Technological advances in molecular biology and genetics have allowed neuroscientists to reduce the study of the brain to individual proteins or genes of interest. The challenge of making the jump from such detailed molecular study to an integrated understanding of brain structure or function is highlighted by the large number of genes and combinations of genes and the time necessary to study the function of any one gene product individually. These same advances in molecular biology that have propelled our knowledge of neuronal development and functioning over the last decade have also widened an implicit division between molecular and systems neuroscientists. cDNA microarray technology offers the exciting potential to bridge the molecular-systems gap by allowing for a systems-level molecular or “genomic” approach to neurobiology because of its unprecedented capacity to monitor simultaneously widespread changes in gene expression. The parallel monitoring of thousands of genes provides a more global view of the system being studied than classic reductionist techniques and requires sophisticated quantitative analytic and computational approaches that are not currently available at the bench of the typical molecular neurobiology lab. Despite some of the barriers to entry, microarray expression profiling is one of several genetic approaches that will have a broad and lasting impact on our molecular and systems understanding of brain development and functioning.
Two basic platforms for fluorescent microarray expression studies currently exist—oligonucleotide arrays and spotted cDNA arrays (Fig. 1; refs. 1 and 2). In this issue of PNAS, Sandberg et al. (3) have elegantly applied microarray expression profiling by using oligonucleotide arrays (4, 5) to begin to map out the baseline differences in gene expression that may underlie structural and behavioral differences between inbred laboratory mouse strains. Over 400 inbred mouse strains with known genealogy exist, and their power for the dissection of complex behavioral and quantitative trait analysis is gaining more widespread appreciation (6–8). The synergistic combination of mouse genetics and microarray expression profiling presented here is paradigmatic for future work in this area.
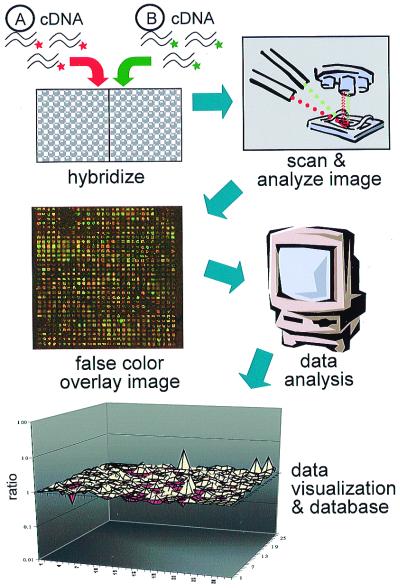
Figure 1.
Microarray experimental schema. cDNA-synthesized RNA from two samples, A and B, is simultaneously hybridized onto a spotted ordered array of cDNA clones. Each sample is labeled with fluors that emit at different wavelengths so that signal from each sample can be distinguished. On the false color image depicted, red signals represent stronger Sample A hybridization, and green signals represent stronger Sample B hybridization or relative enrichment. Yellow spots indicate equal abundance in both samples, and a yellow signal is observed in control nondifferentially expressed spots (group of four in lower left-hand corner). A three-dimensional matrix plot of gene expression data is depicted simply to illustrate one form of analysis and data visualization. The highest peaks represent the most differentially expressed genes. Sandberg et al. (3) use the genechip oligonucleotide array system (Affymetrix, Santa Clara, CA), in which samples are hybridized individually onto separate arrays, and interarray comparisons are made by using a global scaling factor that normalizes by scaling to the average signal intensity on each array. The term “probe” refers to the DNA attached to the array surface and the term “target” to the cDNA that is hybridized onto the array. Radioactive targets may be used as well, and each system has advantages and disadvantages in terms of cost, sample preparation, ease of use, and reproducibility.
Sandberg et al. study baseline gene expression differences in six brain regions (cortex, hippocampus, amygdala, entorhinal cortex, midbrain, and cerebellum) in two mouse strains—C57BL/6 (BL6) and 129SvEv (129)—that have been shown previously to have major differences in behavior and disease susceptibility (9, 10). Furthermore, embryonic stem cells used to generate knockouts are typically derived from 129 strains, and the C57BL/6 is commonly used in pharmacology and quantitative trait mapping. Thus, appreciation of the influence of genetic background on gene expression in these strains has significant practical implications. Comparisons are made between gene expression in the two mouse strains and between different regions within each strain. Mouse embryonic fibroblasts are used as a non-central nervous system comparison sample. Gene expression differences after a drug-induced seizure are determined as an example of the molecular phenotypes underlying the differential disease susceptibility of the two strains.
Several aspects of the approach that make this a well-designed microarray experiment deserve highlighting. The first is the determination of experimental reliability and reproducibility. Small changes in experimental conditions can alter gene expression significantly in small numbers of genes, a problem magnified when thousands of genes are queried in parallel. To assess variability because of dissection methods and other nongenetic factors, the authors (3) compared duplicate brain samples from the same region from the same strain and demonstrated that fewer than 0.02% (2/13,000) of probe sets on the array were differentially expressed. This verifies a high level of experimental reproducibility that reflects careful experimental methods and stringent analytic criteria that accepted only the most robust changes. The use of these analytic criteria is supported further by the identification of changes previously shown to be differentially expressed between the two strains. This demonstration of reproducibility and low noise level in these control samples is critical for the further interpretation of data generated in the experimental conditions and lends significant credence to the results. Another strength is the use of alternative methods to confirm a subset of the differences observed on the microarrays. Sandberg et al. use reverse transcription–PCR and Northern blotting to confirm qualitatively the differential expression of several of the genes identified (3). Although only one Northern blot is shown, care was taken to compare the consistency of the microarray data with previously published regional expression patterns. One related issue that has not been discussed adequately in the microarray literature is to what extent microarray expression data compare quantitatively with more standard methods such as Northern blotting. Anecdotal experience suggests that low-stringency microarray hybridizations have higher background, blunting the observed mRNA expression differences relative to other techniques such as Northern blotting. This does not detract from the utility of microarray technology but highlights an area where care and caution have to be taken in the interpretation of results. Sandberg et al. (3) and others (4, 11–15) demonstrate that microarray expression profiling has great utility for screening thousands of mRNAs in parallel to identify those with the largest expression changes between comparison samples.
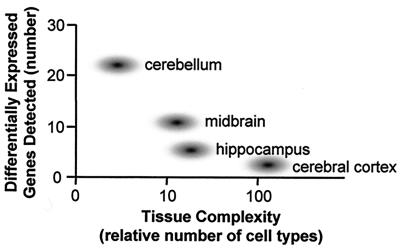
In this regard, it is remarkable that of more than 7,000 expressed genes detected on the array by Sandberg et al. (3), only 24 genes were identified that were differentially expressed in all six brain regions between the two strains. Seventy-three genes were differentially expressed in at least one of the six brain regions between the two strains, or approximately 1% of the genes showing hybridization signals on the array. A similar number of genes were detected as differentially expressed between different brain regions within the same strain. On the basis of this analysis, Sandberg et al. postulate that the cerebellum is the most molecularly distinct region, with 23 unique genes, followed by the midbrain (n = 10), hippocampus (n = 4), and cortex (n = 2). Furthermore, the percent of genes detected that are differentially expressed between fibroblasts from the two strains (1%) is the same as that detected between the brains of the two strains. These results bear careful scrutiny. Are the molecular distinctions between fibroblasts from 129SvEv and C57BL/6 equal in magnitude to those between their brains? How can the histologically less complex cerebellum show many more uniquely expressed genes than the cerebral cortex or hippocampus? It is counterintuitive that a tissue comprised of two major neuronal phenotypes appears the most molecularly complex of all brain regions sampled.
These unexpected results likely reflect methodological limitations rather than a complete catalogue of gene expression differences. cDNA microarray analysis is limited currently to the detection of genes expressed at a relative abundance of about 1/100,000 mRNAs, although the sensitivity continues to improve (1, 5, 14). Additionally, because stringent analytic criteria were used by Sandberg et al., genes expressed at the low end of detection limits that may yield more variable signals would be at a further disadvantage. Because most arrays, including those used here, are based on known genes and expressed sequence tags, which are currently biased toward more abundant genes, their utility for detecting changes in low-abundance genes may be further restricted.
Current models of central nervous system (CNS) development postulate that specific aspects of neuronal phenotype are dictated by low-abundance mRNAs (16) or by those expressed at very specific times in development (17, 18). Differences in these low-abundance species are probably at the detection limit of current microarray experiments. This problem is magnified further in complex CNS tissues such as the cerebral cortex, where hundreds of different cell types may reside within a few square millimeters of tissue (17–19), further diluting low-abundance RNAs by another two orders of magnitude. Because fibroblasts are homogeneous cell populations, even relatively rare mRNAs expressed at 1 to 5 copies per 100,000 may be detected. This is in contrast to brain tissue, which shows marked cellular heterogeneity (17–19). In the same vein, the simplest of the CNS tissues studied, the cerebellum, appears to show more genetic variability than the cerebral cortex. Thus, another interpretation of the data presented is that the less complex the tissue studied, the more likely expression differences will be identified should they be present (Fig. 2). This dilution of detected molecular differences by increasing tissue complexity is emphasized further by the finding that 7,169 of the 13,069 genes represented on the array (55%) were detected by this microarray system as expressed in the adult mouse brain. This is in contrast to the approximately 9,500 genes detected as expressed by fibroblasts. At face value, this suggests the implausible conclusion that more genes are expressed in fibroblasts than in the mouse CNS. However, if the alternative interpretation that the critical low-abundance genes have not been queried is correct, the data in this paper highlight the potential importance of simplified systems or detailed single-cell analysis (20, 21) in fully categorizing gene expression in complex tissues. Further expression analyses of individual cell types or laminae within the brain regions studied are needed to test this premise. This does not mean that studies using CNS tissue are not valuable, nor does it limit the value of expression changes that were detected by this analysis. It merely indicates that an incomplete subset of total expression differences will be detected in complex tissues by using current technologies.
Figure 2.
Molecular diversity and tissue complexity. There is a rough inverse correlation between the number of uniquely expressed genes in a region and the number of cell types. The cell numbers on the x axis are approximations included to facilitate comparison.
One advantage of these data for neuroscientists is that the relatively small number of genes identified as differing between brain regions at baseline or after a seizure facilitates the in-depth study of how these genes may contribute to specific phenotypes. Do genes that are enriched in one particular brain region reflect differences in cell number or the specific novel molecular phenotype of a particular cell type? Followup study with higher-resolution techniques, such as in situ hybridization, is likely to lead to a structural and molecular understanding of the observed strain differences as well as of the potential role of each gene product in the brain's vulnerability to environmental insults that provoke seizures. Functional analysis is facilitated, because many of the differentially regulated genes have previously defined roles (3). The presented comparison of only two mouse strains does not yet allow for strong correlations with seizure susceptibility but serves as a template and proof of principle that should guide future studies. The data generated by Sandberg et al. begin to address the issue of how much genetic variability might contribute to observed experimental variability. The availability of these data on the Internet makes them an accessible resource that others can use, analyze, and contribute to.
When compared with other model organisms, inbred mouse strains currently offer considerable power to the neuroscientist who wishes to exploit genetic methods to study neurobehavioral phenotypes or other quantitative traits (6–8). As Sandberg et al. emphasize (3), inbred mouse strains exhibit significant variation in a wide range of neurological phenotypes, including susceptibility to neurodegeneration, seizure threshold, and cytotoxic cell death, as well as behavioral phenotypes, such as spatial memory or fear conditioning (6). Also, quantitative anatomical traits such as the size of various brain regions, neuronal types, or even neuron number show significant variability between mouse strains that are caused by heritable factors and thus provide an avenue for identifying the structural components that underlie behaviors (22). The combination of microarray expression analysis with traditional mapping and positional cloning approaches in mice (23) is likely to provide an efficient route to disease gene or quantitative trait (QTL) identification. Multiple gene–gene interactions and gene–environment contributions to complex neurobehavioral phenotypes constitute one area where the use of microarray gene expression analysis may have considerable advantages over classic QTL analysis. Instead of genotyping, quantitative trait analysis could be conducted by using cDNA expression data across a range of recombinant inbred strains. cDNA expression probing would consider simultaneously the effects of environment and epistasis and would provide a more direct functional assay than genotyping. Inbred mouse strains also may offer the most powerful route to understanding the influences of the environment on gene expression, because genetic variability is controlled (24). Additionally, the gene expression pattern of particular brain regions can be treated as phenotypic assays with which to identify mutant animals of interest in mutagenesis screens. Appreciation of baseline gene expression differences in inbred mouse strains that are the staples of these approaches and developing databases that catalogue these differences are the necessary preludes for these experiments, as Sandberg et al. describe (3).
In contrast to those in experimental animals, cDNA expression studies aimed at understanding the molecular genetic basis for human phenotypes face the confounding variables of tissue preservation, postmortem interval, and other environmental variables. The present findings in mice suggest that differing genetic backgrounds among human subjects may contribute significantly to variability in gene expression. However, this source of variation has not been dealt with in any published study using human brain tissue. This does not alter the utility of microarrays for expression studies in humans but rather highlights important variables that should be considered when interpreting experimental results. In fact, cDNA microarrays could provide an efficient functionally relevant method for the analysis of individual variation in brain gene expression in humans. These variations, coupled with environmental factors, form the basis for individual variability in behavior, personality, and cognition and the brain structures that underlie them.
Acknowledgments
I thank Ms. Bonita Porch for her editorial assistance; Ms. Carol Gray of the University of California, Los Angeles, Mental Retardation Research Center's Media Core for her help with the figures; and Harley Kornblum, M.D., Ph.D., for his critical reading of the manuscript. Acknowledgement is also given to the National Institute of Mental Health (Grant no. MH60233) for their support of our microarray research.
Footnotes
See companion article on page 11038.
References
- 1.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 3.Sandberg R, Yasuda R, Pankratz D G, Carter T A, Del Rio J A, Wodicka L, Mayford M, Lockhart D J, Barlow C. Proc Natl Acad Sci USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipshutz R J, Fodor S P, Gingeras T R, Lockhart D J. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart D J, Winzeler E A. Nature (London) 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 6.Frankel W N. Trends Genet. 1995;11:471–477. doi: 10.1016/s0168-9525(00)89155-6. [DOI] [PubMed] [Google Scholar]
- 7.Beck J A, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig J T, Festing M F, Fisher E M. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 8.Blake J A, Eppig J T, Richardson J E, Davisson M T. Nucleic Acids Res. 2000;28:108–111. doi: 10.1093/nar/28.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawley J N, Belknap J K, Collins A, Crabbe J C, Frankel W, Henderson N, Hitzemann R J, Maxson S C, Miner L L, Silva A J, et al. Psychopharmacology. 1997;132:107–124. doi: 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- 10.Ingram D K, Jucker M. Neurobiol Aging. 1999;20:137–145. doi: 10.1016/s0197-4580(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 11.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 13.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 14.Brown P O, Botstein D. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 15.Diehn M, Eisen M B, Botstein D, Brown P O. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhari N, Hahn W E. Science. 1983;220:924–928. doi: 10.1126/science.6189184. [DOI] [PubMed] [Google Scholar]
- 17.Levitt P, Barbe M F, Eagleson K L. Annu Rev Neurosci. 1997;20:1–24. doi: 10.1146/annurev.neuro.20.1.1. [DOI] [PubMed] [Google Scholar]
- 18.McConnell S K. J Neurosci. 1995;15:6987–6998. doi: 10.1523/JNEUROSCI.15-11-06987.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawatari A, Callaway E M. Neuron. 2000;25:459–471. doi: 10.1016/s0896-6273(00)80908-3. [DOI] [PubMed] [Google Scholar]
- 20.Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Proc Natl Acad Sci USA. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo L, Salunga R C, Guo H, Bittner A, Joy K C, Galindo J E, Xiao H, Rogers K E, Wan J S, Jackson M R, Erlander M G. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 22.Williams R W, Strom R C, Goldowitz D. J Neurosci. 1998;18:138–146. doi: 10.1523/JNEUROSCI.18-01-00138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikke B A, Johnson T E. Mamm Genome. 1998;9:963–968. doi: 10.1007/s003359900907. [DOI] [PubMed] [Google Scholar]
- 24.Lee C K, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]