Abstract
Background
Natrialba magadii is an aerobic chemoorganotrophic member of the Euryarchaeota and is a dual extremophile requiring alkaline conditions and hypersalinity for optimal growth. The genome sequence of Nab. magadii type strain ATCC 43099 was deciphered to obtain a comprehensive insight into the genetic content of this haloarchaeon and to understand the basis of some of the cellular functions necessary for its survival.
Results
The genome of Nab. magadii consists of four replicons with a total sequence of 4,443,643 bp and encodes 4,212 putative proteins, some of which contain peptide repeats of various lengths. Comparative genome analyses facilitated the identification of genes encoding putative proteins involved in adaptation to hypersalinity, stress response, glycosylation, and polysaccharide biosynthesis. A proton-driven ATP synthase and a variety of putative cytochromes and other proteins supporting aerobic respiration and electron transfer were encoded by one or more of Nab. magadii replicons. The genome encodes a number of putative proteases/peptidases as well as protein secretion functions. Genes encoding putative transcriptional regulators, basal transcription factors, signal perception/transduction proteins, and chemotaxis/phototaxis proteins were abundant in the genome. Pathways for the biosynthesis of thiamine, riboflavin, heme, cobalamin, coenzyme F420 and other essential co-factors were deduced by in depth sequence analyses. However, approximately 36% of Nab. magadii protein coding genes could not be assigned a function based on Blast analysis and have been annotated as encoding hypothetical or conserved hypothetical proteins. Furthermore, despite extensive comparative genomic analyses, genes necessary for survival in alkaline conditions could not be identified in Nab. magadii.
Conclusions
Based on genomic analyses, Nab. magadii is predicted to be metabolically versatile and it could use different carbon and energy sources to sustain growth. Nab. magadii has the genetic potential to adapt to its milieu by intracellular accumulation of inorganic cations and/or neutral organic compounds. The identification of Nab. magadii genes involved in coenzyme biosynthesis is a necessary step toward further reconstruction of the metabolic pathways in halophilic archaea and other extremophiles. The knowledge gained from the genome sequence of this haloalkaliphilic archaeon is highly valuable in advancing the applications of extremophiles and their enzymes.
Background
Archaea are the least well-characterized members among the extant three domains of life, and recent genome sequencing efforts have facilitated our understanding of these unusual microbes [1]. The phylum Euryarchaeota contains a diverse array of archaea currently classified under eight named classes (Archaeoglobi, Halobacteria, Methanobacteria, Methanococci, Methanomicrobia, Methanopyri, Thermococci, and Thermoplasmata) and ten orders [2]. Members of the Euryarchaeota, particularly those of Halobacteria, have received attention because of their ecological and evolutionary importance. Halophilic archaea are physiologically and phylogenetically diverse and occur in a wide variety of environments [3,4]. Most halophilic archaea thrive in hypersaline environments (>15% NaCl). To survive in such extreme conditions, these organisms have evolved strategies to cope with not only osmotic stress and desiccation, but also oxygen limitation and the damaging effects of UV light [5]. The haloalkaliphiles constitute a distinct group of microorganisms since they survive in two extremes: high pH and hypersalinity [6,7]. In addition, haloalkaliphilic archaea have an asymmetric C20 C25 diether isoprenoid core lipid that is uncommon among neutrophilic halophilic archaea [8].
The genus Natrialba within Halobacteria is a heterogeneous group of halophiles including those that thrive in neutral as well as alkaline environments [9]. The type species Natrialba asiatica, which was isolated from a beach in Japan, is non-alkaliphilic and requires a pH of 6.6 to 7.0 for optimum growth [10]. Natrialba magadii (formerly Natronobacterium magadii, a strictly aerobic chemoorganotroph isolated from Lake Magadi in Kenya) is an obligately haloalkaliphilic archaeon that requires 20% (3.5 M) NaCl, pH 9.5, and 37 to 40°C for optimum growth [9,11]. In contrast to the white-yellow color of Nab. asiaticaNab. magadii is red-orange colored due to the presence of carotenoid pigments in the cell membrane [12]. Furthermore, Nab. magadii lacks glycolipids, whereas Nab. asiatica contains bis-sulfated glycolipid S2-DGD {2, 3-diphytanyl- or phytanyl-sesterterpenyl-1-[2, 6-(HSO3)2-α-Manp-1 → 2-Glcp]-sn-glycerol} [9,13]. Previous work has demonstrated that Nab. magadii synthesizes and accumulates 2-sulfotrahalose as an osmolyte under hypersaline conditions [14]. The biochemical features of the flagellar apparatus, a nucleoside diphosphate kinase, a leucine dehydrogenase, and an extracellular serine protease of Nab. magadi have also been characterized since its discovery [15-18].
Although knowledge on the biology of halophilic archaea has greatly advanced during the last decade, attempts to understand the physiology and genetics of the haloalkaliphilic archaea are scarce [19]. The study of haloalkaliphilic archaea is interesting from several perspectives because these are among the most alkaliphilic microorganisms reported to date [20]. Of the halophilic archaea related to Nab. magadii, a low pass genomic sequence of Nab. asiatica strain ATCC 700177 has been reported [21]. The genome of Haloterrigena turkmenica strain DSM 5511, a halophilic archaeon isolated from sulfate saline soil in Turkmenistan, is complete and contains 5,287 protein coding genes [22]. Furthermore, the complete genome of Natronomonas pharaonis DSM 2160, a haloalkaliphilic archaeon isolated from a soda lake in Egypt, contains 2,843 protein coding genes [23]. In addition, a detailed analysis of the metabolic pathways of halophilic archaea has been described [24]. The goal of the current study was to explore the physiology of Nab. magadii at the whole genome level and perform comparative genomic studies with other halophilic as well as haloalkaliphilic archaea. An exploration of the pathways of coenzyme biosynthesis and proteolysis within Nab. magadii was also envisaged.
Methods
Nab. magadii was grown at 37°C aerobically in liquid medium containing 20 g/L yeast extract using the method of Tindall et al. [11], and genomic DNA was extracted from the pelleted cells using the procedure described by Ng et al. for Halobacterium halobium (salinarum) [25]. Genomic library construction, sequencing, and finishing were performed at the Joint Genome Institute (JGI) facilities at Walnut Creek and the Genome Science facilities at Los Alamos National Laboratory. Briefly, the draft genome of Nab. magadii was sequenced using a combination of both Sanger and 454 technologies. A Sanger whole genome shotgun library, which produced 26,484 reads with an average insert size of 6.5 kb, and a 454 FLX standard library, which generated 96.3 Mbp of data, were constructed for this genome. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/. The Phred/Phrap/Consed software package (http://www.phrap.com) was used for sequence assembly and quality assessment. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, custom primer walk, or PCR amplification (Roche Applied Science, Indianapolis, IN). A total of 594 additional custom primer reactions were necessary to close all gaps and raise the quality of the finished sequence. The estimated error rate for the completed genome of Nab. magadii was less than 1 in 100,000. The final assembly was based on 19.1 Mbp of Sanger draft data, which provided 4.3x coverage of the genome, and 96.3 Mbp of 454 draft data, which provided 21.7x coverage of the genome.
Preliminary automated annotation, prediction of the number of subsystems, and pairwise BLAST comparisons of protein sets within different strains were performed using the Rapid Annotation using Subsystems Technology (RAST), which is a fully automated, prokaryotic genome annotation service [26]. Subsequently, a detailed manual curation was performed to ensure consistency with the annotation of other halophilic archaea. Annotation of genes involved in coenzyme biosynthesis was based on the information available in recent literature and/or their relatedness to functionally characterized homologs present in other organisms. These annotation details are provided at the web site http://wiki.rzg.mpg.de/HaloferaxWiki. Proteins deemed to be specific to Nab. magadii were compared against the NCBI non-redundant protein database to determine whether they were hypothetical or conserved hypothetical. If there was no adequate alignment with any protein (less than 25% identity or aligned region is less than 25% of the predicted protein length), the translated ORF was named a hypothetical protein.
Multiple genome comparisons were performed using the ‘progressive alignment’ option available in the program MAUVE version 2.3.0 [27,28]. Default scoring and parameters were used for generating the alignment. Prior to the alignment, the Nab. magadii genome sequence was rearranged to facilitate visual comparison. This was accomplished using the Artemis Comparison Tool to identify a coordinate where the sequence was shifted relative to that of Htg. turkmenica. The coordinate was located at 1961610 bp and the Nab. magadii sequence was cut starting at this coordinate until the end of the sequence and placed at the beginning of the fasta file so that the genome start was near the major origin of replication.
A synteny plot was generated using the program NUCmer, which uses exact matching, clustering, and alignment extension strategies to create a dot plot based on the number of identical alignments between two genomes [29]. NUCmer was used with the maxmatch argument and, to be consistent with the MAUVE comparison, the rearranged Nab. magadii sequence was aligned with that of Htg. turkmenica. The Nab. magadii genome project is deposited in the Genomes OnLine Database (GOLD) and the complete genome sequence is available from GenBank/EMBL/DDBJ with accession numbers CP001932, CP001933, CP001934, and CP001935. The genome of Nab. magadii is also accessible through HaloLex (http://www.halolex.mpg.de) and the UCSC Archaeal genome browser (http://archaea.ucsc.edu/).
Results and discussion
Nab. magadii genome features and comparison with the genomes of other halophilic archaea
The complete genome sequence of Nab. magadii consisted of four replicons (total size 4,443,643 bp). Three of these elements had a GC content of ~61% whereas pNMAG02 had a GC content of 56.82%. A comparison of some of the relevant features of these four elements is shown in Table 1. A BLASTN analysis of pNMAG03 on the NCBI database revealed 99% identity to halovirus φCh1 (58,498 bp; GenBank accession number NC_004084), a bacteriophage-like element isolated from Nab. magadii. Since halovirus φCh1 has already been described elsewhere [30-32], the analysis of pNMAG03 was excluded from the scope of the current work. The large chromosome of Nab. magadii contained two genes encoding putative replication factor C-like proteins (Nmag_1868 and 1910). The large chromosome, pNMAG01, and pNMAG02 were predicted to replicate using a conserved archaeal mechanism [33], since each of these replicons contained at least one gene encoding an Orc1/Cdc6 family replication initiation protein. For the large chromosome, the major replication origin was predicted to be at ca 1.9 Mb, located between Orc1 (Nmag_1930) on the forward strand and a three-gene operon on the reverse strand (Nmag_1927-1929). This set of four highly conserved genes was found adjacent to the replication origin in almost all halophilic archaea.
Table 1.
Characteristics of the replicons ofNatrialba magadiiATCC 43099
| Replicon | Large chromosome | Small chromosome | Large plasmid | Virus φCh1 |
|---|---|---|---|---|
| Annotation |
None |
pNMAG01 |
pNMAG02 |
pNMAG03 |
| Topology |
Circular |
Circular |
Circular |
Linear/Circular |
| Size |
3,751,858 bp |
378,348 bp |
254,950 bp |
58,487 bp |
| GC content |
61.42% |
60.09% |
56.82% |
61.90% |
| Number of RNA genes |
53 |
7 |
None |
None |
| Number of protein-coding genes |
3559 (+1790/-1769) |
340 (+183/-157) |
219 (+110/-109) |
94 (+11/-83) |
| Number of hypothetical proteins |
1278 |
96 |
69 |
75 |
| Full length orc1/cdc6 homologs |
5 |
1 |
1 |
None |
| Glycosyltransferase genes |
19 |
3 |
1 |
None |
| IS elements |
21 |
2 |
13 |
None |
| Overall coding density |
83% |
80% |
76% |
93% |
| GenBank accession number | CP001932 | CP001933 | CP001934 | CP001935 |
Archaeal genomes can contain a large number of transposable elements and the variety of archaeal insertion sequences is thought to approximate that of bacteria [34]. However, most archaeal genomes lack prophage elements [35]. Manual curation indicated that the genome of Nab. magadii contained ~36 full-length or truncated genes encoding putative transposases. These insertion sequence elements were scattered throughout the chromosomes and about 20 of these belong to the IS605 OrfB family. The IS605 OrfB transposase (also called IS1341-type transposase) genes were highly diverse, as is typical of halophilic archaea [36]. A single IS605 OrfA (Nmag_4105, also called IS200-type transposase) was identified in the genome. Other transposase genes in Nab. magadii include 7 of the broad category IS4 (3 IS7-type and 4 IS9-type), a single IS240-type, and 4 related to ISSod10. The small number of transposase genes and their heterogeneity may indicate that Nab. magadii is only minimally affected by these elements. The genome also contained several genes related to bacteriophage elements (e.g., PhiH1 repressor protein, phage tail proteins, and phage protein D) and a vgr-like gene related to recombination hot spot elements. In addition, there were 13 genes encoding integrase/recombinase-like proteins (Additional file 1: Table S1).
Archaeal genomes generally have 1–4 rRNA operons consisting of the 16 S, 23 S, and 5 S rRNA genes with a tRNAAla gene located in the internal transcribed spacer [37]. The large chromosome of Nab. magadii contained two copies of 16 S rRNA-tRNAAla-23 S rRNA-5 S rRNA sequences, one each on the plus and minus strands, as well as two genes encoding components of the RNA guide machinery (Nmag_0693-0694) with fibrillarin-like RNA methyltransferase as the catalytic component. The small chromosome pNMAG01 contained a copy of 16 S rRNA-tRNAAla-23 S rRNA-5 S rRNA sequence on the minus strand and a copy of 23 S rRNA-5 S rRNA sequence on the plus strand. The three 16 S rRNA-tRNAAla-23 S rRNA-5 S rRNA sequences of Nab. magadii had 99% nucleotide identity to each other. The small chromosome pNMAG01 also contained an orphan 5 S rRNA sequence that had 89% nucleotide identity to the other four 5 S rRNA genes of Nab. magadii. Since pNMAG02 lacked rRNA operons and had a lesser GC content than the large and small chromosomes, this self replicating element could be considered a large plasmid. The heterogeneity of the rRNA operons within Nab. magadii is not a unique feature and the occurrence of such rRNA operons among halobacterial genomes is thought to be due to recombination between rRNA genes of different strains or species [38]. The 16 S rRNA genes of Nab. magadii were closely related to those of Nab. asiatica (97% identity), Htg. turkmenica (96% identity), and Nmn. pharaonis (90% identity). Furthermore, the genome of Nab. magadii was compared to 17 complete haloarchaeal genomes available in the public databases (Additional file 2: Table S2). Based on this analysis, Htg. turkmenica contained the highest number of orthologs (2601 symmetrical hits), followed by Halopiger xanaduensis strain SH-6 (2533 symmetrical hits). There were lesser number of orthologs (1805 symmetrical hits) in Nmn. pharaonis, which has a relatively smaller genome. However, when the data for the percentage of proteins having a bidirectional best blast hit in Nab. magadii was computed (Additional file 2: Table S2), Nmn. pharaonis was the top (63% of the proteins having a bidirectional best blast pair), followed by Hpg. xanaduensis (60%) and Htg. turkmenica (51%). Results from Nmn. pharaonis and Htg. turkmenica are emphasized in this paper since the former was the only other haloalkaliphilic archaeon with a complete genome sequence and the latter contained the highest number of orthologs.
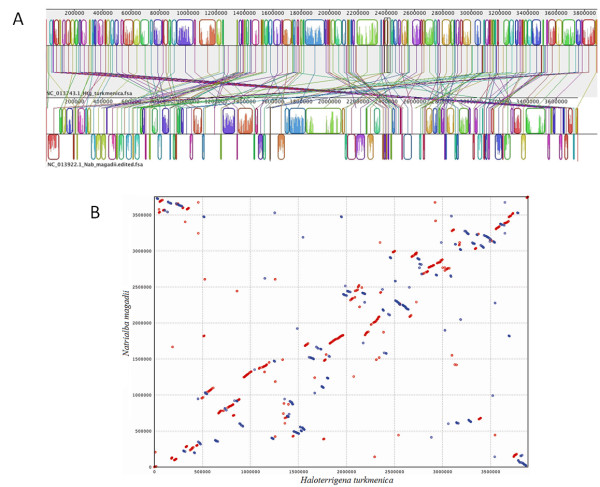
The combined size of the complete genome of Nab. magadii was 1.7 Mb larger than the complete genome of Nmn. pharaonis, which consists of three replicons (total size 2,749,696 bp). However, Nab. magadii genome was 1 Mb smaller than the complete genome of Htg. turkmenica, which consists of seven replicons (total size 5,440,782 bp). The GC content (61.42%) of the large chromosome of Nab. magadii was slightly lesser than that of the large chromosomes of Htg. turkmenica (~66% GC) and Nmn. pharaonis (~63% GC). Alignment of the large chromosome of Nmn. pharaonis (2,595,221 bp) with that of Nab. magadii using MAUVE showed the presence of very few short syntenic regions (data not shown), whereas a similar alignment using the large chromosome of Htg. turkmenica (3,889,038 bp) showed the presence of numerous short syntenic regions (Figure 1A). To further dissect this co-linearity, a BLASTN comparison of the large chromosomes of Nab. magadii and Htg. turkmenica was performed. This analysis revealed the presence of 400 homologous regions (226 plus/plus and 174 plus/minus, sequence range >300 bp, but <2,000 bp) with an average nucleic acid identity of 89% (the identity range was 84%-98%, E-value = 0). The plus and minus strand matches among the chromosomes of Htg. turkmenica and Nab. magadii generated by NUCmer are shown in Figure 1B.
Figure 1.
A. Alignment of the large chromosomes ofNatrialba magadiiATCC 43099 andHaloterrigena turkmenicaDSM 5511 using MAUVE 2. Prior to the alignment, the Nab. magadii genome sequence was rearranged to facilitate visual comparison. The Nab. magadii sequence was cut starting at 1961610 bp (located between Nmag_1929 and Nmag_1930, encoding putative GTP-binding protein and ORC1 replication initiation protein, respectively) until the end of the sequence and placed at the beginning of the fasta file so that the genome start was near the major origin of replication. Identically colored boxes, known as locally collinear blocks (LCBs), depict homologous regions in the two chromosomes. The edges of LCBs indicate chromosome rearrangements due to recombination, insertions, and/or inversions. Sequences of Nab. magadii inverted in relation to those of Htg. turkmenica are shown as blocks below the horizontal line. The vertical lines connecting the LCBs point to regions of homology among the two chromosomes. Numbers above the maps indicate nucleotide positions within the respective chromosomes. B. Synteny plot of the large chromosomes of Natrialba magadii ATCC 43099 and Haloterrigena turkmenica DSM 5511 generated by NUCmer. NUCmer was used with the maxmatch argument and the Nab. magadii genome sequence was rearranged as in Figure 1A to facilitate visual comparison. Regions of identity between the two chromosomes were plotted based on pair-wise alignments. Numbers indicate nucleotide positions within the respective chromosomes. Plus strand matches are slanted from the bottom left to the upper right corner and are shown in red. Minus strand matches are slanted from the upper left to the lower right corner and are shown in blue. The number of dots/lines shown in the plot is the same as the number of exact matches found by NUCmer.
A three-way comparison of all predicted protein-coding genes of Nab. magadii using the TaxPlot tool of NCBI revealed that Htg. turkmenica contained 2387 orthologs, whereas Nmn. pharaonis contained only 426 orthologs (symmetrical hits). These analyses further confirmed that Nab. magadii was more closely related to Htg. turkmenica than to Nmn. pharaonis. In Nab. magadii, 1518 genes could not be assigned a function based on BLAST analysis and were annotated as encoding hypothetical or conserved hypothetical proteins. The isoelectric point (pI) of most of the predicted proteins of Nab. magadii was in the 3–5 range, indicating that the general proteome is acidic, which is typical of most halophilic archaea. A two-way comparison of the large chromosomes revealed that Nab. magadii contained ~945 putative protein-coding genes that had no homologs in Htg. turkmenica. A vast majority (~75%) of these Nab. magadii-specific genes encoded hypothetical proteins. Other genome-specific genes in Nab. magadii encoded ABC-type transporters, ATPases, kinases, phosphatases, proteases, and oxidoreductases. The genome of Nab. magadii also contained a variety of simple sequence repeats encoding characteristic peptide repeat patterns.
General adaptive features
In addition to maintaining an acidic proteome and a cell wall composed of acidic glycoproteins, haloalkaliphilic species appear to have evolved several other mechanisms of adaptation to their niche [39-41]. These include, but are not limited to, intracellular accumulation of inorganic cations and/or neutral organic compounds [42-44]. Halophilic archaea maintain the necessary water balance and osmotic pressure even when the extracellular Na+ concentration exceeds 5 M by pumping Na+ out and K+ into the cell using a variety of cation/proton antiporters [12,45,46]. The genome of Nab. magadii contained an operon of nine genes encoding a putative pH adaptation K+ efflux system (Nmag_3445-3453). Genes related to this operon were present in several halophilic archaea, indicating that they may not encode a specialized system involved in stress response to alkaline growth conditions. Apart from this operon, the genome contained three other genes encoding putative cation/proton antiporters and a gene encoding a putative OsmC family protein (Additional file 1: Table S1).
Low molecular weight organic compounds such as amino acids, polyols, and sugars facilitate cellular adaptation to high-osmolarity and are referred to as osmoprotectants or compatible solutes [47]. Halophilic species also accumulate neutral organic compounds as a means of adaptation to their niche [14,43,44]. The large chromosome of Nab. magadii contained a locus encoding a putative trehalose-phosphate synthase and a trehalose-phosphatase, which may be involved in the biosynthesis of the osmoprotectant 2-sulfotrehalose. The orthologs of these genes were found in few other halophilic archaea and the osmolyte has been detected by nuclear magnetic resonance spectroscopic analysis in Nab. magadii[14]. Nab. magadii also contained genes encoding the biosynthesis of spermine as well as transporters for the uptake of choline/carnitine/betaine and spermidine/putrescine, which may also provide protection at high-osmolarity (Additional file 1: Table S1). Therefore, it appeared that Nab. magadii had multiple mechanisms (e.g., intracellular accumulation of inorganic cations as well as neutral and/or charged organic compounds) for osmotic adaptation.
The scarcity of molecular oxygen in a hypersaline milieu could be a growth-limiting factor for aerobic chemoorganotrophic prokaryotes [48-50]. It has been proposed that some archaeal species accumulate intracellular gas vesicles that help them float on the surface and perform oxidative respiration in their native saturated saltwater habitats [12]. The large chromosome of Nab. magadii contained a cluster of 11 genes encoding putative gas vesicle synthesis family proteins, which were related to the gas vesicle family proteins of Hbt. salinarum. However, Nab. magadii lacked genes related to those encoding the minor gas vesicle protein (GvpC) and the regulators (GvpD and GvpE). The gas vesicle gene clusters of Nab. magadii and Htg. turkmenica were highly similar to each other and appeared to contain a distant homolog of the Hbt. salinarum gvpI gene (Nmag_0338 and Htur_2370, respectively). Nevertheless, these genes encode putative proteins with an N-terminal extension of more than 200 residues not found in GvpI of Hbt. salinarum. Furthermore, the gas vesicle clusters of Nab. magadii and Htg. turkmenica contained an additional gene (Nmag_0337 and Htur_2371, respectively) that was absent in Hbt. salinarum. Nab. magadii also contained a gene encoding a hemAT-type aerotactic transducer with a putative globin-coupled sensor protein comprising of a globin fold domain and a methyl-accepting chemotaxis transducer domain (Additional file 1: Table S1). These traits, in addition to the osmotic adaptation mechanisms discussed above, may play a role in the survival of Nab. magadii in its natural environment.
Other features likely to facilitate the adaptation of Nab. magadii to its niche included genes encoding putative mechanosensitive ion channels (MscS) that afford protection against hypoosmotic shock, chaperone proteins DnaJ and DnaK, a thermosome, and heat shock proteins that may participate in protein quality control and cellular response to stress. Nab. magadii also contained 47 genes (26 on the large chromosome, 18 on pNMAG01, and 3 on pNMAG02) encoding putative proteins of various sizes with a universal stress protein (UspA) domain. One of these genes (Nmag_1302) appeared to form an operon with a gene encoding a putative GCN5-related N-acetyltransferase (GNAT, Nmag_1303), and a similar gene pair was found in Htg. turkmenica (Htur_3429-3430, 80% identity, E-value = 2e-82 to 1e-52) and Nmn. pharaonis (NP1710A-NP1712A, 49% identity, E-value = 2e-41 to 4e-28). It is possible that the GNAT is involved in the acetylation of the linked universal stress protein in these species. In addition, the large chromosome of Nab. magadii contained genes encoding a superoxide dismutase (sodA), two catalases (katG, which is common to most halophiles, and katE, which is closely related to katE of methanogens, also present in Htg. turkmenica and Hpg. xanaduensis, but not in other halophiles), two alkyl-hydroperoxidase-like proteins, a carbonic anhydrase, and methionine sulfoxide reductases (msrA and msrB; Additional file 1: Table S1). It is possible that these enzymes have a role in the adaptation of this haloarchaeon to various oxidative stresses associated with energy metabolism.
Furthermore, Nab. magadii contained genes encoding metal transport proteins and a putative copper resistance protein. Nab. magadii copper resistance protein appears to contain fused CopC-CopD domains and a distant homolog of this protein occurs in Nmn. pharaonis (NP4610A, 39% identity, E-value = 2e-105), but not in other archaea. These genes may be involved in metal homeostasis in the hypersaline environment that Nab. magadii inhabits. Nab. magadii also encoded DNA methylases, DNA damage repair excinuclease ABC subunits, DNA mismatch repair proteins, and DNA repair/recombination proteins RadA and RadB (Additional file 1: Table S1). Homologs of these genes are found in several other archaea and they are predicted to be involved in stress response and maintaining genetic integrity.
Proteases, peptidases, protease inhibitors, and protein translocation
At least 83 genes encoding various types of peptidases/proteases were identified in the genome of Nab. magadii by manual curation (~1.8% of all protein-coding genes; Additional file 3: Table S3). Interestingly, Nab. magadii appears to encode a larger set of proteolytic enzymes compared to most halophilic archaea, including Nmn. pharaonis, Hfx. volcanii and Hbt. salinarum. This suggests that the natural environment inhabited by Nab. magadii contains an ample supply of protein debris, which could be used as a major carbon and nitrogen source. The closest homologs of the vast majority of Nab. magadii genes (~50%) encoding putative peptidases/proteases were found in Htg. turkmenica (Additional file 3: Table S3). Most of the Nab. magadii predicted proteases belong to the catalytic type of metallo- and serine proteases. Other proteases include various amino- and carboxypeptidases, oligopeptidases, signal peptidases, ATP-dependent proteases, and intramembrane cleaving proteases (I-CLiPs).
Subtilases (COG1404; subtilisin-like serine proteases) are a large superfamily of functionally diverse endo- and exo-peptidases that occur in prokaryotes and eukaryotes [51]. Nab. magadii contained nine genes encoding putative S8 and S53 subtilisin kexin sedolisins (Additional file 3: Table S3). Although the predicted subtilisins of Nab. magadii had diverse sizes (ranging from 402 to 1710 aa), the amino acid motifs containing the catalytic triad (Asp-His-Ser) were conserved in all of them. Six of the predicted subtilisins (Nmag_0073, 0714, 0715, 1249, 1874, and 3633) of Nab. magadii contained putative targeting signals for translocation through the twin-arginine transport (Tat) pathway, suggesting that these proteases are most likely exported out of the cell. Within this group, Nmag_0715 has been biochemically characterized and designated as the Natrialba extracellular protease (Nep) [16]. Nep was demonstrated to be alkali-resistant, a feature that correlates with the conditions that predominate in the natural environment of Nab. magadii[16]. Interestingly, the C-terminal domain of Nep contains an acidic patch composed of 12 amino acid residues that is absent in the subtilases of neutrophilic organisms [52]. This distinctive feature of Nep may be involved in its stability at high salt and/or high pH. In addition, pNMAG01 contained a gene encoding a putative microcystin LR degradation protein (Nmag_3774, MlrC-like-protein). MlrC peptidases, initially isolated from the bacterium Sphingomonas, are a specialized group of metalloproteases assigned to M81 family and they participate in the last step of the degradation pathway of microcystin LR [53]. These enzymes rarely occur in the archaeal domain and the homologs of Nmag_3774 were not found in Nmn. pharaonis and Htg. turkmenica (Additional file 3: Table S3).
All archaeal genomes studied to date are predicted to encode self-compartmentalized proteases (20 S proteasomes and Lon-type proteases) likely to function in energy-dependent proteolysis and an ubiquitin-type mechanism for targeting proteins to proteasomes termed sampylation [54,55]. In archaea, 20 S proteasomes of α- and β-type subunits are thought to function with AAA-ATPases such as the proteasome-activating nucleotidase (PAN) in degrading folded proteins [54]. In addition, ubiquitin-like small archaeal modifier proteins (SAMPs) appear to be conjugated to protein targets by an E1-like enzyme termed ubiquitin-like conjugating enzyme of archaea or UbaA (based on study of Hfx. volcanii[56]). The genome of Nab. magadii contained an operon encoding putative 20 S proteasome α and β subunits (Nmag_0515-0514, respectively). Apart from this operon, the genome contained separate genes encoding 20 S proteasome α and β subunit homologs (Nmag_3313 and Nmag_3351, respectively). Nab. magadii was also predicted to encode homologs of PAN (Nmag_1362 and 2440) and ubiquitin-like small archaeal modifier proteins (SAMPs; Nmag_0567, 1914, 2668, and 2971). The genome of Nab. magadi contained two genes encoding putative ubiquitin-like activating enzymes of archaea (UbaA; Nmag_1394 and 3812). Furthermore, it also encoded a distant homolog of UbaA (Nmag_0356) containing a C-terminal JAB1/MPN/Mov34 metalloenzyme (JAMM) domain that was predicted to remove SAMPs from target proteins. In contrast, Hfx. volcanii encodes only a single UbaA-type protein that functions in both protein conjugation (sampylation) and sulfur mobilization [56]. Nab. magadii also encoded an archaeal-type LonB protease (Nmag_2822), which was demonstrated in its cell membranes [57]. While LonB homologs are conserved and likely act as key energy-dependent proteases in archaea, the physiological significance of these enzymes has not been addressed.
The tetrahedral aminopeptidase (TET protease) is an energy-independent protein complex (with a peptidase domain of the clan MH, family M42, according to the MEROPS database) that was isolated from the neutrophilic haloarchaeon Har. marismortui[58]. It has been suggested that TET degrades oligopeptides released by ATP-dependent proteases such as the proteasome and LonB. Nab. magadii encodes a homolog of TET (Nmag_1335, peptidase M42 family protein), which, in combination with the energy-dependent proteases, may participate in the intracellular protein turnover in this extremophile. Furthermore, similar to the majority of haloarchaea, Nab. magadii appears to encode homologs of the three families of membrane-embedded regulatory proteases denoted as I-CLiPs. These include sppA-type signal peptide peptidases (SPPs, Nmag_2612 and 2635), site-2 protease class of zinc metalloproteases that cleave transmembrane domains (S2P type peptidases, Nmag_1508, 1514, 2136, and 3494), and rhomboids (Rho, Nmag_1128, 1636, 2518, and 3579). Furthermore, Nab. magadii contained genes encoding type I signal peptidases (sec11-type, Nmag_1326, 1932, 1944, 3375, 3743, and 4175) and a type IV prepilin peptidase (Nmag_1752). The type I signal peptidases and the type IV prepilin peptidase are predicted to be involved in the processing of N-terminal signal peptides of exported proteins and flagellin precursors, respectively.
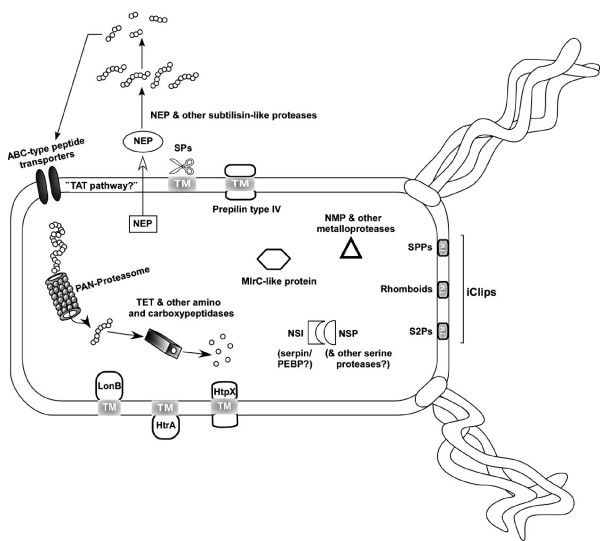
Cellular protease activity is frequently controlled by endogenous protease inhibitors [59]. Genes encoding putative homologs of protease inhibitors of the serpin (Nmag_2110) and phosphatidylethanolamine-binding protein (Nmag_0329) types were present in Nab. magadii. A subtilisin protease inhibitor from this archaeon, denoted NSI, was previously purified and biochemically characterized [60]. This protease inhibitor remains to be investigated at the molecular level and the availability of its gene sequence could facilitate cloning and expression of the recombinant protein for further analysis. A representation of the major proteolytic systems (predicted and/or validated by detecting mRNA and/or assaying protein activity) of Nab. magadii is presented in Figure 2. Although this depiction assumes that the proteolytic systems of Nab. magadii are independent of each other, their synergistic action in vivo cannot be ruled out.
Figure 2.
Schematic representation ofNatrialba magadiiATCC 43099 proteolytic systems. The major proteases and protease inhibitor proteins predicted from the analyses of Nab. magadii genome are depicted. NSP (Nab. magadii serine protease), NMP (Nab. magadii metalloprotease), NSI (Nab. magadii subtilisin inhibitor), NEP (Nab. magadii extracellular protease), and LonB have been described in the literature. SPs, Signal peptidases; I-CLiPs, Intramembrane-Cleaving Proteases; SPPs, Signal peptide peptidases; S2Ps, Site-2 proteases; HtrA, serine protease HtrA; HtpX, putative membrane-bound zinc metalloprotease; TET, Tetrahedral aminopeptidase (Peptidase M42 family); MlrC-like protein, Microcystin LR degradation protein; PEBP, Phosphatidylethanolamine-binding protein (putative protease inhibitor); TM, Transmembrane domain; Tat, Twin arginine translocation pathway.
Protein translocation across the cell membrane in prokaryotes is facilitated by at least three mechanisms including the general secretion (Sec) system, the specialized Tat system, and the highly ornate, substrate-specific secretion systems for delivering effector proteins to target sites [61-63]. Nab. magadii contained genes that encoded putative components of the Sec system (secYEGDF, Nmag_0233, 1140, 1564, 2707, and 2708; srp19, Nmag_3604; srp54, Nmag_1802; ftsY, Nmag_0182) and of the Sec-independent Tat protein translocase complex (tatC, Nmag_2050-2051; tatA, Nmag_3135). While the Tat pathway is commonly used for a small subset of exported proteins in bacteria, it is a dominant export route in halophilic archaea. Many of the exported proteins are subsequently attached to the cell membrane by a lipid anchor and Nab. magadii has 119 genes encoding lipid-modified Tat target proteins, as detected by TatLipo analysis [64]. Furthermore, Nab. magadii contained genes encoding putative components of a type II secretion system (Nmag_3137-3138) and an archaeosortase (Nmag_2750) for which 17 targets with PGF_CTERM motif were identified [65].
N-glycosylation, glycosyltransferases, and polysaccharide biosynthesis
N-glycosylation in archaea and eukaryotes uses dolichol phosphate as the lipid base for the assembly of oligosaccharides [66,67]. Glycosyltransferases (GTs) are key components of N-glycosylation in all three domains of life, and the genome of Nab. magadii contained 23 genes encoding putative GTs. Based on BLASTP analysis on the NCBI database and the presence of conserved domains, these genes were assigned into the GTA (Nmag_0916, 0926, 1132, 1200, 2046, 2620, 2830, 3015, 3273, 3275, 3807, and 4148) and GTB (Nmag_0132, 0135, 0432, 0925, 2541, 3017, 3285, 3431, 3512, 3832, and 3843) superfamilies. One of these genes (Nmag_3015) is in an operon with Nmag_3011 (hexapeptide repeat-containing transferase, 192 aa), Nmag_3012 (aminotransferase, 481 aa), Nmag_3013 (oxidoreductase, 340 aa), and Nmag_3014 (nucleotide sugar dehydrogenase, 544 aa). Nab. magadii also contained genes encoding a putative oligosaccharyltransferase subunit (Nmag_0927, aglB homolog) and a dolichol kinase-like protein (Nmag_1986). Therefore, Nab. magadii appears to have the genetic potential for N-glycosylation.
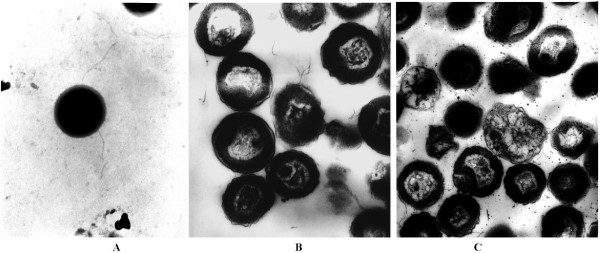
Several species of halophilic archaea are known to produce copious amounts of extracellular polysaccharides [68]. Although transmission electron microscopic (TEM) images show the presence of an exopolysaccharide-like material around Nab. magadii cells (Figure 3A), purification and biochemical analyses of this material are yet to be accomplished. Nab. magadii contained six genes encoding putative polysaccharide biosynthesis proteins (Nmag_0147, 0922, 2457, 3122, 3272, and 3437). Other genes in the genome that encoded putative enzymes involved in polysaccharide biosynthesis included six polysaccharide deacetylases (Nmag_1899, 2045, 2647, 3024, 3271, and 3278), two polyprenyl glycosyl-phosphotransferases (Nmag_0111 and 1184, 65% identity at the predicted protein level), an O-antigen polymerase (Nmag_0143), two UDP-N-acetylglucosamine 2-epimerases (Nmag_0149 and Nmag_0676), an acylneuraminate cytidylyltransferase (Nmag_0148), an O-acetyltransferase (Nmag_0150), a N-acylneuraminate-9-phosphate synthase (Nmag_0151), and two capsule synthesis proteins (Nmag_1511 and 3999). It is possible that some of these genes are involved in the biosynthesis of Nab. magadii exopolysaccharide- or capsule-like material identified in the TEM images.
Figure 3.
Transmission electron microscopic images ofNatrialba magadiiATCC 43099 stationary phase cultures.A. Negative stain of a single cell with 2% phosphotungstic acid. B and C. Ultrathin sections stained with uranyl acetate and lead citrate, respectively. The thread shaped appendages in all three images are most likely flagella. The lightly stained material around the cell in panel A is probably an exopolysaccharide.
Coenzyme biosynthesis
Archaeal metabolic pathways are unique and diverse, in comparison to those of eubacteria [24]. However, the biosynthesis of several coenzymes involved in archaeal metabolism has only been partially understood. Recent advances in this area include the discovery of a new heme biosynthetic pathway [69], further understanding of the pathway of cobalamin biosynthesis, and the reconstruction of a complete pathway for coenzyme F420 biosynthesis in haloarchaea. Analyses of genes putatively involved in coenzyme biosynthesis in Nab. magadii was performed in light of these new discoveries. This section describes the genes related to the biosynthesis of thiamine, riboflavin, NAD, coenzyme F420, folate, heme, and cobalamin.
Vitamin B1 (thiamine pyrophosphate) is involved in several microbial metabolic functions [70]. Prokaryotes have evolved elaborate mechanisms to either synthesize this important co-factor de novo or acquire it from their niche [71]. Thiamine biosynthetic pathways among prokaryotes are very diverse [70]. Thiamine biosynthesis is accomplished by joining two intermediate molecules that are synthesized separately. One of these molecules is hydroxymethylpyrimidine pyrophospate (HMP-PP), which is made from aminoimidazole ribotide (AIR, an intermediate of purine biosynthesis) using ThiC and ThiD. The other molecule is hydroxyethylthiazole phosphate (HET-P), which in bacteria is generated by ThiGH and TenI and involves the sulfur carrier ThiS. The sulfur carrier is activated for thiolylation via C-terminal adenylation catalyzed by the N-terminal E1-like domain of ThiI. Nab. magadii contained a gene (Nmag_3460) encoding a putative ThiI and several ubiquitin-like β-grasp fold proteins (Nmag_0567, 1914, 2668, and 2971). However, β-grasp fold proteins have multiple functions in halophilic archaea, being involved in sulfur chemistry as well as ubiquitin-like protein modification by SAMPylation [56]. The specific β-grasp fold protein likely to participate in thiamine biosynthesis in Nab. magadii remains unidentified.
Furthermore, Nab. magadii lacked homologs of ThiG, ThiH, and TenI involved in HET-P biosynthesis in bacteria. Interestingly, Nab. magadii encoded a homolog of the yeast HET-P synthase THI4 (Nmag_2419). However, Nmag_2419 is currently annotated as ribose-1,5-bisphosphate isomerase based on the functional characterization of the ortholog MJ0601 from Methanocaldococcus jannaschii. In contrast, the ortholog of Nmag_2419 in Pyrococcus kodokaraensis (TK0434) does not have ribose-1,5-bisphosphate isomerase activity [72]. Biochemical characterization is required to ascertain the potential role of Nmag_2419 in thiamine biosynthesis. The genome of Nab. magadii contained purM (Nmag_1281) and thiC (Nmag_2593) homologs, which were predicted to be involved in AIR and HMP biosynthesis, respectively (Table 2).
Two distinct proteins, ThiE and ThiN, are known to join HMP-PP and HET-P to generate thiamine phosphate. Nab. magadii and other halophilic archaea contain both enzymes. Whereas ThiE (Nmag_1811) is a monofunctional protein, ThiN (Nmag_1282) exists as a C-terminal domain in a ThiDN fusion protein. At the last step, thiamine phosphate is predicted to be further phosphorylated to thiamine pyrophosphate by ThiL (Nmag_1515). Therefore, the conversion of AIR to HMP-PP in Nab. magadii appears to be similar to the bacterial pathway and may involve ThiC and ThiD, whereas HET-P biosynthesis in this haloarchaeon appears to be similar to the eukaryotic pathway and may involve Nmag_2419. In addition, Nab. magadii contained genes encoding a HET kinase (ThiM, Nmag_1810, predicted to be involved in thiamine salvage) and a thiamine transporter (ThiBPQ, Nmag_460-462; ThiB2, Nmag_1940).
Vitamin B2 (riboflavin) is the precursor of coenzymes flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), which are cofactors for several biochemical reactions [73]. Most bacteria, fungi, and plants can synthesize riboflavin de novo using one molecule of GTP and two molecules of ribulose 5-phosphate as substrates [74]. Riboflavin biosynthesis has been described in M jannaschii[75]. In general, reduction precedes deamination in the archaeal riboflavin biosynthesis pathway, which appears to be similar to the fungal pathway. In M. jannaschii, GTP cyclohydrolase III, the first enzyme of the riboflavin biosynthesis pathway, produces an archaeal-specific formylated intermediate that requires a subsequent deformylation step [75]. However, the haloarchaeal homolog of GTP cyclohydrolase III has not been identified thus far. Furthermore, riboflavin kinases of halophilic archaea are homologous to those of bacteria, but are unrelated to M. jannaschii riboflavin kinase. Conversely, riboflavin synthases of halophilic archaea are related to those of M. jannaschii, but are unrelated to bacterial riboflavin synthases. Overall, six genes encoding putative enzymes of the riboflavin biosynthesis pathway were identified in Nab. magadii and only two of these (Nmag_0941 and 0942) were clustered together (Table 2).
Vitamin B3 (nicotinic acid) is the central component of coenzymes NAD and NADP, which are essential redox cofactors in metabolism. Like most bacteria, halophilic archaea synthesize NAD from aspartate via quinolinate. Nab. magadii contained 7 genes (Nmag_2920-2922 form an operon and encode nadABC, Nmag_2823 encodes nadM, and Nmag_1544, 2475, and 2846 encode nadE) that were predicted to be involved in NAD biosynthesis.
Coenzyme F420 is involved in methanogenesis [76,77] and other metabolic pathways (e.g., aflatoxin reduction in mycobacteria [78,79]) that require hydride transfer from the low-potential reduced deazaflavin F420 to substrates with electron-deficient ring systems. Furthermore, 5-amino-6-(D-ribitylamino)uracil is an intermediate of coenzyme F420 and riboflavin biosynthesis pathways [76,77]. Although coenyzme F420 has been detected in some halophilic archaea [80], and coenyzme F420 biosynthesis genes have been identified among the methanogens [81], its precise function in halophilic archaea is unknown. Nab. magadii and other halophilic archaea contained several genes encoding putative enzymes of the coenyzme F420 biosynthesis pathway. These genes were identified based on the presence of their homologs among the methanogens. Furthermore, using SIMBAL analysis [82], coenyzme F420 dependent enzymes have been predicted in halophilic archaea (e.g., NP1902A, TIGRFam 04024, D. Haft, personal communication). Nab. magadii predicted proteins with an assigned F420-related TIGRFam are shown in Table 2. A possible function of coenzyme F420 in the respiratory chain of Nab. magadii is discussed later in this paper.
Tetrahydrofolate participates in a number of biochemical reactions and reduced folate cofactors are required for the biosynthesis of a variety of molecules in both prokaryotes and eukaryotes [83,84]. The production of folate involves several enzymes catalyzing the pterin and para-aminobenzoic acid branches of the pathway [85]. Nab. magadii genes putatively involved in folate biosynthesis were generally similar to those described in Nmn. pharaonis, including the archaeal-type GTP cyclohydrolase (Nmag_2853). Nab. magadii contained a pabABC operon (Nmag_2792-2794) and the homologs of these genes were predicted to be involved in para-aminobenzoate biosynthesis in Nmn, pharaonis[24]. Nab. magadii also contained genes encoding a dihydropteroate synthase, a fused dihydropteroate-dihydrofolate synthase, a dihydrofolate reductase, and a methenyltetrahydrofolate cyclohydrolase (Table 2). The latter enzyme is predicted to participate in the conversion of the C1 metabolite attached to tetrahydrofolate. However, none of the other genes encoding C1-converting enzymes identified in Haloquadratum or Haloarcula were found in Nab. magadii and Nmn. pharaonis. Furthermore, Nab. magadii contained three genes encoding putative enzymes of the later stages of the folate biosynthesis pathway (FolP, Nmag_0002; FolC-Prd-FolP, Nmag_2554; FolA, Nmag_2988). Nevertheless, genes encoding the bacterial homologs of FolQBK, the atypical FolQ described in Desulfovibrio, or the alternative pathway bypassing FolQB (described in Plasmodium and several bacteria) were absent in Nab. magadii and Nmn. pharaonis. Therefore, precise mechanisms of folate biosynthesis in these species remain to be discovered.
Environmental bacteria utilize a variety of redox molecules such as porphyrins and other modified tetrapyrroles like heme, siroheme, and adenosylcobalamin for catalysis, energy transfer, and signal transduction [86]. These tetrapyrroles are synthesized de novo using a branched pathway and aminolevulinic acid as the precursor [87,88]. In most prokaryotes, the conversion of glutamate to aminolevulinic acid is catalyzed by glutamyl-tRNA synthetase, glutamyl-tRNA reductase, and glutamate-1-semialdehyde aminotransferase. Two molecules of aminolevulinic acid are condensed by the action porphobilinogen synthase to form porphobilinogen. Four molecules of porphobilinogen are polymerized by the action of the porphobilinogen deaminase to form the tetrapyrrole hydroxymethylbilane. Uroporphyrinogen III methyltransferase cyclizes hydroxymethylbilane to produce uroporphyrinogen III. Uroporphyrinogen III is converted to precorrin-2 in the biosynthetic pathway of adenosylcobalamin and siroheme, which was recently found to be an intermediate of heme biosynthesis (see below).
The complete pathway for the biosynthesis of adenosylcobalamin from precorrin-2 involves two major branches and several enzymes [89,90], some of which are archaea-specific (e.g.cobYcbiZ) [91,92]. Halophilic archaea (e.g.Nmn. pharaonis) use the “anaerobic” branch, which is characterized by an oxygen-independent ring contraction process (cibG) [93]. However, it has been shown that Halobacterium synthesizes cobalamin de novo under aerobic conditions [91]. The “anaerobic” branch is also characterized by early cobalt insertion and Nmn. pharaonis has homologs of the ATP-independent early cobalt chelatase (CbiX) from Bacillus halodurans and Archaeoglobus fulgidus[94,95]. In the “anaerobic” branch, seven archaeal enzymes are known to be involved in the conversion of precorrin-2 into cobyrinic acid (cbiLcbiHcbiFcbiGcbiEcbiT, and cbiC), but two pathway gaps (corresponding to cbiD/cobF and cbiJ/cobK) still remain. A set of 11 genes is known to be involved in conversion of cobyrinic acid into adenosylcobalamin (cbiAcobA, pduO, cbiPcbiZcbiBcobYcobScobCcobD, and cobT).
Based on genome analyses, it appeared that Nab. magadii was incapable of de novo cobalamin biosynthesis since it lacked the genes encoding enzymes for conversion of precorrin-2 into cobyrinic acid. This is in contrast to Htg. turkmenica, which was predicted to be capable of de novo cobalamin biosynthesis since it contained the corresponding genes. However, Nab. magadii was predicted to be capable of corrinoid salvage since it contained a gene encoding a putative corrinoid transporter. Nab. magadii also contained a set of genes that were predicted to be involved in the conversion of cobyrinic acid into adenosylcobalamin, including a gene (cbiZ) that is specific to the archaeal corrinoid salvage pathway (Table 2).
The heme biosynthesis pathway in archaea involving uroporphyrinogen III, precorrin-2, and siroheme appears to be similar to that of Desulfovibrio[69]. Conversion of uroporphyrinogen III into siroheme requires three functions (methylation by SirA, iron chelation, and oxidation by SirC). The enzyme catalyzing iron chelation is unknown since the haloarchaeal precorrin-2 dehydrogenase might be monofunctional (as in Bacillus megaterium SirC) or might also be a ferrochelatase (as in trifunctional Escherichia coli CysG or the bifunctional yeast MET8). From comparison of Nab. magadii with other halophilic archaea, another possibility emerges: iron chelation may be performed by one of the proteins annotated as CbiX-type cobalt chelatase. Nmn. pharaonis has three cbiX paralogs (cbiX1, NP1108A; cbiX2, NP1588A; cbiX3, NP0734A), two of which have closely related orthologs in nearly all other halophilic archaea. Htg. turkmenika, probably capable of de novo cobalamin biosynthesis, has orthologs of both cbiX1 and cbiX2. However, Nab. magadii, which is predicted to be incapable of de novo cobalamin biosynthesis, and therefore expected to lack these proposed early cobalt chelatases, surprisingly contained a cbiX2 ortholog (Nmag_3212). It is possible that cbiX2 functions as a ferrochelatase during siroheme/heme biosynthesis rather than as a cobaltochelatase during de novo cobalamin biosynthesis.
Didecarboxysiroheme, a common intermediate of heme and heme d1 biosynthesis, is generated by the decarboxylation of siroheme on the C12 and C18 acetyl groups [69]. Siroheme decarboxylase activity is attributed to the nirDLGH gene set, which is represented by a pair of two-domain proteins in halophilic archaea (AhbA/NirDL, Nmag_2894; AhbB/NirGH, Nmag_1221). Heme d1 is a coenzyme of dissimilatory nitrite reductase (also called cytochrome cd1) and is not required by organisms lacking this enzyme. The last steps of heme biosynthesis include the removal of acetyl side chains of Fe-coproporphyrin by AhbC (encoded by Mbar_A1793 in Methanosarcina barkeri) and the oxidative decarboxylation of heme by AhbD (encoded by DVU_0855 in Desulfovibrio) [69] [gene assignments: M. Warren, personal communication, including the information that Supplemental Figure 3 shows the sequence of ahbC and not ahbD. Orthologs encoding AhbC and AhbD were present in Nmn. pharaonis (NP1542A and NP1546A) and Htg. turkmenica (Htur_1726 and 1728), but not in Nab. magadii. The presence of ahbC and ahbD in some halophilic archaea but not in others is believed to be due to metabolic heterogeneity rather than incomplete heme biosynthesis. Conversion of heme (also called heme B) into heme A in Nab. magadii was predicted to be catalyzed by CtaA and CtaB homologs (ctaA, Nmag_0636; ctaB, Nmag_2302).
Vitamin H, commonly known as biotin, acts as a coenzyme in several enzyme-catalyzed carboxylation and decarboxylation reactions [96]. Most bacteria can synthesize biotin de novo using pimelic acid as a precursor, and some others have evolved mechanisms for importing this essential cofactor from their natural environments [97,98]. Whereas Nab. magadii is a biotin auxotroph, Nmn. pharaonis is a biotin prototroph and the genome of this haloalkaliphilic archaeon has been shown to contain at least three genes putatively involved in the biosynthesis of biotin [24]. The absence of genes for the biosynthesis of biotin in Nab. magadii was apparent from the analyses of its genome sequence. However, the large chromosome of Nab. magadii contained a locus encoding a putative biotin transporter (BioYMN, Nmag_0886-0888), which may facilitate the uptake of biotin from the environment.
Metabolic and co-factor competency
Nutritional requirements of halophilic archaea in the laboratory are as diverse as their observed phenotypes, suggesting that the metabolic pathways in these organisms are quite intricate [24,99]. The analysis of the genome sequence provided an unprecedented opportunity to comprehend the metabolic versatility of Nab. magadii. Additional file 4: Table S4 contains a comprehensive list of genes predicted to be involved in a diverse array of functions. Furthermore, genes encoding putative enzymes for archaeal modified pathways of gluconeogenesis and glycolysis as well as those of ribose metabolism and the tricarboxylic acid cycle were present in Nab. magadii. Genes that encoded putative enzymes for glycerol utilization, aromatic amino acid catabolism, ureagenesis, and urea degradation were also identified in Nab. magadii. Other putative metabolic features of Nab. magadii included xylose isomerases, an alpha amylase, a methylglyoxal synthase, sulfatases, a chlorite dismutase, sarcosine oxidases, and aldehyde dehydrogenases (Additional file 4: Table S4).
Molybdenum cofactor (MoCo) is essential for the functioning of molybdoenzymes such as dimethylsulfoxide and trimethylamine-N-oxide reductases, formate dehydrogenases, and nitrate reductases [24,100]. Molybdopterin is the dithiolene-containing tricyclic moiety found within MoCo of all molybdoenzymes except nitrogenases [101]. In bacteria, genes of the moa, mob, mod, moe, and mog loci have been implicated in the biosynthesis of MoCo using GTP as the substrate [102]. The large chromosome of Nab. magadii contained 9 genes encoding MoCo biosynthesis functions (Additional file 4: Table S4). It is uncertain if this subset of genes is sufficient for MoCo biosynthesis in Nab. magadii and biochemical studies are required to test whether this haloarchaeon is molybdenum-dependent.
The haloarchaeon Haloarcula marismortui converts acetyl-CoA to glyoxylate via the key intermediate methylaspartate [103]. Glyoxylate is condensed with a second molecule of acetyl-CoA to form malate, which is an intermediate of the tricarboxylic acid cycle. Malate can subsequently be converted to oxaloacetate, which is used by phosphoenolpyruvate carboxykinase for gluconeogenesis. In Nab. magadii, activities of the enzymes of the methylaspartate cycle, but not those of the key enzymes of the glyoxylate cycle, were detected [103]. An operon (Nmag_3333-3338) encoding putative homologs of the methylaspartate cycle and a gene encoding a putative phosphoenolpyruvate carboxykinase (Nmag_3507) were present in Nab. magadii. The square archaeon Haloquadratum walsbyi contains a gene (HQ2709A) encoding a phosphoenolpyruvate-dependent phosphotransferase system (PTS) that is predicted to be involved in the phosphorylation of dihydroxyacetone [104]. Homologs of HQ2709A and genes encoding additional PTS components were present in Htg. turkmenicaHfx. volcanii, and several other haloarchaeal genomes. However, Nab. magadii and Nmn. pharaonis lacked homologs of these genes encoding PTS components.
Respiratory chain and ATP synthesis
Running a proton-driven, energy-conserving ATP synthase at high extracellular pH is an obvious challenge. Energy coupling of sodium ions instead of protons was proposed to be an adaptation to alkaliphilic growth conditions and an ATP synthase driven by Na+ is the hallmark of such an adaptation. Nab. magadii had a cluster of eight genes that form the atpHIKECFAB operon encoding putative ATP synthase subunits (Nmag_1370-1377) and an unlinked atpD homolog (Nmag_1366). Similar gene clusters were found in several halophilic archaea. Ion specificity of the ATP synthase is determined by the c-ring, which is encoded by the atpK gene (Nmag_1375) for A-type ATP synthases. Nab. magadii may have a proton-driven ATP synthase since its predicted AtpK lacks the sequence signature of Na+-dependent ATP synthases [105]. Instead, within the ion-determining region of AtpK, the sequence (PETLVIL) is identical to that of the proton-driven ATP synthases from Hfx. volcanii[106], Hbt. salinarum[107], and Nmn. pharaonis[23].
Reduction of oxygen and the associated proton-coupled electron transfer (respiration) is the primary source of energy among aerobic organisms. Respiratory complexes, which include a variety of cytochromes and terminal oxidases, are essential components of this process. Biochemical and comparative genomic analyses of the electron transport chain of Nmn. pharaonis have revealed several novel features, including a gene encoding a type II NADH dehydrogenase (NP3508A) [23,108]. A homolog of NP3508A in Acidianus ambivalens was proposed to be involved in NADH reoxidation, feeding into the lipid-soluble quinone pool [109]. A homolog of NP3508A was also present in Nab. magadii (Nmag_0301) and several other halophilic archaea. Nab. magadii also contained genes encoding a putative nuo complex (Nmag_3245-3255), which was similar to the mitochrondrial NADH dehydrogenase (complex I). Although 13 nuo cluster subunits were conserved among halophilic archaea and E. coli, the nuoEFG subcomplex, which is involved in accepting NADH, was missing in halophilic archaea [23]. Furthermore, involvement of a type I complex in NADH reoxidation has been ruled out in Hbt. salinarum[110]. It is speculated that reduced coenzyme F420, which is similar to NADH in its redox potential, may interact with the nuo complex in halophilic archaea. In addition to the NADH dehydrogenases, Nab. magadii and other halophilic archaea are predicted to encode a succinate dehydrogenase that may oxidize succinate and reduce the quinone pool of the electron transport chain.
A number of cytochromes involved in respiratory electron transport have been characterized among the archaea [12,111]. Terminal oxidases, also known as oxygen reductases, can accept electrons from a variety of donors and reduce dioxygen to water. The large chromosome of Nab. magadii contained loci encoding putative cytochrome c-type terminal oxidase subunits I and II (Nmag_0263-0264) and cytochrome ubiquinol oxidase subunits I and II (Nmag_1036-1037). Furthermore, pNMAG02 contained an operon encoding putative cytochrome ubiquinol oxidase subunits I and II (Nmag_4038-4039) that were related to the proteins encoded by Nmag_1036-1037 (46% identity at the protein level). The homologs of Nmag_0263-0264 and Nmag_1036-1037 were present in Htg. turkmenica (Htur_2248- 2249 and Htur_4570- 4571, respectively), but not in Nmn. pharaonis. Two cbaDBAC operons (plus a monocistronic cbaE) encoding putative cytochrome ba3 terminal oxidase complexes were identified in pNMAG01 (Nmag_3754-3758 and 3802–3805), and these operons appeared to be related to each other (38-65% identity at the predicted protein level). The homologs of these ORFs were present in Htg. turkmenica (Htur_0462- 0466) and Nmn. pharaonis (NP2960A-NP2968A), the latter of which have been functionally characterized [112]. Halocyanins are predicted to act as one-electron carriers to the terminal oxidases in halophilic archaea [23,108]. This prediction is supported by the observation that cbaD is fused to halocyanin in Hbt. salinarum and Har. marismortui[113]. Nab. magadii contained several genes encoding putative halocyanin-like proteins (Nmag_2576, 2424, 0446, 0741, 3725, 1878, 3525, 1774, and 3800). Halocyanins are coupled to the reduced quinone pool by the cytochrome bc1 complex. Although genes encoding a cytochrome bc1 complex are present in Hbt. salinarumHfx. volcanii, and several other halophilic archaea, they were absent in Htg. turkmenicaNmn. pharaonis, and Nab. magadii. The electron transfer from the reduced lipid-soluble quinone pool to halocyanin remains unresolved in species that lack a cytochrome bc1 complex. Based on genome comparisons, it appeared that the respiratory chain of Nab. magadii was more similar to that of Htg. turkmenica than to Nmn. pharaonis.
Other genes encoding putative cytochrome-related proteins in the large chromosome of Nab. magadii included Nmag_0636 (cytochrome oxidase assembly protein), Nmag_1972 (cytochrome P450), and Nmag_3057 (cytochrome c biogenesis protein). The small chromosome pNMAG01 contained a gene encoding a putative cytochrome c biogenesis protein (Nmag_3708) that had 64% identity to the protein encoded by Nmag_3057. Nab. magadii also contained genes (Nmag_2430-2432) encoding a putative sulfur utilization factor (SUF) system, which was shown to be important for Fe-S cluster biogenesis during stress in E. coli[114]. Other genes predicted to participate in bioenergetic conversion in Nab. magadii include those encoding electron transfer flavoprotein subunits (Nmag_0482-0483 and 1388–1389), SCO1/SenC electron transport proteins (Nmag_0793, 3059, and 3710), and a redoxin domain protein (Nmag_3709).
Signal transduction, motility, and transcriptional regulation
Two-component signal transduction systems consisting of a histidine kinase (HK) and a response regulator (RR) constitute one of the most frequently encountered bacterial and archaeal communication circuits [115]. Nab. magadii contained a pair of genes (Nmag_3535-3536) encoding putative HK-RR proteins. Nab. magadii also contained two pairs of genes (Nmag_2042-2043 and Nmag_3740-3741) encoding putative HK-RR proteins with an additional RR domain in the N-terminus of the predicted HK protein. Furthermore, another set of genes (Nmag_1129-1130) encoding a HK-like protein, which was distantly related to CheA of Nmn. pharaonis, and a putative protein containing a RR domain was also predicted in Nab. magadii. Interestingly, Nab. magadii also contained 15 genes encoding putative HK proteins without a cognate RR gene (Nmag_0168, 0323, 0435, 1296, 1909, 2062, 3106, 3188, 3216, 3297, 3954, 4064, 4101, 4114, and 4119) and 11 genes encoding putative RR proteins without a cognate HK gene (Nmag_0951, 1095, 1797, 2147, 2692, 2693, 2877, 3151, 3379, 3389, and 3679).
Halobacterial perception of and response to physical stimulus such as light (phototaxis) is mediated by photoreceptors [116]. The genome of Nab. magadii contained a single rhodopsin gene (Nmag_2582, the chloride pump halorhodopsin). Nab. magadii also contained 3 genes (Nmag_1701, 2879, and 2881) encoding distant homologs of rhodopsins that were related to each other (36-49% sequence identity). They were also related to NP1758A from Nmn. pharaonis (33%-42% sequence identity) and predicted to encode distant rhodopsin homologs that lack the Lys residue involved in covalent retinal attachment. Whether these rhodopsin homologs interact with retinal noncovalently, or if they interact with retinal at all, is unknown. Two of the three genes encoding retinal homologs in Nab. magadii (Nmag_2879 and 2881) were located adjacent to genes (Nmag_2880 and 2882) encoding putative methyl-accepting chemotactic transducer proteins. In Nmn. pharaonis, NP1758A and NP3132A, which are homologs of Nmag_2879 and 2881, were also found adjacent to genes (NP1756A and NP3134A, respectively) encoding putative methyl-accepting chemotactic transducer proteins [23]. Therefore, this group of distant rhodopsin homologs may be involved in perception of external stimuli, although it remains to be determined if they are involved in light perception. Although Htg. turkmenica lacked genes encoding bacteriorhodopsin and halorhodopsin, it contained a single gene (Htur_3663) that appeared to be a distant homolog of Nmag_2879 and 2881. Furthermore, similar to Nmn. pharaonisNab. magadii lacked a gene encoding a proton pump bacteriorhodopsin. However, Nab. magadii contained a locus encoding putative phytoene desaturase, UbiA prenyltransferase, carotene biosynthesis protein, and phytoene synthase (Nmag_1001-1004) as well as a gene encoding a putative squalene/phytoene synthase (Nmag_2309, unrelated to Nmag_1004).
Two unrelated enzyme families are used for the cleavage of β-carotene into retinal in halophilic archaea. Distant paralogs, which belong to one of these β-carotenase enzyme families, have been identified in Hbt. salinarum and designated Brp and Blh [117]. Although homologs of brp and blh were present in Nmn. pharaonis (NP0650A and NP0206A, respectively), they were absent in Nab. magadii. However, Nab. magadii contained a homolog (Nmag_4083) of Hqr. walsbyi HQ2020A, which was predicted to encode a distinct β-carotenase unrelated to those mentioned above [104]. Interestingly, Htg. turkmenica lacked homologs of brp and blh as well as HQ2020A, which is consistent with the absence of all canonical rhodopsins in this organism.
Microbial response to chemical stimulus (chemotaxis, movement toward nutrients or away from stressors) is mediated by chemoreceptors [118]. The large chromosome of Nab. magadii contained two loci encoding putative motility and signal transduction functions. One of them contained exclusively “che genes” in a cheYBACCDR operon (Nmag_3145-3151), which is preceded by two divergently transcribed and distantly related cheW genes (Nmag_3152-3153). The cheYBACCDR operon encodes a very long signal transduction histidine kinase (CheA, 1576 amino acids), a response regulator receiver protein (CheY), a CheR-type MCP methyltransferase, and a response regulator receiver-modulated methylesterase (CheB). While CheD has been reported to function as glutamine deamidase in some organisms or as methylesterase in others, CheB functions as both [119]. A similar locus was also present in Htg. turkmenica (Htur_0954-0962) and based on predicted protein homology, it appeared that the two loci were evolutionarily very closely linked. Highly similar gene clusters were also found in the genomes of Halopiger xanaduensisNatrinema pellirubrum, and Natronobacterium gregoryi. The second locus (Nmag_2859-2889) contained “che genes” along with “fla genes” encoding flagellin biosynthesis and assembly functions (see below). The “che genes” in this locus encode putative CheA, CheY, CheR, CheB, CheD, a two-domain CheC, and two CheW proteins. Nab. magadii contained two cheF genes within this locus and homologs of these genes were shown to be involved in chemotaxis in Halobacterium[120]. This gene cluster also encoded three methyl-accepting chemotaxis sensory transducers (Nmag_2880, 2882, 2884), two of which were adjacent to genes encoding distant rhodopsin homologs (Nmag_2879 and 2881). Other genes encoding putative methyl-accepting chemotaxis sensory transducers in Nab. magadii include Nmag_0478, 0937, 1253, 1386, 1542, 2639, 3325, 3638, and 3856). Among these, two (Nmag_1253, 1386) were adjacent to genes encoding periplasmic ligand-binding proteins.
Archaeal flagella are very different in composition and assembly in comparison to bacterial flagella [121]. In contrast to the bacterial flagellar motor, which is driven by an ion gradient, the archaeal flagellar motor is driven by ATP, as shown in Hbt. salinarum[122]. Within the second motility and signal transduction gene cluster (Nmag_2859-2889) of Nab. magadii is a region (14,766 bp, 58.66% GC) with 13 predicted ORFs encoding putative flagellin biosynthesis and assembly proteins (Nmag_2862-2874). Except Nmag_2871, which encoded a protein of unknown function, all other ORFs were located on the plus strand. This region contains 4 flagellin genes (Nmag_2862-2865), which encode the flagella proteins previously identified [123] and characterized [17]. Furthermore, Nab. magadii contained homologs of flaF (Nmag_2866), flaG (Nmag_2867), flaH (Nmag_2873), flaI (Nmag_2874), and flaJ (Nmag_2869). The latter two genes (flaI and flaJ) encode putative proteins homologous to the type II secretion system proteins E and F, respectively. In several archaea, FlaI has been shown to be involved in flagellin assembly [124,125], and was recently proposed as a motor component [126]. The motility gene clusters of halophilic archaea are generally polymorphic, probably due to divergence of genome organization and deletion/duplication of the accessory genes [23]. Nevertheless, flaHflaI, and flaJ represent a core set of highly conserved genes presumably crucial for archaeal motility.
Since previous electron microscopic analyses have demonstrated that Nab. magadii contains distinctive flagella [127-129], and structures resembling flagella are also visible in the TEM images in Figure 3, it is likely that the fla locus (Nmag_2862-2874) of Nab. magadii was involved in flagellin biosynthesis and motility. In addition, the large chromosome of Nab. magadii contained genes encoding a putative full-length PilT protein (Nmag_1543) and a prepilin peptidase (Nmag_1752), whose homologs were found in Nmn. pharaonis (NP0198A and NP1276A) and Htg. turkmenica (Htur_3514 and Htur_0098).
Archaeal basal transcription machinery has many similarities to the eukaryotic RNA polymerase II apparatus. However, the mechanisms of transcription regulation and the transcriptional regulators among archaea are distinct from those of eukaryotes [130,131]. Nab. magadii contained 90 genes (~2.2% of all protein-coding genes) encoding putative transcriptional regulators (TRs). BLASTP analyses indicated that most of these predicted proteins were related to bacterial TRs. These TRs were categorized into the following families based on their helix-turn-helix (HTH) motifs (numbers in parenthesis indicate the number of proteins in each family): AsnC (16), IclR (13), PadR (10), ArsR/GntR/Fur (11), XRE (6), TrmB (8), AbrB (3), TetR (3), MarR (3), CopG/Arc/MetJ/NikR (10), HxlR (2), TenA (1), ModE (1), and unassigned (3). Apart from these TRs, Nab. magadii also contained 27 genes encoding TRs with an HTH-10 domain, which was also found in bacterio-opsin activators . In addition, Nab. magadii encoded a two-domain archaeal histone (Nmag_2205), a single TATA-binding transcription initiation factor (Nmag_1863, 93% protein sequence identity to TbpE from Hbt. salinarum), a single transcription initiation factor TFE (Nmag_3157), and a set of seven transcription initiation factors TFB (Nmag_0308, 0527, 2197, 2805, 3037, 3548, and 4179).
Conclusions
This report describes the genome sequence of Nab. magadii, a haloalkaliphilic archaeon that belongs to a physiologically distinct subgroup of halophilic archaea. Although Nab. magadii appears to have developed strategies similar to Nmn. pharaonis to optimally thrive in low water activity and high pH habitats, the genetic architecture of Nab. magadii is more similar to that of Htg. turkmenica than to Nmn. pharaonis. The presence of genes encoding the biosynthesis of the osmoprotectant 2-sulfotrehalose is an uncommon feature among halophilic archaea and this may have contributed to the evolution of Nab. magadii in its natural environment. Nab. magadii has genes encoding a number of cation/proton antiporters as well as pathways for the biosynthesis and/or transport of various cofactors and vitamins. The occurrence of genes encoding enzymes involved in glycolysis, gluconeogenesis, and glycerol utilization suggests that Nab. magadii is metabolically versatile and can use different carbon and energy sources to sustain growth. Furthermore, the large repertoire of genes encoding putative proteases/peptidases and peptide transport systems is indicative of the protein/peptide catabolic potential of Nab. magadii. It also appears that Nab. magadii can perceive and process physical and chemical stimuli, and respond appropriately by moving toward or away from those stimuli using the flagellar apparatus. The information obtained from this comparative genomic analysis contributes to our overall understanding of the biology and diversity of halophilic archaea. In particular, it will guide current and future research on the genetics and physiology of Nab. magadii. Such studies are expected to facilitate the manipulation of this archaeon as a model for haloalkalphilic metabolism and its optimization for biotechnological applications.
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
SS performed the annotation using RAST, planned the comparative analysis, and drafted most of the manuscript. JFC contributed to whole genome comparisons and generated Figure 1. JCD, KWD, LAG, NK, RT, SP, SL, and TW contributed to project planning, project management, genome sequencing, and genome annotation. DES and MIG performed the manual annotation of proteolytic enzymes and protease inhibitors and generated the schematic representation of Nab. magadii proteases/inhibitors (Figure 2). RAP prepared Nab. magadii samples used for TEM analysis. FP participated in the manual curation of Nab. magadii genome, reconstructed several coenzyme biosynthetic pathways, and helped draft the manuscript. RDC and JAM conceived the study, participated in genome analyses, and helped draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table S1.Natrialba magadii ATCC 43099 genes discussed in the text. This table lists Nab. magadii ATCC 43099 genes related to bacteriophage and recombination elements, rRNA genes, and genes encoding adaptive features.
Table S2. Bidirectional best blast pairs among proteins fromNatrialba magadii and 17 other haloarchaeal genomes. This table lists the number of bidirectional best blast pairs among proteins from Nab. magadii and 17 other halophilic archaea. The first column is the number of total proteins in each genome, the second column is the number of bidirectional best blast pairs, and the third column is the percentage of proteins having a bidirectional best blast hit in Nab. magadii.
Table S3.Natrialba magadii ATCC 43099 genes encoding putative peptidases/proteases, protease inhibitors, and regulatory proteins. This table lists Nab. magadii ATCC 43099 genes encoding various types of proteases and peptidases as well as protease inhibitors and regulatory proteins.
Table S4. Natrialba magadii ATCC 43099 genes involved in metabolism. This table lists Nab. magadii ATCC 43099 genes encoding molybdenum cofactor biosynthesis and other metabolic functions.
Contributor Information
Shivakumara Siddaramappa, Email: ssiddara@vt.edu.
Jean F Challacombe, Email: jchalla@lanl.gov.
Rosana E DeCastro, Email: decastro@mdp.edu.ar.
Friedhelm Pfeiffer, Email: fpf@biochem.mpg.de.
Diego E Sastre, Email: sastre@mdp.edu.ar.
María I Giménez, Email: migimen@mdp.edu.ar.
Roberto A Paggi, Email: paggi@mdp.edu.ar.
John C Detter, Email: cdetter@lanl.gov.
Karen W Davenport, Email: kwdavenport@lanl.gov.
Lynne A Goodwin, Email: lynneg@lanl.gov.
Nikos Kyrpides, Email: nckyrpides@lbl.gov.
Roxanne Tapia, Email: rox@lanl.gov.
Samuel Pitluck, Email: s_pitluck@lbl.gov.
Susan Lucas, Email: lucas11@llnl.gov.
Tanja Woyke, Email: twoyke@lbl.gov.
Julie A Maupin-Furlow, Email: jmaupin@ufl.edu.
Acknowledgments
This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. Research in the laboratory of JMF was funded in part by grants from the National Institutes of Health (R01 GM057498) and the Department of Energy (DE-FG02-05ER15650). Research in the laboratory of RDC was supported by grants from CONICET (PIP-1783) and UNMDP (EXA 542–2011), Argentina.
References
- Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross ST. In: Practical Handbook of Microbiology. 2. Goldman E, Green LH, editor. CRC Press, Taylor & Francis Group, LLC, Boca Raton, FL; 2009. Introduction to Archaea; pp. 663–674. [Google Scholar]
- Ventosa A. In: Prokaryotic Diversity: Mechanisms and Significance. Logan NA, Lappin-Scott HM, Oyston PCF, editor. Cambridge University Press, New York; 2006. Unusual micro-organisms from unusual habitats: hypersaline environments; pp. 223–254. [Google Scholar]
- Cuadros-Orellana S, Pohlschröder M, Durrant LR. Isolation and characterization of halophilic archaea able to grow in aromatic compounds. Int Biodeterior Biodegradation. 2006;57:151–154. doi: 10.1016/j.ibiod.2005.04.005. [DOI] [Google Scholar]
- Soppa J. From genomes to function: haloarchaea as model organisms. Microbiology. 2006;152:585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- Grant WD, Ross HNM. The ecology and taxonomy of halobacteria. FEMS Microbiol Lett. 1986;39:9–15. doi: 10.1111/j.1574-6968.1986.tb01836.x. [DOI] [Google Scholar]
- Grant WD. Cultivation of aerobic alkaliphiles. Method Microbiol. 2006;35:439–449. [Google Scholar]
- DeRosa M, Gambacorta A, Grant WD, Lanzotti V, Nicolaus B. Polar lipids and glycine betaine from haloalkaliphilic archaebacteria. J Gen Microbiol. 1988;134:205–211. doi: 10.1099/00221287-134-12-3159. [DOI] [PubMed] [Google Scholar]
- Kamekura M, Dyall-Smith ML, Upasani V, Ventosa A, Kates M. Diversity of alkaliphilic halobacteria: proposals for transfer ofNatronobacterium vacuolatum,Natronobacterium magadii, andNatronobacterium pharaonistoHalorubrum,Natrialba, andNatronomonasgen. nov., respectively, asHalorubrum vacuolatumcomb. nov.,Natrialba magadiicomb. nov., andNatronomonas pharaoniscomb. nov., respectively. Int J Syst Bacteriol. 1997;47:853–857. doi: 10.1099/00207713-47-3-853. [DOI] [PubMed] [Google Scholar]
- Kamekura MD-S, Dyallsmith ML. Taxonomy of the familyHalobacteriaceaeand the description of two new generaHalorubrobacteriumandNatrialba. J Gen Microbiol. 1995;41:333–350. doi: 10.2323/jgam.41.333. [DOI] [Google Scholar]
- Tindall BJ, Ross HNM, Grant WD. Natronobacteriumgen. nov. andNatronococcusgen. nov., two new genera of haloalkaliphilic archaebacteria. Syst Appl Microbiol. 1984;5:41–57. doi: 10.1016/S0723-2020(84)80050-8. [DOI] [Google Scholar]
- Oren A. In: The Prokaryotes. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editor. Springer, New York; 2006. The OrderHalobacteriales; pp. 113–164. [Google Scholar]
- Kamekura M. In: Microbiology and Biogeochemistry of Hypersaline Environments. Oren A, editor. CRC Press, Boca Raton, FL; 1999. Diversity of members of the familyHalobacteriaceae; pp. 13–26. [Google Scholar]
- Desmarais D, Jablonski PE, Fedarko NS, Roberts MF. 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J Bacteriol. 1997;179:3146–3153. doi: 10.1128/jb.179.10.3146-3153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polosina Y, Jarrell KF, Fedorov OV, Kostyukova AS. Nucleoside diphosphate kinase from haloalkaliphilic archaeonNatronobacterium magadii: purification and characterization. Extremophiles. 1998;2:333–338. doi: 10.1007/s007920050076. [DOI] [PubMed] [Google Scholar]
- Giménez MI, Studdert CA, Sánchez JJ, De Castro RE. Extracellular protease ofNatrialba magadii: purification and biochemical characterization. Extremophiles. 2000;4:181–188. doi: 10.1007/s007920070033. [DOI] [PubMed] [Google Scholar]
- Serganova I, Ksenzenko V, Serganov A, Meshcheryakova I, Pyatibratov M, Vakhrusheva O, Metlina A, Fedorov O. Sequencing of flagellin genes fromNatrialba magadiiprovides new insight into evolutionary aspects of archaeal flagellins. J Bacteriol. 2002;184:318–322. doi: 10.1128/JB.184.1.318-322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh R, Ngata S, Ozawa A, Ohshima T, Kamekura M, Misono H. Purification and characterization of leucine dehydrogenase from an alkaliphilic halophile,Natronobacterium magadiiMS-3. J Mol Catal B: Enzym. 2003;23:231–238. doi: 10.1016/S1381-1177(03)00085-7. [DOI] [Google Scholar]
- Kamekura M. Diversity of extremely halophilic bacteria. Extremophiles. 1998;2:289–295. doi: 10.1007/s007920050071. [DOI] [PubMed] [Google Scholar]
- Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YA, Roach J, Glusman G, Baliga NS, Deutsch K, Pan M, Kennedy S, DasSarma S, Ng WV, Hood L. Low-pass sequencing for microbial comparative genomics. BMC Genomics. 2004;5:3. doi: 10.1186/1471-2164-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders E, Tindall BJ, Fahnrich R, Lapidus A, Copeland A, Del Rio TG, Lucas S, Chen F, Tice H, Cheng JF. et al. Complete genome sequence ofHaloterrigena turkmenicatype strain (4 k) Stand Genomic Sci. 2010;2:107–116. doi: 10.4056/sigs.681272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D. Living with two extremes: conclusions from the genome sequence ofNatronomonas pharaonis. Genome Res. 2005;15:1336–1343. doi: 10.1101/gr.3952905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falb M, Muller K, Konigsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzalez O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D. Metabolism of halophilic archaea. Extremophiles. 2008;12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Yang C-F, Halladay JT, Arora P, DasSarma S. In: Archaea, a Laboratory Manual: Halophiles. DasSarma S, Fleischmann EM, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1995. Isolation of genomic and plasmid DNAs fromHalobacterium halobium; pp. 179–184. [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Treangen TJ, Messeguer X, Perna NT. Analyzing patterns of microbial evolution using the mauve genome alignment system. Methods Mol Biol. 2007;396:135–152. doi: 10.1007/978-1-59745-515-2_10. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte A, Baranyi U, Klein R, Sulzner M, Luo C, Wanner G, Kruger DH, Lubitz W. Characterization ofNatronobacterium magadiiphage φCh1, a unique archaeal phage containing DNA and RNA. Mol Microbiol. 1997;23:603–616. doi: 10.1046/j.1365-2958.1997.d01-1879.x. [DOI] [PubMed] [Google Scholar]
- Klein R, Greineder B, Baranyi U, Witte A. The structural protein E of the archaeal virus φCh1: evidence for processing inNatrialba magadiiduring virus maturation. Virology. 2000;276:376–387. doi: 10.1006/viro.2000.0565. [DOI] [PubMed] [Google Scholar]
- Klein R, Baranyi U, Rossler N, Greineder B, Scholz H, Witte A. Natrialba magadiivirus φCh1: first complete nucleotide sequence and functional organization of a virus infecting a haloalkaliphilic archaeon. Mol Microbiol. 2002;45:851–863. doi: 10.1046/j.1365-2958.2002.03064.x. [DOI] [PubMed] [Google Scholar]
- Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. Genetic and physical mapping of DNA replication origins inHaloferax volcanii. PLoS Genet. 2007;3:e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filee J, Siguier P, Chandler M. Insertion sequence diversity in archaea. Microbiol Mol Biol Rev. 2007;71:121–157. doi: 10.1128/MMBR.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H. In: The bacteriophages. Calendar R, editor. Oxford University Press, New York; 2006. Prophage genomics; pp. 17–25. [Google Scholar]
- Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One. 2011;6:e20968. doi: 10.1371/journal.pone.0020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug G, Evguenieva-Hackenberg E, Omer AD, Dennis PP, Marchfelder A. In: Archaea: Molecular and Cellular Biology. Cavicchioli R, editor. ASM Press, Washington, DC; 2007. RNA processing; pp. 158–174. [Google Scholar]
- Boucher Y, Douady CJ, Sharma AK, Kamekura M, Doolittle WF. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J Bacteriol. 2004;186:3980–3990. doi: 10.1128/JB.186.12.3980-3990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich TA. In: The Prokaryotes. 3. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editor. Springer, New York; 2006. Alkaliphilic prokaryotes; pp. 283–308. [Google Scholar]
- Konstantinidis K, Tebbe A, Klein C, Scheffer B, Aivaliotis M, Bisle B, Falb M, Pfeiffer F, Siedler F, Oesterhelt D. Genome-wide proteomics ofNatronomonas pharaonis. J Proteome Res. 2007;6:185–193. doi: 10.1021/pr060352q. [DOI] [PubMed] [Google Scholar]
- Paul S, Bag SK, Das S, Harvill ET, Dutta C. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008;9:R70. doi: 10.1186/gb-2008-9-4-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. In: Halophilic Microorganisms and their Environments. Oren A, editor. Kluwer Academic, Dordrecht, The Netherlands; 2002. Intracellular salt concentrations and ion metabolism in halophilic microorganisms; pp. 207–232. [Google Scholar]
- Roberts MF. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 2005;1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DD, Ciulla RA, Roberts MF. Osmoadaptation in archaea. Appl Environ Microbiol. 1999;65:1815–1825. doi: 10.1128/aem.65.5.1815-1825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Liu J, Ma Y, Krulwich TA. Three putative cation/proton antiporters from the soda lake alkaliphileAlkalimonas amylolyticaN10 complement an alkali-sensitiveEscherichia colimutant. Microbiology. 2007;153:2168–2179. doi: 10.1099/mic.0.2007/007450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz M. Compatible solute influence on nucleic acids: many questions but few answers. Saline Syst. 2008;4:6. doi: 10.1186/1746-1448-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood JE, Stagnitti F, Kokkinn MJ, Williams WD. Dissolved oxygen concentrations in hypersaline waters. Limnol Oceanogr. 1991;36:235–250. doi: 10.4319/lo.1991.36.2.0235. [DOI] [Google Scholar]
- Oren A. The ecology of the extremely halophilic archaea. FEMS Microbiol Rev. 1994;13:415–439. doi: 10.1111/j.1574-6976.1994.tb00060.x. [DOI] [PubMed] [Google Scholar]
- Ochsenreiter T, Pfeifer F, Schleper C. Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles. 2002;6:267–274. doi: 10.1007/s00792-001-0253-4. [DOI] [PubMed] [Google Scholar]
- Siezen RJ, Leunissen JA. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro RE, Ruiz DM, Giménez MI, Silveyra MX, Paggi RA, Maupin-Furlow JA. Gene cloning and heterologous synthesis of a haloalkaliphilic extracellular protease ofNatrialba magadii(Nep) Extremophiles. 2008;12:677–687. doi: 10.1007/s00792-008-0174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, Riddles P, Jones GJ, Smith W, Blakeley RL. Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ Toxicol. 2001;16:523–534. doi: 10.1002/tox.10013. [DOI] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Humbard MA, Kirkland PA, Li W, Reuter CJ, Wright AJ, Zhou G. Proteasomes from structure to function: perspectives from Archaea. Curr Top Dev Biol. 2006;75:125–169. doi: 10.1016/S0070-2153(06)75005-0. [DOI] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. Ubiquitin-like small archaeal modifier proteins (SAMPs) inHaloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Soll D, Maupin-Furlow JA. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci U S A. 2011;108:4417–4422. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre DE, Paggi RA, De Castro RE. The Lon protease from the haloalkaliphilic archaeonNatrialba magadiiis transcriptionally linked to a cluster of putative membrane proteases and displays DNA-binding activity. Microbiol Res. 2011;166:304–313. doi: 10.1016/j.micres.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Franzetti B, Schoehn G, Hernandez JF, Jaquinod M, Ruigrok RW, Zaccai G. Tetrahedral aminopeptidase: a novel large protease complex from archaea. EMBO J. 2002;21:2132–2138. doi: 10.1093/emboj/21.9.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konaklieva MI, Plotkin BJ. The relationship between inhibitors of eukaryotic and prokaryotic serine proteases. Mini Rev Med Chem. 2004;4:721–739. [PubMed] [Google Scholar]
- Giménez MI, Sánchez JJ, De Castro RE. Detection of an intracellular protease inhibitor in archaea. Curr Microbiol. 2003;46:334–339. doi: 10.1007/s00284-002-3861-z. [DOI] [PubMed] [Google Scholar]
- Bolhuis A. The archaeal Sec-dependent protein translocation pathway. Philos Trans R Soc Lond B Biol Sci. 2004;359:919–927. doi: 10.1098/rstb.2003.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschröder M, Dilks KC. In: Archaea: Molecular and Cellular Biology. Cavicchioli R, editor. ASM Press, Washington, DC; 2007. Protein translocation into and across archaeal cytoplasmic membranes; pp. 369–384. [Google Scholar]
- Ellen AF, Zolghadr B, Driessen AM, Albers SV. Shaping the archaeal cell envelope. Archaea. 2010;2010:608243. doi: 10.1155/2010/608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storf S, Pfeiffer F, Dilks K, Chen ZQ, Imam S, Pohlschröder M. Mutational and bioinformatic analysis of haloarchaeal lipobox-containing proteins. Archaea. 2010;2010:410975. doi: 10.1155/2010/410975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Payne SH, Selengut JD. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol. 2012;194:36–48. doi: 10.1128/JB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426:239–257. doi: 10.1016/S0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Magidovich H, Eichler J. Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol Lett. 2009;300:122–130. doi: 10.1111/j.1574-6968.2009.01775.x. [DOI] [PubMed] [Google Scholar]
- Oren A. In: Halophilic Microorganisms and their Environments. Oren A, editor. Kluwer Academic Publishers, Dordrecht, The Netherlands; 2002. Biotechnological applications and potentials of halophilic microorganisms; pp. 357–390. [Google Scholar]
- Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, Howard MJ, Ferguson SJ, Warren MJ. Molecular hijacking of siroheme for the synthesis of heme andd1 heme. Proc Natl Acad Sci U S A. 2011;108:18260–18265. doi: 10.1073/pnas.1108228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- Begley TP, Downs DM, Ealick SE, McLafferty FW, Van Loon AP, Taylor S, Campobasso N, Chiu HJ, Kinsland C, Reddick JJ, Xi J. Thiamin biosynthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science. 2007;315:1003–1006. doi: 10.1126/science.1135999. [DOI] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Bacher A. Biosynthesis of vitamin B2: structure and mechanism of riboflavin synthase. Arch Biochem Biophys. 2008;474:252–265. doi: 10.1016/j.abb.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Fischer M, Bacher A. Biosynthesis of vitamin B2: a unique way to assemble a xylene ring. Chembiochem. 2011;12:670–680. doi: 10.1002/cbic.201000681. [DOI] [PubMed] [Google Scholar]
- Graham DE, White RH. Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat Prod Rep. 2002;19:133–147. doi: 10.1039/b103714p. [DOI] [PubMed] [Google Scholar]
- White RH. Biosynthesis of the methanogenic cofactors. Vitam Horm. 2001;61:299–337. doi: 10.1016/s0083-6729(01)61010-0. [DOI] [PubMed] [Google Scholar]
- Graham DE. A new role for coenzyme F420in aflatoxin reduction by soil mycobacteria. Mol Microbiol. 2010;78:533–536. doi: 10.1111/j.1365-2958.2010.07358.x. [DOI] [PubMed] [Google Scholar]
- Taylor MC, Jackson CJ, Tattersall DB, French N, Peat TS, Newman J, Briggs LJ, Lapalikar GV, Campbell PM, Scott C. et al. Identification and characterization of two families of F420H2-dependent reductases fromMycobacteriathat catalyse aflatoxin degradation. Mol Microbiol. 2010;78:561–575. doi: 10.1111/j.1365-2958.2010.07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XL, White RH. Occurrence of coenzyme F420and its γ-monoglutamyl derivative in nonmethanogenic archaebacteria. J Bacteriol. 1986;168:444–448. doi: 10.1128/jb.168.1.444-448.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selengut JD, Haft DH. Unexpected abundance of coenzyme F420-dependent enzymes inMycobacterium tuberculosisand other actinobacteria. J Bacteriol. 2010;192:5788–5798. doi: 10.1128/JB.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selengut JD, Rusch DB, Haft DH. Sites Inferred by Metabolic Background Assertion Labeling (SIMBAL): adapting the Partial Phylogenetic Profiling algorithm to scan sequences for signatures that predict protein function. BMC Bioinformatics. 2010;11:52. doi: 10.1186/1471-2105-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JM, Nichols BP, Matthews RG. In: Escherichia coli and Salmonella: cellular and molecular biology. 2. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley MA, Schaechter M, Umbarger HE, editor. ASM Press, Washington, D.C; 1996. Folate biosynthesis, reduction, and polyglutamylation; pp. 665–673. [Google Scholar]
- Lucock M. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol Genet Metab. 2000;71:121–138. doi: 10.1006/mgme.2000.3027. [DOI] [PubMed] [Google Scholar]
- de Crécy-Lagard V, El Yacoubi B, de la Garza RD, Noiriel A, Hanson AD. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics. 2007;8:245. doi: 10.1186/1471-2164-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascelles J. Synthesis of tetrapyrroles by microorganisms. Physiol Rev. 1961;41:417–441. doi: 10.1152/physrev.1961.41.2.417. [DOI] [PubMed] [Google Scholar]
- Warren MJ, Smith AG, Deery E, Rose R-S. In: Tetrapyrroles: Birth, Life and Death. Warren MJ, Smith AG, editor. Springer, New York; 2009. Biosynthesis of siroheme and coenzyme F430; pp. 344–351. [Google Scholar]
- Zappa S, Li K, Bauer CE. The tetrapyrrole biosynthetic pathway and its regulation inRhodobacter capsulatus. Adv Exp Med Biol. 2010;675:229–250. doi: 10.1007/978-1-4419-1528-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio-Hall LA, Escalante-Semerena JC. In vitrosynthesis of the nucleotide loop of cobalamin bySalmonella typhimuriumenzymes. Proc Natl Acad Sci U S A. 1999;96:11798–11803. doi: 10.1073/pnas.96.21.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Peck RF, Krebs MP, Escalante-Semerena JC. ThecobYgene of the archaeonHalobacteriumsp. strain NRC-1 is required for de novo cobamide synthesis. J Bacteriol. 2003;185:311–316. doi: 10.1128/JB.185.1.311-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Escalante-Semerena JC. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12precursor cobinamide in archaea. Proc Natl Acad Sci U S A. 2004;101:3591–3596. doi: 10.1073/pnas.0305939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara Y, Santander PJ, Roessner CA, Perez LM, Scott AI. Genetically engineered synthesis and structural characterization of cobalt-precorrin 5A and -5B, two new intermediates on the anaerobic pathway to vitamin B12: definition of the roles of the CbiF and CbiG enzymes. J Am Chem Soc. 2006;128:9971–9978. doi: 10.1021/ja062940a. [DOI] [PubMed] [Google Scholar]
- Raux E, Leech HK, Beck R, Schubert HL, Santander PJ, Roessner CA, Scott AI, Martens JH, Jahn D, Thermes C. et al. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin inBacillus megaterium. Biochem J. 2003;370:505–516. doi: 10.1042/BJ20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Xu LX, Cherney MM, Raux-Deery E, Bindley AA, Savchenko A, Walker JR, Cuff ME, Warren MJ, James MN. Crystal structure of the vitamin B12biosynthetic cobaltochelatase, CbiXS, fromArchaeoglobus fulgidus. J Struct Funct Genomics. 2006;7:37–50. doi: 10.1007/s10969-006-9008-x. [DOI] [PubMed] [Google Scholar]
- Moss J, Lane MD. The biotin-dependent enzymes. Adv Enzymol Relat Areas Mol Biol. 1971;35:321–442. doi: 10.1002/9780470122808.ch7. [DOI] [PubMed] [Google Scholar]
- Schneider G, Lindqvist Y. Structural enzymology of biotin biosynthesis. FEBS Lett. 2001;495:7–11. doi: 10.1016/S0014-5793(01)02325-0. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D, Dyall-Smith ML. Cultivation of haloarchaea. Methods Microbiol. 2006;35:535–552. [Google Scholar]
- Kisker C, Schindelin H, Rees DC. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu Rev Plant Biol. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- Magalon A, Frixon C, Pommier J, Giordano G, Blasco F. In vivointeractions between gene products involved in the final stages of molybdenum cofactor biosynthesis inEscherichia coli. J Biol Chem. 2002;277:48199–48204. doi: 10.1074/jbc.M205806200. [DOI] [PubMed] [Google Scholar]
- Khomyakova M, Bukmez O, Thomas LK, Erb TJ, Berg IA. A methylaspartate cycle in haloarchaea. Science. 2011;331:334–337. doi: 10.1126/science.1196544. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, Pfeiffer F, Oesterhelt D. The genome of the square archaeonHaloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 2006;7:169. doi: 10.1186/1471-2164-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Galperin MY, Makarova KS, Wolf YI, Koonin EV. Evolutionary primacy of sodium bioenergetics. Biol Direct. 2008;3:13. doi: 10.1186/1745-6150-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert K, Wagner V, Kroth-Pancic PG, Bickel-Sandkotter S. Characterization and subunit structure of the ATP synthase of the halophilic archaeonHaloferax volcaniiand organization of the ATP synthase genes. J Biol Chem. 1997;272:6261–6269. doi: 10.1074/jbc.272.10.6261. [DOI] [PubMed] [Google Scholar]
- Mukohata Y, Yoshida M. The H+-translocating ATP synthase inHalobacterium halobiumdiffers from F0F1-ATPase/synthase. J Biochem. 1987;102:797–802. doi: 10.1093/oxfordjournals.jbchem.a122118. [DOI] [PubMed] [Google Scholar]
- Scharf B, Wittenberg R, Engelhard M. Electron transfer proteins from the haloalkaliphilic archaeonNatronobacterium pharaonis: possible components of the respiratory chain include cytochromebcand a terminal oxidase cytochromeba3. Biochemistry. 1997;36:4471–4479. doi: 10.1021/bi962312d. [DOI] [PubMed] [Google Scholar]
- Gomes CM, Bandeiras TM, Teixeira M. A new type-II NADH dehydrogenase from the archaeonAcidianus ambivalens: characterization and in vitro reconstitution of the respiratory chain. J Bioenerg Biomembr. 2001;33:1–8. doi: 10.1023/a:1005630221892. [DOI] [PubMed] [Google Scholar]
- Sreeramulu K, Schmidt CL, Schafer G, Anemuller S. Studies of the electron transport chain of the euryarcheonHalobacterium salinarum: indications for a type II NADH dehydrogenase and a complex III analog. J Bioenerg Biomembr. 1998;30:443–453. doi: 10.1023/A:1020538129400. [DOI] [PubMed] [Google Scholar]
- DiSpirito AA, Kunz RC, Choi D-W, Zahn JA. In: Respiration in Archaea and Bacteria: Diversity of Prokaryotic Respiratory Systems. Zannoni D, editor. Springer, Dordrecht, The Netherlands; 2004. Respiration in methanotrophs; pp. 149–168. [Google Scholar]
- Mattar S, Engelhard M. Cytochromeba3fromNatronobacterium pharaonis-an archaeal four-subunit cytochrome-c-type oxidase. Eur J Biochem. 1997;250:332–341. doi: 10.1111/j.1432-1033.1997.0332a.x. [DOI] [PubMed] [Google Scholar]
- Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D. Evolution in the laboratory: the genome ofHalobacterium salinarumstrain R1 compared to that of strain NRC-1. Genomics. 2008;91:335–346. doi: 10.1016/j.ygeno.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Xu XM, Moller SG. Iron-sulfur clusters: biogenesis, molecular mechanisms, and their functional significance. Antioxid Redox Signal. 2011;15:271–307. doi: 10.1089/ars.2010.3259. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Engelhard M, Schmies G, Wegener AA. In: Photoreceptors and Light Signalling. Batschauer A, editor. The Royal Society of Chemistry, Cambridge, U.K; 2003. Archeabacterial phototaxis; pp. 1–39. [Google Scholar]
- Peck RF, Echavarri-Erasun C, Johnson EA, Ng WV, Kennedy SP, Hood L, DasSarma S, Krebs MP. brpandblhare required for synthesis of the retinal cofactor of bacteriorhodopsin inHalobacterium salinarum. J Biol Chem. 2001;276:5739–5744. doi: 10.1074/jbc.M009492200. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MK, Staudinger WF, Siedler F, Oesterhelt D. Physiological sites of deamidation and methyl esterification in sensory transducers ofHalobacterium salinarum. J Mol Biol. 2008;380:285–302. doi: 10.1016/j.jmb.2008.04.063. [DOI] [PubMed] [Google Scholar]
- Schlesner M, Miller A, Streif S, Staudinger WF, Muller J, Scheffer B, Siedler F, Oesterhelt D. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 2009;9:56. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NA, Bardy SL, Jarrell KF. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol Rev. 2001;25:147–174. doi: 10.1111/j.1574-6976.2001.tb00575.x. [DOI] [PubMed] [Google Scholar]
- Streif S, Staudinger WF, Marwan W, Oesterhelt D. Flagellar rotation in the archaeonHalobacterium salinarumdepends on ATP. J Mol Biol. 2008;384:1–8. doi: 10.1016/j.jmb.2008.08.057. [DOI] [PubMed] [Google Scholar]
- Fedorov OV, Pyatibratov MG, Kostyukova AS, Osina NK, Tarasov VY. Protofilament as a structural element of flagella of haloalkalophilic archaebacteria. Can J Microbiol. 1994;40:45–53. doi: 10.1139/m94-007. [DOI] [Google Scholar]
- Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D. Theflagene cluster is involved in the biogenesis of flagella inHalobacterium salinarum. Mol Microbiol. 2001;41:653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa S, Jarrell KF. Systematic deletion analyses of theflagenes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon,Methanococcus maripaludis. Mol Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Hartung S, van der Does C, Tainer JA, Albers SV. Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem J. 2011;437:43–52. doi: 10.1042/BJ20110410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatibratov MG, Leonard K, Tarasov VY, Fedorov OV. Two immunologically distinct types of protofilaments can be identified inNatrialba magadiiflagella. FEMS Microbiol Lett. 2002;212:23–27. doi: 10.1111/j.1574-6968.2002.tb11239.x. [DOI] [PubMed] [Google Scholar]
- Selb RM. Construction of mutants ofNatrialba magadiiand φCh1. PhD thesis. Universität Wien, Molecular Biology Department, ; 2010. [Google Scholar]
- Reiter M. Gene regulation of φCh1: characterization of ORF49 and further characterization of the origin of replication of the halophage φCh1. PhD thesis. Universität Wien, Molecular Biology Department, ; 2010. [Google Scholar]
- Berquist BR, Soneja Ja, DasSarma S. In: Adaptation to Life at High Salt Concentrations in Archaea, Bacteria, and Eukarya. Gunde-Cimerman N, Oren A, Plemenitaš A, editor. Springer, Dordrecht, The Netherlands; 2005. Comparative genomic survey of information transfer systems in two diverse extremely halophilic Archaea,Halobacteriumsp. strain NRC-1 andHaloarcula marismortui; pp. 148–182. [Google Scholar]
- Thomm M. In: Archaea: Molecular and Cellular Biology. Cavicchioli R, editor. ASM Press, Washington, DC; 2007. Transcription: mechanism and regulation; pp. 139–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.Natrialba magadii ATCC 43099 genes discussed in the text. This table lists Nab. magadii ATCC 43099 genes related to bacteriophage and recombination elements, rRNA genes, and genes encoding adaptive features.
Table S2. Bidirectional best blast pairs among proteins fromNatrialba magadii and 17 other haloarchaeal genomes. This table lists the number of bidirectional best blast pairs among proteins from Nab. magadii and 17 other halophilic archaea. The first column is the number of total proteins in each genome, the second column is the number of bidirectional best blast pairs, and the third column is the percentage of proteins having a bidirectional best blast hit in Nab. magadii.
Table S3.Natrialba magadii ATCC 43099 genes encoding putative peptidases/proteases, protease inhibitors, and regulatory proteins. This table lists Nab. magadii ATCC 43099 genes encoding various types of proteases and peptidases as well as protease inhibitors and regulatory proteins.
Table S4. Natrialba magadii ATCC 43099 genes involved in metabolism. This table lists Nab. magadii ATCC 43099 genes encoding molybdenum cofactor biosynthesis and other metabolic functions.