Abstract
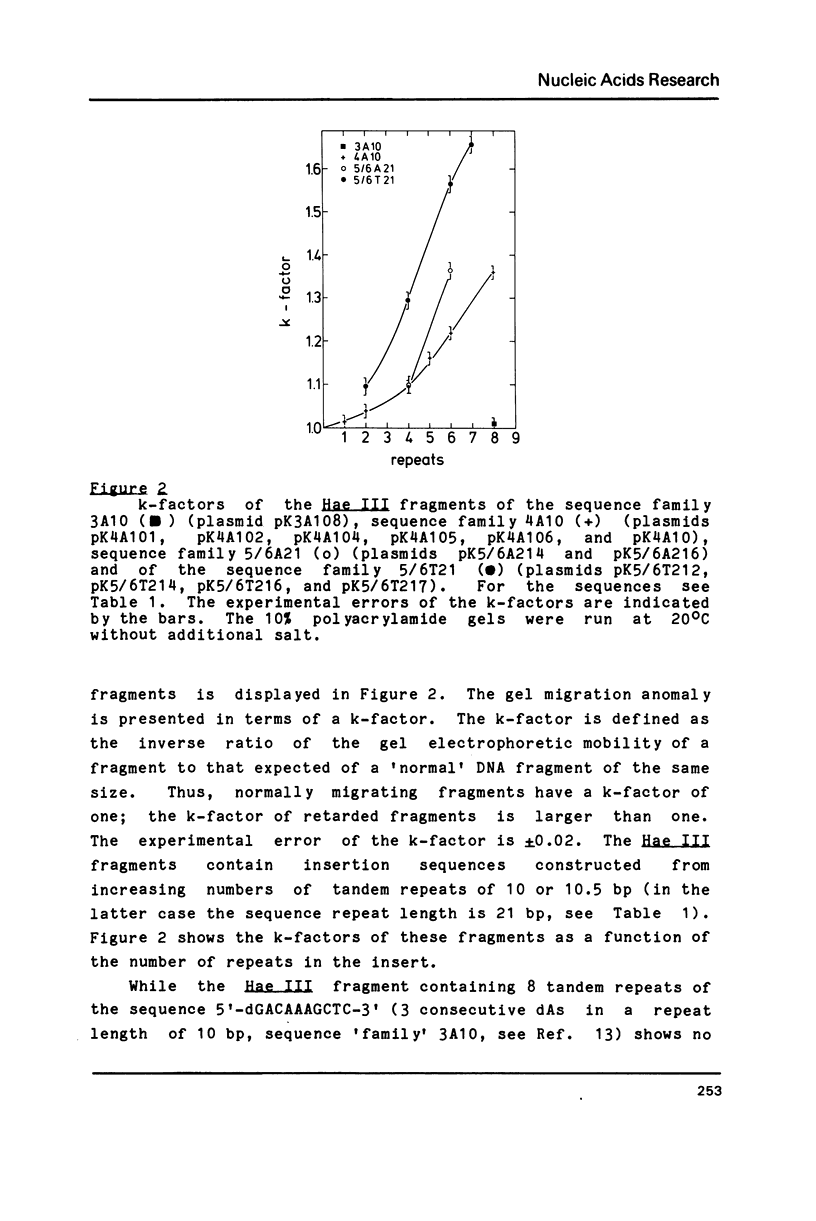
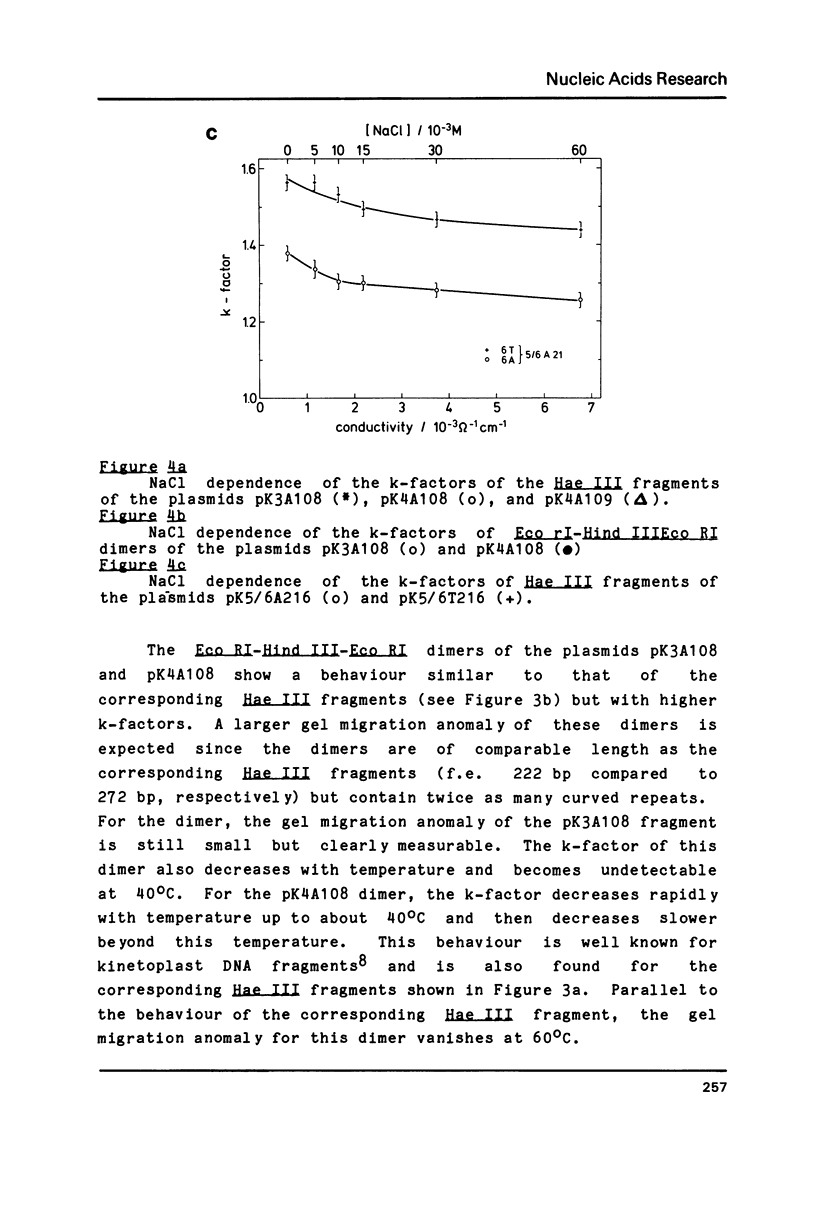
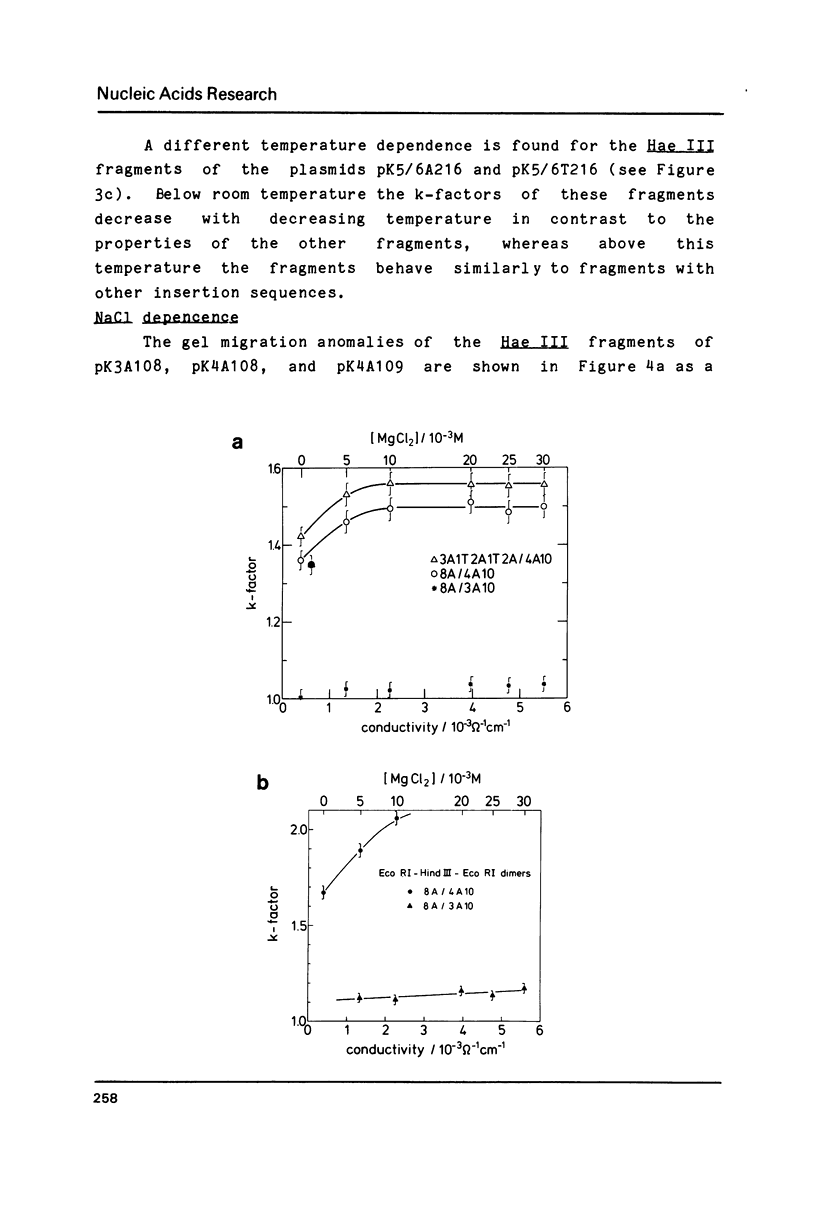
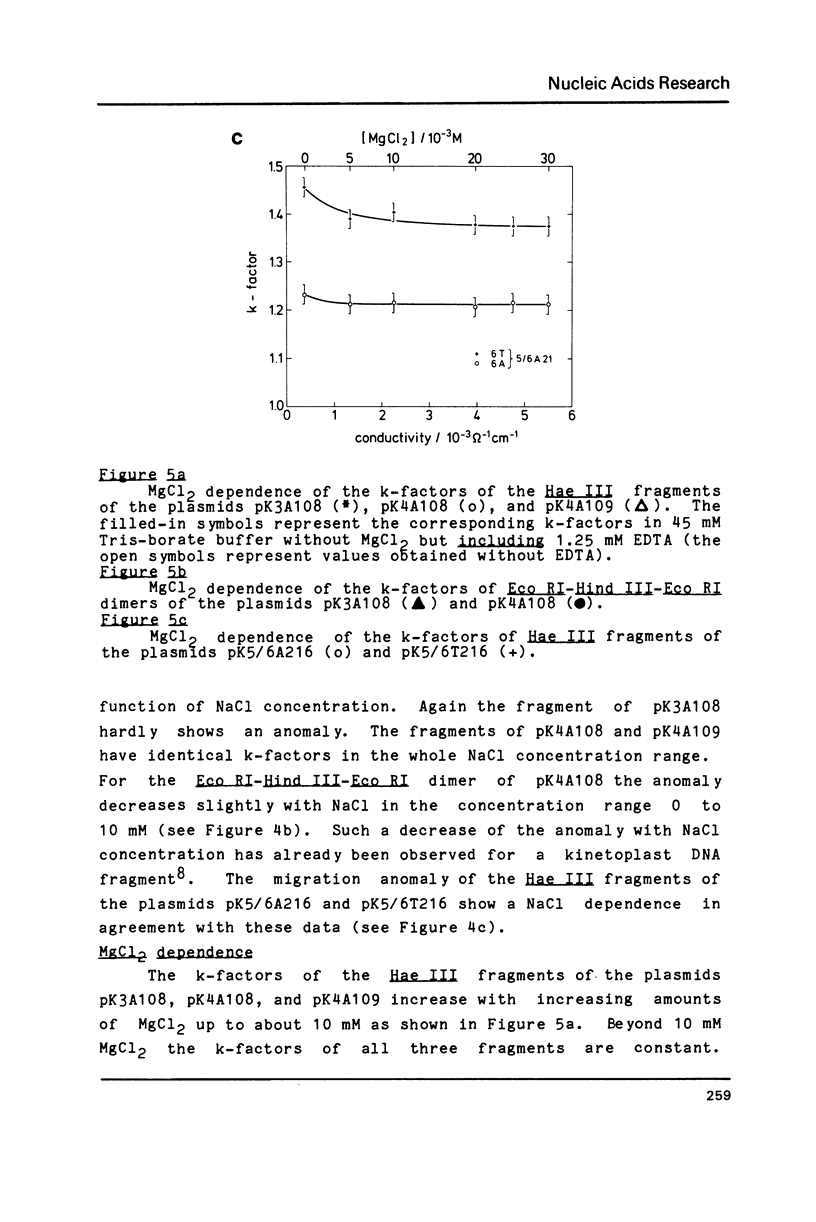
A series of oligonucleotides of different sequences have been cloned to study DNA curvature. Several DNA fragments containing these oligonucleotides in various numbers of repeats were analyzed in 10% polyacrylamide gels. A strong gel migration anomaly was found for dA4 sequences; a comparably very small but clearly detectable anomaly was observed for dA3 (both in a repeat length of 10 base-pairs). The temperature and salt (NaCl, MgCl2) dependence of the gel migration anomaly of these DNA fragments was measured. While a similar behaviour of all sequences is observed for the addition of NaCl, the temperature and MgCl2 dependence of the anomaly varies with the oligonucleotide sequence. These data are interpreted in terms of local DNA structure changes induced by changes in the temperature and the MgCl2 concentration which affect the planarity of the curved DNA fragments.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. Sequence specificity of curved DNA. FEBS Lett. 1986 Jan 20;195(1-2):53–56. doi: 10.1016/0014-5793(86)80128-4. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bleyman M., Rauch C. A., Kitchin P. A., Englund P. T. Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell. 1986 Aug 29;46(5):717–724. doi: 10.1016/0092-8674(86)90347-8. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 1985 Dec 3;24(25):7033–7037. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Nature. 1986 May 22;321(6068):449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- Kitchin P. A., Klein V. A., Ryan K. A., Gann K. L., Rauch C. A., Kang D. S., Wells R. D., Englund P. T. A highly bent fragment of Crithidia fasciculata kinetoplast DNA. J Biol Chem. 1986 Aug 25;261(24):11302–11309. [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Wu H. M., Crothers D. M. Bending and flexibility of kinetoplast DNA. Biochemistry. 1986 Jul 15;25(14):3988–3995. doi: 10.1021/bi00362a003. [DOI] [PubMed] [Google Scholar]
- Lilley D. DNA structure. Bent molecules--how and why? Nature. 1986 Apr 10;320(6062):487–488. doi: 10.1038/320487a0. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ott J., Eckstein F. Filter disc supported oligonucleotide synthesis by the phosphite method. Nucleic Acids Res. 1984 Dec 11;12(23):9137–9142. doi: 10.1093/nar/12.23.9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]



