coreceptor PD-1 is a sensitive indicator of low-level replication of SIV and HIV-1. The Journal of Immunology. 2010;184:476. doi: 10.4049/jimmunol.0902781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lichterfeld M, Yu XG, Mui SK, Williams KL, Trocha A, Brockman MA, Allgaier RL, Waring MT, Koibuchi T, Johnston MN, Cohen D, Allen TM, Rosenberg ES, Walker BD, Altfeld M. Selective depletion of high-avidity human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells after early HIV-1 infection. J Virol. 2007;81:4199–4214. doi: 10.1128/JVI.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage PA, Davis MM. A kinetic window constricts the T cell receptor repertoire in the thymus. Immunity. 2001;14:243–252. doi: 10.1016/s1074-7613(01)00106-6. [DOI] [PubMed] [Google Scholar]
- 37.Paterson A, Sharpe A. Taming tissue-specific T cells: CTLA-4 reins in self-reactive T cells. Nature immunology. 2010;11:109–111. doi: 10.1038/ni0210-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munz C, Lunemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol. 2009;9:246–258. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Bockel DJ, Price DA, Munier ML, Venturi V, Asher TE, Ladell K, Greenaway HY, Zaunders J, Douek DC, Cooper DA, Davenport MP, Kelleher AD. Persistent survival of prevalent clonotypes within an immunodominant HIV gag-specific CD8+ T cell response. J Immunol. 2011;186:359–371. doi: 10.4049/jimmunol.1001807. [DOI] [PubMed] [Google Scholar]
- 40.Kedzierska K, La Gruta NL, Davenport MP, Turner SJ, Doherty PC. Contribution of T cell receptor affinity to overall avidity for virus-specific CD8+ T cell responses. Proc Natl Acad Sci U S A. 2005;102:11432–11437. doi: 10.1073/pnas.0504851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.