Abstract
Limited proteolysis, accomplished by endopeptidases, is a ubiquitous phenomenon underlying the regulation and activation of many enzymes, receptors and other proteins synthesized as inactive precursors. Serine proteases are one of the largest and conserved families of endopeptidases involved in diverse cellular activities including wound healing, blood coagulation and immune responses. Heteromeric α,β,γ-epithelial sodium channels (ENaC) associated with diseases like cystic fibrosis and Liddle’s syndrome, are irreversibly stimulated by membrane-anchored proteases (MAPs) and furin-like convertases. Matriptase/Channel activating protease-3 (CAP3) is one of the several MAPs that potently activate ENaC. Despite identification of protease cleavage sites, the basis for enhanced susceptibility of α- and γ-ENaC to proteases remains elusive. Here, we elucidate the energetic and structural bases for activation of ENaC by CAP3. We find a region near the γ-ENaC furin site that is previously unidentified as a critical cleavage site for CAP3-mediated stimulation. We also report that CAP3 mediates cleavage of ENaC at basic residues downstream of the furin site. Our results indicate that surface proteases alone are sufficient to fully activate uncleaved ENaC, and explain how ENaC in epithelia expressing surface-active proteases can appear refractory to soluble proteases. Our results support a model in which proteases prime ENaC for activation by cleaving at the furin site, and cleavage at downstream sites is accomplished by membrane surface proteases or extracellular soluble proteases. Based on our results, we propose a dynamics-driven “anglerfish” mechanism that explains less stringent sequence requirements for substrate recognition and cleavage by matriptase compared to furin.
Keywords: ENaC, serine endopeptidase, Xenopus, voltage clamp, discrete molecular dynamics
Limited proteolysis is the last step in the attainment of a functional form of many proteins of biological significance and perhaps the first step in protein degradation (1). This important regulatory phenomenon is frequently observed in activation of many enzymes, hormones, receptors, and other biologically active proteins and is conserved through evolution (2). Early structural studies on limited proteolysis led to two opposing theories that argued whether it is segmental mobility or surface accessibility of the proteolytic fragment that determines protease susceptibility (3, 4). Following these hypotheses, others have performed systematic computational analyses concluding that local unfolding, in addition to segmental mobility and surface accessibility, guides protease susceptibility (5, 6). Although the structural requirements for cleavage by serine proteases are relatively well characterized (7), the energetics of enzyme-substrate interactions remains unclear. Determining the energetics of enzyme-substrate interactions is critical to understand the subtle differences in substrate specificities featured by serine proteases. The ENaC family of ion channels offers a unique platform to study limited proteolysis due to the uniqueness of the mode of their regulation via cleavage by proteases, resulting in constitutive channel activation.
ENaC is considered the rate-limiting step in electrogenic Na+ absorption from luminal compartments lined by epithelia. Na+ absorption by ENaC is critical in many biological processes including renal and intestinal control of salt balance and blood pressure and affects the functioning of many organs including airways, sweat ducts, cornea and inner ear (8–11). Over the last decade it has become clear that an important determinant of ENaC activity is the extent of partial proteolysis of the channel subunits (12, 13). Proteolytic regulation of ENaC includes selective cleavage by furin-like proteases during biosynthetic maturation as well as cleavage at the cell surface by proteases that can be membrane-associated or soluble. The current model for proteolytic regulation of ENaC is that the open probability (PO) of the heteromultimer is determined by cleavage events, so far confined to its α and γ subunits. In general, channels made of uncleaved subunits exhibit a very low PO and a range of cleavage events increases PO. Two specific gaps in understanding proteolytic activation of ENaC involve the dominant role of γ-ENaC in proteolytic regulation of the channel. First, studies of at least five ENaC activating proteases identify essential sites in γ-ENaC, despite accepted importance of furin-like cleavage sites in α-ENaC. Second, the structural and/or energetic features of the region of γ-ENaC (residues 130–200), that renders it susceptible to cleavage by multiple proteases, have not been investigated.
Previous studies on MAPs including prostasin/CAP1 and TMPRSS4/CAP2 revealed an important role for γ-ENaC in channel regulation via limited proteolysis (14–16). Additionally, the extracellular domain of γ-ENaC has been hypothesized to harbor an allosteric regulatory subdomain with an important role in channel function (17). Interestingly, proteolytic activity of CAP1 is not required for its stimulation of ENaC. This observation led to a suggestion that CAP1 may play a critical non-catalytic role in ENaC regulation, perhaps as part of a cascade of surface associated proteases (18). It was recently demonstrated that matriptase/CAP3 is a critical activator of CAP1 (19). In this study we focus on the activation of rat ENaC by CAP3-mediated cleavage of the γ subunit.
To uncover the energetic basis for activation of ENaC upon cleavage, we performed computational analyses on peptides from ENaC that are susceptible to proteolysis by serine proteases. We performed discrete molecular dynamics (DMD) simulations to elucidate the structural and energetic bases for ENaC peptide recognition by CAP3. Our computational studies of various peptide-binding configurations to CAP3 establish the structural and energetic bases for CAP3 activity. Using potential of mean force (PMF) analyses, we determined the energetic basis for substrate recognition and cleavage by CAP3. We compared the results to those obtained via similar analyses for furin and elucidate the energetic basis for the lower sequence specificity of CAP3 compared to furin. We designed experimental studies to identify CAP3-mediated cleavage sites in ENaC that lead to channel activation. We conclude that CAP3, with less stringent sequence requirements than furin, robustly activates ENaC by cleaving at multiple basic residues in the extracellular domain. Using computational and experimental studies, we show that a site upstream of the traditional furin site in γ-ENaC is a potential substrate for CAP3. Based on our findings, we propose an “anglerfish” mechanism driven by dynamics of the enzyme whereby the peptide substrate interacts with and is stabilized by the loops enveloping the active site of the enzyme and is subsequently subject to proteolysis. This two-step mechanism underscores the importance of the energetic contribution of surface loops in serine proteases to their proteolytic activity. Furthermore, this mechanism provides a broad outlook for understanding stringency in sequence requirements for cleavage by serine proteases.
Materials and Methods
Enzyme-peptide Docking
We represented γ-ENaC cleavage sites tentatively identified by mutagenesis by 8-mer peptides with the putative wild type P4-P1 cleavage sequence contained in the first four residues. We constructed three linear 8-mer peptides from rat γ-ENaC (Seq1: 135-RKRREAGS; Seq2: 178-RKRKISGK; Seq3: 132-KESRKRRE) such that the P1 site is between the fourth and fifth residues. We chose the initial configuration for the peptide by random placement at sites distant from the active site of either enzyme. We imposed two distance constraints – one between the epsilon nitrogen atom (NE2) of active site histidine and backbone amine of the P1’ site on the peptide and the other between the gamma oxygen atom (OG) of active site serine and carbonyl oxygen of the P1 site – to draw the peptide close to the active pocket. In order not to bias the configuration of the peptide in the active site of the enzyme, we placed the peptide at ten different, randomly chosen starting positions and with different orientations with respect to one another (Figure S2A, B). We performed replica exchange DMD simulations of each such initial configuration with eight replicas in a temperature range of 0.35 to 0.75 reduced units at increments of 0.035 units (Figure S2C) (20). DMD uses Medusa force field to treat interactions between atoms in the macromolecule (21). We used EEF1, an implicit solvation model to treat the solvation of the simulation system (22). Energy minimization of the crystal structure of furin and matriptase was performed using Chiron prior to replica exchange simulations (23). The total simulation time was 106 DMD time units for each replica. Each DMD time unit is approximately 50 fs in real time, accounting for a total simulation time of 50 ns per replica. The relationship between DMD time unit and real time and their inter-conversion is discussed elsewhere (24). Proteases were maintained static during simulations allowing movement of only the loops surrounding the active site. We selected snapshots across simulation trajectories of all replicas that satisfied both the distance constraints and clustered them based on pair wise root mean square deviation (RMSD) of Cα atoms. We selected the representative structures from five such clusters and performed side-chain optimization using the fixed backbone custom design protocol from the Medusa suite (21).
PMF Calculations
To estimate the free energy of peptide binding for each enzyme-peptide combination, we computed 2D-PMF using distance from the active site and energy as reaction coordinates. To obtain an accurate estimate of free energy, we removed the bias from the simulation trajectories by subtracting the appropriate value of constraint potential as a function of distance from the active site. We used the multiscale modeling tools for structural biology (MMTSB) toolkit (25) to perform WHAM (weighted histogram analysis method) analysis with replica-exchange simulation trajectories and compute PMFs at a given temperature. Using WHAM analysis, we self-consistently computed the density of states by combining overlapping histograms from different simulation trajectories (26). We generated 2D-contour plots of the 2D-PMF values using gnuplot (http://www.gnuplot.info).
Peptide Disorder Prediction
We used Disopred2 (http://bioinf.cs.ucl.ac.uk/disopred) to analyze the peptide sequences of alpha, beta and gamma subunits of rat ENaC. We used the sequences L150–L290 from rat α-ENaC, K117-P240 from rat β-ENaC and K91-S222 from rat γ-ENaC.
Plasmid Preparation
For biochemical analyses of ENaC subunit proteolysis, cDNAs encoding rat α,β and γ-ENaC with HA-N-terminal (HA-NT) and V5-C-terminal (V5-CT) epitope tags were generated. Wild type and mutant constructs (α-ENaC; R205A/R231A, β-ENaC, γ-ENaC; R135Q/K136Q/R137Q/R138Q, R135Q/K136Q/R137Q/R138;R135Q/K136Q/R137/R138Q;R135Q/K136/R137Q/R138Q/R135/K136Q/R137Q/R138Q, hepatocyte growth factor activator inhibitor 1 and 2 (HAI-1, HAI2) and CAP3 were generated by PCR and cloned into pCR-BluntII-TOPO (Invitrogen), linearized (HindIII) and in vitro transcribed using T7 RNA polymerase. A PolyA tail was added after transcription (Ambion). Mutations were done with the Quikchange multi site-directed mutagenesis kit (Stratagene). The WT ENaC plasmids were generously provided by Dr. Bernard Rossier. The sequence of all plasmids was verified at the University of North Carolina sequencing facility.
Western Blot Analysis
Proteins were extracted from oocytes as described above. Biotinylated and total proteins were solubilized by boiling in Laemmli sample buffer for 10 min prior to loading onto 4–12% SDS-PAGE gels. Western blots were performed with anti-V5 (Invitrogen), anti-HA (Covance), anti-CAP3 (Bethyl, Laboratories, Inc.) and anti-actin (Chemicon International) antibodies.
Functional Studies of ENaC in Xenopus oocytes
V–VI stage healthy oocytes were harvested as described previously (27) and maintained in modified Barth’s solution (MBS) at 18°C. Animals were maintained and studied under protocols approved by the University of North Carolina Institutional Animal Care and Use Committee. Oocytes expressing the desired combinations of ENaC subunits and CAP3 were obtained as before (14). Briefly, cRNAs encoding wild-type (WT) of both untagged and HA-NT/V5-CT epitope tagged subunits or mutant HA-NT/V5-CT tagged subunits of rat αβ and γ-ENaC (0.3 ng each) and CAP3 cRNA (typically 1 ng) were co-injected into oocytes. Twenty-four hr after injection, two-electrode voltage clamping was performed using a Genclamp amplifier (Axon Instruments) in a constant perfusion system. Currents were measured in the presence and absence of 10 µM amiloride, with membrane voltage clamped to −100 mV. Currents were digitized and recorded using a Digidata 1200 A/D converter (Axon Instruments) and Axoscope software. After basal amiloride sensitive current (INa) was recorded by washing out amiloride, oocytes were superfused with amiloride containing buffer and trypsin or hNE (2–20 µg/ml) for 5 m, followed by a second determination of INa. All results are expressed as the mean ± S.E. or as fold stimulation by CAP3 or hNE. The means of two groups were tested for significant difference using an unpaired Student’s t test, differences between three or more groups were evaluated using ANOVA analysis (GraphPad Prism software). Proteins extracted from control and injected oocytes were analyzed by Western blots to verify expression of ENaC and actin.
Surface labeling
Xenopus oocytes were injected with desired combinations of WT or mutant double epitope (HA/V5) tagged αβ and γ rat ENaC subunits (0.3 ng each) and with or without CAP3 cRNA (1 ng). After 24 hrs, 70 oocytes per experimental condition were pre-chilled on ice for 30 minutes and labeled with 0.7 mg/ml sulfo-NHS-biotin in MBS-Ca++ (mM), 85 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.41 CaCl, 0.33 Ca(NO3), 16.3 hepes titrated to pH 8.0 with NaOH, while tumbling gently for 20 min at 4°C. Oocytes were washed twice with chilled MBS-Ca++ buffer and incubated in MBS-Ca2+ buffer with 100 mM glycine for 10 min at 4°C to quench free biotin. Oocytes were washed again three times with chilled MBS-Ca++ buffer, then lysed with lysis buffer (in mM; 20 Tris, 50 NaCl, 50 NaF, 10 β-glycerophosphate, 5 Na4P2O7 pyrophosphate, 1 EDTA, pH 7.5 containing protease inhibitors (complete, Roche), aprotinin (Sigma)). Cell lysates were prepared by passing oocytes through a 27G1/2 needle twice and by centrifugation at 3,600 rpm for 10 minutes at 4°C. Supernatants were transferred to new tubes and samples were spun at 14,000 rpm for 20 minutes at 4°C. Supernatants were discarded and pellets were solubilized in solubilization buffer (in mM; 50 Tris, 100 NaCl, 1% triton X-100, 1% NP-40, 0.2% SDS, 0.1% Na deoxycholate, 20 NaF, 10 Na4P2O7 pyrophosphate, 10 EDTA + protease inhibitor cocktail, pH 7.5). Total inputs were taken from whole cell samples representing 4% of total protein. Solubilized proteins were incubated with 100 µl of neutravidin beads (Pierce) overnight while tumbling at 4°C. Samples were washed twice with (mM) 500 NaCl 50 Tris pH 7.5 buffer and once with 150 NaCl 50 Tris pH 7.5 buffer. Laemmli buffer was added and samples were loaded on a 4–12% gradient Tris-glycine gel after incubation for 10 minutes at 96°C. Samples were transferred to 0.45 µm polyvinylidene difluoride (PVDF) membranes (Millipore) and Western blot analysis was performed using an anti-V5 (Invitrogen), anti-HA (Covance) and anti-actin (Chemicon International) monoclonal antibodies. Surface ENaC biotinylated fragments were quantified using the metamorph imaging 4.5 program (Hooker Microscopy Facility, University of North Carolina). Densitometry of selected bands was performed, using uninjected oocyte samples as background signal.
Results
CAP3 has less stringent sequence requirement for cleavage than furin
To assess the accessibility of the known protease cleavage tracts of different subunits of ENaC, we computed residue-wise disorder probability in the respective segments using Disopred2 (Figure S1) (28). To eliminate any bias in the prediction, we considered segments (α: V151-L290; β: K117-P240; γ: K91-S222) such that most of the cleavage sites are enclosed but are not near either end of the segment. Interestingly, the regions susceptible to cleavage by furin-like convertases in α- and γ-subunits (α: 202-RSSR, 228-RTAR; γ: 135-RKRR, 178-RKRK) of rat ENaC are intrinsically disordered (Figure S1A, C). The two peaks in the disorder plot for γ-ENaC correspond to the traditional furin site (135-RKRR-138) and the polybasic tract (178-RKRK-181) identified as cleavage sites for complete activation by furin and CAP1 (16) (Figure S1C). This observation is in agreement with previous computational analyses reporting preference for intrinsic structural disorder in cleavage by serine proteases (5).
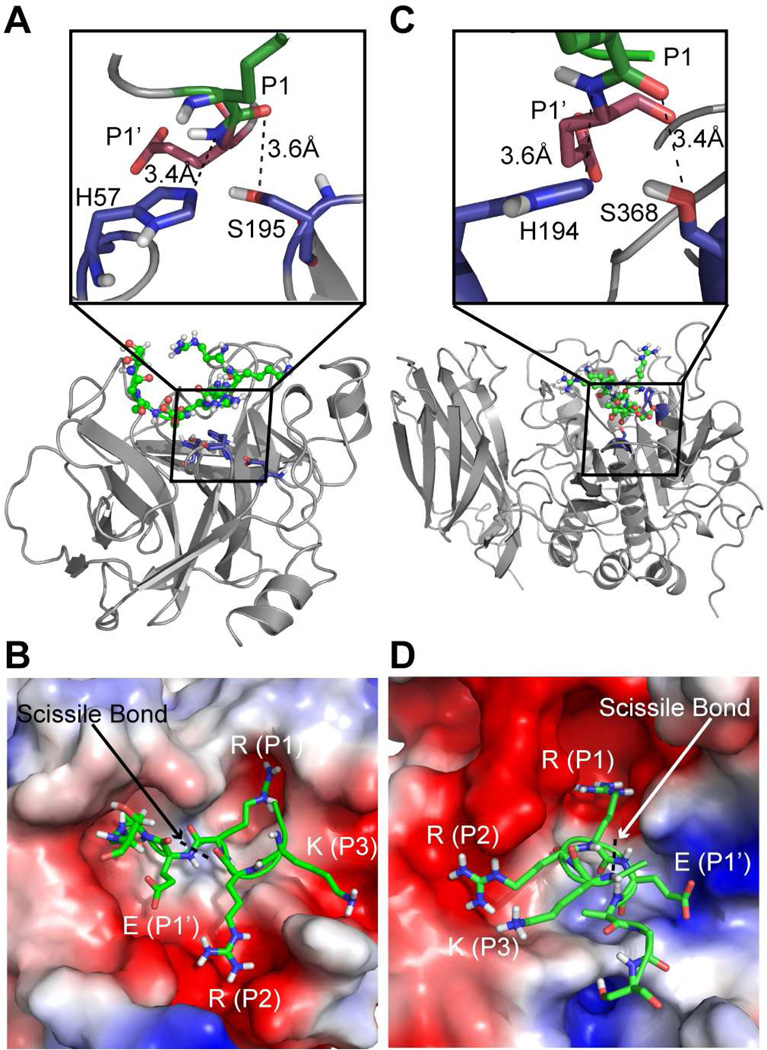
To elucidate the structural basis for substrate recognition and activation of ENaC by CAP3, we performed replica exchange DMD simulations (20, 29) of three peptide sequences (Seq1, 2, 3) from rat γ-ENaC (Materials and Methods) to study peptide binding to the active pocket of furin and CAP3 starting with the unbound state (Figure S2A, B). Using RMSD as the clustering criterion, we selected five final structures for each enzyme-substrate combination (Figure S2C). We found that the maximum RMSD between structures of peptide complexes with either furin or CAP3 is 1.5 Å. The final substrate-bound configurations of both furin and CAP3 satisfy the distance constraints imposed during simulations (Materials and Methods, Figure 1A, C). The peptide-bound configurations also portray the differences in the size, shape and charge distribution of the active sites of furin and CAP3 (Figure 1B, D). In our models, the residue at the P1 site of Seq1 is positioned in charge complementary pockets of both furin and CAP3 as observed in the corresponding crystal structures with bound inhibitors (Figure 1B, D) (30, 31).
Figure 1. Structural models of peptides from γ-ENaC bound to furin and CAP3.
A) Final docked configuration of peptide Seq1 in the active pocket of CAP3. Inset shows the distances between amine of the P1’ site from the NE2 of active site histidine and that of the carbonyl oxygen of the P1 site from the OG of active site serine. B) Electrostatic surface representation of CAP3 with the side chains of residues in peptide Seq1 shown as sticks. The guanidium group of the arginine at the P1 site docks into a negatively charged groove in the enzyme. C) Final docked configuration of the peptide Seq1 in the active pocket of furin. Inset shows the distances between amine of the P1’ site from the NE2 of the active site histidine and that of the carbonyl oxygen of the P1 site from the OG site of active site serine. D) Electrostatic surface representation of furin with the side chains of residues in peptide Seq1 shown as sticks. The guanidium group of arginine at P1 site docks into a negatively charged groove in the enzyme.
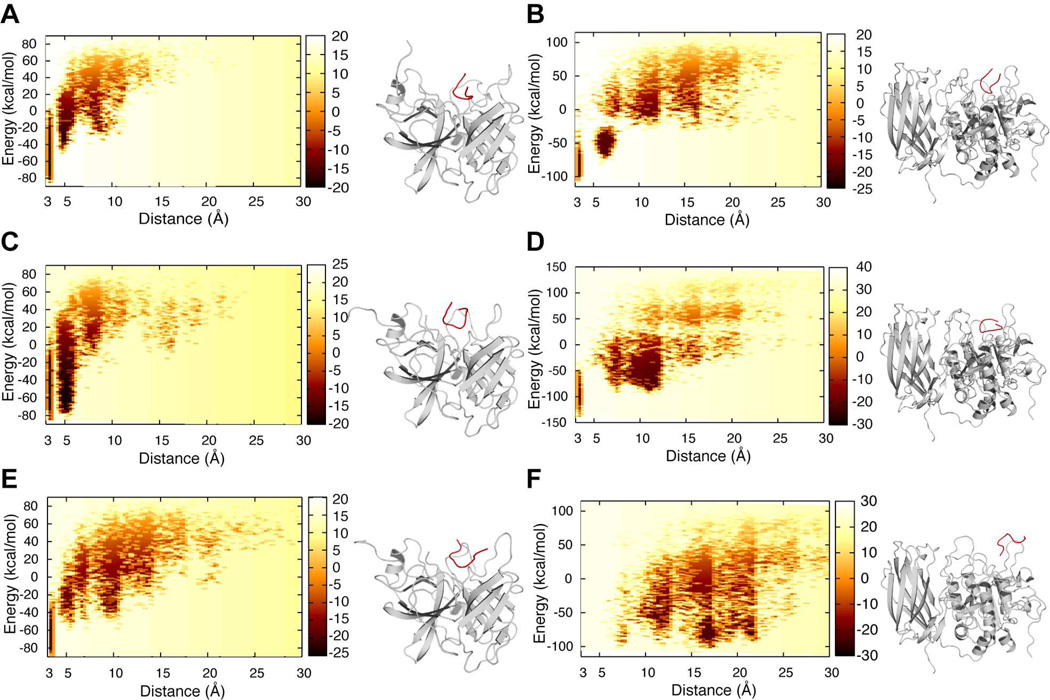
To establish the energetic basis for substrate recognition, we computed the two-dimensional potential of mean force (2D-PMF) with E (normalized energy) and d (distance in Å between active site residues and the P1, P1’ sites on the peptide) as the two reaction coordinates. We have unbiased the constrained replica exchange simulations to obtain an accurate measure of free energy of peptide binding to a given enzyme (Materials and methods). We observe from the 2D-PMF contours that Seq1 features low energy basins at a distance of 3–5 Å from the active site of both CAP3 and furin (Figure 2A, B). Similarly, the polybasic tract consisting of the alternate furin cleavage site (178-RKRK) presents a low energy basin at a distance of 3–5 Å or less from the active site of both furin and CAP3 suggesting that binding of Seq2 to either enzyme is energetically favorable (Figure 2C, D; black arrows). Takeuchi et al. proposed that the lysine residue (K132) upstream of the traditional furin site could be the P4 site of a bona fide consensus cleavage motif (132-KESR) for CAP3 (32). To determine whether a peptide containing this amino acid sequence is energetically compatible to be in the vicinity of the active site of CAP3, we performed simulations with Seq3 approaching the active sites of CAP3 and furin. We observe low energy configurations for Seq3 in the active site of CAP3 in simulations (Figure 2E, Supplementary video 1). Surprisingly, none of the configurations from the simulation with Seq3 in presence of furin satisfied the distance constraints for recognition, suggesting that binding of Seq3 to furin is energetically less favorable (Figure 2F, Supplementary video 2). Such differences are reflected in the contour plot for furin and CAP3 with Seq3 where a low energy basin at a distance of 3–5 Å from the active site of furin is absent while a low energy basin is observed in case of CAP3 (compare Figure 2E, F). These results indicate that CAP3 is more effective than furin at stimulating ENaC containing γ-ENaC 132KESR. Our computational results suggest low stringency sequence requirements for CAP3-mediated cleavage and presence of less ideal furin substrates in this region of γ-ENaC.
Figure 2. Energetic basis for peptide binding to furin and CAP3.
2D-PMF with normalized energy of enzyme-substrate complex and distance between the active site and respective peptides as reaction coordinates. The color on the 2D-PMF plot is indicative of the free energy of peptide binding to the enzyme. Darker the color, lower the free energy in kcal/mol. The peptide is colored red and the enzyme is colored gray in the accompanying models representing the minimum energy configurations for the corresponding enzyme-peptide combinations. A, B) 2D-PMF of Seq1 binding to CAP3 and furin respectively. C, D) 2D-PMF of Seq3 binding to CAP3 and furin respectively. E, F) 2D-PMF of Seq3 binding to CAP3 (video 1) and furin (video 2) respectively. Seq3 is energetically incompatible to be in the vicinity of the active site of furin while it reaches the active site of CAP3.
Catalytic activity of matriptase is required for activation of ENaC
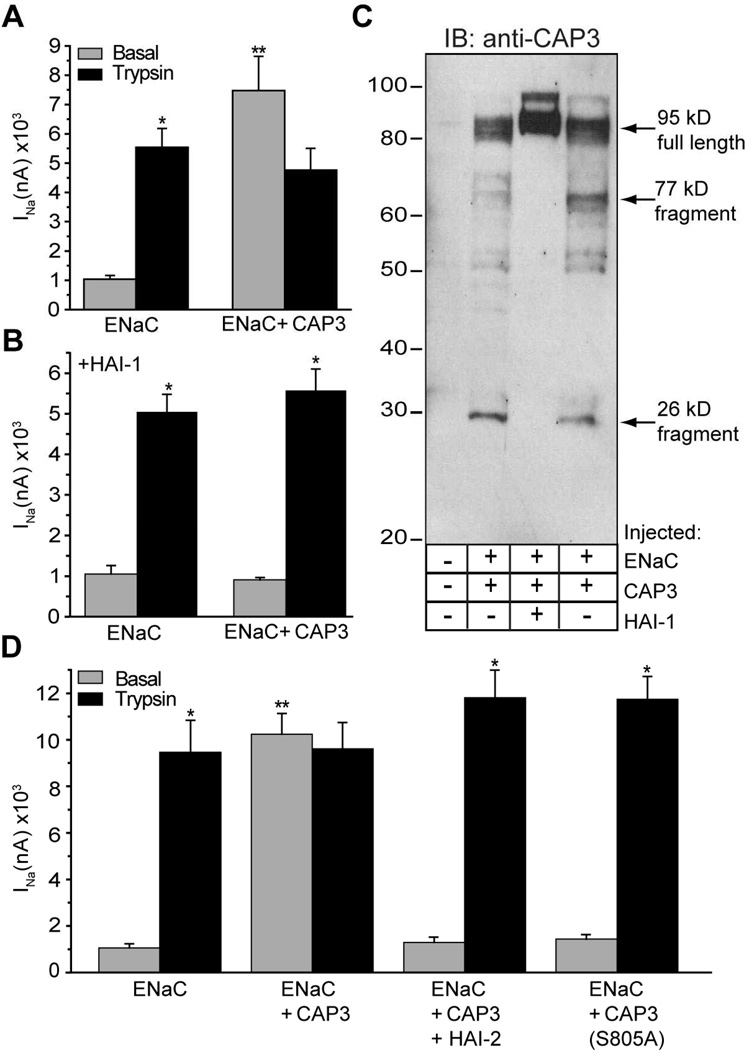
To biochemically characterize the sequence requirements of CAP3, we first established that the catalytic activity of CAP3 is required for activation of ENaC. The motivation for this study arose from the fact that the catalytic activity of CAP1, a GPI-anchored membrane serine protease, is not required for its regulation of ENaC (18). We found that co-expression of CAP3 with ENaC for 24 h robustly increased basal amiloride-sensitive sodium current (INa), typically in the range of 3–5 fold (Figure 3A). Moreover, unlike the basal INa generated by ENaC alone, the larger basal INa with CAP3 co-expression was not further increased by application of trypsin (Figure 3A) or hNE. The decrement of INa sometimes seen following exposure of CAP3-expressing oocytes to exogenous protease (Figure 3A, ENaC+CAP3, black bar) reflects run down of the stimulated INa. We observed that CAP3 stimulation of ENaC is inhibited by co-expressed hepatocyte activator inhibitor-1 (HAI-1) (Figure 3B). HAI-1 is a Kunitz-type serine protease inhibitor identified as the physiologic cognate inhibitor of CAP3 catalytic activity (33). Western blots of CAP3 in lysates of the injected oocytes show that CAP3 was robustly expressed in its active form, as indicated by the expected fragmentation pattern of this self-activating protease (Figure 3C) (34). Furthermore, co-expression of HAI-1 with CAP3 prevented cleavage associated with CAP3 activation. Co-expression of HAI-2, a related Kunitz-type inhibitor, also completely prevented CAP3 stimulation of ENaC (Figure 3D) (35). Finally, CAP3 inactivated by mutation of S805 of the catlaytic triad had no effect on INa of co-expressed ENaC (Figure 3D) (36). Thus, ENaC co-expressed with CAP3 is fully activated through a mechanism that requires catalytic activity of CAP3.
Figure 3. Catalytic activity of matriptase/CAP3 is required for stimulation of ENaC.
A, B) Coexpression of the Kunitz domain containing inhibitor, HAI-1 blocks stimulation of ENaC. Oocytes were co-injected with 0.3 cRNA of α, β and γ-ENaC alone (Panel A), or in combination with 1 ng cRNA of HAI-1 (Panel B). After 24 h incubation, amiloride-sensitive current (INa) was recorded before (gray bars) and following (black bars) 5 min exposure to 2 µg/ml trypsin. C) Expression and autocleavage of matriptase/CAP3, and effect of HAI-1. Oocyte lysates from (A) and (B). were studied by Western blotting using anti-CAP3 antibody. D) CAP3 stimulation of ENaC was blocked by co-expressed HAI-2 or by mutating serine 805 of the catalytic triad. Experiments were repeated on 2–4 batches of oocytes, with a total of 12–26 oocytes per condition. *Trypsin stimulated INa different from basal INa. **Basal INa different from ENaC alone. ANOVA, p < 0.05.
CAP3 cleaves γENaC at an alternate site N-terminal to the furin site
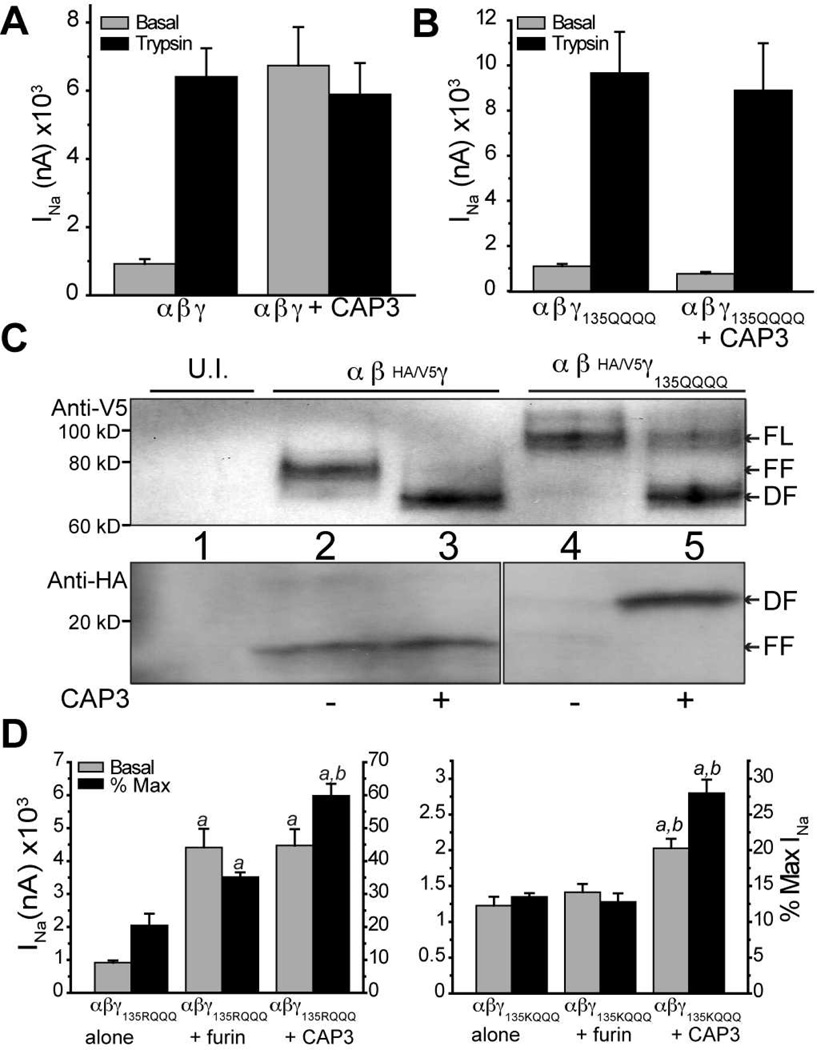
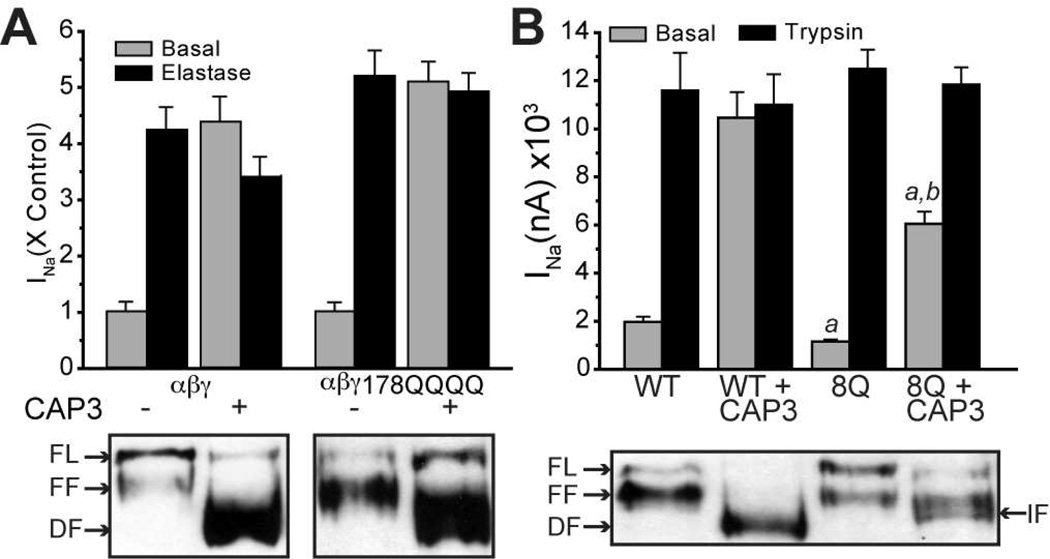
We studied the role of γ-ENaC in stimulation by CAP3, by co-expressing the furin site mutants of α-ENaC with WT β- and γ-ENaC in oocytes. CAP3 robustly activated ENaC without the furin sites in the α-subunit suggesting predominant cleavage of the γ-subunit (Figure S3A, B). In γ-ENaC, we specifically examined the importance of the basic P1 residue (R138) in the 135-RKRR tract recognized as the furin site. Co-expressed CAP3 robustly stimulated ENaC containing γ-subunit furin resistant mutant R138K and the CAP2 insenstive furin mutant R138A (Figure S3C, D) (14) indicating that CAP3 activates ENaC by a mechanism involving cleavage at a site distinct from γ-R138. Although the consensus sequences for convertases (R/K-X-X-R) and CAPs (R/K-X-X-R/K) overlap, some studies have reported distinct preferences at individual residue positions (37, 38). Therefore, we asked if extensive mutagenesis of the 135-RKRR tract in γ-ENaC to 135-QQQQ would affect the action of CAP3 toward ENaC. Co-expressed CAP3 did not stimulate this mutant ENaC, even though subsequent trypsin exposure lead to significant stimulation (Figure 4B). Further mutagenesis revealed that the presence of R135 is sufficient for CAP3-mediated cleavage of γ-ENaC (Figure S4).
Figure 4. CAP3 mediates neither activation nor cleavage of γ-135QQQQ ENaC.
A) WT ENaC was expressed alone or with CAP3. INa was recorded 24 hr after cRNA injection, before (gray bars) and after (black bars) a 5 min trypsin exposure. B) ENaC containing the mutant γ135QQQQ was expressed alone or with CAP3. INa was recorded as in A. C) ENaC made of WT subunits or containing the mutant γ-135QQQQ were expressed alone or with CAP3. After 24 h, uninjected (U.I.) oocytes and oocytes from each treatment group were surface biotinylated and the surface protein pool was captured on streptavidin beads. Full length and fragments of HA/V5 γ-ENaC were recognized by blotting for V5 (C-terminal epitope) or HA (N-terminal epitope). FL = full length; FF = furin fragment; DF =distal fragment. D) Oocytes were injected with αFM,β and either γ135RQQQ (left) or γ135KQQQ (right). Each of these ENaC combinations was expressed alone or co-expressed with furin or CAP3, and studied after 24 h. Basal INa was recorded before and following 5 min exposure to 2 µg/ml hNE. Basal INa as raw current (left ordinate) or as a percent of maximum INa following elastase (right ordinate) is shown. Mean values were compared by ANOVA and Tukey’s test applied. a = different from mutant γ alone, b = different from mutant γ + furin, p < 0.01.
To biochemically characterize CAP3-mediated cleavage of γENaC, we performed Western blot analysis of biotinylated surface protein pool captured on streptavidin beads (Materials and Methods). We characterized the HA/V5 double epitope labeled γ-ENaC by the pattern of anti-V5 (C-terminal tag) and anti-HA (N-terminal tag) staining on Western blot (Figure 4C). In oocytes expressing WT ENaC subunits, V5-tagged γ-ENaC at the cell surface exists as a mixture of full length (“FL”, ~93 kD band) and furin fragments (“FF”, ~75 kD band) (Figure 4C, top panel). Under basal conditions, HA-label was found in a complementary ~18 kD band (Figure 4C, lower panel) and in a FL band (not shown). With co-expression of CAP3, a more rapidly migrating fragment of ~70 kD (“DF”, distal fragment) replaces the FF band, a result now seen as characteristic of proteolysis of γ-ENaC at a site 20–40 residues downstream of the furin site (16, 39, 40). The 18 kD anti-HA reactive (N-terminal) fragment indicates that cleavage at the traditional furin site is not affected. As predicted, mutant γ-ENaC containing the 135-QQQQ tract shows no FF, either V5- or HA-labeled, when co-expressed with α- and β-ENaC alone (Figure 4C, lane 4). However, when co-expressed with CAP3, the proportion of FL V5-labeled γ-135-QQQQ at the cell surface decreases, with a significant increase in the intensity of the DF fragment (Figure 4C, lane 5). A complementary N-terminal HA-labeled mutant γ-ENaC fragment of ~23 kD appears at the surface of CAP3 co-expressing cells (Figure 4C, lower panel, lane 5). These results indicate that cleavage within the 135–138 furin site is blocked in γ-135-QQQQ while CAP3 induces cleavage C-terminal to the furin site. In addition, we conclude that cleavage within residues 135–138 is essential for CAP3 stimulation of ENaC.
Based on previous reports and our computational analyses, we hypothesized that K132 could be part of a bona fide consensus motif (132-KESR) targeted by transmembrane serine proteases (TSPs) or furin-like convertases (32, 41). To test this hypothesis, we generated γ-ENaC with 132-HESRQQQ, which associates with WT α- and β-ENaC to produce reduced INa that responds briskly to hNE (2 µg/ml) or trypsin (20 µg/ml) (Figure S4, black bars). Although the presence of 132-KESR is evidently not optimal for endogenous convertases, we reasoned that over-expressed furin might recognize this site. Because the preference of furin at the P1 residue is strong for arginine over lysine (42), while CAP3 is reported to tolerate lysine at P1 (32), we compared the ability of co-expressed human furin and CAP3 to stimulate the γ-ENaC mutants 135-RQQQ and 135-KQQQ. To simplify interpretation, we expressed these mutant γ-subunits with WT β-ENaC and with α-ENaC furin site mutant (Figure 4D). Interestingly, co-expressed human furin partially activated ENaC containing γ-132-KESRQQQ, to 35% of maximum stimulated INa, albeit less efficiently than CAP3 which led to 60% of maximum stimulated INa (Figure 4D, left panel). Co-expressed furin did not stimulate ENaC with a lysine at position 135 (Figure 4D, right panel) while CAP3 stimulated this mutant, albeit to a lesser extent than the mutant with R135 preserved. These results suggest that CAP3, due to its less stringent sequence requirements, can target basic residues in the 132–138 tract of γ-ENaC that are more resistant to endogenous convertases and over-expressed furin.
CAP3 cleaves ENaC at multiple sites C-terminal to the furin site
A candidate site for cleavage events responsible for the broadly staining DF (~70 kD) (Figure 4C) is the tract 178-RKRK, as this polybasic region is required for cleavage of γ-ENaC by CAP1 (16), and shares the same minimal sequence requirements for cleavage with CAP3 (43). DMD simulations revealed that CAP3/furin binding at this site is energetically favorable. Surprisingly, however, γ-ENaC with 178-QQQQ was stimulated by co-expressed CAP3 to about the same extent as ENaC containing WT γ-subunit (Figure 5A). The CAP3-stimulated basal current in either WT or mutant channel groups was not further increased by hNE, indicating that CAP3 attained full proteolytic stimulation of mutant ENaC at the surface. In addition, the patterns of C-terminal V5-labeled fragments of WT or mutant γ-ENaC contained in the cell surface pool of each expression group were affected similarly by co-expressed CAP3, each showing a shift from a mixture of FL or FF to a population dominated by DFs, consistent with CAP3-induced cleavage at sites downstream from the furin site (Figure 5A, lower panel). While these results do not refute the fact that 178-RKRK is a potential cleavage site, the data indicate that other potential cleavage sites exist in the vicinity of this basic tract.
Figure 5. The basic tract 178-RKRK in γ-ENaC is not essential for CAP3 stimulation of INa.
A) WT HA/V5-γ-ENaC or HA/V5-γ-RKRK(178–181)QQQQ cRNA was co-injected with WT α- and β-ENaC cRNA (0.3 ng/subunit). Half of each group was also injected with 1 ng cRNA for CAP3. INa was recorded after 24 h, before and following 5 min of exposure to 2 µg/ml hNE (upper panel, N = 10–12 oocytes per condition from two batches). 30–60 oocytes per condition were surface biotinylated, as described elsewhere. WT and mutant γ-ENaC fragments present in the cell surface pool were analyzed by Western blot (lower panel). B) WT α- and β-ENaC subunits were expressed for 24 h with WT or mutant (8Q) HA/V5 γ-ENaC, with or without CAP3 (see text for description of mutant 8Q). INa was recorded before and following 5 min exposure to 20 µg/ml trypsin (a = different from WT basal; b = different from 8Q basal, p < 0.01) Surface biotinylated proteins from the same injection groups were analyzed by anti-V5 Western blotting are shown in lower panel. FL = Full Length; FF + Furin Fragment; DF = Distal Fragment; IF = Intermediate Fragment.
As mutation of the polybasic tract 178-RKRK has no effect on CAP3-mediated stimulation or banding pattern of γ-ENaC fragments, we tested the contribution of flanking basic residues in the region from residues 172 to 202. We investigated the importance of 172R, 185K, 189K and 201K, 202K, individually, and in various combinations with 178-QQQQ. We observed no significant effects of mutating any single basic residue in this region on CAP3 cleavage and stimulation of ENaC. However, from a threshold of mutating 6–7 basic residues, up to 9 basic residues in this region replaced by glutamine, we observed progressively decreased CAP3-mediated stimulation of INa, staining density of DF, and resting whole cell Po. Particularly, ENaC containing mutant γ-subunits with eight glutamines substituted for basic residues within the 172–202 tract (“8Q”) was only partially stimulated by co-expressed CAP3 (Figure 5B). Western blot analysis of the C-terminal V5-tagged fragments of 8Q γ-ENaC on the cell surface suggests that in this extensively mutated channel, CAP3 generates an intermediate fragment (IF) that migrates between FF and DF, characteristic of WT γ-ENaC (Figure 5B). Thus, CAP3 cleaves ENaC at multiple basic sites including those that do not conform to the furin consensus sequence requirements. These results are in agreement with our conclusion that CAP3 has less stringent sequence requirements for cleavage than furin. It is likely that the local structure of the protein is altered upon extensive mutagenesis, thereby hampering cleavage. Assuming that the structure of ENaC is intact, our results suggest that CAP3 cleaves γ-ENaC at multiple sites C-terminal to the furin site resulting in robust channel activation.
Discussion
Here, we report the energetic, structural and sequence requirements for cleavage of ENaC by CAP3 and furin. While we focus our attention on activation of ENaC by proteases, the results of our computational analyses are broadly applicable to catalysis by serine proteases. Limited endoproteolysis of ENaC is a complex mechanism for regulating Na+ absorption by epithelia. Numerous proteases that stimulate ENaC have been identified, alongside endogenous protease inhibitors that oppose this effect (12, 13). Thus, ENaC may be controlled in a tissue specific manner by the complement of endogenous proteases and anti-proteinases expressed in a given cell type. We experimentally characterized the effects of CAP3 co-expression on proteolysis and stimulation of ENaC in Xenopus oocytes. We identified two regions of γ-ENaC susceptible to direct cleavage by CAP3. Cleavage after any of several basic residues within each region is necessary and sufficient for full stimulation of ENaC. These observations support three main conclusions. First, cleavage of the γ-subunit alone accounts for CAP3 stimulation of ENaC. Second, though limited in extent, the specificity of γ-ENaC cleavage is not based on stringent docking of extended segments of γ-ENaC within the active site in CAP3. Instead, multiple basic residues comprising the γ-ENaC furin site, along with those including and flanking 178-RKRK are exposed for proteolysis. Finally, CAP3 alone can achieve full proteolytic stimulation of ENaC.
Structural studies of ENaC are set back due to the unavailability of a complete atomistic structure of the heteromultimer. Recently, different ENaC subunits were threaded onto the structure of a homologous acid-sensing ion channel (ASIC1) (44, 45). In the sequence alignment used for model building, the region in γ-ENaC containing the putative cleavage sites for CAP3 and furin was designated the “hypervariable region” (44). Consequently, the structural features of this region are unavailable despite the modeling efforts. The observation that the protease cleavage sites in ENaC are intrinsically disordered is in line with the proposed structural requirement for serine protease activity (5, 6). To study the energetic basis for substrate recognition by CAP3 and furin, we performed DMD simulations of both enzymes with different candidate peptides. Although crystal structures of matriptase/CAP3 (PDB: 1EAW) and furin (PDB: 1P8J) with bound inhibitors are available for this effort (30, 31, 46), we reasoned that the static orientation of an inhibitor in the catalytic pocket does not effectively capture the dynamics of enzyme-substrate complexes. Therefore, we resorted to DMD simulations and we observed from the low energy configurations that a charge complementary pocket in both furin and CAP3 mediates critical enzyme-peptide interactions (Figure 1B, D). While these simulations are not intended to model the hydrolysis reaction carried out by either enzyme, they provide insight into the binding properties of a given enzyme-peptide combination. Given that substrate recognition is the first step in hydrolysis, we consider a peptide that can reach the vicinity of the enzyme (3–5 Å) as a potential substrate for cleavage. By maintaining most of the enzyme static during simulations, we assume that furin and matriptase do not undergo significant conformational changes during peptide recognition. Furthermore, by using an implicit solvent model (EEF1) for treating solvent interactions, we disregard specific water-mediated hydrogen bond formation involved in hydrolysis. Therefore, our simulation studies are only aimed at determining whether a given peptide is energetically suitable for presenting itself in the proximity (within 3–5 Å) of the active site of furin or matriptase, and not to quantify the rate of hydrolysis of a given peptide by a protease. PMF analysis with Seq1 features low energy basins at the active sites of both furin and CAP3 suggesting that Seq1 can be a substrate for both enzymes. Interestingly, analogous computational study of Seq3 indicates that 132KESR can be a substrate for CAP3 but not for furin. This result is consistent with our observation that CAP3 cleaves and stimulates ENaC effectively when 132KESR are the only potentially susceptible residues available near the furin site, whereas exogenous furin is much less effective at stimulating ENaC in this context (Figure 3D). These computational analyses of WT sequences, with our functional and biochemical analyses of WT and mutant γ-ENaC clearly indicate that the γ-ENaC sequence 132-KESRKRR is highly susceptible to cleavage by CAP3.
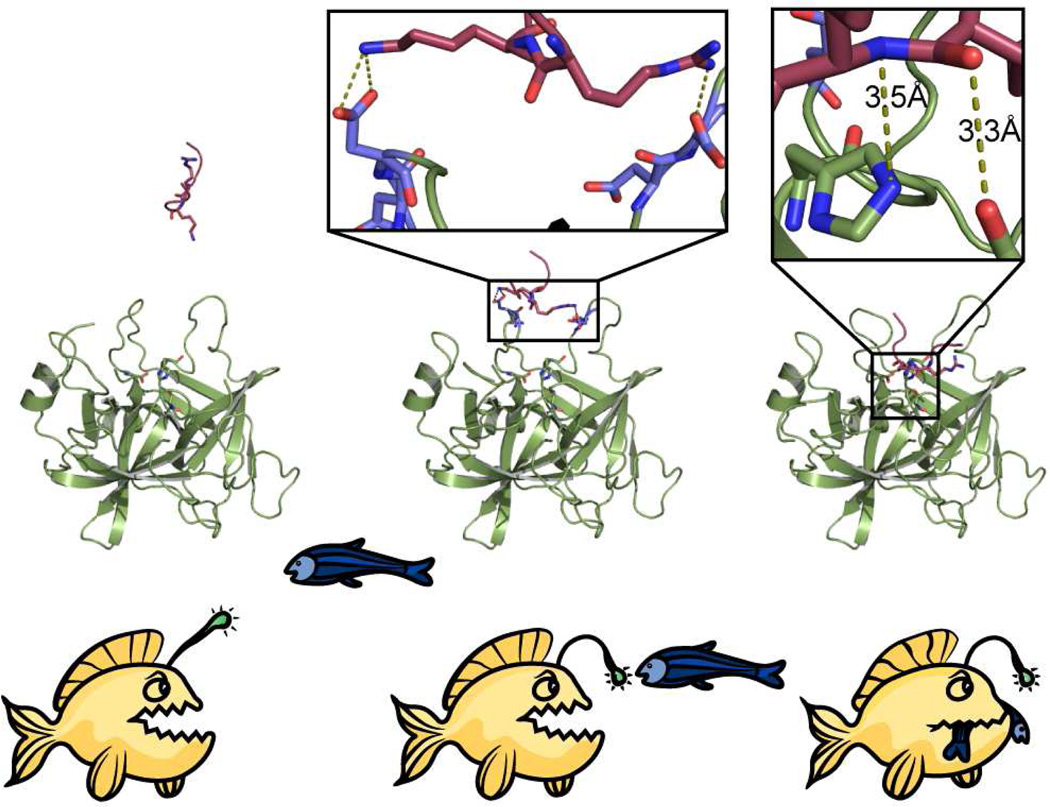
CAP3, unlike furin in this study, or CAP2 in our previous work (14), appears to cleave γ-ENaC after several basic residues within 132-KESRKRR, including 135R, 136K and 138R. Such greater susceptibility of γ-ENaC to cleavage may reflect CAP3’s low substrate stringency coupled with favorable substrate access due to local structural disorder. 2D-PMF analyses of all peptide-catalytic site combinations considered in this study broadly support this notion. Energy minima at 3–5 Å indicate close contact between peptide and catalytic site required for proteolysis (Figure 2), whereas low energy basins beyond 5 Å are suggestive of electrostatic attraction between the net positively charged peptides and the negatively charged surface loops of the enzyme. The absence of a low energy basin at a distance of 3–5 Å from the active site reflects the fact that Seq3 is a less suitable substrate for furin than CAP3 (Figure 2E, F). Seq1 and Seq2 feature energy minima at 3–5 Å from the active site of both CAP3 and furin (compare Figure 2A, B and C, D). Low energy basins corresponding to interaction between the peptide and the surface loops of the enzyme are observed beyond 5 Å for all peptides in simulations. However, only Seq1 and Seq2 are energetically compatible to be in the proximity of the active of CAP3 and furin, as evidenced from the PMF analysis (Figure 2A–D). PMF analysis indicates that it is favorable for Seq3 to be in the vicinity of CAP3, but not furin. In light of these observations, we propose an “anglerfish” mechanism analogous to the characteristic mode of predation by the anglerfish where the fish represents the enzyme, with its illicium being the surface loops that capture the peptide to be cleaved and proteolysis being the eventual irreversible event (Figure 6). The realization of this mechanism is confined by the assumption that proteolysis is guaranteed if a peptide is energetically compatible to be in the vicinity (3–5 Å) of the active site of a hydrolytic enzyme.
Figure 6. “Anglerfish” mechanism of peptide recognition and cleavage by MAPs.
A two-step mechanism depicting the importance of surface loops in proteolysis by MAPs. The incoming peptide interacts with the surface loops and is ultimately delivered to the active site resulting in irreversible cleavage. 2D-PMF analyses reveal the energetic basis for this mechanism.
Our examination of CAP3 activity suggests that several different events underlie stimulation of ENaC. Mutation analysis suggests that 132K participates in a sequence (132-KESR) predicted to be optimal for cleavage by CAP3 (32). Residual cleavage and stimulation of ENaC containing γ-135-RQQQ requires the intact sequence 132-KESR. Mutagenesis of the furin sites in α-ENaC has no effect on CAP3 stimulation of ENaC, even though CAP3 converts all FL WT α-ENaC to fragments expected from cleavage at the two furin sites, similar to our previous study with CAP2 (14). The stimulation of ENaC by CAP1 was also linked by mutagenesis to γ-ENaC (16). Complete proteolytic stimulation of ENaC by soluble serine proteases, including trypsin, hNE and plasmin is also exerted by cleavage of the γ-subunit (40, 47, 48). Thus, our results demonstrating that CAP3-mediated stimulation of ENaC can be blocked by mutations in γ-ENaC add to a growing body of data indicating that γ-ENaC plays a central role in the proteolytic regulation of ENaC.
Based on our results, cleavage of γ-ENaC mediated by CAP3 occurs in two regions: the first is discretely defined by the furin site and the second includes basic residues centered on and surrounding the 178-RKRK tract. CAP3’s low stringency requirements for cleavage are consistent with its cleavage of γ-ENaC at multiple basic residues downstream of the furin cleavage site. Our results provide a compelling hypothesis that recognition and/or cleavage of γ-ENaC by proteases is substantially determined by accessibility of key sites rather than solely by a stringent structural requirement. The ability of certain proteases to activate ENaC has been linked to a specific residue or tract within 172–202 (16, 40, 47, 48). Based on such work for prostasin (CAP1), the initial expectation was that CAP3 would cleave at the 178-RKRK tract. However, when 178-RKRK is unavailable, both CAP2 and CAP3 readily cleave at multiple basic residues in the region 172–202. This phenomenon is consistent with the concept that this region is highly accessible to proteases due to intrinsic structural disorder. The low stringency of CAP3 explains why we could not identify a single residue or short tract that can be mutated to block CAP3 stimulation of ENaC. We expect the accessibility of either region to increase with time and extent of proteolysis, leading to further cleavage and consequent activation by relatively non-specific proteases.
Our results indicate that CAP3 potently stimulates ENaC in an in vitro setting, which is dependent on direct catalytic attack of the protease on extracellular segments of γ-ENaC. It has been suggested that CAP3 completes its auto-activation at the cell surface (49, 50). As a step towards understanding the colocalization of ENaC and CAP3 in vivo, we performed immunofluorescence microscopy to confirm that matriptase localizes to the subapical pools near the plasma membrane of human bronchial epithelial (HBE) cells derived from different individuals (Figure S5). ENaC can be detected in apical and subapical compartments of HBE cultures and is processed by limited cleavage in these cells (51, 52). Thus, it appears highly likely that ENaC is cleaved by matriptase in HBE cells. Additional work is needed to confirm that ENaC and active CAP3 occupy positions at the cell surface that enable ENaC cleavage, but our results clearly imply that CAP3 is capable of activating ENaC independent of furinlike convertases. CAP3 also activates CAP1 (53), which stimulates ENaC regardless of its catalytic activity (54). These and other recent observations suggest a possibility for a cascade of interactions at the cell surface involving CAP3 and other proteases like CAP1, exercising dynamic control over proteolytic regulation of ENaC (55).
We conclude that CAP3, a MAP, can independently activate ENaC without contributions from furinlike convertases or soluble proteases, without a requirement for specific structural motifs for recognition and/or cleavage. Our computational analyses give rise to a potentially generic “anglerfish” mechanism applicable broadly to serine protease mediated recognition and cleavage.
Supplementary Material
Acknowledgments
We acknowledge the expert technical contributions of Yan Dang and Hong He.
Footnotes
This work was supported by the National Institutes of Health Grants 5P01HL034322 and 5R01HL080561 (to MJS) and R01GM080742 and the ARRA supplements GM080742-03S1 and GM066940-06S1 (to NVD).
Supporting Information Available. Figures supporting the results obtained in this study are provided as a pdf file. Videos representing computational trajectories are provided as quicktime movies. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation (a review) Proc Natl Acad Sci U S A. 1976;73:3825–3832. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neurath H. Evolution of proteolytic enzymes. Science. 1984;224:350–357. doi: 10.1126/science.6369538. [DOI] [PubMed] [Google Scholar]
- 3.Neurath H. Proteolytic processing and physiological regulation. Trends Biochem Sci. 1989;14:268–271. doi: 10.1016/0968-0004(89)90061-3. [DOI] [PubMed] [Google Scholar]
- 4.Novotny J, Bruccoleri RE. Correlation among sites of limited proteolysis, enzyme accessibility and segmental mobility. FEBS Lett. 1987;211:185–189. doi: 10.1016/0014-5793(87)81433-3. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard SJ, Beynon RJ, Thornton JM. Assessment of conformational parameters as predictors of limited proteolytic sites in native protein structures. Protein Eng. 1998;11:349–359. doi: 10.1093/protein/11.5.349. [DOI] [PubMed] [Google Scholar]
- 7.Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellenberger S, Gautschi I, Schild L. An external site controls closing of the epithelial Na+ channel ENaC. J Physiol (Lond) 2002;543:413–424. doi: 10.1113/jphysiol.2002.022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy M, Wang X, Quinton P. Effect of Cytosolic pH on Epithelial Na+ Channel in Normal and Cystic Fibrosis Sweat Ducts. Journal of Membrane Biology. 2008;225:1–11. doi: 10.1007/s00232-008-9126-4. [DOI] [PubMed] [Google Scholar]
- 10.Kim SH, Kim KX, Raveendran NN, Wu T, Pondugula SR, Marcus DC. Regulation of ENaC-mediated sodium transport by glucocorticoids in Reissner's membrane epithelium. Am J Physiol Cell Physiol. 2009;296:C544–C557. doi: 10.1152/ajpcell.00338.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauz S, Walker EA, Murray PI, Stewart PM. Expression and distribution of the serum and glucocorticoid regulated kinase and the epithelial sodium channel subunits in the human cornea. Experimental Eye Research. 2003;77:101–108. doi: 10.1016/s0014-4835(03)00088-5. [DOI] [PubMed] [Google Scholar]
- 12.Rossier BC, Stutts MJ. Activation of the Epithelial Sodium Channel (ENaC) by Serine Proteases. Annual Review of Physiology. 2009;71:361–379. doi: 10.1146/annurev.physiol.010908.163108. [DOI] [PubMed] [Google Scholar]
- 13.Kleyman TR, Carattino MD, Hughey RP. ENaC at the Cutting Edge: Regulation of Epithelial Sodium Channels by Proteases. Journal of Biological Chemistry. 2009;284:20447–20451. doi: 10.1074/jbc.R800083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Caballero A, Dang Y, He H, Stutts MJ. ENaC Proteolytic Regulation by Channel-activating Protease 2. J. Gen. Physiol. 2008;132:521–535. doi: 10.1085/jgp.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adachi M, Kitamura K, Miyoshi T, Narikiyo T, Iwashita K, Shiraishi N, Nonoguchi H, Tomita K. Activation of Epithelial Sodium Channels by Prostasin in Xenopus Oocytes. J Am Soc Nephrol. 2001;12:1114–1121. doi: 10.1681/ASN.V1261114. [DOI] [PubMed] [Google Scholar]
- 16.Bruns JB, Carattino MD, Sheng S, Maarouf AB, Weisz OA, Pilewski JM, Hughey RP, Kleyman TR. Epithelial Na+ Channels Are Fully Activated by Furin- and Prostasin-dependent Release of an Inhibitory Peptide from the {gamma}-Subunit. J. Biol. Chem. 2007;282:6153–6160. doi: 10.1074/jbc.M610636200. [DOI] [PubMed] [Google Scholar]
- 17.Winarski KL, Sheng N, Chen J, Kleyman TR, Sheng S. Extracellular allosteric regulatory subdomain within the gamma subunit of the epithelial Na+ channel. J Biol Chem. 2010;285:26088–26096. doi: 10.1074/jbc.M110.149963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuagniaux G, Vallet V, Jaeger NF, Hummler E, Rossier BC. Synergistic activation of ENaC by three membrane-bound channel- activating serine proteases (mCAP1, mCAP2, and mCAP3) and serum- and glucocorticoid-regulated kinase (Sgk1) in Xenopus Oocytes. J Gen Physiol. 2002;120:191–201. doi: 10.1085/jgp.20028598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.List K, Hobson John P, Molinolo Alfredo, Bugge Thomas H. Co-localization of the channel activating protease prostasin/(CAP1/PRSS8) with its candidate activator, matriptase. Journal of Cellular Physiology. 2007;213:237–245. doi: 10.1002/jcp.21115. [DOI] [PubMed] [Google Scholar]
- 20.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 23.Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins. 79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S, Ding F, Dokholyan NV. Multiscale modeling of nucleosome dynamics. Biophys J. 2007;92:1457–1470. doi: 10.1529/biophysj.106.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feig M, Karanicolas J, Brooks CL., 3rd MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations of biomolecules. 1. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 27.Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC, Gabriel SE. Regulation of the Epithelial Sodium Channel by Serine Proteases in Human Airways. J. Biol. Chem. 2002;277:8338–8345. doi: 10.1074/jbc.M105044200. [DOI] [PubMed] [Google Scholar]
- 28.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 30.Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I, Bode W, Than ME. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10:520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 31.Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, Madison EL. Catalytic Domain Structures of MTSP1/Matriptase, a Matrix-degrading Transmembrane Serine Proteinase. J. Biol. Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 33.Szabo R, Molinolo A, List K, Bugge TH. Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene. 2007;26:1546–1556. doi: 10.1038/sj.onc.1209966. [DOI] [PubMed] [Google Scholar]
- 34.Benaud C, Dickson RB, Lin CY. Regulation of the activity of matriptase on epithelial cell surfaces by a blood-derived factor. Eur J Biochem. 2001;268:1439–1447. doi: 10.1046/j.1432-1327.2001.02016.x. [DOI] [PubMed] [Google Scholar]
- 35.Szabo R, Hobson JP, List K, Molinolo A, Lin CY, Bugge TH. Potent inhibition and global co-localization implicate the transmembrane Kunitz-type serine protease inhibitor hepatocyte growth factor activator inhibitor-2 in the regulation of epithelial matriptase activity. J Biol Chem. 2008;283:29495–29504. doi: 10.1074/jbc.M801970200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyake Y, Yasumoto M, Tsuzuki S, Fushiki T, Inouye K. Activation of a Membrane-Bound Serine Protease Matriptase on the Cell Surface. J Biochem. 2009;146:273–282. doi: 10.1093/jb/mvp066. [DOI] [PubMed] [Google Scholar]
- 37.Bugge TH, Antalis TM, Wu Q. Type II Transmembrane Serine Proteases. Journal of Biological Chemistry. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris M, Firsov D, Vuagniaux G, Stutts MJ, Rossier BC. A Novel Neutrophil Elastase Inhibitor Prevents Elastase Activation and Surface Cleavage of the Epithelial Sodium Channel Expressed in Xenopus laevis Oocytes. J. Biol. Chem. 2007;282:58–64. doi: 10.1074/jbc.M605125200. [DOI] [PubMed] [Google Scholar]
- 40.Passero CJ, Mueller GM, Rondon-Berrios H, Tofovic SP, Hughey RP, Kleyman TR. Plasmin Activates Epithelial Na+ Channels by Cleaving the {gamma} Subunit. J. Biol. Chem. 2008;283:36586–36591. doi: 10.1074/jbc.M805676200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishi K, Yamazaki K, Yasuda I, Yahagi N, Ichinose M, Tsuchiya Y, Athauda SBP, Inoue H, Takahashi K. Characterization of a Membrane-Bound Arginine-Specific Serine Protease from Rat Intestinal Mucosa. J Biochem. 2001;130:425–430. doi: 10.1093/oxfordjournals.jbchem.a003002. [DOI] [PubMed] [Google Scholar]
- 42.Matthews DJ, Goodman LJ, Gorman CM, Wells JA. A survey of furin substrate specificity using substrate phage display. Protein Sci. 1994;3:1197–1205. doi: 10.1002/pro.5560030805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shipway A, Danahay H, Williams JA, Tully DC, Backes BJ, Harris JL. Biochemical characterization of prostasin, a channel activating protease. Biochemical and Biophysical Research Communications. 2004;324:953–963. doi: 10.1016/j.bbrc.2004.09.123. [DOI] [PubMed] [Google Scholar]
- 44.Stockand JD, Staruschenko A, Pochynyuk O, Booth RE, Silverthorn DU. Insight toward epithelial Na+ channel mechanism revealed by the acid-sensing ion channel 1 structure. IUBMB Life. 2008;60:620–628. doi: 10.1002/iub.89. [DOI] [PubMed] [Google Scholar]
- 45.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel-1 at 1.9[thinsp]A resolution and low pH. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 46.Henrich S, Lindberg I, Bode W, Than ME. Proprotein Convertase Models based on the Crystal Structures of Furin and Kexin: Explanation of their Specificity. Journal of Molecular Biology. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 47.Diakov A, Bera K, Mokrushina M, Krueger B, Korbmacher C. Cleavage in the {gamma}-subunit of the epithelial sodium channel (ENaC) plays an important role in the proteolytic activation of near-silent channels. J Physiol. 2008 doi: 10.1113/jphysiol.2008.154435. jphysiol.2008.154435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adebamiro A, Cheng Y, Rao US, Danahay H, Bridges RJ. A Segment of {gamma} ENaC Mediates Elastase Activation of Na+ Transport. J. Gen. Physiol. 2007;130:611–629. doi: 10.1085/jgp.200709781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szabo R, Bugge TH. Type II transmembrane serine proteases in development and disease. The International Journal of Biochemistry & Cell Biology Directed Issue: Proteases and Antiproteases in Development, Homeostasis and Disease. 2008;40:1297–1316. doi: 10.1016/j.biocel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 50.Lee M-S, Tseng I-C, Wang Y, Kiyomiya K-i, Johnson MD, Dickson RB, Lin C-Y. Autoactivation of matriptase in vitro: requirement for biomembrane and LDL receptor domain. Am J Physiol Cell Physiol. 2007;293:C95–C105. doi: 10.1152/ajpcell.00611.2006. [DOI] [PubMed] [Google Scholar]
- 51.Gentzsch M, Dang H, Dang Y, Garcia-Caballero A, Suchindran H, Boucher RC, Stutts MJ. The cystic fibrosis transmembrane conductance regulator impedes proteolytic stimulation of the epithelial Na+ channel. J Biol Chem. 2010;285:32227–32232. doi: 10.1074/jbc.M110.155259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaillard EA, Kota P, Gentzsch M, Dokholyan NV, Stutts MJ, Tarran R. Regulation of the epithelial Na+ channel and airway surface liquid volume by serine proteases. Pflugers Arch. 2010;460:1–17. doi: 10.1007/s00424-010-0827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Netzel-Arnett S, Currie BM, Szabo R, Lin C-Y, Chen L-M, Chai KX, Antalis TM, Bugge TH, List K. Evidence for a Matriptase-Prostasin Proteolytic Cascade Regulating Terminal Epidermal Differentiation. J. Biol. Chem. 2006;281:32941–32945. doi: 10.1074/jbc.C600208200. [DOI] [PubMed] [Google Scholar]
- 54.Vallet V, Pfister C, Loffing J, Rossier BC. Cell-Surface Expression of the Channel Activating Protease xCAP-1 is Required for Activation of ENaC in the Xenopus Oocyte. J Am Soc Nephrol. 2002;13:588–594. doi: 10.1681/ASN.V133588. [DOI] [PubMed] [Google Scholar]
- 55.Svenningsen P, Uhrenholt TR, Palarasah Y, Skjodt K, Jensen BL, Skott O. Prostasin-dependent activation of epithelial Na+ channels by low plasmin concentrations. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1733–R1741. doi: 10.1152/ajpregu.00321.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.