Abstract
Prognosis in colorectal cancer patients is quite variable, even after adjustment for clinical parameters such as disease stage and microsatellite instability status. It is possible that the psychological distress experienced by patients, including anxiety and depression, may be correlated with poor prognosis. In the present study, we hypothesize that genetic variations within three genes biologically linked to the stress response, namely serotonin transporter (SLC6A4), brain-derived neurotrophic factor (BDNF), and arginine vasopressin receptor (AVPR1B) genes are associated with prognosis in colorectal cancer patients. We used a population-based cohort of 280 patients who were followed for up to 12.5 years after diagnosis. Our multivariate analysis showed that a tagSNP in the SLC6A4 gene (rs12150214) was a predictor of shorter overall survival (HR: 1.572, 95%CI: 1.142–2.164, p = 0.005) independent of stage, age, grade and MSI status. Additionally, a multivariate analysis using the combined genotypes of three polymorphisms in this gene demonstrated that the presence of any of the minor alleles at these polymorphic loci was an independent predictor of both shorter overall survival (HR: 1.631, 95%CI: 1.190–2.236, p = 0.002) and shorter disease specific survival (HR: 1.691, 95%CI: 1.138–2.512, p = 0.009). The 5-HTT protein coded by the SLC6A4 gene has also been implicated in inflammation. While our results remain to be replicated in other patient cohorts, we suggest that the genetic variations in the SLC6A4 gene contribute to poor survival in colorectal cancer patients.
Introduction
Colorectal cancer is a common disease with millions of new cases worldwide each year. Although outcomes in patients have improved in the recent decades, because of earlier detection, improvement in diagnostic procedures, and development of better treatment options, current 5-years survival rates for the patients are quite disappointing: ∼60–65% in North America [1] and much lower in developing countries [2]. Therefore, similar to other cancers, there is a need to develop better strategies to clinically manage this disease.
Clinical outcomes are associated with disease stage, age, and co-morbidities [3]. But, patient outcome may be modified by other factors, such as emotional health and quality of life. An important determinant of the emotional health and quality of life in cancer patients is the patients’ effectiveness of coping with the psychological distress caused by the cancer diagnosis and the undesirable consequences of treatment. In patients with advanced stage, especially in palliative care, coping with the prospect of death is also a serious emotional burden. These challenges often surface as depression or anxiety in cancer patients, which are observed in up to 50% of cancer patients [4]. Some studies have suggested an association between distress coping effectiveness and survival in cancer patients [5], [6], although conflicting results have also been reported [7].
Susceptibility to depression or anxiety has been linked to several genes, including, SLC6A4, which codes for the serotonin transporter protein (also known as 5-HTT and SERT). Serotonin is a neurotransmitter, deficiency of which leads to clinical depression in susceptible individuals [8]. By regulating the serotonin uptake and its synaptic availability, 5-HTT has an important role in serotonin metabolism. Various observational and experimental studies have shown that SLC6A4 and its variations are involved in susceptibility to several psychiatric conditions in humans and mice, including depression and anxiety [8]–[10]. The BDNF gene codes for a protein (brain-derived neurotrophic factor) involved in survival, growth, and differentiation of neurons [11], and, like SLC6A4, is involved in the response to stress. Several studies, including those in BDNF-mutant mice, have suggested that alterations in expression or function of this gene are linked to anxiety, depression and post-traumatic disorder [12], [13]. Another gene involved in stress response is the AVPR1B gene coding for the arginine vasopressin receptor [14]. Functional and epidemiological studies have shown that this gene and its ligand vasopressin are also linked to anxiety and depression in humans and in model species [15], [16].
In this study, we hypothesized that genetic variants located within the SLC6A4, BDNF, and AVPR1B genes are associated with prognosis in colorectal cancer patients. Therefore, we tested the association of five such variations from these genes with survival in a colorectal cancer cohort from Newfoundland, Canada.
Materials and Methods
Ethics Statement
This study was approved by the Human Investigation Committee of the Memorial University of Newfoundland as well as the Regional Health Boards. Informed consent was not required by the local ethics board as the study was considered an anonymous chart review. Patient data was analyzed anonymously.
Patient Cohort
This is a population based and retrospective study. From the Avalon Peninsula of Newfoundland, new cases (diagnosed at the Health Sciences Centre, Grace General Hospital, St. Clare’s Mercy Hospital (all in St. John’s), and Carbonear General Hospital) were ascertained over a two-year period between January 1, 1997 and December 31, 1998. Surgical specimens were available for 280 of the 292 patients identified. DNA was extracted from formalin-fixed paraffin-embedded non-tumor colon and rectum tissues. Outcomes were ascertained using medical records until July 2009.
Selection and Genotyping of Polymorphisms
The three genes (SLC6A4, BDNF and AVPR1B) were prioritized and investigated in this study because of their biological roles in depression or anxiety. We identified tagSNPs with a minor allele frequencies >10% in these genes using the genotype information for Caucasians posted in the HapMap database, phase I & II (http://hapmap.ncbi.nlm.nih.gov/) and using the pair-wise tagger function implemented in the Haploview program [17]. Following this approach, we initially selected five tagSNPs in SLC6A4 (rs11080121, rs12150214, rs2066713, rs4251417, and rs140700) and two tagSNPs in BDNF (rs7124442 and rs6265). For AVPR1B we analyzed a previously reported polymorphism (rs35369693) [18] since there was no HapMap data available. The genotypes were obtained at an outsourced genotyping facility (The Centre for Applied Genomics Facility, Toronto, Canada) using the Sequenom MassArray® technology. Of the SNPs selected, rs11080121 and rs2066713 in SLC6A4 as well as rs7124442 in BDNF could not be genotyped with this method. As a result, data from five SNPs were included in this study. At least 5% of the samples were genotyped twice and all duplicated genotypes were concordant. DNA samples of 8 patients had failed to be genotyped for all five polymorphisms investigated. Overall at least 94% of the patients were genotyped for each polymorphism. These polymorphisms, their minor allele frequencies (MAFs), and the successful genotyping rates are summarized in Table 1 .
Table 1. Genes and SNPs investigated in this study.
| Gene | SNP ID | Polymorphism | MAF | Patients with available genotypes |
| SLC6A4 | rs4251417 | G>A (non-coding) | 8% | 272 (97.0%) |
| SLC6A4 | rs12150214 | G>C (non-coding) | 39.30% | 271 (96.8%) |
| SLC6A4 | rs140700 | G>A (non-coding) | 9.30% | 268 (95.7%) |
| BDNF | rs6265 | G>A (Val66Met) | 19.40% | 271 (96.8%) |
| AVPR1B | rs35369693 | G>C (Lys65Asn) | 6.80% | 264 (94.3%) |
Summary of the genetic variations included in this study. MAF: minor allele frequency, SNP ID: dbSNP database [41] SNP identifiers.
Linkage Disequilibrium (LD) Map
The LD map of the SLC6A4 gene (Figure S1) was constructed using the HapMap (phase I & II) genotype data obtained from the Caucasian samples (http://hapmap.ncbi.nlm.nih.gov/) using the Haploview program [17].
Outcomes Investigated
The primary endpoint analyzed was overall survival (OS) defined as the time from date of diagnosis until the date of death from any cause. Our secondary endpoints were disease-free survival (DFS) and disease-specific survival (DSS). DFS is the time from diagnosis until the occurrence of first recurrence, metastasis or death from any cause. DSS is defined as the time from diagnosis until death due to colorectal cancer (or due to post-operative complications within the 30 days after surgery, n = 17). Patients who did not experience the event of interest were censored at the date of last follow up. Number of events for OS, DFS, and DSS are 162, 171, and 107, respectively.
Demographic and Clinicopathological Variables
Disease stage, grade (poorly differentiated or undifferentiated vs well or moderately differentiated), location (rectum vs colon), age, sex (male vs female), histology (mucinous vs non-mucinous), and microsatellite stability status (microsatellite stable or microsatellite instability-low (MSS/MSI-L) vs microsatellite-instability high (MSI-H)) were investigated.
Statistical Analysis
All genotypes were first manually analyzed to identify the major and the minor alleles and then were grouped together assuming the dominant inheritance model (aa+Aa vs AA, where a is the minor allele and A is the major allele). Deviation of the obtained genotype data from Hardy-Weinberg Equilibrium (HWE) was tested using an online tool described by Rodriguez and colleagues [19]. Age was analyzed as a continuous variable where as the remaining variables were categorized. We estimated the survival curves using the Kaplan-Meier method, which also provided the log-rank p-values. Differences between the survival times of different patient groups were examined using the Cox regression method, which calculated the p-values, hazard ratios (HRs), and 95% confidence intervals. Those variables that had p-values less than 0.05 in the univariate Cox regression analysis were entered into a multivariate analysis using the same method. In the genotype combination analysis for the SLC6A4 SNPs, we combined patients with at least one minor allele (AG+AA genotypes in SLC6A4-rs4251417, CG+CC genotypes in SLC6A4-rs12150214, or AG+AA genotypes in SLC6A4-rs140700) and compared them with the patients homozygous for the major alleles of these polymorphisms.
In order to test whether the inclusion of the SLC6A4 genotypes as a variable improved the predictive accuracy of the multivariate models, we calculated and compared the Akaike Information Criterion (AIC) [20] for three OS and DSS models. For both OS and DSS, Model 1 included age, stage, grade, and MSI status; Model 2 included the SLC6A4-rs12150214 genotype in addition to the variables in Model 1, and Model 3 included the combined SLC6A4 genotypes in addition to the variables in Model 1. For each model, the AIC was calculated by the following formula: -2 log likelihood +2K, where -2 log likelihood is the likelihood of the Cox regression model and K is the number of parameters in the model for which an HR was calculated. The model with the smallest AIC represents the best informative model fitting the data.
Correlation between SLC6A4-combined genotypes and patient demographic and clinical parameters were tested using the Chi-square test. Statistical significance was set at p<0.05, all statistical tests were two-sided and were performed using the IBM-SPSS software package (version 19). Because of the hypothesis-generating nature of this study, correction for multiple testing was not done.
Power Calculations
Power calculations for randomly selected hazard ratios of 2 and 1.5 were performed for each polymorphism using the Power and Sample Size program [21] assuming a follow up time of 11 years, accrual time of 2 years, and a type-I error probability (α) of 0.05. As a result, a minimum 80% study power was reached for each SNP to detect a hazard ratio of 2, but not 1.5. As expected, insufficient study power was more profound in less frequent polymorphisms (rs4251417 and rs140700 in SLC6A4 and rs35369693 in AVPR1B genes) when compared to other two common polymorphisms (rs12150214 in SLC6A4 and rs6265 in BDNF).
Results
The baseline characteristics of this patient cohort are summarized in Table 2 . As expected, the majority of the cases had well or moderately differentiated (i.e. low grade; 83.6%), non-mucinous (84.6%), colon (79.6%), and MSS/MSI-L (87.9%) tumors. The median age at diagnosis was 68.4 years. The five-year and ten-year overall survival rates for this cohort were 50% and 40%, respectively. Almost two-thirds of the patients had died (61.4%) or experienced disease progression (i.e. recurrence, metastasis, or death; 65.7%) by the last follow-up date. In addition, 40.4% of the cases had died of colorectal cancer ( Table 2 ).
Table 2. Baseline characteristics of the patient cohort.
| Variables | n | % |
| Sex | ||
| male | 150 | 53.57 |
| female | 130 | 46.43 |
| Median age at diagnosis | 68.4 years,range (25.3–91.6) | |
| Grade | ||
| poorly differentiated/undifferentiated | 42 | 15.0 |
| well/moderately differentiated | 234 | 83.6 |
| unknown | 4 | 1.4 |
| Histology | ||
| mucinous | 43 | 15.4 |
| non-mucinous | 237 | 84.6 |
| Location | ||
| rectum | 57 | 20.4 |
| colon | 223 | 79.6 |
| Stage | ||
| I | 54 | 19.3 |
| II | 94 | 33.6 |
| III | 76 | 27.1 |
| IV | 47 | 16.8 |
| unknown | 9 | 3.2 |
| MSI status | ||
| MSI-H | 34 | 12.1 |
| MSI-L/MSS | 246 | 87.9 |
| OS status | ||
| dead | 172 | 61.4 |
| alive | 108 | 38.6 |
| Median OS and DSS (follow up) time | 5.30 years,range (0–12.5 years) | |
| DFS status | ||
| recurrence/metastasis/death (+) | 184 | 65.7 |
| recurrence/metastasis/death (−) | 96 | 34.3 |
| DFS (follow up) time | 3.37 years,range (0–12.5 years) | |
| DSS status | ||
| death from colorectal cancer | 113 | 40.4 |
| death from other causes/alive | 167 | 59.6 |
Summary of the baseline characteristics of the study cohort. (+): present, (−): absent, DFS: disease-free survival, DSS: disease-specific survival, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, OS: overall survival.
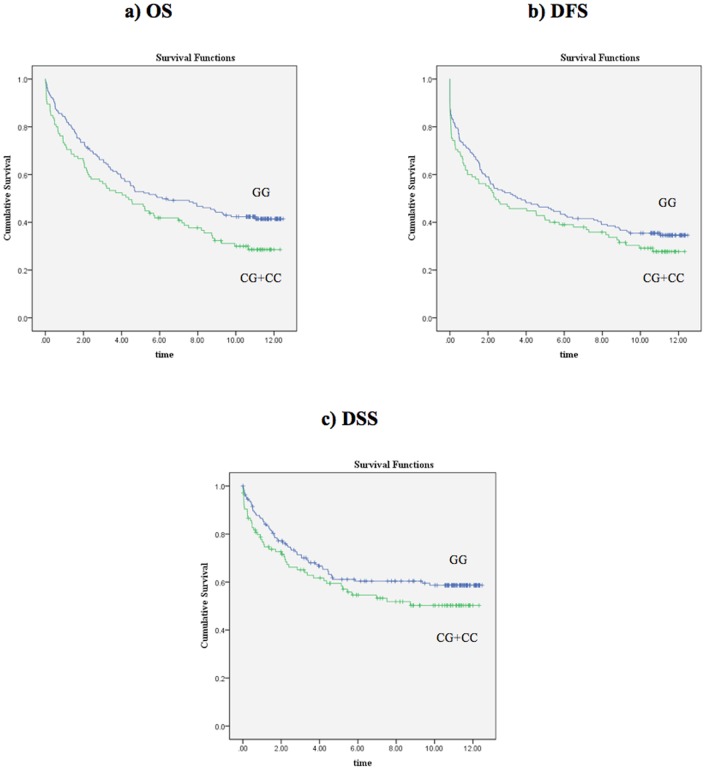
The minor allele frequencies (MAFs) of the polymorphisms are shown in Table 1 . All polymorphisms were in Hardy-Weinberg Equilibrium. The univariate Cox regression analysis results for OS, DFS, and DSS are summarized in Tables S1, S2, S3. For the SLC6A4-rs12150214 polymorphism, patients carrying the variant C allele (CG or CC genotypes) were at increased risk of death from any cause (HR: 1.399, 95%CI: 1.031–1.897, p = 0.031) when compared to patients homozygous for the major allele (GG genotype) ( Figure 1 ). However, a significant association of this SNP was not observed in either the DFS or the DSS analyses ( Figure 1 ). Increasing age, disease stage, high tumor grade (poorly differentiated or undifferentiated), and MSS/MSI-L phenotype were also associated with worse OS, DFS, and DSS. Significant associations of the remaining four polymorphisms in SLC6A4, BDNF, and AVPR1B with OS, DFS, or DSS were not detected.
Figure 1. Kaplan-Meier survival curves for patients grouped based on the SLC6A4-rs12150214 genotype data.
a) OS (p = 0.030, log-rank test), b) DFS (p = 0.225, log-rank test), and c) DSS (p = 0.159, log-rank test). Time is shown in years.
The results of the multivariate analysis for OS are shown in Table 3 . Similar to the univariate analysis, for the SLC6A4-rs12150214 polymorphism, when compared to the patients with major homozygote genotype (GG), patients carrying the variant C allele (CG or CC genotypes) were at increased risk of death from any cause (HR: 1.572, 95%CI: 1.142–2.164, p = 0.005), after adjustment for sex, age, grade, stage and MSI status. Increasing age, grade, stage, and MSI status were also independent predictors of OS in this cohort ( Table 3 ). Additional multivariate analyses also showed the independent association of age, grade, stage, and MSI status with both DFS and DSS (data not shown).
Table 3. Multivariate analysis results for OS.
| 95% CI | |||||
| Variable | p-value | HR | Lower | Upper | n |
| SLC6A4-rs12150214 (CG+CC vs GG) | .005 | 1.572 | 1.142 | 2.164 | 259 |
| Age | <.001 | 1.049 | 1.034 | 1.064 | |
| Grade (poorly diff./undiff. vs well/moderately diff.) | <.001 | 2.372 | 1.566 | 3.592 | |
| Stage | <.001 | ||||
| Stage (II vs I) | .715 | 1.103 | .651 | 1.87 | |
| Stage (IIIvs I) | .001 | 2.410 | 1.419 | 4.093 | |
| Stage (IV vs I) | <.001 | 11.083 | 6.267 | 19.603 | |
| MSI status (MSI-H vs MSS/MSI-L) | .002 | .357 | .186 | .686 | |
Only those variables with a p<0.05 in the univariate analysis are included in the multivariate analysis. Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus. Number of events for OS is 162.
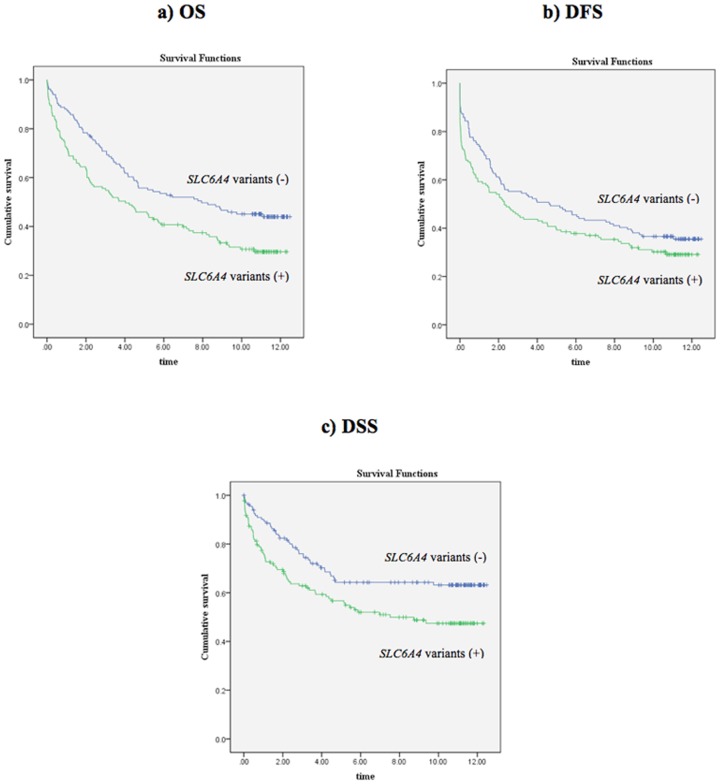
We also performed a genotype combination analysis for the three SNPs from the SLC6A4 gene. Patients with genotypes with at least one minor allele in any of the three SNPs were compared with patients who were homozygous for major alleles. Univariate analysis showed that the presence of at least one minor allele was associated with worse OS and DSS, but not with DFS in our cohort ( Table 4 ; Figure 2 ). More interestingly, when adjusted for other variables, the presence of at least one minor allele was independently associated with both worse OS (HR: 1.631, 95%CI: 1.190–2.236, p = 0.002) and worse DSS (HR: 1.691, 95%CI: 1.138–2.512, p = 0.009) ( Tables 5 and 6 ). These associations were stronger than those detected when SLC6A4-rs12150214 was analyzed alone ( Table 3 ; Figures 1 and 2 ). The AIC calculations indicated that the addition of the SLC6A4-rs12150214 genotype into the multivariate models (both for OS and DSS) also containing age, stage, grade, and MSI status variables improved the model predictions ( Table 7 ). However, the best improvement was detected when the model contained the combined SLC6A4 genotypes in addition to the clinical variables ( Table 7 ). In addition, when compared to the DSS model, the model improvement was more profound in the OS model.
Table 4. Univariate analysis results for the combined genotypes of three SNPs within the SLC6A4 gene.
| 95% CI | ||||||
| Variable | p-value | HR | Lower | Upper | n | Endpoint |
| SLC6A4 variants (+ vs -) | .005 | 1.544 | 1.137 | 2.097 | 269 | OS |
| SLC6A4 variants (+ vs -) | .108 | 1.271 | .949 | 1.704 | 269 | DFS |
| SLC6A4 variants (+ vs -) | .009 | 1.656 | 1.133 | 2.419 | 269 | DSS |
In the combined analysis, patients with at least one minor (variant) allele in any of the three SNPs in SLC6A4 were categorized together (+) and were compared with the patients who did not have the variant alleles at these polymorphic loci (−). OS: overall survival, DFS: disease-free survival, DSS: disease-specific survival.
Figure 2. Kaplan-Meier survival curves for the combined genotypes of three SLC6A4 polymorphisms.
a) OS (p = 0.005, log-rank test), b) DFS (p = 0.104, log-rank test), and c) DSS (p = 0.008, log-rank test). Time is shown in years.
Table 5. Multivariate analysis results for the combined genotypes of three SNPs within the SLC6A4 gene (OS).
| 95% CI | |||||
| Variable | p-value | HR | Lower | Upper | n |
| SLC6A4 variants (+ vs -) | .002 | 1.631 | 1.190 | 2.236 | 257 |
| Age | <.001 | 1.049 | 1.034 | 1.065 | |
| Grade (poorly diff./undiff. vs well/moderately diff.) | <.001 | 2.364 | 1.556 | 3.589 | |
| Stage | <.001 | ||||
| Stage (II vs I) | .698 | 1.111 | .654 | 1.886 | |
| Stage (IIIvs I) | .001 | 2.476 | 1.456 | 4.211 | |
| Stage (IV vs I) | <.001 | 11.244 | 6.335 | 19.957 | |
| MSI status (MSI-H vs MSS/MSI-L) | .001 | .331 | .172 | .640 | |
Multivariate analysis results for overall survival. (+): patients with at least one minor (variant) allele in any of the three SLC6A4 SNPs, (−): patients who did not have the variant allele in any of the three SLC6A4 SNPs, OS: overall survival.
Table 6. Multivariate analysis results for the combined genotypes of three SNPs within the SLC6A4 gene (DSS).
| 95% CI | |||||
| Variable | p-value | HR | Lower | Upper | n |
| SLC6A4 variants (+ vs -) | .009 | 1.691 | 1.138 | 2.512 | 257 |
| Age | <.001 | 1.038 | 1.020 | 1.056 | |
| Grade (poorly diff./undiff. vs well/moderately diff.) | <.001 | 2.917 | 1.798 | 4.733 | |
| Stage | <.001 | ||||
| Stage (II vs I) | .154 | 2.040 | .766 | 5.436 | |
| Stage (IIIvs I) | <.001 | 6.630 | 2.573 | 17.083 | |
| Stage (IV vs I) | <.001 | 35.993 | 13.77 | 94.097 | |
| MSI status (MSI-H vs MSS/MSI-L) | .002 | .207 | .075 | .574 | |
Multivariate analysis results for disease-specific survival. (+): patients with at least one minor (variant) allele in any of the three SLC6A4 SNPs, (−): patients who did not have the variant allele in any of the three SLC6A4 SNPs, DSS: disease-specific survival.
Table 7. Akaike information criterion (AIC) calculations for the OS and DSS multivariate models with or without the SLC6A4 genotypes.
| Endpoint | −2 log likelihood | Number of parameters | AIC | ΔAIC |
| OS | ||||
| Model 1 | 1545.859 | 6 | 1557.859 | 53.792 |
| Model 2 | 1514.252 | 7 | 1528.252 | 24.185 |
| Model 3 | 1490.067 | 7 | 1504.067 | 0 |
| DSS | ||||
| Model 1 | 977.655 | 6 | 989.655 | 30.834 |
| Model 2 | 962.69 | 7 | 976.69 | 17.869 |
| Model 3 | 944.821 | 7 | 958.821 | 0 |
Model 1 contains the clinical variables that were significant in the multivariate analyses; namely age, stage, grade, and MSI-H status (for both OS and DSS); Model 2 contains the SLC6A4-rs12150214 genotype as a variable in addition to the clinical variables in Model 1, and Model 3 contains the combined SLC6A4 genotypes (for rs4251417, rs12150214, rs140700) in addition to the variables in Model 1. ΔAIC is the difference in AIC values of a model and the best model, which is Model 3 for both OS and DSS.
There was no correlation between the combined genotype status and the baseline demographic and clinical characteristics listed in Table 2 (data not shown).
Discussion
While well-established prognostic markers such as disease stage are used in prognostication, an inter-patient prognostic variability remains [3]. Better prognostic performance may come from the identification of yet-to-be recognized prognostic indicators. For example, several epidemiological studies have suggested an effect of life-style factors (such as level of physical activity) on prognosis [22], [23]. Although not well studied in colorectal cancer, emotional distress and psychological interventions after diagnosis have been implicated in modification of outcome [24]–[27]. Consequently, in the present study, we have aimed to test the associations of five genetic variations in stress response genes (SLC6A4, BDNF, and AVPR1B) with clinical outcomes in a population-based cohort of 280 colorectal cancer patients from Newfoundland and Labrador (NL). NL is one of the 13 provinces/territories in Canada and has the highest incidence of colorectal cancer as well as one of the highest cancer mortality rates in Canada [1], [28]. This population is also characterized by a high incidence rate of familial colorectal cancer [29], [30]. A strength of our study is the long period of time that patients have been followed (up to 12.5 years after cancer diagnosis). As a consequence we have documented many events, which increased our study power. A weakness of our study is that the results obtained remain to be replicated in other cohorts. In addition, serum biomarkers with prognostic potentials, such as interleukin-6 (IL-6), carcinoembryonic antigen (CEA), and carbohydrate antigen 19–9 (CA 19–9) were not evaluated in our study.
Our results showed that the polymorphisms in neither the BDNF nor the AVPR1B genes were correlated with outcome in our patient cohort, though we cannot rule out the false-negative findings due to lack of adequate study power, especially in the case of the relatively rare AVPR1B-rs35369693 polymorphism. However, we found that the rs12150214 SNP, which is a relatively common G>C substitution in the non-coding region of the SLC6A4 gene, was associated with poor OS in a univariate as well as in a multivariate analysis. Specifically, when adjusted for sex, stage, grade, age and MSI status, the patients carrying the variant C allele (both CC homozygotes and CG heterozygotes) were at 57% increased risk of death (95%CI: 1.142–2.164; Table 3 ) when compared to patients with the GG genotype. When the genotypes containing the minor alleles were combined for the three SLC6A4 SNPs, we detected a significant correlation with not only OS, but also with DSS ( Tables 5 and 6 ). These associations were independent of other clinically important variables including age, stage, grade, and MSI status. Although our approach did not include all variations in the SLC6A4 gene, these results suggest the association of multiple SLC6A4 variations with prognosis in colorectal cancer patients. In addition, our results show that inclusion of the SLC6A4 genotypes, particularly the combined genotypes, as a variable improves the predictive accuracy of both the OS and DSS multivariate models ( Table 7 ). Figures 1 and 2 also show that the combined SLC6A4 genotypes, when compared to rs12150214 genotypes alone, results in a better separation of patient survival curves.
The linkage disequilibrium (LD) map of the SLC6A4 gene and the relative positions of the three SNPs investigated in the present study (rs4251417, rs12150214, and rs140700) are shown in Figure S1. These three SNPs are all non-coding polymorphisms and are located in non-linked regions of the SLC6A4 gene (i.e. they are not correlated with each other; r2<0.8). Specifically, rs4251417 and rs12150214 are located in the 5′-end of the gene (in intron 1) and rs140700 is located in intron 6. At the time being, it is not clear how these polymorphisms may affect the SLC6A4 gene expression or function; however, it is also likely that other functional polymorphisms that are highly correlated with them in fact might be the variants biologically related to altered gene function and thus prognosis (Figure S1).
To our knowledge, there is no report linking rs4251417 to stress response or other human disease. However, previously, the rs12150214 polymorphism (located in the LD block-2 in Figure S1) was reported to be borderline associated with the Beck Depression Inventory scores in a study investigating 360 male twins [31]. These authors also performed a haplotype analysis with the polymorphisms linked to rs12150214 and found that this region of the SLC6A4 gene was associated with not only depression but also elevated IL-6 levels in the study subjects. Interestingly, IL-6 is a pro-inflammatory cytokine (see below). In another study involving 567 subjects, Lazary et al. [32] suggested that the rs140700 polymorphism (or other variations linked to it) acts as a modifier of depressive symptoms (this polymorphism is located in the LD block-1 in Figure S1). These previously reported findings suggest that multiple variations within the SLC6A4 gene are likely to be involved in altering gene function with adverse consequences for stress response and depression. This can also explain why we have detected a stronger association with both OS and DSS in our patient cohort when the genotype data for the three SLC6A4 SNPs were combined and analyzed in the multivariate analyses. However, in the absence of the psychological health data in our cohort, we cannot determine whether the observed association of the SLC6A4 gene with shorter survival times is due to its role in psychological distress or due to other biological roles of the SLC6A4.
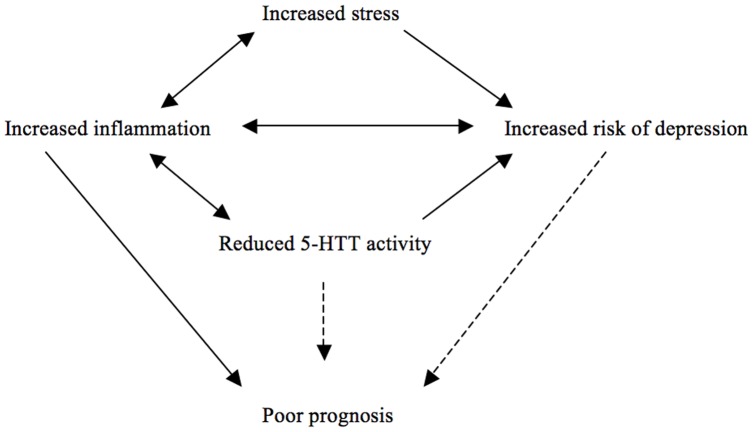
In fact, in addition to its role in psychological stress response, the 5-HTT protein coded by the SLC6A4 gene as well as its ligand (serotonin) function in the immune response and inflammation, including in gut. For example, serotonin signaling regulates stool transition in the gut and the serotonin levels in this organ may be altered (either by altered rates of serotonin secretion or function of the 5-HTT protein) as a result of local infection and inflammation [33]. Interestingly, a recent study using a mouse model of colitis showed that the loss of 5-HTT activity enhances the severity of inflammation in the colon [34]. These findings suggest the presence of a two-way interaction between 5-HTT and inflammation. A similar pattern is also observed in inflammation and depression, where the one seems to induce or exacerbate the other [35], [36]. Increased or persistent inflammation is also linked to tumor progression, including colorectal cancers [37], [38]. Interestingly IL-6 was previously reported to be associated with variations in the SLC6A4 gene [31] (see the previous paragraph), which also seems to be a mediator of tumor progression in colorectal cancer [37], [39], [40]. Therefore, we suggest that emotional or physiological stress (i.e. inflammation), and their associated neurological and immune responses can be modified by the activity of the 5-HTT protein (either by environmental factors or by the genetic variations in the SLC6A4 gene), which can explain the poor survival observed in our cohort of colorectal cancer patients ( Figure 3 ). Further studies on the potential roles of the SLC6A4 gene variations and the depression in survival of colorectal cancer patients are therefore warranted.
Figure 3. The 5-HTT activity may be altered as a response to changing cellular environment (such as inflammation) or by the genetic variations in the SLC6A4 gene.
The direct links between the altered 5-HTT activity as well as the depression and prognosis in colorectal cancer patients (arrows with broken lines) remain to be established by further studies.
Supporting Information
The three SLC6A4 SNPs investigated in this study (rs4251417, rs12150214, and rs140700) are circled. The black triangles designate the LD blocks. The red squares are where the correlation between markers is the strongest. Please note that the gene is shown in a 3′ to 5′ orientation in this figure. According to the Haploview (pairwise tagger) [17] results, rs12150214 is highly correlated with the following polymorphisms along the SLC6A4 gene with a correlation coefficient (r2) of >0.85: rs8076005, rs11080122, rs6354, rs25528, rs2020936, and rs8071667 (annotated with stars on the Figure S1).
(TIFF)
Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus.
(DOC)
Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus.
(DOC)
Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus.
(DOC)
Acknowledgments
The authors thank Payal Sipahimalani for technical assistance and Dr. Veeresh Gadag for his help with the AIC calculations.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Canadian Institute of Health Research Institute of Genetics and the Canadian Gene Cure Foundation, Memorial University of Newfoundland, Research and Development Corporation (RDC) of Newfoundland, Interdisciplinary Team in Colorectal Cancer funded by CIHR, and Interdisciplinary Team in Human Genetics funded by the Atlantic Innovation Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Canadian Cancer Society's Steering Committee. Canadian Cancer Statistics, 2010. Toronto: Canadian Cancer Society. 2010.
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Savas S, Liu G. Genetic variations as cancer prognostic factors: review and update. Hum Mutat. 2009;30:1369–1377. doi: 10.1002/humu.21078. [DOI] [PubMed] [Google Scholar]
- 4.Wein S, Sulkes A, Stemmer S. The oncologist's role in managing depression, anxiety, and demoralization with advanced cancer. Cancer J. 2010;16(5):493–499. doi: 10.1097/PPO.0b013e3181f28b64. [DOI] [PubMed] [Google Scholar]
- 5.Johansson M, Ryden A, Finizia C. Mental adjustment to cancer and its relation to anxiety, depression, HRQL and survival in patients with laryngeal cancer - a longitudinal study. BMC Cancer. 2011;11:283. doi: 10.1186/1471-2407-11-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamer M, Chida Y, Molloy GJ. Psychological distress and cancer mortality. J Psychosom Res. 2009;66(3):255–258. doi: 10.1016/j.jpsychores.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Phillips KA, Osborne RH, Giles GG, Dite GS, Apicella C, et al. Psychosocial factors and survival of young women with breast cancer: a population-based prospective cohort study. J Clin Oncol. 2008;26(28):4666–4671. doi: 10.1200/JCO.2007.14.8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp T, Cowen PJ. 5-HT and depression: is the glass half-full? Curr Opin Pharmacol. 2011;11(1):45–51. doi: 10.1016/j.coph.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Murphy DL, Moya PR. Human serotonin transporter gene (SLC6A4) variants: their contributions to understanding pharmacogenomic and other functional GxG and GxE differences in health and disease. Curr Opin Pharmacol. 2011;11(1):3–10. doi: 10.1016/j.coph.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167(5):509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirstein M, Farinas I. Sensing life: regulation of sensory neuron survival by neurotrophins. Cell Mol Life Sci. 2002;59(11):1787–1802. doi: 10.1007/PL00012506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chourbaji S, Brandwein C, Gass P. Altering BDNF expression by genetics and/or environment: impact for emotional and depression-like behaviour in laboratory mice. Neurosci Biobehav Rev. 2011;35(3):599–611. doi: 10.1016/j.neubiorev.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Martinowich K, Lu B. Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology. 2008;33(1):73–83. doi: 10.1038/sj.npp.1301571. [DOI] [PubMed] [Google Scholar]
- 14.Griebel G, Simiand J, Stemmelin J, Gal CS, Steinberg R. The vasopressin V1b receptor as a therapeutic target in stress-related disorders. Curr Drug Targets CNS Neurol Disord. 2003;2(3):191–200. doi: 10.2174/1568007033482850. [DOI] [PubMed] [Google Scholar]
- 15.Surget A, Belzung C. Involvement of vasopressin in affective disorders. Eur J Pharmacol. 2008;583(2–3):340–349. doi: 10.1016/j.ejphar.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 16.Landgraf R. The involvement of the vasopressin system in stress-related disorders. CNS Neurol Disord Drug Targets. 2006;5(2):167–179. doi: 10.2174/187152706776359664. [DOI] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Dempster EL, Burcescu I, Wigg K, Kiss E, Baji I, et al. Evidence of an association between the vasopressin V1b receptor gene (AVPR1B) and childhood-onset mood disorders. Arch Gen Psychiatry. 2007;64(10):1189–1195. doi: 10.1001/archpsyc.64.10.1189. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169(4):505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akaike H. Information theory and extension of the maximum likelihood principle. In Second International Symposium on Information Theory, Petrov B, Csaki F, editors. Budapest, Hungary: Akademiai Kiado, pp 267–281. 1973.
- 21.Dupont WD, Plummer WD., Jr Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11(2):116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 22.Morikawa T, Kuchiba A, Yamauchi M, Meyerhardt JA, Shima K, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305(16):1685–1694. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huxley R, Asia Pacific Cohort Studies Collaboration. The role of lifestyle risk factors on mortality from colorectal cancer in populations of the Asia-Pacific region. Asian Pac J Cancer Prev. 2007;8(2):191–198. [PubMed] [Google Scholar]
- 24.Sharma A, Sharp DM, Walker LG, Monson JR. Predictors of early postoperative quality of life after elective resection for colorectal cancer. Ann Surg Oncol. 2007;14(12):3435–3442. doi: 10.1245/s10434-007-9554-x. [DOI] [PubMed] [Google Scholar]
- 25.Gripp S, Moeller S, Bolke E, Schmitt G, Matuschek C, et al. Survival prediction in terminally ill cancer patients by clinical estimates, laboratory tests, and self-rated anxiety and depression. J Clin Oncol. 2007;25(22):3313–3320. doi: 10.1200/JCO.2006.10.5411. [DOI] [PubMed] [Google Scholar]
- 26.Weinberger T, Forrester A, Markov D, Chism K, Kunkel EJ. Women at a dangerous intersection: diagnosis and treatment of depression and related disorders in patients with breast cancer. Psychiatr Clin North Am. 2010;33(2):409–422. doi: 10.1016/j.psc.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Kuchler T, Bestmann B, Rappat S, Henne-Bruns D, Wood-Dauphinee S. Impact of psychotherapeutic support for patients with gastrointestinal cancer undergoing surgery: 10-year survival results of a randomized trial. J Clin Oncol. 2007;25(19):2702–2708. doi: 10.1200/JCO.2006.08.2883. [DOI] [PubMed] [Google Scholar]
- 28.Ugnat AM, Xie L, Semenciw R, Waters C, Mao Y. Survival patterns for the top four cancers in Canada: the effects of age, region and period. Eur J Cancer Prev. 2005;14(2):91–100. doi: 10.1097/00008469-200504000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Woods MO, Hyde AJ, Curtis FK, Stuckless S, Green JS, et al. High frequency of hereditary colorectal cancer in Newfoundland likely involves novel susceptibility genes. Clin Cancer Res 11(19 Pt. 2005;1):6853–6861. doi: 10.1158/1078-0432.CCR-05-0726. [DOI] [PubMed] [Google Scholar]
- 30.Green RC, Green JS, Buehler SK, Robb JD, Daftary D, et al. Very high incidence of familial colorectal cancer in Newfoundland: a comparison with Ontario and 13 other population-based studies. Fam Cancer. 2007;6(1):53–62. doi: 10.1007/s10689-006-9104-x. [DOI] [PubMed] [Google Scholar]
- 31.Su S, Zhao J, Bremner JD, Miller AH, Tang W, et al. Serotonin transporter gene, depressive symptoms, and interleukin-6. Circ Cardiovasc Genet. 2009;2(6):614–620. doi: 10.1161/CIRCGENETICS.109.870386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazary J, Lazary A, Gonda X, Benko A, Molnar E, et al. New evidence for the association of the serotonin transporter gene (SLC6A4) haplotypes, threatening life events, and depressive phenotype. Biol Psychiatry. 2008;64(6):498–504. doi: 10.1016/j.biopsych.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 33.Spiller R. Serotonin and GI clinical disorders. Neuropharmacology. 2008;55(6):1072–1080. doi: 10.1016/j.neuropharm.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Haub S, Ritze Y, Bergheim I, Pabst O, Gershon MD, et al. Enhancement of intestinal inflammation in mice lacking interleukin 10 by deletion of the serotonin reuptake transporter. Neurogastroenterol Motil 22(7): 826–34, e229. 2010. [DOI] [PMC free article] [PubMed]
- 35.Anisman H. Cascading effects of stressors and inflammatory immune system activation: implications for major depressive disorder. J Psychiatry Neurosci. 2009;34(1):4–20. [PMC free article] [PubMed] [Google Scholar]
- 36.Hansel A, Hong S, Camara RJ, von Kanel R. Inflammation as a psychophysiological biomarker in chronic psychosocial stress. Neurosci Biobehav Rev. 2010;35(1):115–121. doi: 10.1016/j.neubiorev.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Ferrone C, Dranoff G. Dual roles for immunity in gastrointestinal cancers. J Clin Oncol. 2010;28(26):4045–4051. doi: 10.1200/JCO.2010.27.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamarron BF, Chen W. Dual roles of immune cells and their factors in cancer development and progression. Int J Biol Sci. 2011;7(5):651–658. doi: 10.7150/ijbs.7.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pucci S, Mazzarelli P, Sesti F, Boothman DA, Spagnoli LG. Interleukin-6 affects cell death escaping mechanisms acting on Bax-Ku70-Clusterin interactions in human colon cancer progression. Cell Cycle. 2009;8(3):473–481. doi: 10.4161/cc.8.3.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker C, Fantini MC, Wirtz S, Nikolaev A, Lehr HA, et al. IL-6 signaling promotes tumor growth in colorectal cancer. Cell Cycle. 2005;4(2):217–220. [PubMed] [Google Scholar]
- 41.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The three SLC6A4 SNPs investigated in this study (rs4251417, rs12150214, and rs140700) are circled. The black triangles designate the LD blocks. The red squares are where the correlation between markers is the strongest. Please note that the gene is shown in a 3′ to 5′ orientation in this figure. According to the Haploview (pairwise tagger) [17] results, rs12150214 is highly correlated with the following polymorphisms along the SLC6A4 gene with a correlation coefficient (r2) of >0.85: rs8076005, rs11080122, rs6354, rs25528, rs2020936, and rs8071667 (annotated with stars on the Figure S1).
(TIFF)
Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus.
(DOC)
Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus.
(DOC)
Significant results are shown in bold. CI: confidence interval, diff: differentiated, HR: hazards ratio, MSI-H: microsatellite instability-high, MSI-L: microsatellite instability-low, MSS: microsatellite stable, n: number of samples included into the analysis, vs: versus.
(DOC)