Abstract
Purpose
This study evaluates the prognostic performance of a 15 gene expression profiling (GEP) assay that assigns primary posterior uveal melanomas to prognostic subgroups: class 1 (low metastatic risk) and class 2 (high metastatic risk).
Design
Prospective, multicenter study.
Participants
459 patients with posterior uveal melanoma were enrolled from 12 independent centers.
Testing
Tumors were classified by GEP as class 1 or class 2. The first 260 samples were also analyzed for chromosome 3 status using a single nucleotide polymorphism assay. Net reclassification improvement analysis was performed to compare the prognostic accuracy of GEP to the 7th edition clinical Tumor-Node-Metastasis (TNM) classification and to chromosome 3 status.
Main Outcome Measures
Patients were managed for their primary tumor and monitored for metastasis.
Results
The GEP assay successfully classified 446/459 (97.2%) cases. The GEP was class 1 in 276 cases (61.9%) and class 2 in 170 cases (38.1%). Median follow-up was 17.4 months (mean, 18.0 months). Metastasis was detected in 3 (1.1%) class 1 cases and 44 (25.9%) class 2 cases (log rank test, P<10−14). Although there was an association between GEP class 2 and monosomy 3 (Fisher exact test, P<0.0001), 54/260 (20.8%) tumors were discordant for GEP and chromosome 3 status, among which GEP demonstrated superior prognostic accuracy (log rank test, P=0.0001). Using multivariate Cox modeling, GEP class had a stronger independent association with metastasis than any other prognostic factor (P<0.0001). Chromosome 3 status did not contribute additional prognostic information that was independent of GEP (P=0.2). At three years follow-up, the net reclassification improvement of GEP over TNM classification was 0.43 (P=0.001) and 0.38 (P=0.004) over chromosome 3 status.
Conclusions
The GEP assay had a high technical success rate and was the most accurate prognostic marker among all of the factors analyzed. GEP provided a highly significant improvement in prognostic accuracy over clinical TNM classification and chromosome 3 status. Chromosome 3 status did not provide prognostic information that was independent of GEP.
Uveal melanoma is the most common primary cancer of the eye and has a propensity for fatal hematogenous metastasis.1 Advances in treatment of the primary tumor have allowed greater preservation of eyes and vision, but they have not led to measurable improvements in patient survival.2 This observation has been attributed to presumed early subclinical micrometastasis coupled with a prolonged latency prior to development of overt metastatic disease.3 Since there are no therapies that have been proven to be effective against advanced metastatic uveal melanoma,4 there has been increasing interest in developing adjuvant therapy for high risk patients who are likely to harbor subclinical micrometastases. An accurate method for identifying these patients is needed as an entry criterion for such clinical trials.
Gene expression profiling (GEP) has revealed that primary uveal melanomas cluster into two highly prognostic molecular subgroups: class 1 (low metastatic risk) and class 2 (high metastatic risk)5 Recently, it was shown that the class 2 GEP correlates with mutations in the BAP1 tumor suppressor gene.6 BAP1 is located on chromosome 3, which is commonly lost in uveal melanoma.7–9 In most metastasizing class 2 tumors, one copy of BAP1 is mutated and the other is absent through loss of the entire chromosome, consistent with the “two hit” model for mutation of tumor suppressor genes. As expected, there is a strong relationship between monosomy 3 and the class 2 GEP.10, 11 However, multiple independent groups have consistently shown that GEP classification is a more accurate prognostic markers in uveal melanoma compared to monosomy 3 and other cytogenetic, clinical and pathologic factors.10, 12–14 Further, GEP classification is the only prognostic method in uveal melanoma to be validated on multiple independent datasets in a manner that was masked to patient outcome.15, 16 Consequently, we developed a GEP assay for routine clinical use based on the expression of 15 carefully selected genes.16 This assay includes a pre-amplification step and is performed on a microfluidics polymerase chain reaction (PCR) platform that together allow it to detect extremely small quantities of tumor RNA from fine needle aspiration biopsy (FNAB) samples, while it can also be used to analyze surgically resected specimens.16
We formed a Collaborative Ocular Oncology Group (COOG), comprising twelve ocular oncology centers in North America, to evaluate prospectively the GEP assay in a large cohort of patients with uveal melanoma. The GEP assay was performed on a sample collected directly from the primary tumor, usually at the time of treatment, and patients were subsequently followed for detection of metastatic disease. This study reports our initial results for patients with posterior (choroidal and ciliary body) uveal melanoma, which represents the most common form of uveal melanoma.
METHODS
Patient data and tumor samples
This study was conducted with the approval of the Institutional Review Board and/or Ethics Committee of each participating institution. Informed consent was obtained from each patient and the study adhered to the tenets of the Declaration of Helsinki. All work using patient information was performed in compliance with the Health Insurance Portability and Accountability Act. Inclusion criteria for the study patients were: (1) clinical diagnosis of uveal melanoma and (2) investigational informed consent. Clinical diagnosis of uveal melanoma was determined by the presence of clinical features typical of uveal melanoma such as thickness ≥ 2.5 mm, orange lipofuscin pigmentation, serous retinal detachment, collar button configuration and/or mushroom shape and growth. 19 tumors with thickness < 2.5 mm were included based on documented growth or ≥2 high risk features listed above. Exclusion criteria were: (1) tumor location limited to the iris and (2) non-melanoma cytopathologic diagnosis. 459 cases were obtained prospectively between June 2006 and November 2010. De-identified patient information was collected from each center, including patient age, gender, tumor thickness (measured by ultrasound), tumor diameter (defined as the largest basal tumor dimension measured by indirect ophthalmoscopy or ultrasound), ciliary body involvement (defined as any portion of the tumor extending anterior to the ora serrata), date tumor sample was obtained, cytopathologic cell type (predominantly spindle, mixed, epithelioid, unspecified melanoma cell type, acellular/quantity not sufficient for diagnosis, or information not available), last known patient survival status (alive with no metastasis, alive with metastasis, dead of metastatic disease, or dead of other causes), presence or absence of metastatic disease, date metastatic disease was first detected, and date of death or most recent follow-up. The 7th edition Tumor-Node-Metastasis (TNM) clinical classification for uveal melanoma was performed using basal tumor diameter, thickness, and ciliary body involvement as described elsewhere17. Following treatment of the primary tumor, patients were monitored for metastasis with a liver function panel every 6 months and liver imaging (usually computed tomography with contrast) once a year or anytime the liver function panel was abnormal or there were symptoms suspicious for metastasis (e.g., right upper quadrant fullness or discomfort, unexplained weight loss). Suspicious findings on imaging were followed up with biopsy confirmation when possible. Patients were coded as having metastasis only if (1) biopsy confirmation was obtained, or (2) imaging studies were highly consistent with uveal melanoma metastasis in the absence of other known primary cancer.
Preparation of RNA Samples
FNAB samples for RNA analysis were expelled into an empty RNase-free tube in the operating room. The empty syringe was filled with 200 μL of extraction buffer (XB) from the PicoPure® RNA isolation kit (Molecular Devices, Sunnyvale, CA), which was flushed through the syringe to collect any additional tumor cells lodged in the needle hub. The tube was then snap frozen in liquid nitrogen or a dry ice-alcohol bath in the operating room prior to transportation. For specimens requiring transport to the Harbour Laboratory at Washington University from other centers, tubes were placed on dry ice and mailed by overnight courier. On arrival in the laboratory, the samples were logged and stored in a freezer at −80°C until they could be processed. RNA was isolated using the PicoPure® kit (including the optional DNase step), which yielded <10 ng to 1.5 μg total RNA per aspirate using the NanoDrop 1000 system (NanoDrop Products, Wilmington, DE). Genomic DNA was prepared using the Wizard Genomic DNA Purification kit (Promega, Madison, WI). RNA quality was assessed by comparing the total RNA yield to the expression of the three endogenous control genes, as previously described.16
Real-Time PCR Analysis
RNA samples were converted to cDNA using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Applied Biosystems Inc., Foster City, CA) following the manufacturer's protocol. Then, the cDNA was pre-amplified for fourteen cycles with pooled TaqMan® primers and Pre-Amp Master Mix, diluted 20-fold into sterile TE buffer, and stored at −20° C. RNA expression for each of the 15 genes was quantified using the 7900HT Real-Time PCR System with Applied Biosystems TaqMan® primers and Gene Expression Master Mix following the manufacturer's protocol. TaqMan® Microfluidics Expression Arrays were custom ordered to include our 12 class discriminating genes and three endogenous control genes in triplicate. The number of PCR cycles required for each gene to reach the expression threshold (Ct) was calculated using the manufacturer's software, and mean Ct values were calculated for all triplicate sets. ΔCt values were calculated by subtracting the mean Ct of each discriminating gene from the geometric mean of the mean Ct values of the three endogenous control genes.
Gene expression profiling and other molecular analyses
The 15-gene classification assay was performed at the Washington University site using GIST 2.3 Support Vector Machine (SVM) (http://chibi.ubc.ca/gist: accessed 10/17/2011) for class assignment, as described in detail in a previous study.16 The assay includes 12 discriminating genes (Table 1) and three control genes (MRPS21, RBM23, and SAP130). A gene was considered undetectable if its amplification product registered no Ct value after 40 cycles of qPCR. A sample was considered a technical failure if one or more endogenous control genes or at least three discriminating genes were undetectable. Chromosome 3 was analyzed for loss of heterozygosity using a validated assay that interrogates SNPs distributed across the chromosome, as previously described.18
Table 1.
Summary of uveal melanoma gene expression profile
| Gene Symbol | Direction of change in class 2 tumors | Gene Name |
|---|---|---|
| CDH1 | Up | E-cadherin |
| ECM1 | Up | Extracellular matrix protein 1 |
| EIF1B | Down | Eukaryotic translation initiation factor 1B |
| FXR1 | Down | Fragile × mental retardation, autosomal homolog 1 |
| HTR2B | Up | 5-hydroxytryptamine (serotonin) receptor 2B |
| ID2 | Down | Inhibitor of DNA binding 2 |
| LMCD1 | Down | LIM and cysteine-rich domains 1 |
| LTA4H | Down | Leukotriene A4 hydrolase |
| MTUS1 | Down | Microtubule associated tumor suppressor 1 |
| RAB31 | Up | RAB31, member RAS oncogene family |
| ROBO1 | Down | Roundabout, axon guidance receptor, 1 |
| SATB1 | Down | SATB homeobox 1 |
Statistical analysis
Cox proportional-hazards regression analysis, Kaplan-Meier survival analysis, and Receiver Operating Characteristic (ROC) curve analysis were performed using Medcalc® version 11.5.1.0. The optimal cut-off values for continuous variables for use in Kaplan-Meier analyses were determined using receiver operating characteristic (ROC) analysis. For Cox proportional hazards modeling, the proportionality of all hazards tested were confirmed over the whole time period using the log(-log(survival)) versus log of survival time method. The net reclassification improvement (NRI) of GEP over TNM classification and chromosome 3 status were calculated by setting cut-offs for disease-free survival at 2 years and 3 years, and then using ROC analysis to determine the sensitivity and specificity of each classification method. The NRI was equivalent to twice the difference in areas under the ROC curves for each method19, using a binary TNM classification based upon the ROC-assigned threshold of Stage > T3a.
RESULTS
Baseline information
The study design is outlined in Figure 1. The gene expression profile (GEP) prognostic assay was performed on 459 posterior uveal melanomas obtained prospectively from twelve participating institutions. 224 patients were female and 235 were male. Mean age was 61.7 years (median, 61.0 years). Mean tumor diameter was 12.8 mm (median, 12.7 mm), and mean tumor thickness was 6.3 mm (median, 5.5 mm). Ciliary body involvement was absent in 308 cases, present in 139 cases and unknown in 12 cases. Tumor samples were obtained by FNAB in 359 cases, post-enucleation FNAB in 92 cases, and local tumor resection in 8 cases. The cytopathologic diagnosis was spindle cell melanoma in 143 cases, mixed cell melanoma in 95 cases, epithelioid cell melanoma in 87 cases, unspecified melanoma cell type in 41 cases, acellular/quantity not sufficient for diagnosis in 60 cases, and sample not obtained for cytopathology in 33 cases. The status of chromosome 3 was assessed by a multi-SNP assay in the first 260 cases. 34 deaths occurred, 28 (82.4%) of which were due to metastatic disease. Another 19 patients had developed metastases but were still alive at the time of last follow-up.
Figure 1.
Schematic of study design. COOG = Collaborative Ocular Oncology Group; GEP = gene expression profiling.
Initial performance of the GEP assay
The GEP assay was successful in rendering a classification in 446/459 (97.2%) cases. Among the 13 samples that failed to yield a GEP result, 5 did not adhere to study protocol (improper buffer, handling or shipping). Of the 446 cases, 276 (61.9%) were class 1 and 170 (38.1%) were class 2. Median follow-up was 17.4 months (mean, 18.0 months). Metastasis was detected in 3 (1.1%) patients with class 1 tumors and 44 (25.9%) patients with class 2 tumors (log rank, P<0.0001). Fifteen samples were obtained from active tumors arising from uveal melanomas that were previously treated with plaque radiotherapy (7 cases), proton beam radiotherapy (4 cases), and laser treatment (4 cases); nine of these were class 1 and six were class 2.
Comparison of GEP to clinical and histopathologic prognostic factors
GEP class 2 showed a significant association with other known prognostic factors, including increased patient age, greater tumor diameter and thickness, ciliary body involvement, and mixed/epithelioid cell type (Table 2). By Kaplan-Meier analysis, GEP class 2 was more strongly associated with metastasis than any of the other prognostic factors that were analyzed, including chromosome 3 status (Figure 2). By univariate Cox proportional hazards analysis (Table 3), factors associated with metastasis included advanced patient age (P=0.02), ciliary body involvement (P=0.03), tumor diameter (P=0.0003), tumor thickness (P=0.006), tumor cell type (P=0.04), chromosome 3 status (P=0.0002) and GEP class (P<10−7). This analysis was performed on all cases with values reported for a given factor, and there was no significant impact on the results when the analysis was restricted to those cases with complete data for all factors. By multivariate Cox modeling, GEP class (P=0.006) was the only variable that contributed independent prognostic information. A significant association was observed between TNM classification and metastasis (P=0.003)(Table 4). Chromosome 3 status did not contribute additional prognostic information that was independent of GEP (P=0.2). The NRI of GEP over TNM classification was 0.37 at 2 years (P=0.008) and 0.43 at 3 years (P=0.001), and the NRI of GEP over chromosome 3 status was 0.36 at 2 years (P=0.006) and 0.38 at 3 years (P=0.004)(Table 5).
Table 2.
Comparison of uveal melanoma molecular classes
| Variable | Class 1 | Class 2 | P value |
|---|---|---|---|
| Age (years) | mean,59.6 median, 59.0 min-max, 13.2–98.4 |
mean,65.3 median, 65.9 min-max, 17.2–91.5 |
0.0001 |
| Gender | female, 130 male, 146 |
female, 89 male, 81 |
0.2 |
| Tumor diameter (mm) | mean, 12.1 median, 12.0 min-max, 4.0–24.0 |
mean, 13.9 median, 14.0 min-max, 1.3–23.0 |
<0.0001 |
| Tumor thickness (mm) | mean, 5.7 median, 5.0 min-max, 1.0–17.5 |
mean, 7.0 median, 6.2 min-max, 1.5–17.5 |
<0.0001 |
| Ciliary body involvement | yes, 61 (22.7%) no, 208 (77.3%) |
yes, 72 (43.6%) no, 93 (56.4%) |
<0.0001 |
| Mixed or epithelioid cytology | yes, 86 (45.5%) no, 103 (54.5%) |
yes, 89 (70.6 %) no, 37 (29.4%) |
<0.0001 |
| Metastasis | yes, 3 (1.1%) no, 273 (98.9%) |
yes, 44 (25.9%) no, 126 (74.1%) |
<0.0001 |
Figure 2.
Comparison of gene expression profile (GEP) classification to other prognostic factors. Kaplan-Meier plots for the indicated prognostic factors. P-values were determined by log rank method. Age indicates patient age at the time of primary tumor diagnosis. D3, disomy 3; M3, monosomy 3. Threshold values for dichotomizing continuous variables (tumor thickness and diameter) were determined by receiver operating characteristic (ROC) analysis.
Table 3.
Cox proportional hazards analysis of prognostic factors
| Variable | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| P value | HR | CI | P value | HR | CI | |
| Increased patient age | 0.02 | 2.2 | 1.2 – 4.0 | 0.2 | 1.9 | 0.7 – 5.0 |
| Ciliary body involvement | 0.03 | 1.9 | 1.1 – 3.4 | 0.9 | 1.1 | 0.4 – 3.3 |
| Increased tumor diameter | 0.0003 | 3.2 | 1.7 – 5.9 | 0.2 | 2.0 | 0.7 – 5.6 |
| Increased tumor thickness | 0.006 | 2.5 | 1.3 – 4.8 | 0.6 | 0.8 | 0.3 – 2.3 |
| Mixed/epithelioid cell type | 0.04 | 2.3 | 1.0 – 5.1 | 0.6 | 1.3 | 0.5 – 3.3 |
| Loss of chromosome 3 | 0.0002 | 4.3 | 2.0 – 9.3 | 0.2 | 2.8 | 0.6 – 13.7 |
| GEP class 2 | < 10−7 | 26.4 | 8.2 – 84.6 | 0.006 | 20.5 | 2.4 – 175.6 |
GEP, gene expression profile; HR, hazard ratio; CI, confidence interval.
Table 4.
Tumor/Node/Metastasis (TNM) staging of the uveal melanomas included in the study based on the guidelines established by the American Joint Committee on Cancer, 7th Ed.
| Number of cases | Cases that Metastasized | Class 2 Cases | |
|---|---|---|---|
| T1 | |||
| a | 77 | 1 (1.3%) | 15 (19%) |
| b | 4 | 1 (25%) | 2 (50%) |
|
| |||
| T2 | |||
| a | 138 | 14 (10%) | 42 (30%) |
| b | 32 | 3 (9.4%) | 14 (44%) |
|
| |||
| T3 | |||
| a | 71 | 5 (7.0%) | 28 (39%) |
| b | 69 | 11 (16%) | 36 (52%) |
|
| |||
| T4 | |||
| a | 12 | 4 (33%) | 6 (50%) |
| b | 22 | 4 (18%) | 18 (82%) |
T indicates tumor sized; a indicates no ciliary body involvement; b indicates ciliary body involvement.
Table 5.
Net Reclassification Improvement of gene expression profile versus other prognostic tools.
| All cases | At 2 years | At 3 years | |
|---|---|---|---|
| TNM Staging | |||
| Sample size | 425 | 146 | 70 |
| Positive for metastasis | 43 (10%) | 32 (22%) | 42 (60%) |
| Net Reclassification Improvement | 0.42 | 0.37 | 0.43 |
| Standard Error | 0.044 | 0.055 | 0.066 |
| Significance level | P < 0.0001 | P = 0.008 | P = 0.001 |
| Monosomy 3 | |||
| Sample size | 260 | 121 | 60 |
| Positive for metastasis | 38 (15%) | 27 (22%) | 37 (62%) |
| Net Reclassification Improvement | 0.28 | 0.36 | 0.38 |
| Standard Error | 0.038 | 0.052 | 0.065 |
| Significance level | P = 0.002 | P = 0.006 | P = 0.004 |
TNM = Tumor/Node/Metastasis: the tumor classification guidelines for uveal melanoma established by the American Joint Committee on Cancer. Two and three year subanalyses represent data known at those time points following primary treatment in evaluable cases.
Comparison of GEP to chromosome 3 status
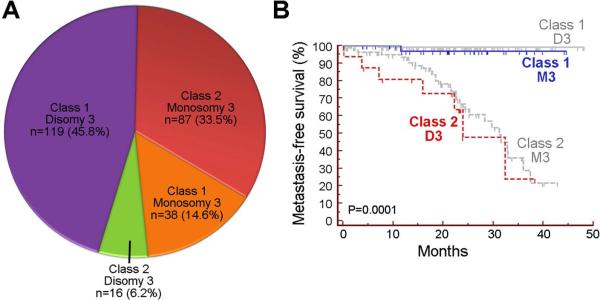
Since monosomy 3 has been widely used as a prognostic marker for uveal melanoma, we wished to study more closely the relationship between GEP and chromosome 3 status. Chromosome 3 status was collected from the first 260 cases, of which, the GEP/chromosome 3 status was class 1/disomy 3 in 119 (45.8%), class 2/monosomy 3 in 87 (33.5%), class 1/monosomy 3 in 38 (14.6%), and class 2/disomy 3 in 16 (6.2%) (Figure 3A). As expected, there was a significant association between class 1 and disomy 3, and between class 2 and monosomy 3 (Chi square, P<0.0001). However, the GEP and chromosome 3 results were discordant in 54 (20.8%) cases. Among 16 cases that were class 2/disomy 3, 7 (43.8%) metastasized, whereas among 38 cases that were class 1/monosomy 3 only one metastasized (2.6%). Thus, GEP was more strongly associated with metastasis than chromosome 3 status among the discordant cases (log rank test, P=0.0001) (Figure 3B). As a result of the inferior prognostic value of chromosome 3 status, we discontinued chromosome 3 testing after the first 260 cases.
Figure 3.
Comparison of gene expression profile (GEP) classification to chromosome 3 status. (A) Pie chart showing relationship between GEP class and chromosome 3 status in 293 patients. (B) Kaplan-Meier plot showing metastasis-free survival after primary tumor diagnosis in cases where GEP class and chromosome 3 status were discordant (class 1/monosomy 3 or class 2/disomy 3). Cases in which GEP and chromosome 3 status were concordant are shown for comparison in gray. P-value was determined by log rank method.
DISCUSSION
In this prospective multicenter collaborative study, the prognostic accuracy of a 15-gene prognostic assay for uveal melanoma was assessed. A strong association was observed between GEP class 2 and other adverse prognostic factors, including increased patient age, ciliary body involvement, larger tumor diameter and thickness, epithelioid cell type and monosomy 3. However, GEP class was more strongly associated with metastasis than these other factors. In multivariate analysis, no combination of other prognostic variables (including chromosome 3 status) was more closely associated with metastasis than GEP alone. Only 6 of 34 deaths were not due to metastasis, such that non-metastatic deaths did not represent a significant competing risk in this data set (data not shown). Fifteen cases were local recurrences that arose following prior treatment, and 9 (60%) of these were GEP class 1. This suggests that recurrent tumors are no more likely to be one class or the other, although more cases are needed to establish statistical significance.
Comparison of GEP and chromosome 3 status
An important goal of this study was to compare the prognostic value of GEP and chromosome 3 status, which has been widely used as a prognostic marker in uveal melanoma. Although there was a strong association between monosomy 3 and GEP class 2, the prognostic accuracy of GEP was superior to monosomy 3. Indeed, by multivariate Cox modeling, chromosome 3 status did not provide prognostic information that was independent of GEP. The technique used for chromosome 3 testing was state of the art for methods applicable to clinical FNAB samples, consisting of a validated assay in which single nucleotide polymorphisms across the chromosome were assessed for loss of heterozygosity.18 This technique has been shown to be more accurate than other commonly used methods such as fluorescence in situ hybridization (FISH) and array-based comparative genomic hybridization (aCGH)18 Other cytogenetic techniques have been extremely valuable for research purposes but are not suitable for routine clinical use on FNAB samples, such as the use of FISH to analyze isolated tumor nuclei from eyes that have been removed.20
This study confirms that GEP is more a more accurate prognostic marker than monosomy 3 for routine clinical use in patients with uveal melanoma, consistent with previous smaller studies from multiple independent institutions.10, 12, 13 Similarly, we previously showed that the inclusion of other commonly used cytogenetic features such as gain of 6p and 8q did not improve the accuracy of GEP alone.21 A likely reason for the superiority of GEP over cytogenetic methods is that the latter are static markers that are often distributed heterogeneously throughout the tumor, and are thus prone to sampling error.20, 22 In contrast, GEP captures a functional “snapshot” of the tumor's microenvironment that does not vary as much across most tumors and is thus amenable to a single pass of a fine needle in most cases.16 Thus, while cytogenetic studies are indispensible for research purposes, they have not performed as well as GEP for routine clinical testing.
Technical performance of the GEP assay on FNAB samples
The combination of pre-amplification and microfluidics in the GEP assay platform has allowed a higher success rate on small FNAB samples compared to cytogenetic testing. The GEP assay produced interpretable results in over 97% of FNAB cases, whereas the failure rate for cytogenetic testing methods has been reported to be as high as 50%,23 owing to the greater tissue requirements and lower sensitivity of these methods. Consequently, one pass of a 25- or 27-gauge needle almost always provides an adequate sample for the GEP assay, in contrast to the more aggressive methods reported for cytogenetic testing, such as the use of a vitreous cutter24 or a high number (up to 12) of needle passes.25 For most cases, a single needle sampling for GEP near the geometric center of the tumor is sufficient. Multiple samples are only taken when there are morphologically distinct tumor components, such as multi-lobulated tumors and those with extraocular extension, in which case we recommend sampling each area separately. This assay has also been optimized for use on formalin fixed paraffin embedded specimens, which will be the subject of another publication.
Clinical applications of GEP testing
The GEP assay represents a prospectively validated tool that can be used for routine clinical prognostic testing and for stratifying patients for entry into clinical trials of adjuvant therapy. About half of patients with a class 2 tumor will develop detectable metastasis by three years after primary tumor diagnosis. Enrolling patients with class 2 tumors into clinical trials at the time of primary tumor diagnosis offers the potential to reduce the number of patients required and length of time needed to detect a difference in outcomes can be greatly reduced relative to other available prognostic markers. The identification of BAP1 mutations associated with class 2 tumors6 may soon lead to new targeted therapeutic agents for such clinical trials.
Acknowledgements
The authors would like to thank Dr. Steve Kymes for statistical consultation.
Financial Support: This work was supported by grants to J.W.H. from the National Cancer Institute (R01 CA125970), Barnes-Jewish Hospital Foundation, Kling Family Foundation, Tumori Foundation, Horncrest Foundation, and a Research to Prevent Blindness David F. Weeks Professorship, and by awards to the Department of Ophthalmology and Visual Sciences at Washington University from a Research to Prevent Blindness, Inc. Unrestricted grant, and the NIH Vision Core Grant P30 EY02687c.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Of Potential Conflicts Of Interest: Dr. Harbour and Washington University may receive royalties based on a license of related technology by the University to Castle Biosciences, Inc. This research was not funded by Castle Biosciences, Inc.
REFERENCES
- 1.Ramaiya KJ, Harbour JW. Current management of uveal melanoma. Expert Rev Ophthalmol. 2007;2:939–46. [Google Scholar]
- 2.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–5. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Eskelin S, Pyrhonen S, Summanen P, et al. Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology. 2000;107:1443–9. doi: 10.1016/s0161-6420(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 4.Augsburger JJ, Correa ZM, Shaikh AH. Effectiveness of treatments for metastatic uveal melanoma. Am J Ophthalmol. 2009;148:119–27. doi: 10.1016/j.ajo.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 5.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horsman DE, Sroka H, Rootman J, White VA. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet Cytogenet. 1990;45:249–53. doi: 10.1016/0165-4608(90)90090-w. [DOI] [PubMed] [Google Scholar]
- 8.Prescher G, Bornfeld N, Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. J Natl Cancer Inst. 1990;82:1765–9. doi: 10.1093/jnci/82.22.1765. [DOI] [PubMed] [Google Scholar]
- 9.Sisley K, Rennie IG, Cottam DW, et al. Cytogenetic findings in six posterior uveal melanomas: involvement of chromosomes 3, 6, and 8. Genes Chromosomes Cancer. 1990;2:205–9. doi: 10.1002/gcc.2870020307. [DOI] [PubMed] [Google Scholar]
- 10.Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007;13:1466–71. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 11.Tschentscher F, Husing J, Holter T, et al. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res. 2003;63:2578–84. [PubMed] [Google Scholar]
- 12.van Gils W, Lodder EM, Mensink HW, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008;49:4254–62. doi: 10.1167/iovs.08-2033. [DOI] [PubMed] [Google Scholar]
- 13.Petrausch U, Martus P, Tonnies H, et al. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye (Lond) 2007;22:997–1007. doi: 10.1038/sj.eye.6702779. [DOI] [PubMed] [Google Scholar]
- 14.Singh AD, Sisley K, Xu Y, et al. Reduced expression of autotaxin predicts survival in uveal melanoma. Br J Ophthalmol. 2007;91:1385–92. doi: 10.1136/bjo.2007.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onken MD, Worley LA, Davila RM, et al. Prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. J Mol Diagn. 2006;8:567–73. doi: 10.2353/jmoldx.2006.060077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finger PT. 7th Edition AJCC-UICC Ophthalmic Oncology Task Force. The 7th edition AJCC staging system for eye cancer: an international language for ophthalmic oncology. Arch Pathol Lab Med. 2009;133:1197–8. doi: 10.5858/133.8.1197. [DOI] [PubMed] [Google Scholar]
- 18.Onken MD, Worley LA, Person E, et al. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clin Cancer Res. 2007;13:2923–7. doi: 10.1158/1078-0432.CCR-06-2383. [DOI] [PubMed] [Google Scholar]
- 19.Pencina MJ, D'Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maat W, Jordanova ES, van Zelderen-Bhola SL, et al. The heterogeneous distribution of monosomy 3 in uveal melanomas: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med. 2007;131:91–6. doi: 10.5858/2007-131-91-THDOMI. [DOI] [PubMed] [Google Scholar]
- 21.Onken MD, Worley LA, Harbour JW. A metastasis modifier locus on human chromosome 8p in uveal melanoma identified by integrative genomic analysis. Clin Cancer Res. 2008;14:3737–45. doi: 10.1158/1078-0432.CCR-07-5144. [DOI] [PubMed] [Google Scholar]
- 22.Dopierala J, Damato BE, Lake SL, et al. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Invest Ophthalmol Vis Sci. 2010;51:4898–905. doi: 10.1167/iovs.09-5004. [DOI] [PubMed] [Google Scholar]
- 23.Young TA, Rao NP, Glasgow BJ, et al. Fluorescent in situ hybridization for monosomy 3 via 30-gauge fine-needle aspiration biopsy of choroidal melanoma in vivo. Ophthalmology. 2007;114:142–6. doi: 10.1016/j.ophtha.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 24.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–92. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 25.Young TA, Burgess BL, Rao NP, et al. Transscleral fine-needle aspiration biopsy of macular choroidal melanoma. Am J Ophthalmol. 2008;145:297–302. doi: 10.1016/j.ajo.2007.09.028. [DOI] [PubMed] [Google Scholar]