Abstract
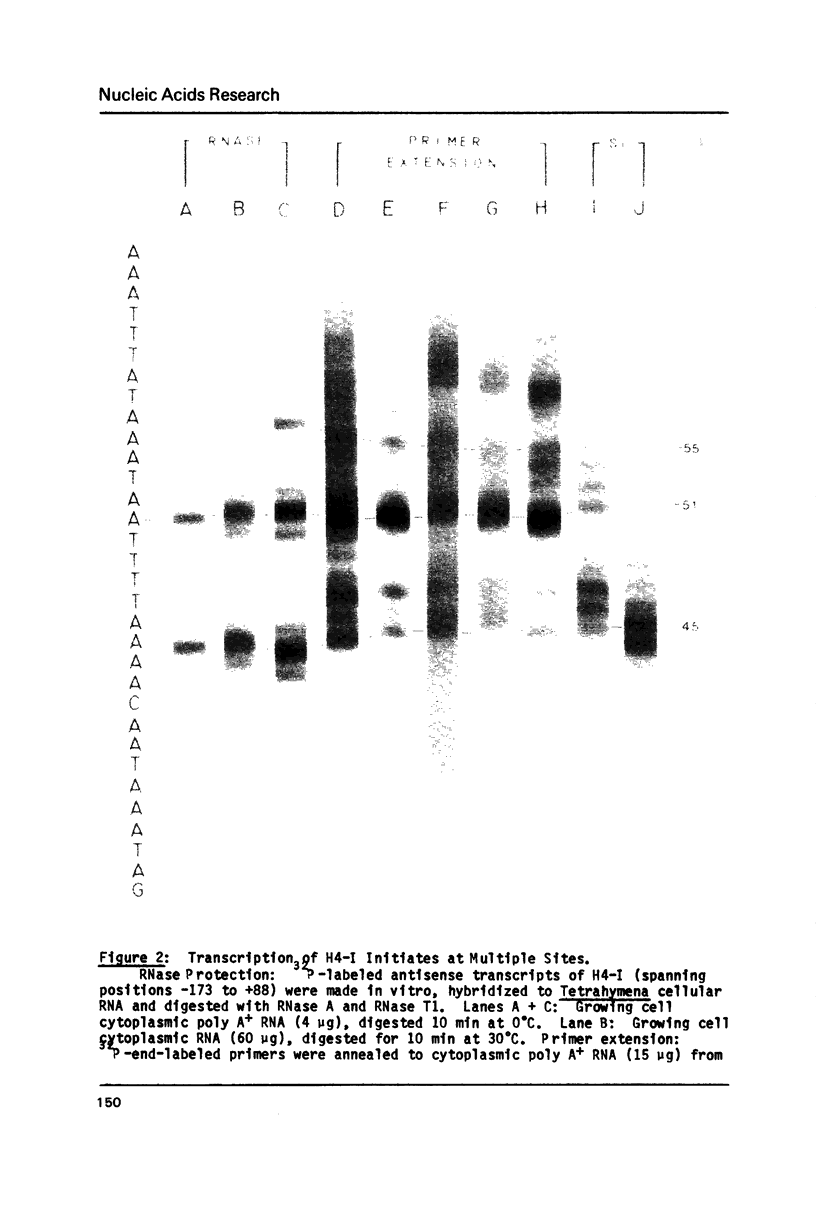
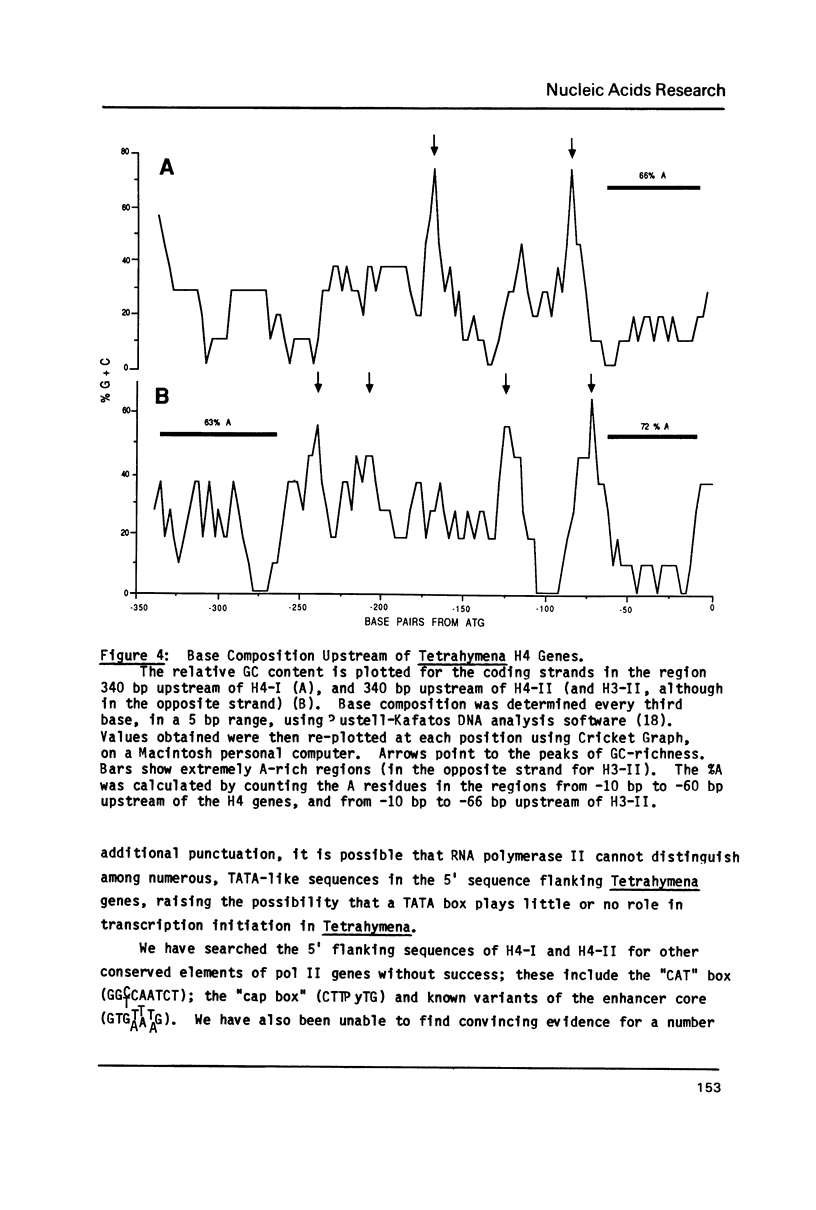
The complete DNA sequence is presented of H4-II, the second of the pair of histone H4 genes of the ciliated protozoan, Tetrahymena thermophila. Both H4 genes code for the same protein. Codon usage in these and other Tetrahymena genes is severely restricted and is similar to that in yeast. Flanking regions are AT-rich (greater than or equal to 75%), relative to coding sequences (approximately 45% GC). Except for small, similarly positioned homologies, flanking sequences of the two genes are different. Canonical sequences in higher eukaryotic promoters are not obvious in these genes. Instead, short, localized, base composition eccentricities characterize the 5' flanking sequences of all Tetrahymena genes analyzed. The consensus, P yP u(A)3-4 ATGG initiates translation in these and all other known Tetrahymena genes. Nuclear transcripts and messages of both growing and starved cells begin at multiple sites, mainly at the first or second A residue following a pyrimidine. The palindrome typical of histone message 3' termini in higher organisms is not present. Downstream of both genes are sequences similar to the processing/polyadenylation signal of higher eukaryotes, although the unique 3' ends are not those predicted by the location of the signals.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannon G. A., Bowen J. K., Yao M. C., Gorovsky M. A. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. doi: 10.1093/nar/12.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon G. A., Calzone F. J., Bowen J. K., Allis C. D., Gorovsky M. A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983 Jun 25;11(12):3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barklis E., Pontius B., Barfield K., Lodish H. F. Structure of the promoter of the Dictyostelium discoideum prespore EB4 gene. Mol Cell Biol. 1985 Jun;5(6):1465–1472. doi: 10.1128/mcb.5.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Dray J. F. Cloned Drosophila alcohol dehydrogenase genes are correctly expressed after transfection into Drosophila cells in culture. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1701–1705. doi: 10.1073/pnas.81.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Calzone F. J., Stathopoulos V. A., Grass D., Gorovsky M. A., Angerer R. C. Regulation of protein synthesis in Tetrahymena. RNA sequence sets of growing and starved cells. J Biol Chem. 1983 Jun 10;258(11):6899–6905. [PubMed] [Google Scholar]
- Caron F., Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985 Mar 14;314(6007):185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- Clerc R. G., Bucher P., Strub K., Birnstiel M. L. Transcription of a cloned Xenopus laevis H4 histone gene in the homologous frog oocyte system depends on an evolutionary conserved sequence motif in the -50 region. Nucleic Acids Res. 1983 Dec 20;11(24):8641–8657. doi: 10.1093/nar/11.24.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J., Din N., Saiga H., Higashinakagawa T. Nucleotide sequence of the 5'-terminal coding region for pre-rRNA and mature 17S rRNA in Tetrahymena thermophila rDNA. Nucleic Acids Res. 1984 Jan 25;12(2):959–972. doi: 10.1093/nar/12.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusauchi Y., Iwai K. Tetrahymena histone H2A. Acetylation in the N-terminal sequence and phosphorylation in the C-terminal sequence. J Biochem. 1984 Jan;95(1):147–154. doi: 10.1093/oxfordjournals.jbchem.a134578. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Piatak M., Lebowitz P., Weissman S. M. Determination of RNA sequences by primer directed synthesis and sequencing of their cDNA transcripts. Methods Enzymol. 1980;65(1):580–595. doi: 10.1016/s0076-6879(80)65061-7. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Nomoto M., Iwai K. Tetrahymena histone H4. Complete amino acid sequences of two variants. J Biochem. 1984 Nov;96(5):1449–1456. [PubMed] [Google Scholar]
- Hayashi T., Hayashi H., Fusauchi Y., Iwai K. Tetrahymena histone H3. Purification and two variant sequences. J Biochem. 1984 Jun;95(6):1741–1749. doi: 10.1093/oxfordjournals.jbchem.a134788. [DOI] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985 Jan 25;13(2):415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Kaine B. P., Spear B. B. Nucleotide sequence of a macronuclear gene for actin in Oxytricha fallax. Nature. 1982 Feb 4;295(5848):430–432. doi: 10.1038/295430a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu N., Tashiro F., Nishimura S. Tetrahymena thermophila glutamine tRNA and its gene that corresponds to UAA termination codon. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4758–4762. doi: 10.1073/pnas.82.14.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Mita T., Nishimura S. Nucleotide sequence of cytoplasmic initiator tRNA from Tetrahymena thermophila. Nucleic Acids Res. 1981 Sep 25;9(18):4557–4562. doi: 10.1093/nar/9.18.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S., Mita T., Higashinakagawa T. The nucleotide sequences of 5S rRNAs from three ciliated protozoa. Nucleic Acids Res. 1982 Jul 24;10(14):4409–4412. doi: 10.1093/nar/10.14.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhausen M., Nelson R. G., Sather S., Selkirk M., Agabian N. Identification of a small RNA containing the trypanosome spliced leader: a donor of shared 5' sequences of trypanosomatid mRNAs? Cell. 1984 Oct;38(3):721–729. doi: 10.1016/0092-8674(84)90267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto M., Hayashi H., Iwai K. Tetrahymena histone H2B. Complete amino acid sequence. J Biochem. 1982 Mar;91(3):897–904. doi: 10.1093/oxfordjournals.jbchem.a133778. [DOI] [PubMed] [Google Scholar]
- Nussinov R. RNA folding is unaffected by the nonrandom degenerate codon choice. Biochim Biophys Acta. 1982 Aug 30;698(2):111–115. doi: 10.1016/0167-4781(82)90125-7. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of DNA sequence analysis programs for microcomputers. Nucleic Acids Res. 1982 Jan 11;10(1):51–59. doi: 10.1093/nar/10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Miura K., Nakaso A., Kimura G. Nucleotide sequence around the replication origin of polyoma virus DNA. FEBS Lett. 1977 Jul 15;79(2):383–389. doi: 10.1016/0014-5793(77)80826-0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]






