Abstract
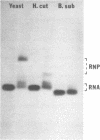
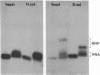
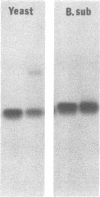
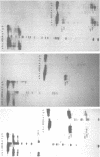
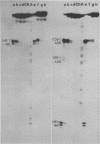
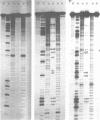
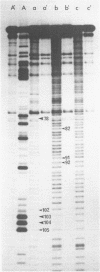
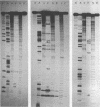
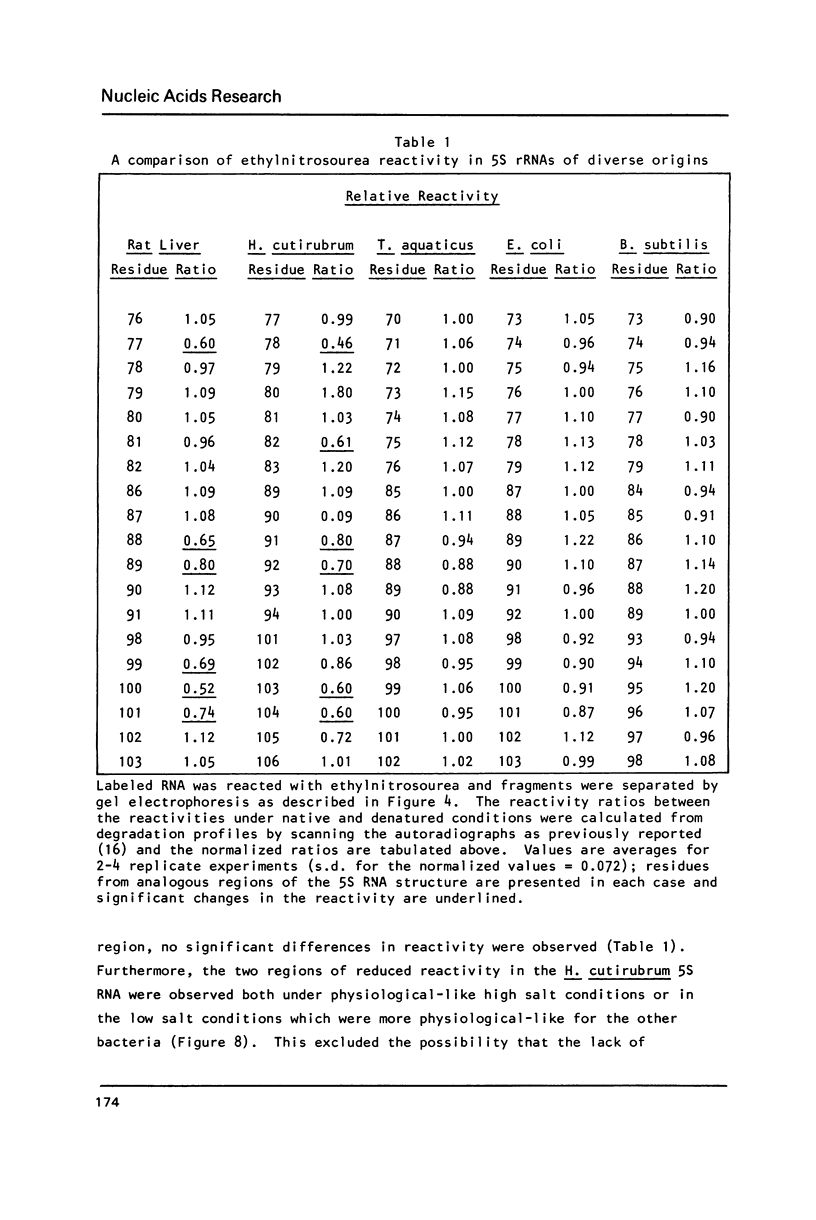
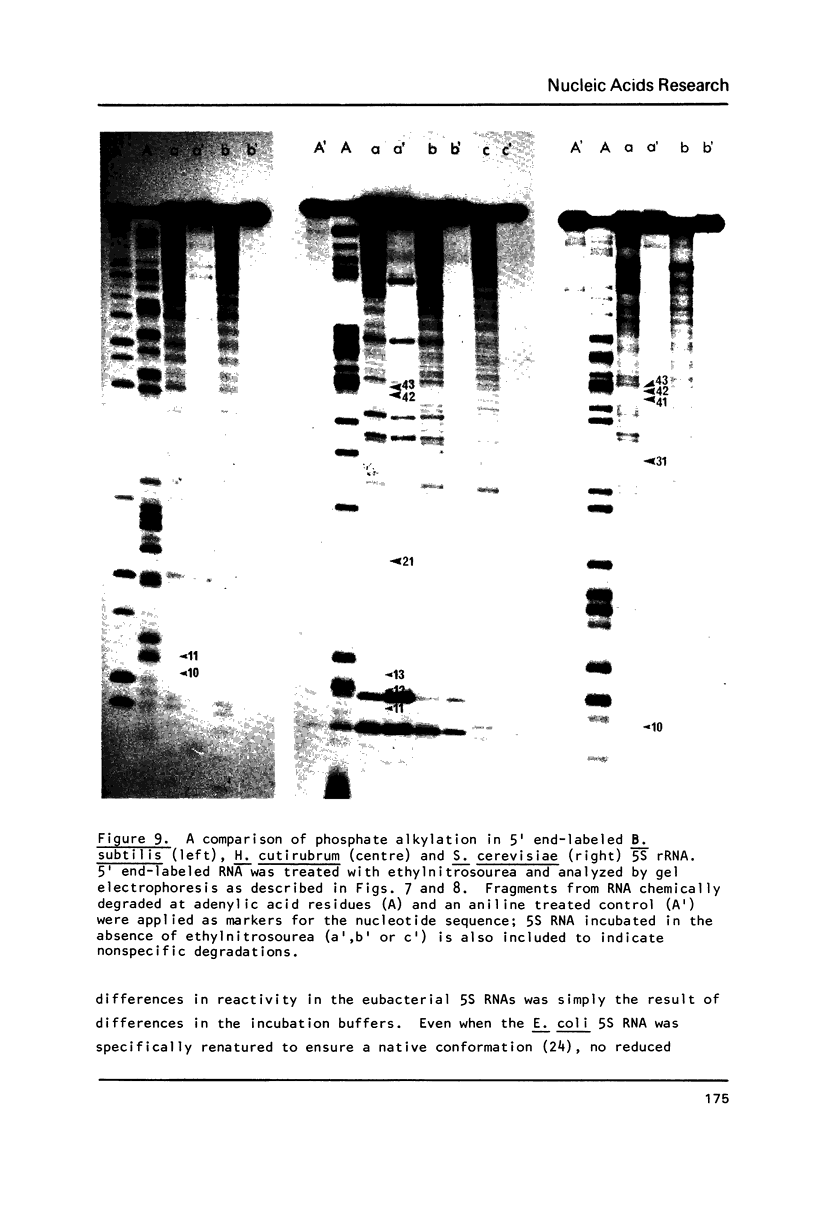
Comparative studies have been undertaken on the higher order structure of ribosomal 5S RNAs from diverse origins. Competitive reassociation studies show that 5S RNA from either a eukaryote or archaebacterium will form a stable ribonucleoprotein complex with the yeast ribosomal 5S RNA binding protein (YL3); in contrast, eubacterial RNAs will not compete in a similar fashion. Partial S1 ribonuclease digestion and ethylnitrosourea reactivity were used to probe the structural differences suggested by the reconstitution experiments. The results indicate a more compact higher order structure in eukaryotic 5S RNAs as compared to eubacteria and suggest that the archaebacterial 5S RNA contains features which are common to either group. The potential significance of these results with respect to a generalized model for the tertiary structure of the ribosomal 5S RNA and to the heterogeneity in the protein components of 5S RNA-protein complexes are discussed.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Bellemare G., Monier R. Selective reaction of glyoxal with guanine residues in native and denatured Escherichia coli 5S RNA. Biochimie. 1973;55(2):135–142. doi: 10.1016/s0300-9084(73)80385-2. [DOI] [PubMed] [Google Scholar]
- Blobel G. Isolation of a 5S RNA-protein complex from mammalian ribosomes. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1881–1885. doi: 10.1073/pnas.68.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. S., Bonen L., Doolittle W. F., Gray M. W. Unique species of 5S, 18S, and 26S ribosomal RNA in wheat mitochondria. FEBS Lett. 1976 Oct 15;69(1):116–122. doi: 10.1016/0014-5793(76)80666-7. [DOI] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Equilibria in 5-S ribosomal RNA secondary structure. Bulges and interior loops in 5-S RNA secondary structure may serve as articulations for a flexible molecule. Eur J Biochem. 1984 Aug 15;143(1):175–182. doi: 10.1111/j.1432-1033.1984.tb08356.x. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Garrett R. A. Secondary structure of prokaryotic 5S ribosomal ribonucleic acids: a study with ribonucleases. Biochemistry. 1981 Dec 8;20(25):7301–7307. doi: 10.1021/bi00528a039. [DOI] [PubMed] [Google Scholar]
- Dyer T. A., Bowman C. M. Nucleotide sequences of chloroplast 5S ribosomal ribonucleic acid in flowering plants. Biochem J. 1979 Dec 1;183(3):595–604. doi: 10.1042/bj1830595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Farber N. M., Cantor C. R. A slow tritium exchange study of the solution structure of Escherichia coli 5 S ribosomal RNA. J Mol Biol. 1981 Feb 25;146(2):223–239. doi: 10.1016/0022-2836(81)90433-2. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Garrett R. A., Olesen S. O. Structure of eukaryotic 5S ribonucleic acid: a study of Saccharomyces cerevisiae 5S ribonucleic acid with ribonucleases. Biochemistry. 1982 Sep 14;21(19):4823–4830. doi: 10.1021/bi00262a047. [DOI] [PubMed] [Google Scholar]
- Hancock J., Wagner R. A structural model of 5S RNA from E. coli based on intramolecular crosslinking evidence. Nucleic Acids Res. 1982 Feb 25;10(4):1257–1269. doi: 10.1093/nar/10.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A. C., Nazar R. N. Topography of 5.8 S rRNA in rat liver ribosomes. Identification of diethyl pyrocarbonate-reactive sites. J Biol Chem. 1982 Apr 10;257(7):3516–3524. [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E., Kilpatrick M. W., Walker R. T., Domdey H., Krupp G., Gross H. J. The nucleotide sequence of the 5S rRNA from the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res. 1981 Feb 25;9(4):965–970. doi: 10.1093/nar/9.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Nicholson D. E., Eubanks D. C., Fox G. E. An archaebacterial 5S rRNA contains a long insertion sequence. Nature. 1981 Oct 29;293(5835):755–756. doi: 10.1038/293755a0. [DOI] [PubMed] [Google Scholar]
- Marotta C. A., Varricchio F., Smith I., Weissman S. M. The primary structure of Bacillus subtilis and Bacillus stearothermophilus 5 S ribonucleic acids. J Biol Chem. 1976 May 25;251(10):3122–3127. [PubMed] [Google Scholar]
- McDougall J., Nazar R. N. Tertiary structure of the eukaryotic ribosomal 5 S RNA. Accessibility of phosphodiester bonds to ethylnitrosourea modification. J Biol Chem. 1983 Apr 25;258(8):5256–5259. [PubMed] [Google Scholar]
- Miyazaki M. Studies on the nucleotide sequence of pseudouridine-containing 5S RNA from Saccharomyces cerevisiae. J Biochem. 1974 Jun;75(6):1407–1410. doi: 10.1093/oxfordjournals.jbchem.a130532. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Matheson A. T., Bellemare G. Nucleotide sequence of Halobacterium cutirubrum ribosomal 5 S ribonucleic acid. An altered secondary structure in halophilic organisms. J Biol Chem. 1978 Aug 10;253(15):5464–5469. [PubMed] [Google Scholar]
- Nazar R. N. The ribosomal protein binding site in Saccharomyces cerevisiae ribosomal 5 S RNA. A conserved protein binding site in 5 S RNA. J Biol Chem. 1979 Aug 25;254(16):7724–7729. [PubMed] [Google Scholar]
- Nazar R. N., Wildeman A. G. Three helical domains form a protein binding site in the 5S RNA-protein complex from eukaryotic ribosomes. Nucleic Acids Res. 1983 May 25;11(10):3155–3168. doi: 10.1093/nar/11.10.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Yaguchi M., Willick G. E., Rollin C. F., Roy C. The 5-S RNA binding protein from yeast (Saccharomyces cerevisiae) ribosomes. Evolution of the eukaryotic 5-S RNA binding protein. Eur J Biochem. 1979 Dec 17;102(2):573–582. doi: 10.1111/j.1432-1033.1979.tb04274.x. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Yaguchi M., Willick G. E. The 5S RNA - protein complex from yeast: a model for the evolution and structure of the eukaryotic ribosome. Can J Biochem. 1982 Apr;60(4):490–496. doi: 10.1139/o82-058. [DOI] [PubMed] [Google Scholar]
- Nichols J. L., Welder L. S1 nuclease as a probe of yeast ribosomal 5 S RNA conformation. Biochim Biophys Acta. 1979 Feb 27;561(2):445–451. doi: 10.1016/0005-2787(79)90152-7. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Nucleotide sequence of 5 S RNA from Torulopsis utilis. FEBS Lett. 1974 Mar 15;40(1):106–109. doi: 10.1016/0014-5793(74)80904-x. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Garrett R. A. Structure of 5 S ribosomal RNA from Escherichia coli: identification of kethoxal-reactive sites in the A and B conformations. J Mol Biol. 1979 Aug 25;132(4):621–636. doi: 10.1016/0022-2836(79)90378-4. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Garrett R. A. Structure of 5 S ribosomal RNA from Escherichia coli: identification of kethoxal-reactive sites in the A and B conformations. J Mol Biol. 1979 Aug 25;132(4):621–636. doi: 10.1016/0022-2836(79)90378-4. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Erdmann V. A. Three-dimensional structural model of eubacterial 5S RNA that has functional implications. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4599–4603. doi: 10.1073/pnas.79.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEELE W. J., OKAMURA N., BUSCH H. EFFECTS OF THIOACETAMIDE ON THE COMPOSITION AND BIOSYNTHESIS OF NUCLEOLAR AND NUCLEAR RIBONUCLEIC ACID IN RAT LIVER. J Biol Chem. 1965 Apr;240:1742–1749. [PubMed] [Google Scholar]
- Smith N., Matheson A. T., Yaguchi M., Willick G. E., Nazar R. N. The 5-S RNA . protein complex from an extreme halophile, Halobacterium cutirubrum. Purification and characterization. Eur J Biochem. 1978 Sep 1;89(2):501–509. doi: 10.1111/j.1432-1033.1978.tb12554.x. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Luehrsen K. R., Woese C. R., Pace N. R. An unusual 5S rRNA, from Sulfolobus acidocaldarius, and its implications for a general 5S rRNA structure. Nucleic Acids Res. 1981 Nov 25;9(22):6129–6137. doi: 10.1093/nar/9.22.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots I., Metspalu A., Villems R., Saarma M. Location of single-stranded and double-stranded regions in rat liver ribosomal 5S RNA and 5.8S RNA. Nucleic Acids Res. 1981 Oct 24;9(20):5331–5343. doi: 10.1093/nar/9.20.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov V. V., Giege R., Ebel J. P. The tertiary structure of yeast tRNAPhe in solution studied by phosphodiester bond modification with ethylnitrosourea. FEBS Lett. 1980 Oct 20;120(1):12–16. doi: 10.1016/0014-5793(80)81034-9. [DOI] [PubMed] [Google Scholar]
- Walker W. F., Doolittle W. F. Redividing the basidiomycetes on the basis of 5S rRNA sequences. Nature. 1982 Oct 21;299(5885):723–724. doi: 10.1038/299723a0. [DOI] [PubMed] [Google Scholar]
- Weidner H., Crothers D. M. Pathway-dependent refolding of E. coli 5S RNA. Nucleic Acids Res. 1977 Oct;4(10):3401–3414. doi: 10.1093/nar/4.10.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner H., Yuan R., Crothers D. M. Does 5S RNA function by a switch between two secondary structures? Nature. 1977 Mar 10;266(5598):193–194. doi: 10.1038/266193a0. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Structural studies of 5 S ribosomal RNAs from a thermophilic fungus, Thermomyces lanuginosus. A comparison of generalized models for eukaryotic 5 S RNAs. J Biol Chem. 1982 Oct 10;257(19):11395–11404. [PubMed] [Google Scholar]
- Willick G. E., Nazar R. N., Van N. T. Physicochemical studies on the 5S ribonucleic acid-protein complex from a eucaryote, Saccharomyces cerevisiae. Biochemistry. 1980 Jun 10;19(12):2738–2742. doi: 10.1021/bi00553a031. [DOI] [PubMed] [Google Scholar]