Abstract
Molecular tools to distinguish strains of Plasmodium vivax are important for studying the epidemiology of malaria transmission. Two sets of markers—tandem repeat (TR) polymorphisms and MSP3α—were used to study Plasmodium vivax in patients in the Peruvian Amazon region of Iquitos. Of 110 patients, 90 distinct haplotypes were distinguished using 9 TR markers. An MSP3α polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) using HhaI and AluI revealed 8 and 9 profiles, respectively, and 36 profiles when analyzed in combination. Combining TR and PCR-RFLP markers, 101 distinct molecular profiles were distinguished among these 110 patients. Nine TR markers arrayed along a 100 kB stretch of a P. vivax chromosome containing the gene for circumsporozoite protein showed non-linear linkage disequilibrium (ISA = 0.03, P = 0.001). These findings demonstrate the potential use of TR markers for molecular epidemiology studies.
Introduction
Malaria caused by Plasmodium vivax generally is not lethal but exacts a substantial human toll throughout Latin America, Asia, Oceania, and parts of Africa and the Middle East. More than two billion people are at risk for P. vivax in malaria-endemic regions, among whom an estimated 70–80 million cases occur annually.1,2 Acute febrile illness caused by primary P. vivax infection or relapse, and chronic illness such as malarial anemia result in substantial economic loss in terms of job loss, economic output, and disability-adjusted life years. Non-lethal malaria impairs early childhood growth and development,3 as well as quality of life, overall health, and robustness in activities of daily living in ways that are difficult to measure. Pregnancies complicated by vivax malaria are associated with low birth weight and increased neonatal morbidity and mortality.4 Several factors combine to increase the priority for translating fundamental, bench-based research on P. vivax malaria into regional and global control efforts for control of this infection: relative neglect of the burden of disease caused by P. vivax infection; its globally widespread distribution; and increased cognizance of the possibility of severe vivax infections.5–10
Infection by P. vivax is often assumed to result inevitably in symptomatic febrile illness,11 being classically associated with 48-hour fever cycles. Yet, in malaria-endemic regions, asymptomatic malaria parasitemia, with degrees of prevalence that are geographically variable, is common.11–18 This observation has raised two important hypotheses: 1) that asymptomatic parasitemia may contribute significantly to maintaining malaria transmission in endemic regions; and 2) different Plasmodium falciparum or P. vivax clones may differ in virulence, some more likely than others to result in symptoms. Substantial data support the former hypothesis; few data to date support the latter. To address questions of mechanisms of immunity, immunity against homologous versus heterologous Plasmodium strains, and delineation of human reservoirs of malaria transmission, it is necessary to be able to precisely identify infecting parasite strains. Recent advances in analyzing the genetic diversity of P. vivax have laid the groundwork for determining relationships of virulence to parasite genotype, mechanisms of strain-specific immunity, and the dynamics of parasite transmission within local communities. Prerequisite to such studies is the development of tools that efficiently discriminate between closely related parasite clones. A number of genetic markers have been assessed for their ability to discriminate between infecting strains of P. vivax.19 Such markers include genes encoding vaccine candidate proteins subject to immune selection such as circumsporozoite protein (PvCSP), merozoite surface protein-1 (PvMSP-1),20 apical membrane protein-1 (PvAMA-1),20 and merozoite surface protein-3α (PvMSP-3α),21 and putatively neutral markers such as recently described tandem repeat (TR) polymorphism markers22 and microsatellites.23
Several genes encoding proteins subject to immune selection—PvMSP-1, PvMSP-3α, and PvCSP—have been suggested as markers that efficiently discriminate between infecting clones of P. vivax in Papua New Guinea, Thailand, and India using polymerase chain reaction (PCR)-restriction fragment polymorphism analysis.21,24,25 Previous work using a PCR-restriction fragment length polymorphism (PCR-RFLP) assay for assessing P. vivax diversity in the field setting includes the use of PvMSP-3α as a marker to distinguish 24 alleles in 74 samples from Papua New Guinea.26 More recently the use of a similar PCR-RFLP approach using PvCSP and PvMSP-1 distinguished 36 and 23 alleles, respectively, among 100 P. vivax infections in Bangkok, Thailand, with about 26 patients having at least two infecting clones of P. vivax discernable.24 Of the remaining 74 individuals infected by a single genotype of P. vivax, 68 genotypes could be distinguished by combining both genetic markers. Thus, the use of PCR-RFLP on P. vivax genes encoding proteins subject to immune selection in the human host27 has been shown to discriminate among infecting malaria parasite strains, at least in Papua New Guinea and Thailand, where the intensity of malaria transmission is relatively high and complex infections that facilitate genetic out-crossing are relatively common.
Malaria in the Iquitos region of the Peruvian Amazon is notable for being hypoendemic.13,28 Plasmodium vivax has been noted to be heterogeneous, highly structured according to microsatellite analysis, and to have a low effective recombination rate.29 Transmission intensity is typically low (entomological inoculation rate < 1 infective bite per year, based on data from the late 1990s30), few Anopheles spp. vector mosquitoes are infected with malaria parasites30,31 and mixed infections with P. falciparum and P. vivax in the Amazon region are uncommon.13 In this study, we hypothesized that immune selection for the genes encoding PvMSP-1, PvMSP-3α, and PvCSP might be less intense in this region than in Oceania and Asia associated with less genetic diversity at these loci as detected by established PCR-RFLP assays. To test this hypothesis, we compared the discriminating ability of a PvMSP-3α PCR-RFLP assay with a set of TR polymorphic genetic markers in acute P. vivax malaria patients in the Peruvian Amazon of Iquitos.22 The analysis of these markers involves a single step PCR reaction analyzed by automated, computerized measurement of band size by agarose gel electrophoresis followed by straightforward computational analysis. The simplicity and superior resolution of this assay has important implications for field studies of drug and vaccine efficacy and for population-based studies of P. vivax transmission.
Materials and Methods
Study sites, patient enrollment, and sample collection and preparation.
Patients with acute vivax malaria were identified by conventional light microscopy among patients presenting to six health centers in the Peruvian Amazon region of Iquitos. Two health centers were within urban zones of Iquitos (Cardoso and the Fever Clinic in the Hospital de Apoyo), one was from a health post located on the Nanay River (Bellavista Nanay), a referral center from up-river rural health posts, and three were in rural areas along the highway from Iquitos to Nauta (Varillal, Moralillo, Villa Buen Pastor).
At the enrollment site, venous blood was collected in EDTA Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ). Samples were aliquoted, frozen at –20°C and shipped on dry ice to the study laboratory in Lima for molecular analysis. The DNA was extracted from 200 μL of thawed anticoagulated whole blood using the Qiagen Blood Kit (Qiagen, Valencia, CA). The diagnosis of non-mixed P. vivax infections was confirmed in all patients using a species-specific nested PCR assay32; no samples contained P. falciparum, Plasmodium malariae, or Plasmodium ovale.
This project was approved by the Ethical Committees of Universidad Peruana Cayetano Heredia, and Asociación Benéfica PRISMA, both in Lima, Peru; by the Directorate of Health, Iquitos, Peru; and by the Institutional Review Boards of the University of California, San Diego and the Johns Hopkins Bloomberg School of Public Health.
Molecular genotyping assays.
Genotyping of P. vivax isolates was performed using two methods: 1) a PCR-based assay based on a set of previously published TR polymorphism markers22; and 2) an assay using PCR-RFLP analysis of the PvMSP-3α locus.21
Tandem repeat polymorphisms in all clinical samples were determined by agarose gel electrophoresis analysis of a single-step PCR reaction using 9 of 33 previously published PCR oligonucleotide primer pairs, according to the published cycling protocol.21 The 9 TR markers were chosen based on a preliminary study of seven patients from different areas of Iquitos, in whom 24 of the TR markers were not found to discriminate between P. vivax strains, but 9 TR markers proved to have at least two alleles each (data not shown).
Briefly, 4 μL of DNA extracted from 200 μL of whole blood was added to 45 μL of PCR mix containing 5 μL of 10×PCR buffer, 1.0 μL of dNTPs (10 mM), and 0.1 μL (5 U/μL) of Taq polymerase (Invitrogen, Carlsbad, CA). A single cycling protocol was used: 947°C for 2 min, 35 cycles of 94°C for 20 sec, 55°C for 10 sec, and 65°C for 2 min, and a final extension time of 5 min at 65°C using a MJR PTC-100 thermal cycler (Bio-Rad, Hercules, CA). Agarose gel electrophoresis with 1% Tris-Acetate-EDTA (TAE) gels was used to analyze PCR products. The TR markers used in this study were as follows: MN3, MN4, MN12, MN17, MN23, MN24, MN25, MN26, and MN29.22 No TR primer pair amplified DNA from either human or P. falciparum (data not shown and ref.22).
To assess allelic types of the MSP-3α gene, the published method was used as described using AluI and HhaI restriction enzymes.21 Briefly, a polymorphic fragment of the PvMSP-3α gene was amplified by nested PCR and 4 μL of the PCR product was analyzed by 1% Tris-Acetate-EDTA (TAE) agarose gel electrophoresis. Approximately 5 μL of the PCR product was digested with each enzyme in separate reactions and analyzed by TAE agarose gel electrophoresis.
Digitized tiff images of ethidium bromide-stained gels were obtained using the KODAK EDAS 120 gel documentation system (Kodak, Rochester, NY). Band sizes of PCR and PCR-RFLP products were calculated using PRO-SCORE Molecular Weight Software Windows PC version 2.39 (DNA Proscan, Nashville, TN), using a 100 bp ladder for calibration (Invitrogen, Carlsbad, CA). Alleles were distinguished based on differences in restriction band patterns and band size differences within 5% were grouped together to be conservative in estimating allelic diversity.
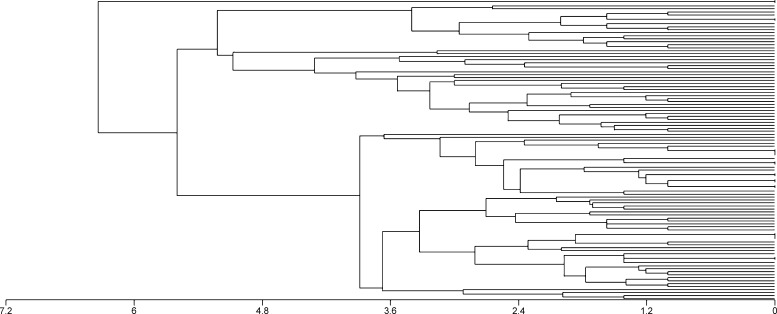
The set of TR alleles from individual patients was determined by creating a dendrogram based on a combination of the 9 markers that allowed us to distinguish individual haplotypes and quantify their relatedness. Alleles (as determined by electrophoretic mobility) were first coded then clustering analysis was performed using the unweighted pair group method with arithmetic means (UPGMA) and the Euclidean distance algorithm, using the Multivariate Statistics Package (MVSP) version 3.13 m (Kovach Computing Services, Anglesey, Wales, UK). The genotypes of isolates from patients with complex infections (> 1 infecting strain) as detected by the TR markers were omitted from the dendrogram.
Sequences of the TR markers were determined by automated cycle sequencing on an ABI PRISM 3300 sequencer (Applied Biosystems, Carlsbad, CA) using Big Dye chemistry. The PCR products determined to be homogeneous by agarose gel electrophoresis were treated with 1 μL of shrimp alkaline phosphatase (ExoSAP-IT, U.S. Biochemical, Cleveland, OH) at 37°C for 15 min and at 80°C for another 15 min. Each sequencing reaction used 2–5 μL of the PCR product. Sequences were determined in both directions using the same oligonucleotide primers used to generate the PCR product. Sequences were assembled and aligned using the DNASTAR analysis suite for Macintosh OS X (version 5.51; Madison, WI).
Statistical analysis.
Multilocus linkage equilibrium was first assessed by calculating a standardized index of association (ISA) was calculated using LIAN v3.5,33 accessed at http://adenine.biz.fh-weihenstephan.de/cgi_bin/lian/lian.cgi.pl). Genetic linkage of the TR markers to assess independence of the allele distribution corresponding to each pair of contiguous markers (MN3-MN4, MN4-MN12, MN12-MN17, etc.) followed by pairwise analysis of MN3 to each of the other 8 loci done was using a Pearson χ2 using Stata v.9 (College Station, TX). Mean genetic diversity and diversity at each locus was calculated using LIAN v3.5.
Results
Molecular confirmation of P. vivax infections.
All acute P. vivax infections of 110 patients were confirmed at the species level by a nested PCR assay. There were no detectable mixed infections with P. falciparum.
Polymorphism analysis of P. vivax using MSP3-α PCR-RFLP assay.
Blood samples from the 110 patients with confirmed P. vivax infections were subjected to PCR analysis for detection of MSP3-α ( Supplementary Figures S1 and S2). Seven allelic forms produced by HhaI digestion of the MSP3-α PCR fragment were discernible (Supplementary Figure S1; Table 1). Allele 6 predominated and defined 28% of all isolates from this population. Eight allelic forms of the MSP3-α PCR fragments were found following digestion with Alul (Table 1). Thirty-nine percent of patient isolates were of allelic haplotype 1. The restriction patterns from nine samples suggested mixed infections because the sum of the restriction fragments exceeded the size of the primary product and so these samples were not analyzed further.
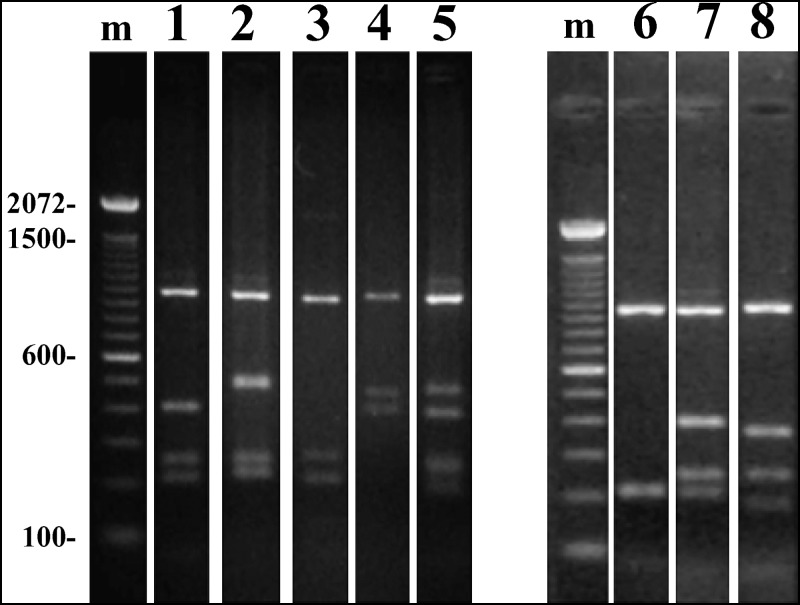
Supplementary Figure S1.
Polymerase chain reaction/restriction fragment polymorphism analysis of Plasmodium vivax MSP3-α using restriction enzyme HhaI ( AluI restriction not shown). Primary amplification products are shown above, digestion products below.
Supplementary Figure S2.
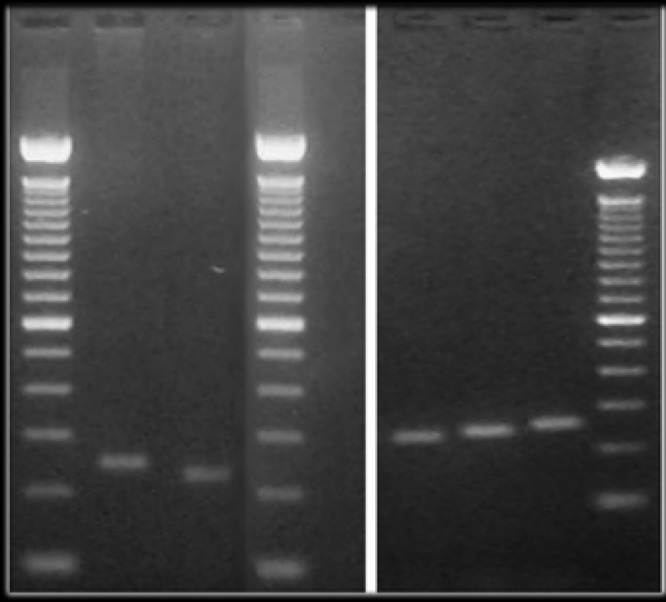
An example (marker MN12, with 5 allelic types) of the products of the tandem repeat polymorphism PCR assay. Band sizes for this marker (shown for each lane on top of the gel), as well as the other MN markers, were assigned by automated computational analysis (see Materials and Methods) using the 100 bp ladders on each side of the gel as standard (200 and 300 bp markers are indicated on the sides of the gel image).
Table 1.
MSP3α polymerase chain reaction (PCR)-restriction fragment length polymorphism alleles
| HhaI Digest Alleles (in bp) | ||||||
|---|---|---|---|---|---|---|
| Allele 1 | 1070 | 431 | 275 | 207 | ||
| Allele 2 | 1070 | 530 | 275 | 236 | ||
| Allele 3 | 1070 | 275 | 223 | |||
| Allele 4 | 1070 | 500 | 431 | |||
| Allele 5 | 1070 | 223 | ||||
| Allele 6 | 1070 | 431 | 275 | 223 | ||
| Allele 7 | 1070 | 385 | 275 | 188 | ||
| AluI Digest Alleles (in bp) | ||||||
| Allele 1 | 551 | 467 | 259 | 185 | 153 | |
| Allele 2 | 551 | 205 | 153 | |||
| Allele 3 | 551 | 447 | 205 | 175 | 153 | |
| Allele 4 | 551 | 398 | 205 | 175 | 153 | |
| Allele 5 | 551 | 259 | 185 | 175 | 153 | |
| Allele 6 | 551 | 447 | 354 | 394 | 165 | 153 |
| Allele 7 | 551 | 354 | 259 | 205 | 173 | 153 |
| Allele 8 | 523 | 253 | 198 | 165 | 144 | 130 |
Genetic diversity of P. vivax using TR Polymorphism and MSP-3α analysis.
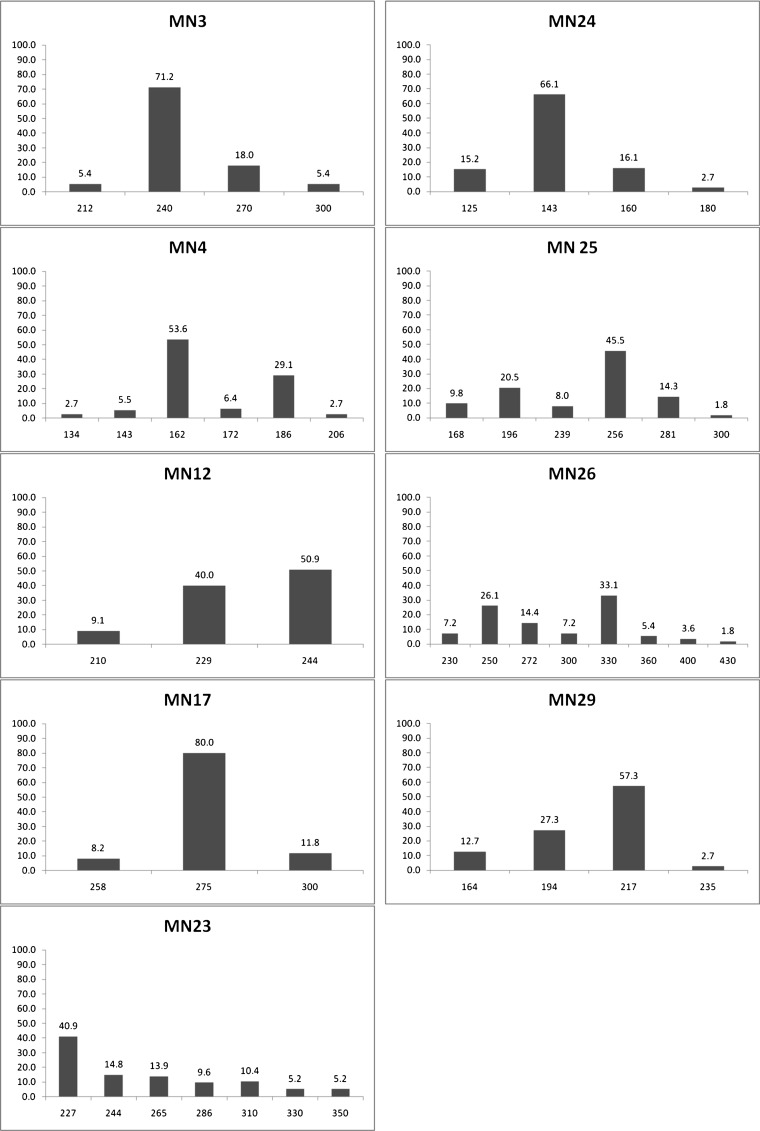
Nine previously published TR primer pairs were used in a PCR assay to generate haplotypes for each P. vivax specimen. The mean genetic diversity for the 9 markers was 0.60 ± 0.05. The genetic diversity of each locus ranged from 0.34 (MN17) to 0.79 (MN23). The allelic frequencies at each locus and the frequencies of MSP3-α types are shown in Figure 1. Combining TR and MSP-3α, 101 of 110 P. vivax samples (92%) could be distinguished (Figure 2 ). Of the remaining 9 patient samples, 7 had two bands detected by a single primer pair, indicating the presence of at least two P. vivax strains in the specimen, and 2 patients had two bands in more than one TR marker. Nine TR haplotypes were common to 20 patient samples; two haplotypes were shared by 3 other patients.
Figure 1.
Frequency of alleles of 9 tandem repeat polymorphism markers in 110 patients with Plasmodium vivax malaria.
Figure 2.
Complexity of Plasmodium vivax populations as demonstrated by the unweighted pair group method with arithmetic mean (UPGMA) dendrograms with Euclidean distance (horizontal axis).
Direct PCR sequencing was successful on 26 of 47 TR alleles (data not shown). All band size polymorphisms were caused by insertion/deletions, with the concomitant presence of single nucleotide polymorphisms and partial TRs. Sequencing also confirmed the automated computational identification of the band sizes even in cases where differences in band size were small (< 10 bp). The DNA sequencing confirmed the specificity of the TR assay, the allele assignments, and ruled out the possibility that artifacts from the PCR assay led to spurious results.
Genetic linkage of TR markers.
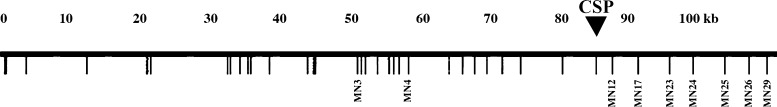
The standard index of association (ISA) for the 9 TR markers for the 101 monoclonal infections was 0.03 (P = 0.001). Because these markers are on a relatively small (100 kB) chromosomal segment we then sought to delineate if the linkage disequilibrium was related to the physical distance along the chromosomal segment. We used a χ2 test to assess the distribution of each pair of contiguous markers (MN3-MN4, MN4-MN12, MN12-MN17, etc. (Figure 3 ; Table 2). In all cases except for the pair of markers MN17-MN23 and MN23-MN24, there was evidence of genetic linkage.
Figure 3.
Map of Plasmodium vivax chromosomal segment containing the 9 tandem repeat polymorphism markers used in the analysis (open reading frames are schematically depicted by locus number in Figure 1 of Reference 22). The approximate location of the gene encoding P. vivax circumsporozoite protein is indicated by CSP.
Table 2.
Pairwise analysis of all 9 tandem repeat polymorphism markers by Chi-square analysis to assess genetic linkage at a distance
| Marker | MN4 | MN12 | MN17 | MN23 | N24 | MN25 | MN26 | MN29 |
|---|---|---|---|---|---|---|---|---|
| MN3 | < 0.0001 | 0.505 | 0.792 | 0.135 | 0.071 | < 0.0001 | < 0.0001 | 0.037 |
| MN4 | 0.007 | 0.406 | 0.001 | < 0.0001 | < 0.0001 | < 0.0001 | 0.031 | |
| MN12 | < 0.0001 | 0.379 | 0.432 | 0.286 | < 0.0001 | < 0.0001 | ||
| MN17 | 0.131 | 0.685 | 0.578 | 0.002 | 0.769 | |||
| MN23 | 0.466 | < 0.0001 | < 0.0001 | 0.032 | ||||
| MN24 | < 0.0001 | < 0.0001 | 0.088 | |||||
| MN25 | < 0.0001 | < 0.0001 | ||||||
| MN26 | < 0.0001 |
It would be expected that distant markers would show less evidence of genetic linkage than contiguous markers. However, we found significant evidence of genetic linkage among distant, non-contiguous markers despite a lack of linkage at intermediate loci (Table 1). In particular, analysis of associations with Marker MN26 demonstrated evidence of distant-marker linkage.
Discussion
The data presented here provide evidence for a remarkably high level of P. vivax genetic diversity in a region of low malaria transmission in the Peruvian Amazon, consistent with previous findings from around the world. We used a simple TR polymorphism assay based on markers arrayed along a previously sequenced ∼100 kB stretch of P. vivax chromosomal DNA to analyze genetic diversity in an unselected group of P. vivax patient samples from Iquitos and surrounding villages. We found that 94 of 101 monoclonal P. vivax infections were distinguishable using TR markers. The MSP-3α PCR-RFLP used alone demonstrated less diversity when compared with TR results.
The TR markers exhibited substantial linkage disequilibrium observed among adjacent markers but the linkage could not be explained by physical linkage alone. Generally, linkage disequilibrium is associated with low levels of diversity and low levels of disease transmission.13 However, in this population linkage disequilibrium coexisted with high levels of diversity; this discordance suggests these markers are both hypervariable and coadaptive. This ∼100 kB locus encodes only one protein (Circumsporozoite Protein) known to be selected for by immune evasion by the parasite.
Although it seems clear that these markers are in a hyperdynamic part of the genome, there are several findings in our study that stand in contrast to descriptions of low genetic diversity reported by others.14 Using only PCR-RFLP using AluI we were able to detect eight P. vivax haplotypes in a limited geographic area and time frame. And, despite hypoendemicity at a population level, our finding that 8.2% of P. vivax infections contained more than one genotype suggests that segments of the population are at a greatly elevated risk compared with the population as a whole, reflecting the heterogeneity of malaria transmission in this region, as is typical elsewhere. This epidemiologic group is likely to be an important factor in the generation of genetic diversity in P. vivax in this geographic area because cross-fertilization and recombination can occur only when more than one clone is present in the mosquito vector—an event greatly facilitated when the human host has a polyclonal infection.
The degree of diversity in P. vivax in the Iquitos region of the Peruvian Amazon is sufficiently high to allow for the differentiation of strains present in human infections using a variety of methods including microsatellites29 and the combination of TRs and MSP3α PCR-RFLP. Furthermore, recent data indicate that the P. vivax population is highly structured in the Iquitos region of the Peruvian Amazon,29 similar to the limited number of haptotypes reported for P. falciparum in the Peruvian Amazon.34 Nonetheless, the large-scale genomic structure underlying the population structure has yet to be determined. Whole genome analysis beyond the currently published data35,36 will be required to further determine the genomic features underlying the population biology and evolution of P. vivax in Peru, which is of particular interest in light of the low malaria transmission intensity in this region.
In this study, we have demonstrated that use of the TR markers in the hypoendemic malaria region of the Amazon effectively and efficiently distinguishes infecting strains of P. vivax. This typing strategy is easy to perform, largely automated, and unambiguous. Allele assignment allows for the rapid classification and easy integration into existing software for genetic analysis. This typing system can be a useful tool in determining the dynamics of parasite transmission within local areas, mechanisms of strain-specific immunity, relationships between parasite genetics and virulence, and potentially distinguishing reintroduction versus continued circulation of P. vivax strains during malaria elimination campaigns.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carlos Vidal Oré and members of the Peruvian Ministry of Health for their assistance and thoughtful input. X. Su of the National Institute of Allergy and Infectious Diseases graciously provided advice early in the design of this study. S. T. Unt provided critical revisions of earlier drafts of the manuscript. We are grateful to Paula Maguina for her expert scientific, logistic, and administrative contributions to the success of this project.
Footnotes
Financial Support: This work was supported by grants from the United States Public Health Service, National Institutes of Health K24AI06803, D43 TW007120, R01067727, 1U19AI089681 (JMV), and K01TW005717 (MK), an Innovations in Clinical Research Award from the Doris Duke Charitable Foundation (JMV), and a pilot project grant from the Johns Hopkins Malaria Institute (RHG).
Authors' addresses: Margaret Kosek, Pablo P. Yori, and Robert H. Gilman, Johns Hopkins Bloomberg School of Public Health, Department of International Health, Baltimore, MD, E-mails: mkosek@jhsph.edu, pyori@jhsph.edu, and gilmanbob@gmail.com. Maritza Calderon, AB Prisma, Lima, Peru, E-mail: mmcalderons@yahoo.es. Mirko Zimic, Raul Chuquiyauri, Cesar Jeri, and Viviana Pinedo-Cancino, Alejandro Llanos-Cuentas, Universidad Cayetano Heredia, Lima, Peru, E-mails: mirko.zimic@upch.edu, raulharo@ucsd.edu, cesajeri@yahoo.es, vivi_cancino@yahoo.com, and elmer.llanos@upch.edu. Michael A. Matthias and Joseph M. Vinetz, University of California San Diego, Department of Medicine, La Jolla, CA, E-mails: mmatthias@ucsd.edu and jvinetz@ucsd.edu.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IR, Baird JK, Snow RW, Hay SI. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis. 2010;4:e774. doi: 10.1371/journal.pntd.0000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee G, Yori P, Olortegui MP, Pan W, Caulfield L, Gilman RH, Sanders JW, Delgado HS, Kosek M. Comparative effects of vivax malaria, fever and diarrhoea on child growth. Int J Epidemiol. 2012 doi: 10.1093/ije/dyr190. doi:10.1093/dyr190. First published online: January 17, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nosten F, McGready R, Simpson JA, Thwai KL, Balkan S, Cho T, Hkirijaroen L, Looareesuwan S, White NJ. Effects of Plasmodium vivax malaria in pregnancy. Lancet. 1999;354:546–549. doi: 10.1016/s0140-6736(98)09247-2. [DOI] [PubMed] [Google Scholar]
- 5.Baird JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007;23:533–539. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Alexandre MA, Ferreira CO, Siqueira AM, Magalhaes BL, Mourao MP, Lacerda MV, Alecrim MG. Severe Plasmodium vivax malaria, Brazilian Amazon. Emerg Infect Dis. 2010;16:1611–1614. doi: 10.3201/eid1610.100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 8.Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, Kochar A, Khatri MP, Gupta V. Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg. 2009;80:194–198. [PubMed] [Google Scholar]
- 9.Baird JK. Severe and fatal vivax malaria challenges ‘benign tertian malaria’ dogma. Ann Trop Paediatr. 2009;29:251–252. doi: 10.1179/027249309X12547917868808. [DOI] [PubMed] [Google Scholar]
- 10.Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–566. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- 11.Vinetz JM, Gilman RH. Asymptomatic Plasmodium parasitemia and the ecology of malaria transmission. Am J Trop Med Hyg. 2002;66:639–640. doi: 10.4269/ajtmh.2002.66.639. [DOI] [PubMed] [Google Scholar]
- 12.Alves FP, Durlacher RR, Menezes MJ, Krieger H, Silva LH, Camargo EP. High prevalence of asymptomatic Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg. 2002;66:641–648. doi: 10.4269/ajtmh.2002.66.641. [DOI] [PubMed] [Google Scholar]
- 13.Roshanravan B, Kari E, Gilman RH, Cabrera L, Lee E, Metcalfe J, Calderon M, Lescano AG, Montenegro-James S, Calampa C, Vinetz JM. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg. 2003;69:45–52. [PubMed] [Google Scholar]
- 14.Harris I, Sharrock WW, Bain LM, Gray KA, Bobogare A, Boaz L, Lilley K, Krause D, Vallely A, Johnson ML, Gatton ML, Shanks GD, Cheng Q. A large proportion of asymptomatic Plasmodium infections with low and sub-microscopic parasite densities in the low transmission setting of Temotu Province, Solomon Islands: challenges for malaria diagnostics in an elimination setting. Malar J. 2010;9:254. doi: 10.1186/1475-2875-9-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steenkeste N, Rogers WO, Okell L, Jeanne I, Incardona S, Duval L, Chy S, Hewitt S, Chou M, Socheat D, Babin FX, Ariey F, Rogier C. Sub-microscopic malaria cases and mixed malaria infection in a remote area of high malaria endemicity in Rattanakiri province, Cambodia: implication for malaria elimination. Malar J. 2010;9:108. doi: 10.1186/1475-2875-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladeia-Andrade S, Ferreira MU, de Carvalho ME, Curado I, Coura JR. Age-dependent acquisition of protective immunity to malaria in riverine populations of the Amazon Basin of Brazil. Am J Trop Med Hyg. 2009;80:452–459. [PubMed] [Google Scholar]
- 17.da Silva NS, da Silva-Nunes M, Malafronte RS, Menezes MJ, D'Arcadia RR, Komatsu NT, Scopel KK, Braga EM, Cavasini CE, Cordeiro JA, Ferreira MU. Epidemiology and control of frontier malaria in Brazil: lessons from community-based studies in rural Amazonia. Trans R Soc Trop Med Hyg. 2010;104:343–350. doi: 10.1016/j.trstmh.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 18.da Silva-Nunes M, Ferreira MU. Clinical spectrum of uncomplicated malaria in semi-immune Amazonians: beyond the “symptomatic” vs “asymptomatic” dichotomy. Mem Inst Oswaldo Cruz. 2007;102:341–347. doi: 10.1590/s0074-02762007005000051. [DOI] [PubMed] [Google Scholar]
- 19.Cui L, Mascorro CN, Fan Q, Rzomp KA, Khuntirat B, Zhou G, Chen H, Yan G, Sattabongkot J. Genetic diversity and multiple infections of Plasmodium vivax malaria in Western Thailand. Am J Trop Med Hyg. 2003;68:613–619. doi: 10.4269/ajtmh.2003.68.613. [DOI] [PubMed] [Google Scholar]
- 20.Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A. Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA 1 and MSP 1 genes. Mol Biochem Parasitol. 2000;108:53–66. doi: 10.1016/s0166-6851(00)00204-8. [DOI] [PubMed] [Google Scholar]
- 21.Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3alpha locus of Plasmodium vivax: global and local diversity. Am J Trop Med Hyg. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Carlton JM, Joy DA, Mu J, Furuya T, Suh BB, Wang Y, Barnwell JW, Su XZ. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc Natl Acad Sci USA. 2003;100:8502–8507. doi: 10.1073/pnas.1232502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imwong M, Sudimack D, Pukrittayakamee S, Osorio L, Carlton JM, Day NP, White NJ, Anderson TJ. Microsatellite variation, repeat array length, and population history of Plasmodium vivax. Mol Biol Evol. 2006;23:1016–1018. doi: 10.1093/molbev/msj116. [DOI] [PubMed] [Google Scholar]
- 24.Imwong M, Pukrittayakamee S, Gruner AC, Renia L, Letourneur F, Looareesuwan S, White NJ, Snounou G. Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar J. 2005;4:20. doi: 10.1186/1475-2875-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prajapati SK, Joshi H, Valecha N. Plasmodium vivax merozoite surface protein-3 alpha: a high-resolution marker for genetic diversity studies. J Vector Borne Dis. 2010;47:85–90. [PubMed] [Google Scholar]
- 26.Bruce MC, Galinski MR, Barnwell JW, Donnelly CA, Walmsley M, Alpers MP, Walliker D, Day KP. Genetic diversity and dynamics of Plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000;121:257–272. doi: 10.1017/s0031182099006356. [DOI] [PubMed] [Google Scholar]
- 27.Kumkhaek C, Phra-Ek K, Renia L, Singhasivanon P, Looareesuwan S, Hirunpetcharat C, White NJ, Brockman A, Gruner AC, Lebrun N, Alloueche A, Nosten F, Khusmith S, Snounou G. Are extensive T cell epitope polymorphisms in the Plasmodium falciparum circumsporozoite antigen, a leading sporozoite vaccine candidate, selected by immune pressure? J Immunol. 2005;175:3935–3939. doi: 10.4049/jimmunol.175.6.3935. [DOI] [PubMed] [Google Scholar]
- 28.Branch O, Casapia WM, Gamboa DV, Hernandez JN, Alava FF, Roncal N, Alvarez E, Perez EJ, Gotuzzo E. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J. 2005;4:27. doi: 10.1186/1475-2875-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van den Eede P, Van der Auwera G, Delgado C, Huyse T, Soto-Calle VE, Gamboa D, Grande T, Rodriguez H, Llanos A, Anne J, Erhart A, D'Alessandro U. Multilocus genotyping reveals high heterogeneity and strong local population structure of the Plasmodium vivax population in the Peruvian Amazon. Malar J. 2010;9:151. doi: 10.1186/1475-2875-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoeler GB, Flores-Mendoza C, Fernandez R, Davila JR, Zyzak M. Geographical distribution of Anopheles darlingi in the Amazon Basin region of Peru. J Am Mosq Control Assoc. 2003;19:286–296. [PubMed] [Google Scholar]
- 31.Flores-Mendoza C, Fernandez R, Escobedo-Vargas KS, Vela-Perez Q, Schoeler GB. Natural Plasmodium infections in Anopheles darlingi and Anopheles benarrochi (Diptera: Culicidae) from eastern Peru. J Med Entomol. 2004;41:489–494. doi: 10.1603/0022-2585-41.3.489. [DOI] [PubMed] [Google Scholar]
- 32.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 33.Haubold B, Hudson RR. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics. 2000;16:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 34.Dharia NV, Plouffe D, Bopp SE, Gonzalez-Paez GE, Lucas C, Salas C, Soberon V, Bursulaya B, Kochel TJ, Bacon DJ, Winzeler EA. Genome scanning of Amazonian Plasmodium falciparum shows subtelomeric instability and clindamycin-resistant parasites. Genome Res. 2010;20:1534–1544. doi: 10.1101/gr.105163.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dharia NV, Bright AT, Westenberger SJ, Barnes SW, Batalov S, Kuhen K, Borboa R, Federe GC, McClean CM, Vinetz JM, Neyra V, Llanos-Cuentas A, Barnwell JW, Walker JR, Winzeler EA. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci USA. 2010;107:20045–20050. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.