Abstract
Breast cancer is the most common tumor among women with inherited mutations in the p53 gene (Li-Fraumeni syndrome). The tumors represent the basal-like subtype which has been suggested to originate from mammary stem/progenitor cells. In mouse mammary epithelium, mammosphere-forming potential was increased with decreased dosage of the gene encoding the p53 tumor suppressor protein (Trp53). Limiting dilution transplantation also showed a 3.3-fold increase in the frequency of long-term regenerative mammary stem cells in Trp53−/− mice. The repression of mammospheres by p53 was apparent despite the absence of apoptotic responses to radiation indicating a dissociation of these two activities of p53. The effects of p53 on progenitor cells were also observed in TM40A cells using both mammosphere-forming assays and the DsRed-let7c-sensor. The frequency of long-term label-retaining epithelial cells (LRECs) was decreased in Trp53−/− mammary glands indicating that asymmetric segregation of DNA is diminished and contributes to the expansion of the mammary stem cells. Treatment with an inhibitor of γ-secretase (DAPT) reduced the number of Trp53−/− mammospheres to the level found in Trp53+/+ cells. These results demonstrate that basal levels of p53 restrict mammary stem/progenitor cells through Notch and that the Notch pathway is a therapeutic target to prevent expansion of this vulnerable pool of cells.
Keywords: Tumor Suppressor Protein p53, Li-Fraumeni Syndrome, Adult Stem Cells, Notch Proteins, apoptosis, Breast Neoplasms
INTRODUCTION
Mammary stem cells have been identified that are capable of regenerating the entire mammary ductal tree and repopulating the mammary fat pad (1, 2). The mechanisms and pathways regulating self-renewal and differentiation of mammary stem cells are of great interest for its potential application in tissue replacement therapies as well as the prevention and treatment of breast cancer. The Wnt pathway was found to regulate the self-renewal of mammary stem/progenitor cells. Ectopic expression of Wnt1 resulted in an increased population of mammary stem/progenitor cells in the mammary gland and eventually induced mammary tumors (3). Another breast cancer related gene, BRCA1, was reported to play a critical role in the differentiation of mammary stem/progenitor cells to luminal cells. Loss of both BRCA1 alleles resulted in expansion of stem/progenitor cells in the breast epithelium of women and increased breast cancer risk (4). The Notch pathway has also been implicated as regulator of mammary stem/progenitor cells self-renewal and differentiation but its function is controversial. Dontu et al reported that the activation of Notch signaling with DSL peptide resulted in 10-fold increase of mammosphere-forming activity (5). Conversely, Bouras et al showed that the inhibition of Notch pathway by knockdown Cbf-1 in CD29hiCD24+ cells resulted in increased transplantation efficiency, suggesting that the Notch pathway may restrict mammary stem/progenitor cells expansion (6).
The p53 protein is a central regulator for multiple tumor suppressor pathways. The role of activated p53 in mediating cell cycle arrest and apoptosis has been studied extensively (7–9). In response to DNA damage, oncogene activation or other stresses, p53 accumulates in nucleus and transactivates downstream genes, such as p21 and PUMA, and directs the fate of damaged cells resulting in repair or elimination (10, 11). In addition to the importance of activated p53 under stress conditions, the basal level p53 under normal conditions may also play an essential role in tumor suppressor function. In both hematopoietic system and neural system, basal levels of p53 were shown to negatively regulate the self-renewal of tissue-specific stem cells (12, 13). The Arf-Trp53 pathway was shown to restrict the efficiency of reprogramming of induced pluripotent stem cells (14, 15).
Disruption of the gene encoding p53 (designated TP53 in human and Trp53 in mouse) predisposes normal mammary epithelium to tumorigenesis. Women with Li-Fraumeni syndrome, which is most commonly associated with germline heterozygous mutations of TP53, have significantly increased risk of breast cancer (16, 17). Mutations and deletions of TP53 are the most common alterations in cancers. The rate of p53 mutation is as high as 82% in the basal-like subtype of breast cancer, whereas in luminal A subtype, p53 mutations are found in only 13% patients suggesting that p53 mutation promotes basal-like breast cancer (18). This class of aggressive tumors express gene signatures enriched in embryonic stem cells, and thus, have been proposed to originate from progenitor cells (19). Mammary tumors from p53 heterozygous mouse models mimic Li-Fraumeni syndrome in women and the tumors share gene expression patterns with tumors from Brca1-deficient and Wnt1 transgenic mice and human basal-like breast cancer, suggesting that mammary tumors from p53-deficient mice may also originate from the stem/progenitor cells (18, 20–22).
In this study, BALB/c-Trp53+/+, Trp53+/− and Trp53−/− mice were used to test the role of p53 in regulating the mammary stem/progenitor cells. We found that decreased p53 dosage resulted in increased frequency of mammary stem/progenitor cells, suggesting that basal levels of p53 inhibited self-renewal of mammary stem/progenitor cells. As the mammosphere-initiating cells of different Trp53 genotypes were resistant to ionizing radiation (IR), p53-mediated apoptosis is comprised in these cells. Therefore, expansion of the mammary stem/progenitor cells population cannot be attributed to differences in apoptosis. Similarly, the decrease in the pool of label-retaining cells in Trp53−/− mammary epithelium also suggest that survival is not increased, but rather asymmetric segregation of DNA is diminished in the absence of p53 leading to dilution of the label during expansion of mammary stem/progenitor cells. We also showed that γ-secretase inhibitors (GSI) can be used to inhibit the expansion of Trp53−/− mammary stem/progenitor cells. The results demonstrate that p53 regulates self-renewal of mammary stem/progenitor cells and that insufficient basal levels of p53 can lead to expansion of the pool of mammary stem/progenitor cells, which are especially vulnerable to tumorigenesis without the proper surveillance of p53. Therefore, the Notch pathway is a potential therapeutic target to inhibit expansion of mammary stem/progenitor cells and reduce breast cancer risk.
MATERIALS AND METHODS
Animals
BALB/c-Trp53+/+, Trp53+/− and Trp53−/− mice were generated by backcrossing (C57BL/c x 129/Sv) Trp53−/− mice onto the BALB/cMed strain as described before (23). Wild type 3 weeks old BALB/c recipient mice for transplantation were purchased from Jackson lab.
Isolation of primary mouse mammary cells
Mammary gland harvested from 8–10 weeks old virgin mice were minced and dissociated in DMEM:F12 (Sigma, St. Louis, MO) supplemented with 5% Fetal Bovine serum (Gibco, Paisley, UK), 2mg/ml collagenase (Worthington Biochemicals, Freehold, NJ), 100u/ml hyaluronidase (Sigma), 100u/ml pen/strep (Gibco) and 100μg/ml gentamicin (Gibco) for 6 hours. The cell pellet was collected and further dissociated with 1ml pre-warmed 0.05% Trypsin-EDTA (Gibco) and 200μl 1mg/ml DNase I (Roche, Mannheim, Germany). Cell suspensions were sieved through a 40μm cell strainer to obtain single cell suspension.
Mammosphere culture
Primary single cells were seeded into ultra-low attachment dishes or plates at a density of 20,000 viable cells/ml. Cells were grown in a serum-free mammary epithelial growth medium (HuMEC, Gibco) supplemented with B27 (Gibco), 20ng/ml EGF (Sigma), 20ng/ml bFGF (Sigma), 4μg/ml heparin (Sigma), 100u/ml Pen/Strep, 5μg/ml gentamicin (24). For the GSI treatment, N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT, Sigma) was added into culture medium to the final concentration of 5μM (final DMSO concentration 0.1%). To passage mammospheres, mammospheres were collected with gentle centrifugation 800rpm for 5min 7 days after culture and dissociated with 1ml pre-warmed 0.05% Trypsin-EDTA and 60μl 1mg/ml DNase I for 5–8min. Cell suspensions obtained from dissociation were sieved through 40μm cell strainer and seeded at a density of 1,000 viable cells/ml. To test the IR responses of mammosphere-initiating cells, single cell suspensions received 0-Gy (control group) or 5-Gy dose (radiation group) of γ-irradiation from a cesium-137 source before being plated.
Limiting dilution and transplantation
Primary mammary epithelial cells (MECs) were freshly isolated as described above and resuspended in DMEM:F12 with 5%FBS. Six different cell concentrations were used: 50,000/10μl, 10,000/10μl, 5,000/10μl, 2,500/10μl, 1,000/10μl, 100/10μl. Trp53+/+ cell suspensions were injected into right side of #4 cleared fat pads of 3 weeks old recipient mice and the same concentration of Trp53−/− cell suspensions were injected into the contralateral left side fat pad. The transplanted fat pads were harvested and stained with Carmine Alum solution 8 weeks after transplantation (25). Outgrowths that occupied >5% of the fat pad were regarded as a successful outgrowth (26). Two methods were used to estimate the frequency of long-term regenerative mammary stem cells. The L-Calc software (Stemcell Tech, Vancouver, Canada) has been reported previously (27). We also used a generalized linear model approach assuming an underlying Poisson distribution of stem cell frequency to model the limiting dilution data using Stata (Stata Corp, College Station, TX). The regression model included a term for the multiplicative effect of Trp53+/+ (relative to Trp53−/−) and model adequacy was assessed using the link test. The Wilcoxon signed-rank test was used to compare the percentage of filled fat pad between Trp53+/+ and Trp53−/− epithelium. The model was not adjusted for the paired design where Trp53+/+ and Trp53−/− transplants are tested within each animal. The paired design would bias results in the direction of the null hypothesis resulting in a conservative estimate of statistically significant findings.
TM40A cell culture and retroviral infection
TM40A cells were maintained in regular MECL media: DMEM:F12 supplemented with 2% adult bovine serum (Gibco), 10μg/ml Insulin (Sigma), 20ng/ml mEGF, and 100u/ml Pen/Strep. Oligos coding for the p53 knockdown or scramble shRNA were annealed and cloned into pSicoR-PGK-puro vector (Addgene, Cambridge, MA). The p53 target sequence was GTACTCTCCTCCCCTCAAT and the scramble sequence was CGCTACACACTTCTTCTCC. The infection of TM40A cells with let-7c sensor plasmid, pSicoR-PGK-puro-p53KD plasmid or the control plasmids were performed as described previously (28, 29).
Flow cytometry and cell sorting
Cells were freshly collected and resuspended in DMEM:F12 supplemented with 1mM EDTA, 25mM HEPES, 1%FBS and 100u/ml Pen/Strep. The FACS data were collected using LSRII (Becton Dickinson, San Jose CA). A total 100,000 events were collected and analyzed using BD FACSDiva software (Becton Dickinson). Cell sorting was performed using a FACSVantage SE (Becton Dickinson).
Western blot
TM40A cell protein lysates were harvested 1 hour after 0-Gy or 10-Gy of γ-irradiation using RIPA buffer (50mM Tris, 150mM NaCl, 1% TritonX-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate, 1x protease inhibitor (Sigma, p8340) and 1x phosphatase inhibitor (Sigma, P5726)). Protein lysates (80μg) were separated by 10% SDS-PAGE and transferred to PVDF membrane (Millipore, Billerica, MA). The membrane was incubated with anti-phospho p53 (1:1000, Cell Signaling, 9284), or anti-β-actin (1:4000, Sigma, A1978), followed by incubation with hoseradish peroxidase conjugated secondary antibodies (1:5000, GE Healthcare, Little Chalfont, Buckinghamshire, UK), and developed using enhanced chemiluminescence (ECL) solution (GE Healthcare) in G:Box imaging system (Syngene, Cambridge, UK).
Label retaining cells
3 weeks old BALB/c-Trp53+/+ and BALB/c-Trp53−/− mice were injected with BrdU (Sigma) 300μg/10g body weight for 7 days. Mammary glands were harvested 9 weeks after the final injection. 5 Trp53+/+ mice and 3 Trp53−/− mice were used in this experiment. BrdU staining was done using the BrdU staining kit (Invitrogen, Carlsbad, CA) and the whole slides were counted for the total epithelial cells and LRECs.
RESULTS
p53 inhibits the expansion of mammary stem/progenitor cells
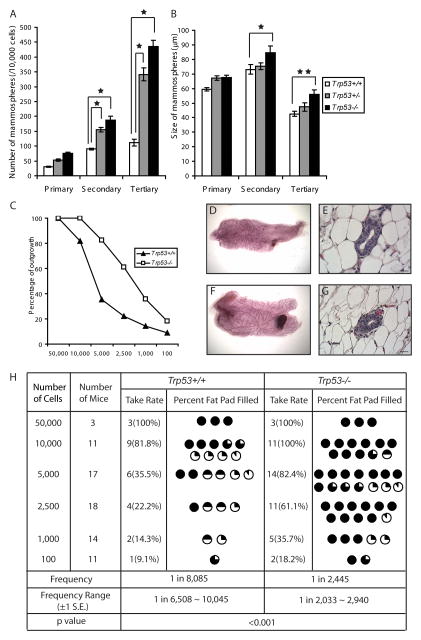
In order to examine the effect of p53 on mammary stem/progenitor cells, mammosphere formation capacity was compared among BALB/c-Trp53+/+, Trp53+/− and Trp53−/−mice. During serial passages, Trp53−/− epithelial cells gave rise to significantly higher numbers of secondary and tertiary mammospheres than wild type epithelium (p<0.01) (Fig. 1A), suggesting that p53 restricts expansion of mammary stem/progenitor cells. Trp53+/− epithelium also gave rise to higher numbers of mammmospheres than Trp53+/+, indicating the importance of p53 dosage with respect to regulation of mammary stem/progenitor cells. Furthermore, the Trp53−/− mammospheres are also larger than Trp53+/+ mammospheres, suggesting more extensive proliferation (Fig. 1B).
Fig. 1.
p53 inhibits the expansion of mammary stem/progenitor cells. (A) Trp53−/− and Trp53+/− mammary epithelial cells (MECs) gave rise to significantly higher number of mammospheres than Trp53+/+ MECs (p<0.01). The data shown represent 12 replicates for each genotype. The results were reproduced in a second independent experiment. (B) Trp53−/− MECs gave rise to significantly larger mammospheres than Trp53+/+ MECs upon serial passages (★p<0.01, ★★p<0.05). (C) Trp53−/− MECs gave a higher outgrowth rate than Trp53+/+ MECs. (D–F) Both Trp53+/+ (D, E) and Trp53−/− (F, G) outgrowths were histologically normal as shown by whole mount and HE staining. (H) The extent of fat pad filled for each successful outgrowth was recorded and the frequency of mammary stem/progenitor cells was estimated for Trp53+/+ and Trp53−/− using L-Calc software. Trp53−/− MECs contained significantly higher frequency of long-term regenerative mammary stem cells than Trp53+/+ MECs (p<0.001). Meanwhile, Trp53−/− outgrowths occupied significantly higher percentage of fat pad than the Trp53+/+ outgrowths (p<0.01).
To estimate the frequency of long-term regenerative mammary stem cells, we performed limiting dilution and transplantation to test the ability of cells to reconstitute the mammary gland. Total mammary cells were isolated from 8–10 week old aged-matched BALB/c-Trp53+/+ and Trp53−/− donor mice. The cells were transplanted into cleared mammary fat pads of 3-week old wild type BALB/c recipients. Both Trp53+/+ and Trp53−/− outgrowths showed normal ductal structure in both whole mounts (Fig.1D, F) and HE staining (Fig.1E, G). UsingL-Calc Software, the frequency of mammary stem cell in BALB/c-Trp53+/+ epithelium was estimated to be 1 in 8,085 (± 1S.E. 6,508 −10,045) compared to 1 in 2,445 (± 1S.E. 2,033 − 2,940) in BALB/c-Trp53−/− epithelium (Fig. 1C, H). The frequency of long-term regenerative mammary stem cells in Trp53−/−epithelium was 3.3-fold higher than the Trp53+/+ epithelium (p<0.001), suggesting that basal levels of p53 inhibits the expansion of mammary stem cells and that insufficient p53 dosage results in increased numbers of mammary stem cells between these genotypes. A generalized linear model approach was also applied and produced similar estimates of the difference in frequency of mammary stem cells. It is also noticeable that Trp53−/−outgrowths occupied a significantly higher percentage of the gland than the Trp53+/+ outgrowth (p<0.01), suggesting increased regenerative capacity of the Trp53−/− mammary stem cells (Fig. 1H).
p53-mediated apoptosis pathway is compromised in mammary stem/progenitor cells
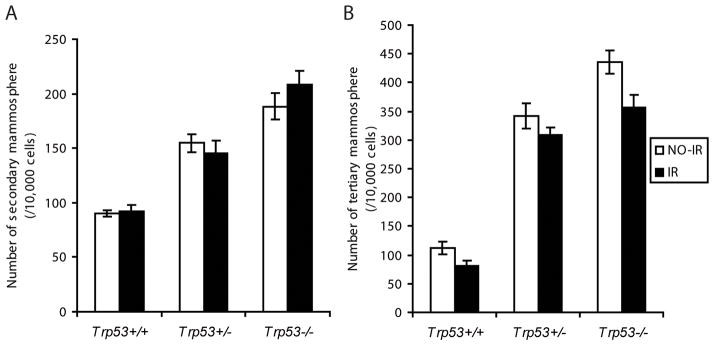
Ionizing radiation (IR) causes DNA double strand breaks, which induces preferentially p53-dependent cell cycle arrest and apoptosis. To test whether the different number of mammospheres may be an artifact of defective apoptosis in Trp53−/− cells, we used IR to trigger DNA damage and apoptosis. Upon serial passages, mammosphere cell suspensions were treated with either 0-Gy or 5-Gy γ-irradiation then seeded in parallel. Surprisingly, the number of secondary or tertiary mammospheres was not affected by IR in any of the genotypes (p>0.05) (Fig. 2A, B), suggesting that the mammosphere-initiating cells are resistant to IR and that the p53-mediated apoptosis pathway is compromised in these cells. These results indicate that the increase in mammary stem/progenitor cells is not attributed to differences in apoptosis or survival in Trp53−/− cells and that p53 acts by a distinct mechanism to limit the mammary stem/progenitor cells.
Fig. 2.
p53-mediated apoptosis is compromised in mammary stem/progenitor cells. Single cell suspensions were treated with 0-Gy or 5-Gyγ-irradiation before being plated and the number of secondary (A) and tertiary (B) mammospheres were compared. The mammospheres of different p53 genotypes showed no difference between control (NO-IR) and irradiated (IR) samples (p>0.05). The data shown represent 12 replicates for each treatment. The results were reproduced in a second independent experiment.
Trp53−/− mammary epithelium contained fewer label retaining epithelial cells (LRECs)
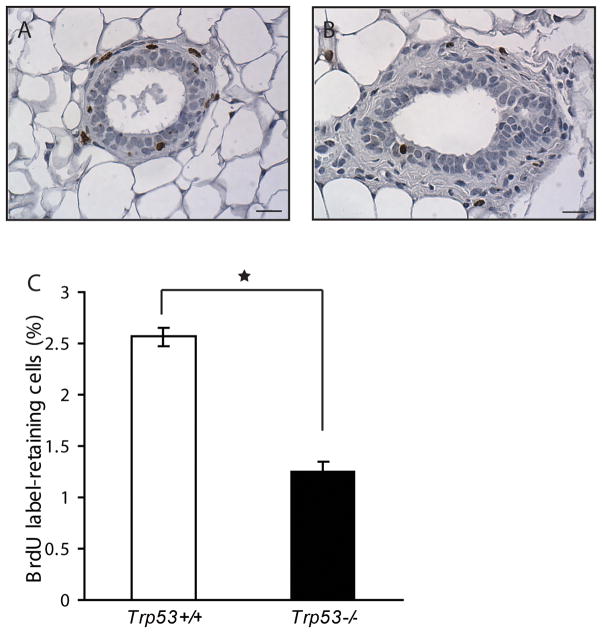
Non-random segregation of chromatids was reported in both embryonic stem cells and multilineage progenitor cells (30). It has been postulated that the tissue specific stem cells maintain their “stemness” and protect themselves from mutation through asymmetric segregation of their template DNA strands (31, 32). LRECs have been reported in mammary gland by using either [3H]-thymidine or BrdU labeling (33). We labeled BALB/c-Trp53+/+ and Trp53−/− mice with BrdU when 3 weeks old and chased for 9 weeks. Both genotypes exhibited similar incorporation of BrdU immediately after the labeling period. After 9 weeks of chasing, the BrdU-retaining epithelial cells were found in both luminal and basal compartments and the distribution of LRECs was similar among both genotypes (Fig. 3A, B). However, Trp53−/− mammary glands contained significantly fewer LRECs (1.26±0.09%) than the Trp53+/+ mammary glands (2.56±0.18%) (p<0.01, Fig. 3C), indicating that asymmetric segregation of DNA is impaired in the absence of p53 resulting in dilution of the BrdU label.
Fig. 3.
Trp53−/− mammary epithelium contained fewer label-retaining epithelial cells (LRECs). LRECs were found in both luminal and basal compartments and the distribution of LRECs were similar among Trp53+/+ (A) and Trp53−/− (B) mammary glands. Quantitative analysis showed that the Trp53−/− glands contained significantly lower number of LRECs than the Trp53+/+ glands (C) (p<0.01).
TM40A cells as an in vitro model to test the function of basal level p53
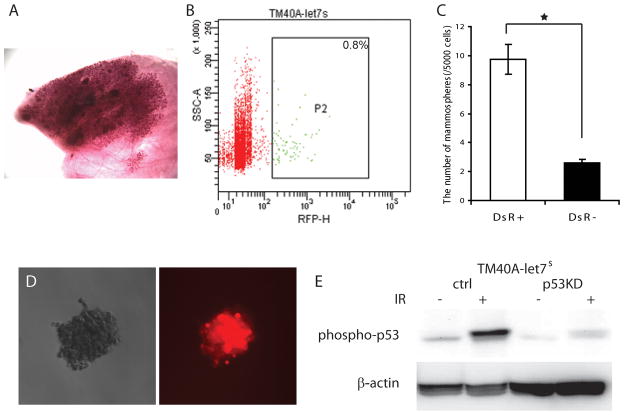
The role of p53 in regulation of mammary stem/progenitor cells was further confirmed in vitro using the TM40A cell line, a mammary epithelial cell line derived from BALB/c mice and retains wild type p53 mRNA (unpublished data, DJJ). TM40A cells form hyperplastic outgrowths when transplanted in vivo (Fig. 4A) but have undetectable tumorigenicity through 20 weeks. The let-7 microRNA family was shown to be depleted in the mammary progenitor cells and highly expressed in the more differentiated cell types and the let-7c sensor plasmid (let7s) has been used to label mammary progenitor population in vitro (28). The TM40A-let7s cells contained 0.8% of DsRed positive (DsR+) progenitor cells (Fig. 4B). The mammosphere-forming capacity of DsR+ cells is 3.8-fold greater than DsR- cells (p<0.01) (Fig. 4C), confirming their progenitor feature. Interestingly, most of the cells in the DsR+ mammospheres remained DsR+ (Fig. 4D).
Fig. 4.
TM40A cells as an in vitro model to test the function of basal level p53. (A) Whole mount of a TM40A outgrowth 20 weeks after transplantation. (B) The let7c-sensor plasmid was introduced into TM40A cells (TM40A-let7s). The TM40A-let7s cells contained 0.8% of DsRed positive (DsR+) progenitor cells. Background levels of fluorescence were determined using control cells. (C) The DsR+ cells gave rise to significantly more mammospheres than DsR− cells (p<0.01). (D) Most cells in the DsR+ mammospheres remained DsRed positive. (E) Western blot showed that phosphorylated p53 (phospho-p53) was reduced in TM40A-let7s-p53KD cells compared to the control cells (TM40A-let7s-ctrl).
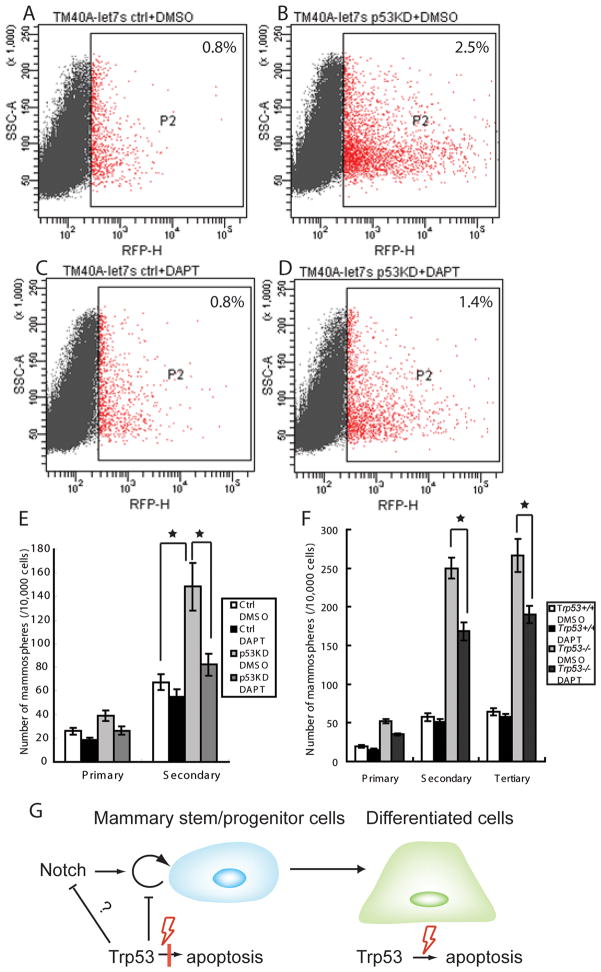
We proceeded to determine whether the knockdown of p53 can change the proportion of DsR+ progenitor cells. TM40A-let7s cells were infected with a p53 shRNA plasmid (TM40A-let7s-p53KD) or control plasmid (TM40A-let7s-ctrl) as described previously (29). The p53 shRNA decreased p53 protein to less than 24% of the original level (Fig. 4E). The TM40A-let7s-p53KD contained increased numbers of DsR+ cells (2.5%) compared to the TM40A-let7s-ctrl cells (0.8%) (Fig. 5A and B). The number of secondary mammospheres formed by TM40A-let7s-p53KD cells was also 2.2-fold higher than the control cells (p<0.01) (Fig. 5E), further proving that the basal level of p53 inhibits the expansion of mammary progenitor cells.
Fig. 5.
Inhibition of mammary stem/progenitor cells with γ-secretase inhibitor. (A-D) TM40A-let7s-p53KD cells contained more DsR+ progenitor cells compared to the TM40A-let7s-ctrl cells. The expansion of DsR+ progenitors in TM40A-let7s-p53KD cells can be inhibited by the treatment of DAPT, while the DsR+ cells in TM40A-let7s-ctrl cells were not affected. (E) TM40A-let7s-p53KD cells gave rise to significantly more secondary mammospheres than control cells (p<0.01). The number of p53KD mammospheres decreased significantly with the treatment of 5μM DAPT (p<0.01), while the number of control mammospheres were not affected (p>0.05). (F) Mammospheres were treated with either 5μM DAPT or DMSO control during serial passages. The number of Trp53−/− mammospheres decreased significantly after the treatment of DAPT (p<0.01). The number of Trp53+/+ mammospheres was not changed with DAPT treatment. (G) Model of p53 tumor suppression function in different cell types. p53 restricts the self-renewal of mammary stem/progenitor cells; however the p53-mediated apoptosis response is compromised in these cells. In the differentiated cells, the p53-mediated apoptosis pathway becomes functional. Notch may be inhibited by basal levels of p53 in mammary stem/progenitor cells. Insufficient p53 can result in increased Notch activity which leads to the expansion of mammary stem/progenitor cells.
Notch inhibitor reduced the number of mammary stem/progenitor cells
Increased mammary stem/progenitor cells could be vulnerable targets for carcinogenesis, especially in Li-Fraumeni patients in whom p53-mediated genome surveillance is compromised. We proceeded to test the potential of pharmacological methods to inhibit the expansion of mammary stem/progenitor cells. The Notch pathway has been reported to both promote and limit progenitor cells (5, 6), therefore we tested whether γ-secretase inhibitors (GSI), could affect the expansion of p53-deficient mammary stem/progenitor cells. The number of TM40A-let7s-p53KD mammospheres decreased to baseline levels after the treatment with DAPT, a GSI (Fig. 5E; p<0.01) indicating that Notch pathway could be a potential therapeutic target for downregulation of mammary stem/progenitor cells. The number of mammospheres in control cells with wild type p53 was not changed with DAPT treatment showing that inhibition of Notch was not a general effect, but specifically reversed the effect of p53-deficiency (Fig. 5E). DAPT treatment also restricted the expansion of progenitors measured by DsR+ sensor after p53 knockdown. In TM40A-let7s-p53KD cells, DAPT decreased the DsR+ cells from 2.5% to 1.4% (compare Fig. 5B and D), whereas the proportion of DsR+ cells was not changed by DAPT in the control group (compare Fig. 5A and C). Trp53−/− and Trp53+/+ primary mammary epithelial cells were also treated with 5μM DAPT or DMSO. Similarly, DAPT significantly inhibited the expansion of Trp53−/− mammospheres. The number of Trp53−/−secondary mammospheres decreased significantly from 250±13/10,000 cells to 168±11/10,000 cells after the DAPT treatment (p<0.01), but the number of Trp53+/+ mammospheres were not affected (Fig. 5F).
DISCUSSION
The importance of p53 in breast cancer is highlighted by the dramatic increase of breast cancer risk among women with Li-Fraumeni syndrome (16, 17). Although the function of activated p53 in mammary epithelium has been extensively studied, its role at basal levels under normal conditions is not fully understood. Both the mammosphere and limiting dilution data showed that insufficient basal levels of p53 resulted in increased numbers of mammary stem/progenitor cells. A gene dosage effect was also detected with the frequency of mammosphere-forming activity being intermediate for Trp53+/− mammary epithelium compared to the Trp53+/+ and Trp53−/−.
Label-retaining cell assays provide a measure of the asymmetric divisions of stem cells. Smith et al reported that by using [3H]-thymidine as the first label for LRECs and BrdU as secondary label for recently proliferating cells, most LRECs were actively synthesizing DNA yet retained their [3H]-thymidine labeled strands, suggesting that asymmetrically dividing cells contribute to most of LRECs (33). Organ specific stem cells could also be static and divide less frequently, which may also contribute to their label-retaining feature (31, 34). Recently, reports showed that p53 is essential for maintaining quiescence of mammary stem cells as well as hematopoietic stem cells (34, 35). In our experiment, the frequency of LRECs in wild type mice was 2.56±0.18%, which is close to that reported by Smith et al using [3H]-thymidine. We showed that Trp53−/− epithelium contained fewer LRECs than wild type epithelium. This could be explained by increased proliferation of Trp53−/− mammary stem cells, which dilute the BrdU after 9 weeks of chasing. Alternatively, p53 may regulate the asymmetric segregation of sister chromatids during mitosis, which could be vital for the fate decision of daughter cells. Loss of p53 may result in the disruption of this asymmetric segregation, leading to the loss of BrdU labeling after several rounds of division.
As p53 plays a prominent role in apoptosis, it was possible that differences in cell survival could contribute to the apparent increase in mammary stem/progenitor cells. However, radiation treatment failed to alter the number of secondary or tertiary mammospheres in any Trp53 genotype suggesting that p53-mediated apoptosis is compromised in the mammary stem/progenitor cell population. Previous studies have also demonstrated the resistance of mammary progenitors to therapeutic doses of ionizing radiation (36, 37). Furthermore, if differences in apoptosis were responsible for the apparent expansion of mammary stem/progenitor cells, the frequency of LRECs would be expected to be increased, but were in fact decreased significantly. Therefore, the expansion of mammary stem/progenitor cells cannot be attributed to altered survival of p53-deficient cells.
These results highlight the disparate functions and roles of p53 in different cell types. It was reported that ES cells could not activate p53-dependent responses to ionizing radiation because p53 protein was sequestered in the cytoplasm (38). Nonetheless, under basal conditions p53 was found to suppress expression of Nanog and induce differentiation of mouse ES cells (39). In mammary gland, irradiation triggers p53-mediated apoptosis in ductal epithelium (40), but this surveillance activity of p53 was not detectable in mammary stem/progenitor cells (Fig. 2). Nonetheless, the ability of basal levels of p53 to restrict the pool of progenitors was retained (Fig. 1). Therefore, the tumor suppressor function of p53 can be divided into two different aspects. In differentiated cells, p53 can be activated due to various genotoxic or cellular stresses so that the damaged cells will either be repaired or eliminated through apoptosis, depending on the extent of damage. In contrast, the apoptosis-inducing function of p53 is compromised in the mammary stem/progenitor cells, which prevents the loss of tissue-specific stem cells and the premature aging process due to DNA damage or other cellular challenges. In these cells, the major tumor suppressor function of p53 is to restrict self-renewal and inhibit inappropriate expansion of mammary stem/progenitor cells, a function that is independent of the pro-apoptotic function of p53 (Fig. 5G).
The elucidation of mammary stem cells and breast cancer stem cells has stimulated greatly the discussion of the cellular origins of breast cancers (21, 41). In small intestine, the deletion of the adenomatosis polyposis coli gene (Apc) in intestinal stem cells showed much higher transformation efficiency than in short-lived transit-amplifying cells, providing direct evidence of the stem cell origin of intestinal cancer (42). In mammary gland, the complexity of breast cancer subtypes and mammary epithelial cell hierarchy makes it hard to identify the cellular origins of breast cancer. Type I human breast epithelial cells (HBECs) express features of luminal stem/progenitor cells and show a greater potential for immortalization and transformation by oncogenes (43–45). The pathologic features of tumors also appear to differ among populations sequentially immortalized and transformed with TERT, SV40-T-antigen and activated Ras (46). These observations suggest that the mammary stem/progenitor cells are sensitive to oncogenic transformation, although the possibility of transformation of differentiated epithelial cells can not be ruled out. Therefore, modest increases in the prevalence of stem/progenitor cells would be anticipated to increase risk of breast cancer and are a likely source of cancer stem cells.
Germline heterozygous mutations in TP53 or BRCA1 significantly increase breast cancer risk. BRCA1 was also shown to regulate self-renewal and cell fate decision of mammary stem/progenitor cells. Loss of heterozygosity (LOH) of BRCA1 resulted in histologically normal lobules, which are comprised of progenitor cells and have higher transformation risk (4). Similarly, loss of p53 function is associated with basal-like breast cancers that express markers of embryonic stem cells (18, 19). Mammary tumors from p53-deficient mouse models also appear to develop from bipotent progenitor cells and have gene expression patterns similar to embryonic stem cells (22, 41). Our lab showed that 62% of spontaneous mammary tumors from Trp53+/− mice contained mixture of cells expressing either K5 or K8/18 (22). It is likely that the expansion of mammary stem/progenitor cells resulting from loss of p53 activity contributes to the great breast cancer risk due to their long life span and the ability to give rise to multiple lineages of differentiated cells. Therefore the inhibition of mammary stem/progenitor cell expansion may be a key target for prevention of hereditary breast cancers.
Expression of Notch pathway members were especially prominent in the gene expression patterns in mammary tumors of p53-deficient mice (22) suggesting that this pathway may contribute to the expansion of mammary stem/progenitor cells. We found that treatment of Trp53−/− primary cells and TM40A-let7s-p53KD cells with DAPT significantly downregulated the mammosphere-forming activity and the number of DsR+ progenitor cells. However DAPT did not change the mammosphere number of Trp53+/+ or TM40A-let7s-ctrl cells. This data agreed with the report of Dontu et al, which demonstrated that the Notch pathway upregulated the number of mammary stem/progenitor cells (5). The p53 protein has been reported to inhibit activation of the Notch pathway at different levels by either inhibiting the transcription of presenilin-1 (PS1) or competing with Notch-1 intracellular domain for co-activator p300/CBP (47–49). Similar mechanisms may lead to the inhibition of Notch pathway by p53 in mammary stem/progenitor cells. In the absence of p53, this inhibition is released, which allows expansion of mammary stem/progenitor cells. While in the presence of p53, the inhibition mechanism is intact and the treatment of DAPT did not affect the self-renewal of mammary stem/progenitor cells (Fig. 5G). Researchers have tried to apply GSI on breast cancer treatment and showed that GSI is effective in suppression of breast cancer stem cells and inhibition of breast cancer growth (50, 51). The GSI-sensitive signature suggested that pathways, including the Notch pathway and chemokine signaling pathway may contribute to the sensitivity of breast cancer to GSI (51). However, it remains possible that other targets of γ-secretase are important. Our results suggest that the Notch pathway is a potential therapeutic target to inhibit the expansion of mammary stem/progenitor cells and GSI may be applied to prevent breast cancer in patients with Li-Fraumeni syndrome.
SUMMARY
Proper regulation of the pool of progenitor cells is increasingly recognized as a factor in determining risk of breast cancer. We show that decreased p53 gene dosage results in increased numbers of progenitor cells through a mechanism that involves loss of asymmetric divisions and apparent increases in Notch activity, but not alterations in apoptosis. Inhibition of Notch signaling with a γ-secretase inhibitor reversed the effect of p53 loss resulting in restriction of the number of mammary stem/progenitor cells. As inhibition of Notch limited the pool of mammary stem/progenitor cells in p53-deficient mammary epithelium but had no effect in Trp53+/+ cells, it appears to be an effective treatment to prevent mammary tumors due to loss of p53 function with minimal consequences to cells with wild type p53.
Acknowledgments
We thank Dr. Daniel Medina for inspiring critiques and background information on TM40A cells, Dr. Gregory Hannon for the gift of let7c-sensor and control plasmids.
Research Support: This work was supported by the Predoctoral Traineeship Award from Department of Defense Breast Cancer Research Program (W81XWH-09-1-0028 awarded to Luwei Tao) and funding from the National Institute of Health (R01-CA095164, R01-CA105452, R01-ES015739 awarded to Dr. D. Joseph Jerry).
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Author Contribution
Luwei Tao: Conception and design, collection and assembly of data, manuscript writing
Amy Roberts: Other (technical support)
Karen Dunphy: Other (technical support)
Carol Bigelow: Data analysis and interpretation
Haoheng Yan: Conception and design, collection and assembly of data
D. Joseph Jerry: Conception and design, manuscript writing, financial support, final approval of manuscript
Reference List
- 1.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. NATURE. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. NATURE. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Welm B, Podsypanina K, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. PROC NATL ACAD SCI U S A. 2003;100(26):15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. PROC NATL ACAD SCI U S A. 2008;105(5):1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dontu G, Jackson KW, McNicholas E, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. BREAST CANCER RES. 2004;6(6):R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. CELL STEM CELL. 2008;3(4):429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Giono LE, Manfredi JJ. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J CELL PHYSIOL. 2006;209(1):13–20. doi: 10.1002/jcp.20689. [DOI] [PubMed] [Google Scholar]
- 8.Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. NAT REV MOL CELL BIOL. 2004;5(9):752–762. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- 9.Rohaly G, Chemnitz J, Dehde S, et al. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. CELL. 2005;122(1):21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez GS, Khan SH, Stommel JM, et al. p53 regulation by post-translational modification and nuclear retention in response to diverse stresses. ONCOGENE. 1999;18(53):7656–7665. doi: 10.1038/sj.onc.1203013. [DOI] [PubMed] [Google Scholar]
- 11.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. NAT REV MOL CELL BIOL. 2008;9(9):702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 12.Dumble M, Moore L, Chambers SM, et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. BLOOD. 2007;109(4):1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meletis K, Wirta V, Hede SM, et al. p53 suppresses the self-renewal of adult neural stem cells. DEVELOPMENT. 2006;133(2):363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Collado M, Villasante A, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. NATURE. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. NATURE. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleihues P, Schauble B, Zur HA, et al. Tumors associated with p53 germline mutations: a synopsis of 91 families. AM J PATHOL. 1997;150(1):1–13. [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols KE, Malkin D, Garber JE, et al. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. CANCER EPIDEMIOL BIOMARKERS PREV. 2001;10(2):83–87. [PubMed] [Google Scholar]
- 18.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PROC NATL ACAD SCI U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. NAT GENET. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. GENOME BIOL. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. NAT REV CANCER. 2007;7(10):791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 22.Yan H, Blackburn AC, McLary SC, et al. Pathways Contributing to Development of Spontaneous Mammary Tumors in BALB/c-Trp53+/− Mice. AM J PATHOL. 2010 doi: 10.2353/ajpath.2010.090438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerry DJ, Kittrell FS, Kuperwasser C, et al. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. ONCOGENE. 2000;19(8):1052–1058. doi: 10.1038/sj.onc.1203270. [DOI] [PubMed] [Google Scholar]
- 24.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. GENES DEV. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen Susan B, Young Lawrence JT, Smith Gilbert H. In: Preparing mammary gland whole mounts from mice. Margot MIp, Bonnie BA., editors. 2000. [Google Scholar]
- 26.Siwko SK, Dong J, Lewis MT, et al. Evidence that an early pregnancy causes a persistent decrease in the number of functional mammary epithelial stem cells--implications for pregnancy-induced protection against breast cancer. STEM CELLS. 2008;26(12):3205–3209. doi: 10.1634/stemcells.2008-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Britt KL, Kendrick H, Regan JL, et al. Pregnancy in the mature adult mouse does not alter the proportion of mammary epithelial stem/progenitor cells. BREAST CANCER RES. 2009;11(2):R20. doi: 10.1186/bcr2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibarra I, Erlich Y, Muthuswamy SK, et al. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. GENES DEV. 2007;21(24):3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura A, Meissner A, Dillon CP, et al. Cre-lox-regulated conditional RNA interference from transgenes. PROC NATL ACAD SCI U S A. 2004;101(28):10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. SCIENCE. 2006;311(5764):1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 31.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. CELL. 2008;135(6):1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 32.Shinin V, Gayraud-Morel B, Gomes D, et al. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. NAT CELL BIOL. 2006;8(7):677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 33.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. DEVELOPMENT. 2005;132(4):681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 34.Cicalese A, Bonizzi G, Pasi CE, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. CELL. 2009;138(6):1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Elf SE, Miyata Y, et al. p53 regulates hematopoietic stem cell quiescence. CELL STEM CELL. 2009;4(1):37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen MS, Woodward WA, Behbod F, et al. Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J CELL SCI. 2007;120(Pt 3):468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 37.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. PROC NATL ACAD SCI U S A. 2007;104(2):618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aladjem MI, Spike BT, Rodewald LW, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. CURR BIOL. 1998;8(3):145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 39.Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. NAT CELL BIOL. 2005;7(2):165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 40.Dunphy KA, Blackburn AC, Yan H, et al. Estrogen and progesterone induce persistent increases in p53-dependent apoptosis and suppress mammary tumors in BALB/c-Trp53+/− mice. BREAST CANCER RES. 2008;10(3):R43. doi: 10.1186/bcr2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M, Behbod F, Atkinson RL, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. CANCER RES. 2008;68(12):4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. NATURE. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 43.Kao CY, Nomata K, Oakley CS, et al. Two types of normal human breast epithelial cells derived from reduction mammoplasty: phenotypic characterization and response to SV40 transfection. CARCINOGENESIS. 1995;16(3):531–538. doi: 10.1093/carcin/16.3.531. [DOI] [PubMed] [Google Scholar]
- 44.Chang CC, Sun W, Cruz A, et al. A human breast epithelial cell type with stem cell characteristics as target cells for carcinogenesis. RADIAT RES. 2001;155(1 Pt 2):201–207. doi: 10.1667/0033-7587(2001)155[0201:ahbect]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 45.Sun W, Kang KS, Morita I, et al. High susceptibility of a human breast epithelial cell type with stem cell characteristics to telomerase activation and immortalization. CANCER RES. 1999;59(24):6118–6123. [PubMed] [Google Scholar]
- 46.Ince TA, Richardson AL, Bell GW, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. CANCER CELL. 2007;12(2):160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Laws AM, Osborne BA. p53 regulates thymic Notch1 activation. EUR J IMMUNOL. 2004;34(3):726–734. doi: 10.1002/eji.200324772. [DOI] [PubMed] [Google Scholar]
- 48.Oswald F, Tauber B, Dobner T, et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. MOL CELL BIOL. 2001;21(22):7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roperch JP, Alvaro V, Prieur S, et al. Inhibition of presenilin 1 expression is promoted by p53 and p21WAF-1 and results in apoptosis and tumor suppression. NAT MED. 1998;4(7):835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- 50.Harrison H, Farnie G, Howell SJ, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. CANCER RES. 2010;70(2):709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watters JW, Cheng C, Majumder PK, et al. De novo discovery of a gamma-secretase inhibitor response signature using a novel in vivo breast tumor model. CANCER RES. 2009;69(23):8949–8957. doi: 10.1158/0008-5472.CAN-09-1544. [DOI] [PubMed] [Google Scholar]