Abstract
Background
Structure-based methods for P450 substrates are commonly used during drug development to identify sites of metabolism. However, docking studies using available x-ray structures for the major drug-metabolizing P450, CYP3A4, do not always identify binding modes supportive of the production of high-energy toxic metabolites. Minor pathways such as P450-catalyzed dehydrogenation have been experimentally shown to produce reactive products capable of forming biomolecular adducts which can lead to increased risk toxicities. 4-hydroxy-tamoxifen (4OHT) is metabolized by CYP3A4 via competing hydroxylation and dehydrogenation reactions.
Methods
Ab initio gas-phase electronic structural characterization of 4OHT was used to develop a docking scoring scheme. Conformational sampling of CYP3A4 with molecular dynamics simulations along multiple trajectories were used to generate representative structures for docking studies using recently published heme parameters. A key predicted binding mode was tested experimentally using site-directed mutagenesis of CYP3A4 and liquid chromatography mass spectroscopy analysis.
Results
Docking with MD-refined CYP3A4 structures incorporating hexacoordinate heme parameters identifies a unique binding mode involving ARG212 and channel 4, unobserved in the starting PDB ID: 1TQN x-ray structure. The models supporting dehydrogenation are consistent with results from in vitro incubations.
Conclusions and General significance
Our models indicate that coupled structural contributions of the ingress, egress and solvent channels to the CYP3A4 active site geometries play key roles in the observed 4OHT binding modes. Thus adequate sampling of the conformational space of these drug-metabolizing promiscuous enzymes is important for substrates that may bind in malleable regions of the enzyme active-site.
Keywords: P450, 4-hydroxytamoxifen, dehydrogenation, molecular dynamics, docking, conformational dynamics
Introduction
Cytochrome P450 enzymes (P450) metabolize a diverse set of substrates via different oxidation reaction mechanisms [1, 2]. As more and more experimental P450 structures with and without substrates become available [3–5], the emerging consensus is that the extent of promiscuity in P450 substrate selectivity has a structural basis which relates to both the varied enzyme active site architectures and their plasticity [6]. These characteristics vary greatly between the different subfamilies of hepatic P450 [7–9]. While hepatic P450 promiscuity in substrate-selectivity favors the elimination of numerous endogenous and exogenous substrates whose accumulation can prove toxic to the organism [10], it presents a significant challenge to structure-based drug design. This promiscuity makes it difficult to a priori predict if a putative lead compound will be adversely metabolized by P450 [11, 12].
The characterization of novel and reactive Phase I drug metabolites, such as those produced by P450, is an area of intensive research efforts during preclinical drug-testing and development to help identify reactive metabolites [13–15]. Of key interest is improving our understanding of factors that contribute to competing Phase I reaction mechanisms, some of which produce stable products that can be further metabolized and/or excreted and others that produce reactive metabolites capable of inducing toxicities [16]. Due to the high energy nature of the catalytic oxyferryl species, Compound I [17], P450s can catalyze a variety of different reaction mechanisms, such as dehydrogenation reactions, that can produce desaturated electrophilic metabolites capable of forming potentially toxic biomolecular adducts [18, 19]. Although a number of tools exist to predict P450 metabolism, they demonstrate a mixed level of success depending on the class of substrate, P450 and reaction mechanism. Since P450 can catalyze a number of different oxidation reactions, including competing reaction mechanisms at the same site of metabolism (SOM), simply identifying the major metabolite is not adequate for drug safety screening.
Computational tools for predicting the site and mechanism of P450-metabolism are commonly used to provide structural alerts for a given set of lead compounds [11], the goal of which is to highlight and identify the potential for the generation of toxic metabolites early in the drug-design process. Ultimately, eliminating potentially reactive lead compounds earlier may reduce drug development costs and timelines [13, 14, 20]. However, due to the diversity of the active site architecture and plasticity of the different subfamilies of hepatic P450, structure-based computational methods face a serious challenge in accurately predicting the SOM and the P450-catalyzed reaction mechanism, especially for CYP3A4 which is the hepatic P450 responsible for the majority of Phase I drug metabolism. Experimental data have demonstrated that CYP3A4 can accommodate a wide variety of structurally diverse ligands, substrates, and cofactors [8, 21, 22]. The currently available x-ray structures of CYP3A4 (PDB ID: 1W0E [23], 1W0F [23], 1W0G [23], 1TQN [24], 2J0D [8], 2V0M [8] and 3NXU [25]) support that this enzyme has a large plastic active site and suggests that the conformational dynamics of the CYP3A4 active site may have a direct impact on drug metabolism by influencing both the substrate selectivity and the specific reaction mechanism.
Our group has focused on dehydrogenation reactions due to their potential to metabolize substrates to reactive metabolites. Dehydrogenation reactions are more difficult to predict than standard P450 reactions and moreover, they can compete with hydroxylation mechanisms. Predicting them is made even more complicated since dehydrogenation may involve the abstraction of two hydrogen atoms from two different SOM, in contrast to the conventional ‘text-book’ rebound mechanism for hydroxylation which only involves one SOM [26]. Although dehydrogenation reactions are not as well studied as other P450-catalyzed reactions, mechanistic studies reveal the requirement for the formation of a stable cationic species and suggest that the enzyme active-site - substrate interactions impose steric hindrance within the P450 active site so as to inhibit the rebound process to form a hydroxylated product [27]. To our knowledge, currently tools do not yet exist for predicting the susceptibility of substrates to be dehydrogenated and to form electrophilic products.
In previous work, we showed that molecular modeling combined with docking methods were able to assess enzyme-substrate interactions important in the P450-metabolism of raloxifene (RALX) by CYP3A4 [28]. It was shown that the reliable prediction of enzyme-substrate interactions was greatly improved by the assignment of an initial set of chemically reasonable atomic partial charges for the resting high-spin ferric state of the heme. These theoretical studies correctly identified the SOM and were able to guide us to experimentally confirm the role of Phe215 in determining the selectivity of dehydrogenation versus oxygenation metabolic pathways. More recently, we have improved our heme models through the development of consistent and AMBER-compatible heme parameters for a number of heme states during the P450 catalytic cycle, including several penta-coordinate and hexa-coordinate states with dioxygen and the catalytically activated oxygen atom bound, Compound I [29]. We validated these molecular mechanics parameters for the modeling of P450-substrate interactions with raloxifene and CYP3A4 [28]. With the new heme parameters, we were able to generate models that further improved our prediction of the SOM through a structure-based modeling approach.
Building on the previous work, here we investigate the P450 metabolism of 4-hydroxy-tamoxifen (4OHT) which, similar to raloxifene, undergoes both hydroxylation and dehydrogenation reactions [30–32]. 4OHT is a primary metabolite of tamoxifen, which is a FDA-approved selective estrogen receptor modulator used extensively in the treatment of breast cancer [33], (Figure 1). 4OHT is a useful molecular probe since it is a pharmacologically active drug metabolite that can undergo P450-oxygenation and dehydrogenation to form reactive products, which is of concern in the MIST guidelines. Moreover, its metabolism is well understood and shown to be dependent on differences in the P450-subclass active-sites [30]. Also, 4OHT provides an additional challenge for improving P450 models since unlike raloxifene, initiation of dehydrogenation reactions competes with hydroxylation reactions at the same SOM. To extend upon our previous approach, we use extensive quantum mechanics calculations to probe substrate reactivity for dehydrogenation, and molecular dynamics simulations to find representative enzyme active-site conformations (of the 1TQN PDB structure). With the incorporation of the recently published heme parameters [29], these CYP3A4 MD –refined structures provide models for the docking experiments that incorporate thermodynamic fluctuations of enzyme structure and changes in the electronic state of the heme.
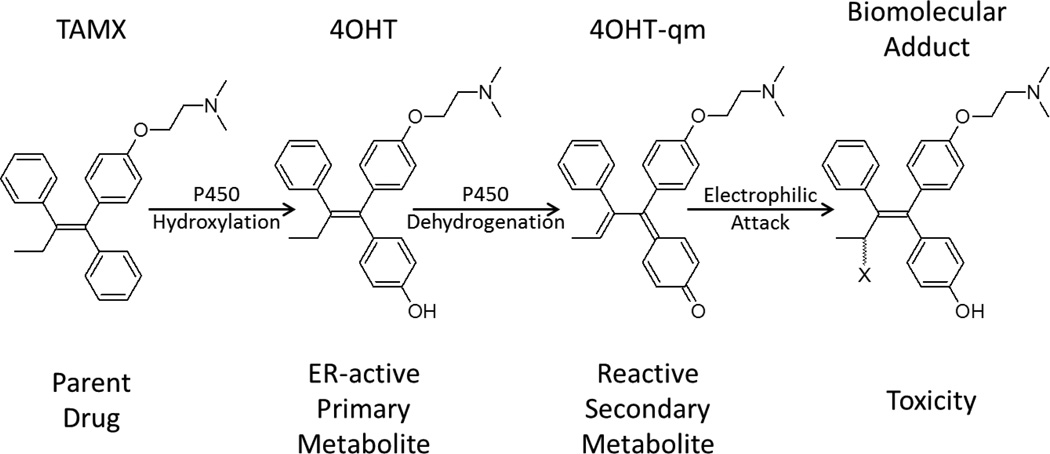
Figure 1.
Scheme for one putative pathway of P450 mediated bioactivation of tamoxifen (TAMX) to estrogen-receptor (ER) active 4-hydroxy-tamoxifen via P450-catalyzed hydroxylation, followed by conversion to 4-hydroxy-tamoxifen quinone methide (4OHT-qm) which then forms an adduct with a biomolecule (X) via an electrophilic attack.
We investigate if gas-phase quantum calculations at the HF/6–31G* and B3LYP/6–31G* level of the substrate structure can identify the dehydrogenation SOM and sequence of hydrogen abstraction. In addition to the gas-phase analysis, since experimental data indicate that P450 structure and dynamics may play a key role in metabolism, we have applied molecular dynamics (MD) methods to full P450 models in an attempt to better model enzyme-substrate interactions. We used clustering analysis to produce representative structures from multiple MD trajectories, modified these MD-refined CYP3A4 structures with new heme parameters for a number of different steps of the P450 catalytic cycle and performed docking with the QM-optimized 4OHT structures. Our results indicate that including representative conformational sampling of enzyme and substrate configurations along with the use of penta- and hexa-coordinate QM-based heme parameters for docking leads to an improved signal-to-noise for identifying the binding modes that are supportive of the entire observed metabolism of 4OHT by CYP3A4. These models are experimentally validated in line with our previous work with in vitro incubations of 4OHT with wild type CYP3A4 and the site-directed point mutant CYP3A4 R212A.
Materials and Methods
Computational
All calculations were performed on the SGI Pople machine at the Pittsburgh Supercomputing Center and at the University of Utah’s Center for High Performance Computing. The calculations involved ab initio QM, homology modeling, current state-of-the-art MD simulations with current force fields and explicit solvent, and molecular docking.
Substrate optimizations
All QM calculations were performed using Gaussian 03 software [34] in the gas-phase. All substrate structures were built, calculations set up, and results analyzed using Gaussview [35]. Initial conformations of 4OHT were intuitively organized to arrange the central ring system in one of two propeller conformations while the other functional groups were arranged to sample the conformational space as completely as possible. These starting structures were all optimized at the HF/6–31G* level of theory. Overlay of optimized structures using UCSF Chimera was used to identify redundant structures which were not used for further refinement. All optimized structures were also subjected to frequency analysis at the same level of theory as the optimization. All species with an amine moiety had optimizations for multiple minima performed with both 0 and +1 formal charges.
Dihedral scans
Scans of all ψ angles defined in Figure 2 were performed at the HF/6–31G* level of theory using one of the two lowest energy conformations of 4OHT, conformation A with a neutral charge. For ψETHYL, additional scans were also performed at the B3LYP/6–31G* and MP2/6–31G* levels of theory.
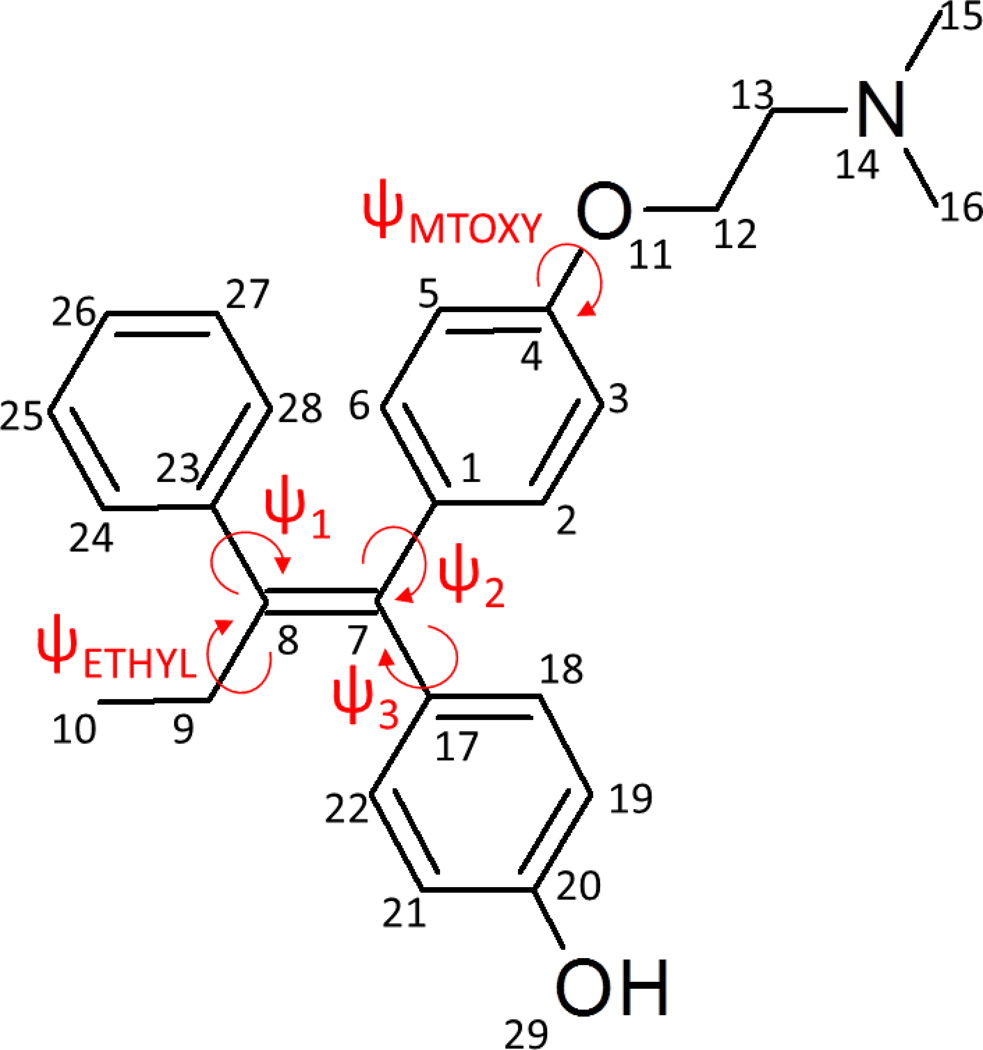
Figure 2.
Atomic numbering and dihedral labeling scheme for 4-hydroxy-tamoxifen.
Substrate bond stretching scans
Since relaxed scans produced nonphysical re-association of the abstracted hydrogen atom with some other portion of the molecule, rigid scans of the three bonds involved in the dehydrogenation of 4OHT to a quinone methide, (Figure 1), were performed for one conformation of each intermediate. Only one conformation was used for these scans and the H9a and H9b naming is arbitrary and was maintained for all intermediates, (Figure 2). Rigid scans of just one conformation may have produced some conformation-dependent differences in the energies of H9a and H9b; hence, we used conformations based on the parent’s A conformation that were identified, as described above, for each intermediate along the dehydrogenation pathway. HF/6–31G*//B3LYP/6–31G* optimized conformations of dehydrogenation reaction coordinate intermediates at stable minima, as determined via frequency analysis, were then subjected to a rigid scan of hydrogen CH or OH bonds involved in the dehydrogenation of TAMX metabolites in 40 × 0.1 Å steps. Initially, relaxed scans were performed, but these were computationally much more intensive and produced spurious results in which the hydrogen atom recombined with another part of the molecule. Certain bonds that could not be successfully scanned as a stretch from the optimized coordinate were performed starting +4.0 Å to the optimized CH bond length of interest followed by 40 × (−)0.1 Å step scans to the return to the optimized structure geometry. The B3LYP/6–31G* scans were performed using the SCF=QC method, NOSYMM constraints and the MAXCYCLE limit extended to 1000.
Substrate charge fitting
Restrained electrostatic potential (RESP) charges [36] were derived using RED-III [37] for the optimized geometries and molecular electrostatic potential (MEP) computations were performed on multiple orientations of four conformations for 4OHT in both charge states, based on atoms C9, O22 and N14 (see Figure 2 for atomic numbering).
Initial P450 loop refinement
For the x-ray structures, (PDB ID: 1TQN [24] & 1W0E [23]) used for docking and molecular dynamics simulations, missing residues in undefined loop regions were identified from the PDB file and added with UCSF MODELLER9.9 [38]. For each missing loop, five models were built with the use of the ‘automodel’ class at the loop refinement level, and ‘refine.fast’ with the ‘env.io.hetatm’ set to ‘True’ to include the heme HETATM from the pdb file. The structure with the lowest Modeller Objective Function value was selected for further refinement.
P450 molecular dynamics
MD was performed as described previously [29] except the current simulations were performed with our preliminary heme parameters [28]. Five independent MD simulations were performed for ~40 ns each with a different starting structure of the loop-refined (from the independent MODELLER loop refinements of) PDB ID: 1TQN [24] due to its higher resolution. The models were built with the ff99SB force field [39] with histidine (δ, ε or doubly protonated) states chosen by visualization (residue 27 HID, other histidines HIE). Heme parameters (shown in the supporting information) were applied and the cysteine to iron bond was manually added in tLEAP (residue 415 atom SG to residue 473 Fe). Charge was neutralized with chloride ions [40] and an additional 70 Na+ and Cl− ions were added to bring the salt concentration to ~200 mM. The models were solvated into a truncated octahedron unit cell with TIP3P water [41] extending at least 12 Å from any of the protein atoms and the positions of the ions randomized by randomly swapping with water molecules such that no ions were within 4 Å of each other and within 6 Å of the protein. All simulations employed AMBER 10.0 or 11.0 with a 9 Å cutoff with an additional 1 Å pairlist buffer, automated pairlist builds, the particle mesh Ewald method with default settings and a homogeneous long-range correction for the van der Waals interactions. MD simulations applied SHAKE [42] on all hydrogen atoms and were integrated with a 2 fs time step. The models were equilibrated with 500 steps each of steepest descent and conjugate gradient minimization with 25.0 kcal/mol-Å2 restraints on the protein Cα atoms. Heating with constant volume MD followed for 50 ps starting at 100 K ramped to 300 K over 5000 steps with 25.0 kcal/mol-Å2 restraints on the Cα atoms. Cycles of 1000 step minimization and 50 ps Berendsen-coupled [43] constant pressure and temperature MD (0.2 coupling times, 300 K) followed as before with Cα positional restraints with gradually reduced force constants at each step of the cycle of 5.0, 4.0, 3.0, 2.0 and 1.0 kcal/mol-Å2. Finally, a 500 ps constant pressure MD simulation was performed with 0.5 kcal/mol-Å2 positional restraints on the protein Cα atoms, followed by production MD with the pressure/temperature coupling times increased to 5.0. Each trajectory was determined to have reached steady state with 2D RMSD plots (shown in supplementary Figure 1). Visual inspection of the 2D RMSD plots was also used to determine the most reasonable number of clusters to search for. Analysis was performed with the ptraj module of AMBER11, each trajectory was searched for 4 clusters using the means clustering algorithm and an average representative structure was generated for each cluster in PDB format. From each trajectory, one representative structure was selected that represented the largest cluster and these were labeled m1–m5.
QM-heme parameters
All P450 models were prepared as described previously [29] with the following modifications; all structures were placed into a common frame of reference with the Matchmaker utility in UCSF Chimera tools [44] and saved. The existing x-ray heme coordinates were modified with published QM-derived coordinates and force field parameters for the resting high-spin and Compound I [29] using LEAP in AMBER tools, and an energy minimization (imin = 1) was performed for 1000 steps (maxcyc = 1000) with electrostatics turned off (type = 'ELEC', value1 = 0.0) and a cut-off of 9.0 Å (cut = 9.0) in implicit solvent was performed to relieve any steric clashes. Overlap check was performed with the check overlap utility in ptraj, and from this final structure a PDB format coordinate file was then generated and prepared for docking with the addition of Autodock3.0 parameters and RESP charges to the heme.
Docking
Four QM energy minimized structures for 4OHT with REDIII-derived RESP charges calculated using multiple conformation and reorientations for the parent molecule were used with each P450 model. The formal charge state was denoted as foht- as a neutral and fohtq- with a formal +1 charge due to protonation of the amine moiety. Then, 256 independent dockings were performed for each of the four conformations, and thus, 1024 dockings for each charge state of 4OHT with each P450 model were performed.
The refinement of the enzyme coordinates involved molecular dynamics simulations of the PDB ID: 1TQN [24] structure with five independent trajectories, and using clustering analysis of all of the structures generated to produce an average structure representative of the largest cluster of each trajectory (m1–m5). All of the MD-refined structures and the x-ray-based PDB ID: 1W0E [23] and 1TQN [24] were then modified with QM-based AMBER compatible heme parameters [29] and energy minimized to remove any steric clashes. These structures were then used with (*-wq) and without (*-00) assignment of atomic partial charges and scored using a distance-based scheme from the iron to heavy atom at a known SOM. The iron atom was used rather than the catalytic oxygen for both penta- and hexa-coordinate states of the heme so as to differentiate the contributions of charges on the heme, sterics due to the presence of the catalytic oxygen for the hexa-coordinate Compound I versus the penta-coordinate ferric and the contributions of changes in the active site architecture to the docking results.
Docking was performed with AutoDock3 [45] and Autodock tools [46] as previously published [28, 29] with the following modifications. The grid box was recentered at −21.545, −20.757 and −11.334 displacement from the default center assigned by Autogrid3.05 with the dimensions of 68 × 72 × 82 and a resolution of 0.375 Å in each dimension. Consistent with our previous work, for each of the four minima of the substrate 4OHT, 256 independent genetic algorithm runs were performed producing a total number of 1024 docked conformations for each substrate and heme charge state. This was performed for the two x-ray based (PDB ID: 1TQN and 1W0E) and the 5 MD-refined (m1–m5) P450 models, and for each P450 model docking studies were performed with charges assigned or not assigned to the heme for each catalytic cycle state. The docking results were analyzed using Autodock tools [46]. The results from each docking were clustered consistent with our previous work [28]. Autodock tools [46] was used to perform conformational clustering of Autodock3 [45] results with the 2 Å RMSD cutoff for conformational similarity of each bin and used the following site of metabolism scoring criteria based on the following distances:
α−OH: Fe-C11 < 8 Å and Fe-R2 > 8 Å
Dehydrogenation: both Fe-C11 and Fe-R2 < 8 Å
N-demethylation: Fe-C27 or C28 < 8 Å.
For each repetition of each substrate in each P450 state, the total number of poses in every cluster above a 5% signal-noise cut-off that represented a particular reaction mechanism was pooled. A number of ambiguous or nonproductive (Non) binding modes were also identified and these were all pooled together.
For each cluster that had more than 52 representative states (>5% cut-off of 1024 total), the lowest energy conformation was saved in PDB format and further analyzed with UCSF CHIMERA [44].
Statistics
Statistical analyses of each reaction mechanism for each substrate-P450 state involved a one-way ANOVA and a post-hoc test using the Tukey test. Analyses were performed using GraphPad Prism version 5.00 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com.
Channel mapping
CAVER [47] was applied to identify the channels present within the different P450 structures via the CAVER add-in to PYMOL [48]. The center selection of the P450 was based on the default setting and all structures were searched for 10 channels. In each of the runs, only 2–4 distinct channels were observed with the remaining channels being redundant to the initial set of 2–4 channels. Nomenclature was assigned consistent with Cojocaru et al. 2007 [49].
Active-site mapping
Active sites were mapped with UCSF HOLLOW [50] using a cylinder type of search with a radius 6.5 Å. The axis for each channel was found for each structure with CAVER and was defined from the activated oxygen of the hexa-coordinate heme (O1) to the Cα of the most appropriate residue deemed by visual inspection. The offset to the Cα was also set at 0.0, −1.0 or −2.0 Å based again on visual inspection.
Molecular graphics
Images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081) [44] and the PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC as noted below.
Experimental
Materials
4OHT, NADPH, silver oxide, glutathione (GSH), N-acetylcysteine (NAC) and raloxifene (internal standard) were obtained from Sigma-Aldrich (St. Louis, MO). All other chemicals were of analytical grade and obtained at the highest quality commercially available.
Synthesis of standards of dehydrogenated 4OHT product adducts
4OHT was oxidized using 500 mg silver oxide in 2 ml anhydrous acetonitrile with constant stirring at 30°C. Production of 4-hydroxy-tamoxifen quinone methide (4OHT-qm), Figure 1, was monitored by UV-vis absorbance at 280 nm. It was determined that 4OHT-qm had reached a steady-state maximum after ~8 min. Reactions were terminated by vacuum filtration of the mixture to remove excess silver oxide and purified by flash chromatography, during which the organic solution was added drop-wise to ½ volume aqueous phase (100 mM KPi, pH 7.4 with 25 mM NAC) to trap the dehydrogenated quinone methides as NAC adducts. These solutions were diluted to <5% organic phase and further purified using the Waters Oasis WCX Cation Exchange SPE column. The eluted products were concentrated under N2, and reconstituted to a final volume 50 µL MeOH prior to analysis by LC/MS.
Incubations with 4-hydroxy-tamoxifen
The CYP3A4 wild type and R212A mutants were expressed, purified and kinetics characterized as described previously [51]. In brief 25 pmol of P450, 50 pmol cytochrome b5 (Invitrogen, Carlsblad, CA) and 50 pmol of recombinant P450 oxido-reductase in a lipid mixture of 20 µg of a lipid mixture composed of equal weights of DLPS, DLPC and DOPC with 0,04% (w/v) sodium cholate were used for each incubation in 50 mM Potassium phosphate puffer (pH 7.4), 15 mM MgCl2, 5 mM NAC and 20 µM 4-hydroxytamoxifen in 1 µL DMSO.. NADPH (5 mM) was used to start the reaction. Reactions were allowed to proceed for 12 min at 37°C and terminated with addition of 200 µL ice-cold acetonitrile with internal standard added (RALX 5 µM). The mixtures were then vortexed and centrifuged at 21,000×g for 5 min. Excess KPi buffer was added to dilute the organic phase to <5%, and products were purified using a Waters Oasis WCX SPE cartridge. The MeOH eluate was then concentrated to dryness under vacuum with a Savant SVC100 Speed Vac and reconstituted in 50 µL MeOH for LC-MS analysis. Negative control incubations were performed without NADPH.
LC-MS analysis of 4OHT metabolites
LC-MS analysis of 4OHT metabolites was performed as described previously with the following [28, 52] modifications: source temperature 385°C and capillary voltage 10V. Chromatography was performed with Phenomenex Jupiter 5µ C4 (150 mm × 2.00 mm) reverse-phase column using a mobile phase consisting of Solvent A: acetonitrile and Solvent B: 20% MeOH, 0.1% formic acid (v/v/v). The analytical gradient was linear from 40% to 95% Solvent A over 15 min at a flow rate of 0.2 ml/min.
4-OHT and its hydroxylated and dehydrogenated metabolites were identified as described previously [53]. Briefly, 4-OHT was identified as the molecular ion at m/z 388.5, with an elution time of 5.6 min. MS2 of this molecular ion with activation energy of 34.0% and an isolation width of 3 amu produced a major daughter ion at m/z 331. Hydroxylated 4OHT was identified as the molecular ion at m/z 404.5, with an elution time of 4.1 min. MS2 of the m/z 404.5 with an activation energy of 42.0% and an isolation width of 3 amu produced a major daughter ion at m/z 386.0 representing the loss of a water. The dehydrogenation product 4OHT + NAC was identified as the molecular ion at m/z 549.0, with an elution time of 4.3 min. MS2 of m/z 549.0 with activation energy of 20.0% and an isolation width of 5 amu, yielded a major daughter ion with m/z 386.0 representing the loss of NAC. MS3 fragmentation of the 386.0 daughter ion with 42% activation energy and isolation width of 3 amu of this molecular ion yielded the major diagnostic ion with m/z 341.1, the diagnostic ion of 4OHT. Raloxifene was identified as m/z of 474.5 with an elution time at 4.4 min.
Results
Substrate geometry optimizations
The structure activity relationships for tamoxifen have been investigated previously in an effort to design the next generation of selective estrogen receptor modulators [54–59]. These studies illustrate the complex interplay of interactions with various estrogen receptors and other targets showing how small differences in the chemical structure can alter the pharmacological activity [60]. Early theoretical investigations at lower levels of theory had also provided insights into the complex conformational landscape produced by the central alkene covalently bound to three aromatic rings, which could occupy multiple minima [61, 62]. However, recent investigations at HF and B3LYP levels of theory revealed one minima existed which was in agreement with experimental x-ray data [63]. Energy minimizations at the HF/6–31G* level-the conventional level of theory applied for molecular mechanical force field parameterization compatible with the nonpolarizable AMBER-based pairwise additive ff9X force fields that produce a slight and beneficial overpolarization-identified multiple minima within ~2 kcal/mol, (Table 1 and Figure 3). The major difference differences in the optimized geometries at the HF/6–31G* level involved two sites: 1) the orientation of the aromatic rings and ethyl group attached via a central double bond from C7 to C8, which compose the central ring system; and 2) the oxy-methyl-amine (MTOXY) ‘tail’ composed of atoms O11, C12, C13, N14, C15 and C16 (refer to Figure 2 for nomenclature), which adopted one of two conformations, (Table 1). The differences with the ring system (dihedral angle ψ1–3) relate to different ‘propellering’ in one of two directions, and the ethyl group is oriented with C10 above or below the plane defined by the central carbons (C1, C7, C8, C9, C17 and C23).
Table 1.
Key dihedral angles of ψA-C, ETHYL & MTOXY and differences in zero-point energies (ΔZPE) and sum of electronic and free energies (ΔG) for four nonredundant optimized geometries of 4-hydroxy-tamoxifen. Conformations labeled (A-D) at results from calculations at (i) HF/6–31G*, (ii) B3LYP/6–31G* levels of theory. Differences in energy are shown in units of kcal/mol. Dihedral angles are shown in degrees.
| (i) | |||||||
|---|---|---|---|---|---|---|---|
| HF/6–31G* | ψETHYL | ψ1 | ψ2 | ψ3 | ψMTOXY | ΔZPE | ΔG |
| A | 114.0 | 119.4 | 127.3 | −64.1 | 1.8 | 0.0 | 0.0 |
| B | 81.2 | −119.8 | −127.5 | 69.1 | 87.3 | 3.2 | 1.9 |
| C | −81.2 | 119.7 | 128.4 | −68.8 | 1.8 | 2.0 | 1.5 |
| D | −113.8 | −119.5 | −126.4 | 64.6 | 87.4 | 1.1 | 0.4 |
| (ii) | |||||||
|---|---|---|---|---|---|---|---|
| B3LYP/6–31G* | ψETHYL | ψ1 | ψ2 | ψ3 | ψMTOXY | ΔZPE | ΔG |
| A | 115.7 | 127.9 | 134.7 | −56.6 | 1.3 | 0.1 | 0.2 |
| B | 76.1 | −136.5 | −137.7 | 63.3 | 1.5 | 1.9 | 2.4 |
| C | −76.1 | 136.0 | 137.1 | −63.0 | 1.5 | 1.8 | 2.1 |
| D | −115.4 | −127.8 | −134.5 | 56.5 | 1.0 | 0.0 | 0.0 |
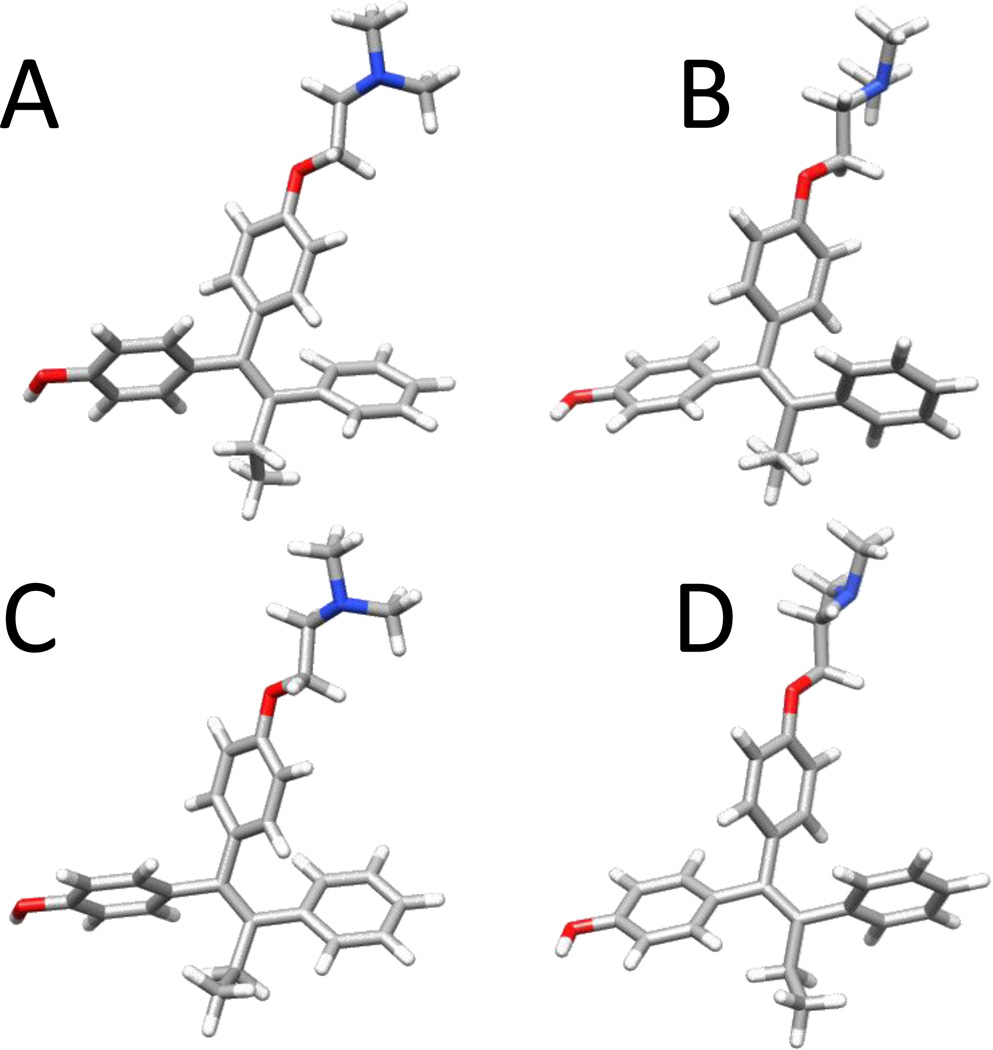
Figure 3.
Four (A–D) different energy minimized geometries of 4-hydroxy tamoxifen identified at the B3LYP/6–31G* level of theory.
Optimizations at the B3LYP/6–31G* level did not show the same difference in orientation of the MTOXY tail. Scans of ψMTOXY at HF/6–31G* and B3LYP/6–31G* level indicate that the minima observed at the HF level at ~87° is not observed at the B3LYP level (supplementary material Figure 2). The B3LYP/6–31G* structures shared all the other trends and showed similar energy differences. Since structures derived at the B3LYP-level of theory have been shown to be in good agreement with experimental structures [63], the DFT-based conformations were used for further modeling. The differences between these four geometries are shown qualitatively in Figure 3. Of the four conformations, denoted A–D, A and C share similar orientations of the ring systems as do B and D. The main differences between these two pairs relate to the orientation of the ethyl group at C9 and C10.
Mulliken population analysis of atomic spin densities for 4OHT open-shell dehydrogenation reaction intermediates
In an attempt to identify sites of metabolism and hydrogen abstraction, atomic spin densities of open shell species were calculated to assess the relative probabilities of unpaired electrons at specific sites on 4OHT. In addition, gas-phase ab initio quantum mechanics calculations probed the sequence of hydrogen abstraction involved in dehydrogenation of 4OHT. Previous gas-phase quantum mechanics-based studies of the desaturation of a radical clock substrate and a truncated thiolate model of the heme/Compound I, concluded that “the oxidase-dehydrogenase mixed activity occurs from the cationic intermediate species and requires electro-steric inhibition of the rebound process” [27]. Based on the hypothesis that the mixed activity occurs with the cationic intermediate species, we evaluated the applicability of this approach for identifying SOM of dehydrogenation reactions that involve two distinct SOM, specifically C9 and O29 for 4OHT, as opposed to other P450-catalyzed reactions that commonly involve one hydroxylation or neighboring atomic centers such as epoxidation [19]. Also, since the major CYP3A4 metabolites of 4OHT have been identified experimentally as α-hydroxylation at C9, and N-demethylation at C15 and C16, and the major CYP2B6 4OHT hydroxylated metabolite at C26 [30], we also evaluated atomic spin densities at these sites.
Gas-phase reaction coordinates for 4OHT dehydrogenation are shown in Figure 4 and for all four conformations of the first cationic species formed, we examined the Mulliken atomic spin densities to see if this approach would identify the site of the first hydrogen abstraction. The highest spin densities were observed at C8, which is not a commonly observed P450 SOM, and C26, the observed SOM for 4’-hydroxylation by both CYP3A4 (minor metabolite) and CYP2B6 (major metabolite) [30]. However, spin densities did not predict metabolism at C9 or at any of the carbon atoms bonded to N14, the SOM for the major metabolites produced by CYP3A4 from 4OHT (supplementary information Table 1). Mulliken atomic spin densities for all three intermediate neutral radical species were also examined for the single lowest energy conformation (A), to determine if these might identify the site of the second hydrogen atom abstraction in the dehydrogenation of 4OHT. The Mulliken atomic spin densities remained high at the site of hydrogen atom abstraction for each species, and at C7 for the radicals formed from aliphathic hydrogen abstraction. Thus, the Mulliken atomic spin densities from gas-phase QM calculations at the 6–31G* level of theory for the open-shell species formed along the putative dehydrogenation reaction coordinate do not identify the SOM for the CYP3A4 metabolism of 4OHT, except for the minor product 4’-hydroxy-tamoxifen. A priori knowledge appears to be required to identify the two dehydrogenation SOM of 4OHT by CYP3A4.
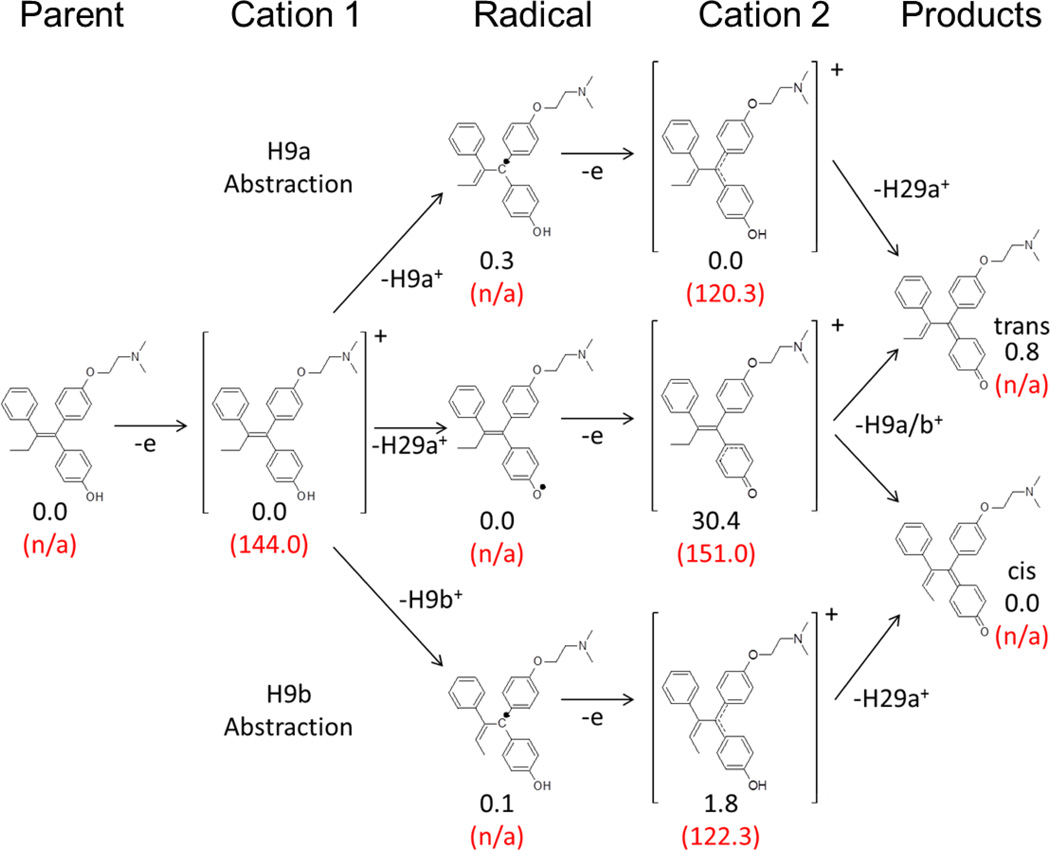
Figure 4.
Scheme for alternate hydrogen abstraction pathways for 4OHT intermediates along the dehydrogenation reaction coordinate. Differences in the sum of electronic and thermal free energies for each species are shown relative to that of the lowest energy equivalent intermediate in black. Electron affinities are shown where applicable as relative energy to previous neutral intermediate in the same pathway in parentheses in red. All energies given in units of kcal/mol.
Investigation of the sequence of hydrogen atom abstraction
Since the SOM for dehydrogenation with gas-phase calculations of the substrate alone were not useful, a priori knowledge of the SOM resulting in the formation of the desaturated quinone methide species identified experimentally [31] was used for further analysis. We selected conformation (A), one of the two lowest energy conformations, to perform analysis of the energy differences of all of the stable species formed along each putative reaction pathway for hydrogen abstraction, (Figure 4). For this study, we utilized two distinct approaches to determine the order of hydrogen abstraction. Since it had been previously proposed that the formation of a stable cation was likely a key factor required to produce dehydrogenation, approach 1 investigated the stability, relative energies and electron affinities of the putative intermediates determined to be involved in the formation of 4OHT quinone methide [31], while approach 2 calculated the bond dissociation energies (BDE) of these bonds.
Approach 1: Stability, relative energies and electron affinities of intermediates along putative l pathways of hydrogen atom abstraction for the formation of quinone methides from 4OHT
We performed geometry optimizations and frequency analyses for all putative gas-phase intermediates that could be formed from abstraction of the three hydrogen atoms putatively involved along the dehydrogenation reaction coordinate, (Figure 4). All species were determined to be at stable minima due to the absence of negative frequencies. The cation 1 intermediate is an open-shell doublet with a +1 charge formed from the parent. The subsequent radical intermediates can be formed from the abstraction of H29a, H9a or H9b, resulting in neutral open-shell doublets. The second cation 2 intermediate can be formed from any of the putative neutral radical intermediate species, has a +1 charge and is a singlet. The parent and product species are neutral closed shell systems. A summary of the difference in the sum of electronic and the thermal free energies relative to the lowest energy equivalent intermediate is given for each species with the corresponding electron affinity, (Figure 4).
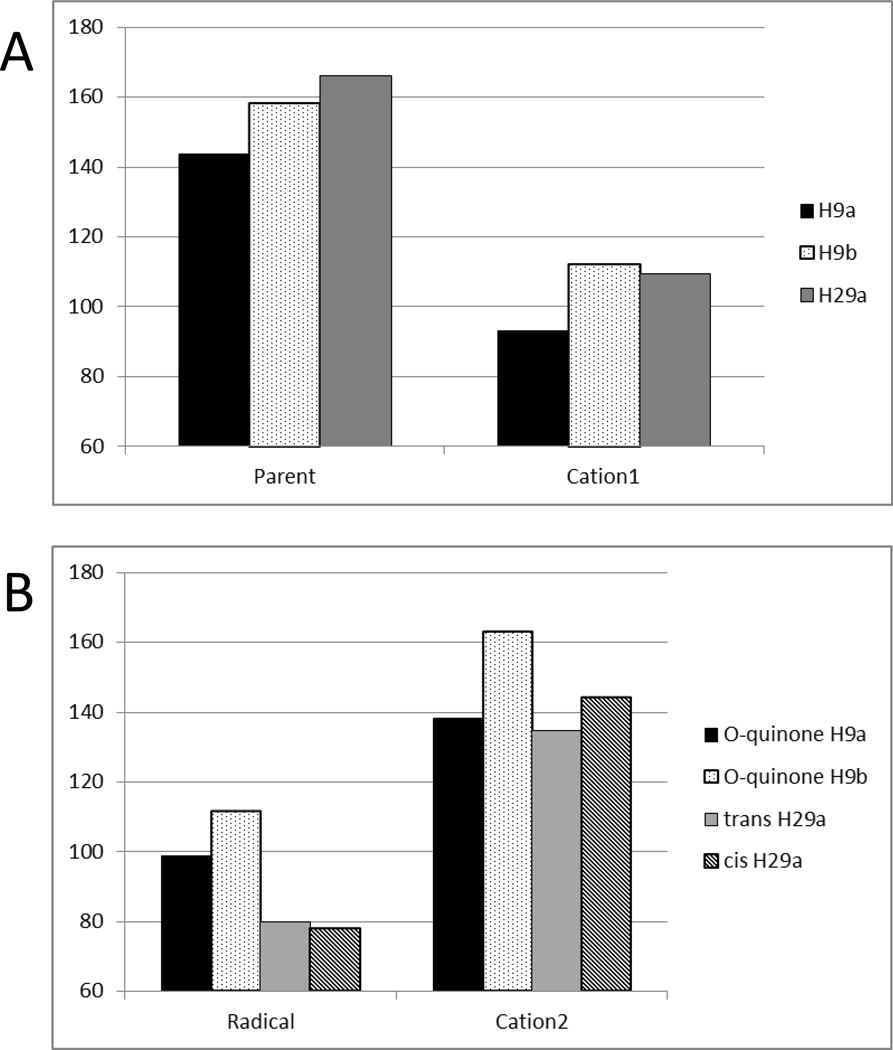
The largest observed energy difference was for the formation of the second cationic species from the abstraction of H29a which was ~30 kcal/mol higher in energy than either of the second cationic species formed from the initial abstraction of either allylic hydrogen atom (H9a or H9b). This species also has a greater electron affinity than either of the two cation 2 equivalents or of cation 1. These data suggest that dehydrogenation would be least favored to proceed via an initial hydrogen abstraction from the hydroxyl group based on the energies of the intermediates along the putative dehydrogenation pathways involving C9 and O29 explored here.
Approach 2: Bond dissociation energies (BDE)
The BDE for the parent and the first cationic species formed show that, for conformation A, the H9a abstraction was the lowest energy, (Figure 5). The BDE for the three neutral radical intermediate species and the second cationic intermediate (Cation 2) indicate that the hydroxyl bond (H29a) is the lowest in energy for the second hydrogen atom abstraction. These results together suggest that the dehydrogenation of 4OHT likely proceeds with the abstraction of the allylic H9a followed by the abstraction of the hydroxyl H29a.
Figure 5.
Final bond dissociation energies of carbon-hydrogen and oxygen-hydrogen bonds involved in the dehydrogenation of 4OHT at 4 Å extension for all intermediates on the dehydrogenation reaction coordinate: (A) first hydrogen abstraction, (B) second hydrogen abstraction. The legend indicates the hydrogen abstracted and for (B), the intermediate and the hydrogen abstracted.
This gas-phase method provides a physical basis to more quantitatively score docking results to accurately identify the site and order of metabolism. These data suggest that 4OHT likely undergoes sequential abstraction of hydrogen atoms beginning with the aliphatic hydrogen. This underscores the importance of sampling and identification of all of the minima available at physiological temperatures. Furthermore, the QM-based reaction coordinate pathway energetic analysis and BDE provide a physical basis to support the postulated dehydrogenation order of 4OHT to be used to score our docking results.
Docking
Molecular docking was performed to investigate putative binding modes of the substrates in the active site of CYP3A4. To provide a more realistic sampling of active site geometries beyond that shown in the available crystal structures, molecular dynamics simulations were employed. The use of ensembles of representative configurations of enzyme-substrate structures sampled with modern MD simulation protocols, when used with computationally affordable tools such as docking, holds great promise for structure-based prediction metabolism of new chemical entities [64, 65]. In spite of sampling limitations and force field inaccuracies [66], MD simulations can fold small proteins [67] and accurately model protein-ligand interactions using modern simulation protocols and balanced force field treatments [68].
To confirm that the energy barriers between the minima were low enough for the minima to be thermally accessible at physiological temperature, scans of all the ψ* dihedral angles as defined in Figure 1 were performed. The results of these scans (supplementary information Figure 3 and supplementary information Table 2) indicated that the energy barriers would indeed allow all of these minima to be occupied at physiological temperatures. Therefore, docking was performed with all four representative structures, and the results from all four dockings were combined.
Single docking runs only provide a “snap-shot” of enzyme- substrate interactions. However, this allows docking programs (e.g. Autodock3) that treat the coordinates for the biomolecule as static and use a united atom treatment to provide for fast sampling of the search space. However, identifying one pose out of many by visually inspecting the docking results can be time consuming, and subject to issues of signal-to-noise, which may not provide statistically significant results. A commonly applied metric for identifying potential SOM after docking is the use of distance-based scoring schemes, such as sites within 5 Å of the heme oxygen [12, 20, 51, 69]. Autodock3 [45] uses a united atom treatment and a static protein model. Moreover, since our heme models now include the catalytic oxygen atom in the hexa-coordinate heme state, the lowest energy docked structure within a family of similar poses may not necessarily correspond to the pose most amenable to catalysis. Therefore, we loosened the distance criteria for scoring the SOM for the lowest energy conformation representative of each cluster of Autodock3 results. Specifically, a cutoff < 8 Å from the iron atom to the heavy atom representing the SOM was used for metabolism involving a single SOM, e.g. hydroxylation and N-demethylation, and for dehydrogenation, one of the two iron-to-SOM distances should be < 5.5 Å and the iron to the second SOM distance should be < 8 Å. Visual inspection of productive clusters showed that within each cluster/bin, structures could be found where the corresponding iron-SOM distance was < 5 Å.
We improved the sampling by performing multiple independent docking runs to permit statistical analysis of the results. We also imposed an arbitrary signal-to-noise cutoff by only evaluating clusters which had >5% of the total population. In most cases, this reduced the number of clusters analyzed per 1024 docking runs to <10 clusters. All of the docking studies were repeated three times, independently, and analyzed for statistically significant differences for each reaction mechanism.
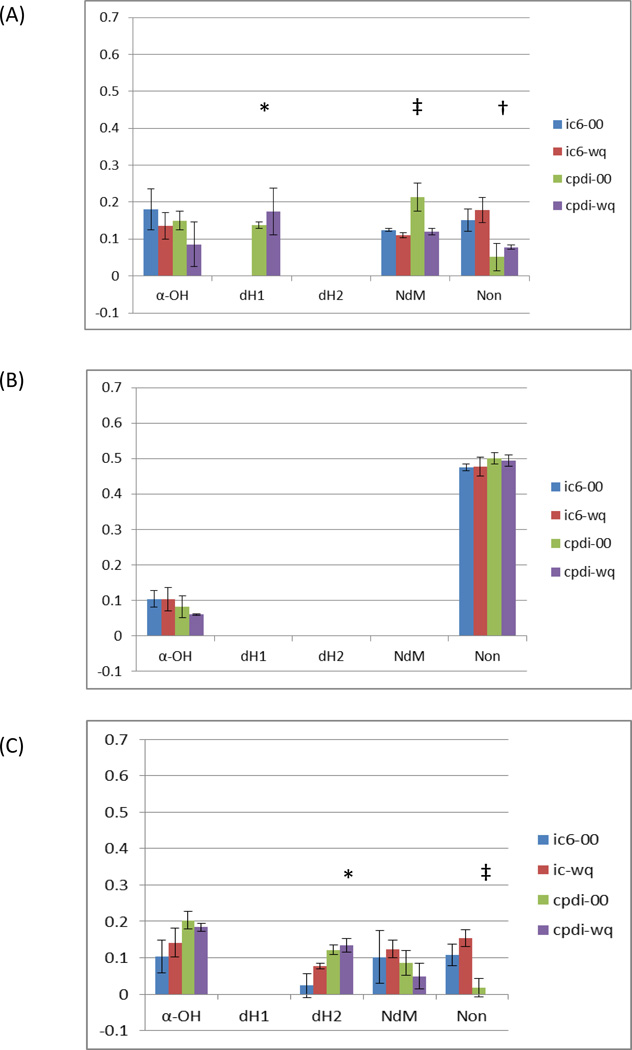
Preliminary docking studies using x-ray structures of CYP3A4 (PDB ID: 1W0E [23] and 1TQN [24]) with a single MM2 energy-minimized structure of 4OHT with Gasteiger charges assigned, no charge assignment to the heme atoms, consistent with our earlier work with RALX, did not identify binding modes that supported the observed metabolism of 4OHT. Conversely, docking studies with multiple conformations of 4OHT with the PDB ID:1W0E-based structure modified with QM-based heme parameters (1w0e-ic6-00/wq and 1w0e-cpdi-00/wq) identified a number of binding modes that were supportive of α-hydroxylation (α-OH) and N-demethylation (NdM) with the penta-coordinate heme, (Figure 6 and supplementary information Figure 4). Docking with this same P450 structure with the hexa-coordinate heme parameters for Compound I identified a binding mode supportive of both α-hydroxylation and dehydrogenation (dH1). The association of this binding mode with a specific dehydrogenation reaction mechanism is ambiguous because no specific residues capable of producing the required steric hindrance that would appear to inhibit the hydroxyl rebound mechanism were observed. Interestingly, the number of configurations supportive of NdM were increased with the presence of the catalytic oxygen without atomic partial charges. This result suggests that, with this enzyme model, the steric effect from the presence of the catalytic oxygen is decreased with the assignment of atomic partial charges which cause a change in the orientation of the flexible MTOXY moiety. Also, the number of non-productive modes in the major clusters significantly decreased with the use of the hexa-coordinate heme parameters, indicating that the presence of the catalytic oxygen atom has a greater steric contribution for 4OHT with this x-ray model.
Figure 6.
Ratio of Autodock3 binding modes supportive of observed metabolism of 4-hydroxy-tamoxifen reaction mechanisms alpha-hydroxylation (α-OH) dehydrogenation modes 1 and 2 (dH1 & dH2), N-demethylation (NdM) and nonproductive and/or ambiguous modes (Non) with CYP3A4 experimentally derived x-ray (A) PDB ID: 1W0E, (B) PDB ID: 1TQN and (C) molecular dynamics refined m2 refined with quantum mechanics based heme parameters for resting high-spin (ic6) and Compound-I (cpdi) with (wq) and without (00) RESP charges assigned to the heme. Statistically significant differences are indicated with symbols: †, ‡ and *.
A similar approach with the PDB ID: 1TQN-based structure modified with QM-based heme parameters (1tqn-ic6-00/wq and 1tqn-cpdi-00/wq), docking studies identified mostly non-productive modes and 1 productive mode supportive of α-hydroxylation, (Figure 6).
To incorporate flexibility and dynamics in the active site structures, five different MD trajectories of the starting 1TQN structure were also evaluated. To choose representative structures for docking, clustering of the structures sampled at 1 ps-intervals by RMSD similarity for each trajectory was performed. The average structure from the largest cluster was chosen for docking in each of the five trajectories. To remove artifacts from averaging, the structure was first minimized with the electrostatic interactions turned off to remove steric clashes.
Docking studies with the MD-based structure m2 modified with QM-based heme parameters (m2-ic6-00/wq and m2-cpdi-00/wq) also predicted α-OH, NdM as well as an alternate binding mode supportive of dehydrogenation (dH2). This binding mode suggested R212 as a single residue that could potentially produce electro-steric inhibition of the hydroxyl-rebound mechanism leading to dehydrogenation. The number of poses in the dH2 binding mode also significantly increased with the inclusion of hexa-coordinate Compound I coordinates and atomic partial charges. For this structure, the number of nonproductive binding modes in the largest cluster also significantly decreased with the incorporation of hexa-coordinate Compound I coordinates.
Analysis of QM-based heme parameters and substrate charge state in docking
Inclusion of the heme charge and coordinate parameters, and alternate charge state for the substrate did not produce a significant “across the board” improvement for modeling 4OHT-CYP3A4 interactions with 1W0E, 1TQN or the MD refined models. Furthermore, significant model-specific differences were observed in binding modes supporting different reaction mechanisms, except α-OH. To ensure that the substrate charge state did not alter the docking results, these studies were repeated with charged 4OHT. Similar results were obtained with neutral, and +1 4OHT with respect to identifying SOM and various reaction mechanisms. Significant differences for the same model, with and without charges, were also observed for a few dockings, but no significant improvement based on the charge state of 4OHT or heme parameters was observed across all CYP3A4 models, supplementary information Figure 4. However, it should be noted that inclusion of the hexa-coordinate state never eliminated a binding mode identifying the SOM for any structure tested. In fact, inclusion of these coordinates was required to identify the dH1 and dH2 binding modes.
4OHT binding modes and CYP3A4 conformational dynamics and active site architecture
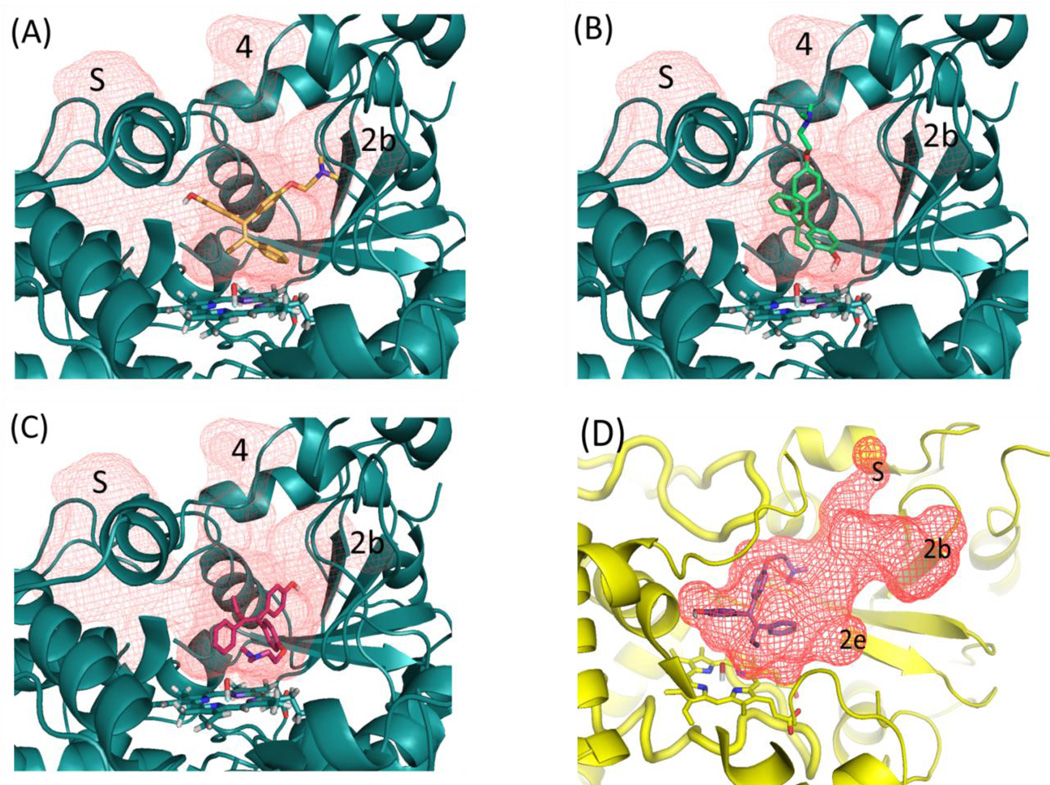
For CYP3A4 models that produced binding modes supportive of dehydrogenation, two modes were observed, labeled dH1 and dH2. The dH1 binding mode was similar in orientation to the binding mode that supported α-OH, except with 4OHT “flipped over”, thus placing O29 in a position within hydrogen abstraction distance to the heme, (Figure 7). However, with the dH1, no residues were observed to intervene between 4OHT and the catalytic center of the heme to provide the necessary steric inhibition of the rebound process. This makes interpretation of dehydrogenation versus hydroxylation ambiguous using this type of MM-based approach. In contrast, the dH2 binding mode clearly positioned Arg212 in an orientation that imposed steric and potentially electro-static inhibition of the rebound process. This was only observed with the m2 model. Further, the m2-based dH2 pose showed significant improvement in the results predicting the involvement of the dehydrogenation with the inclusion of the QM-based heme charges.
Figure 7.
Active site and major channels (red grid) for structure m2-cpdi (cyan) and w0e-cpdi (yellow) shown with orientation of representative poses scored for different reaction mechanisms (A) α-hydroxylation [αOH], (B) dehydrogenation [dH2], (C) N-demethylation [NdM] and (D) dehydrogenation [dH1]. Volumes defining active-site and ingress/egress channels are shown as red mesh and labeled S (solvent), 2b, 2e and 4 channels. The 1w0e-cpdi and m2-cpdi model are shown at slightly different angles, and the peptide backbone in the foreground for all pictures has been hidden to allow the differences in the structure and orientation of channels and substrate to be seen more clearly. Active-site mapping performed with UCSF HOLLOW, figures generated with PYMOL.
Ingress, egress and solvent channels
Examination of the binding modes within the context of the active site of the MD-refined m2-cpdi and w0e-cpdi models show that for 4OHT, the opening and closing of the ingress and egress channel 4 provides a space for the substrate with respect to the catalytic site of the enzyme, (Figure 7). In the m2 structure, channels 2b, 4 and S were determined to be open. The dH2 binding mode is only possible with the opening of channel 4, in such a manner so as to allow 4OHT to partially occupy it. The other major reaction mechanisms (i.e. α-OH, dH1 and NdM), are supported by binding modes that allow 4OHT to occupy a region of the active site directly above the heme, that among the models tested, appears to be less malleable.
Table 3.2 shows the channels that were identified for each model and the reaction mechanisms determined to be supported by our scoring scheme. All of the channels present in the MD-refined structures are present in published CYP3A4 x-ray structures, but in different combinations, including channel 4, which is observed in another x-ray structure (PDB ID: 2V0M supplementary information Table 3).
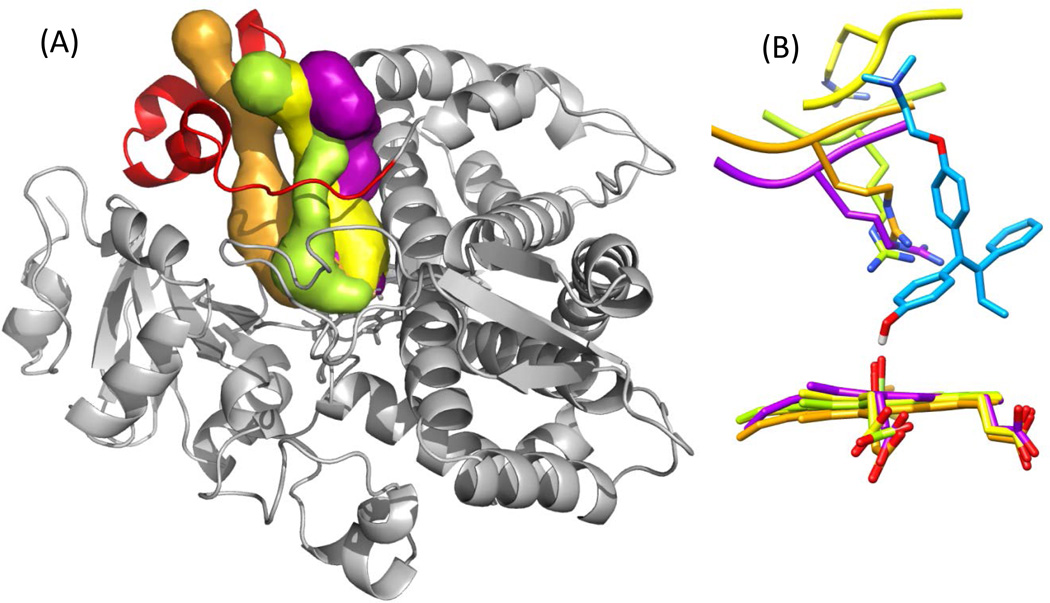
By comparing structure-specific channels and binding modes (Table 3.2), it can be clearly seen that there is no one-to-one relation between channel opening and closing and substrate binding. For structures that do have channel 4 open, specifically m2, m3, m5 and PDB ID: 2V0M, the orientation and structure of channel 4 was examined, (Figure 8). For all four structures different volumes and paths defined for channel 4 were observed. The positioning of Arg212 toward the heme is similar for m2, m3 and m5, but for 2V0M [8], the Arg212 is oriented away from the heme, (Figure 8). This would suggest that the use of this model would not have implicated the potential role of this residue in dehydrogenation. Therefore, both the channel 4 open state and Arg212 oriented toward the heme are required for a binding mode that supports dehydrogenation of 4OHT.
Figure 8.
Differences in channel 4 and position of ARG212. (A) Volume and placement of channel 4 as defined by CAVER shown as a surface for each model m2 (lime green), m3 (purple), m5 (orange) and 2V0M (yellow). The CYP3A4 backbone is shown for the m2 model as a grey ribbon, the F–G loop (which includes the F’ and G’ helices) is shown in red. (B) Position of ARG212 and heme for m2 (lime green), m3 (purple), m5 (orange) and 2V0M (yellow) shown in reference to docked conformation of 4OHT (cyan) in dH2 pose. Hydrogen atoms have been hidden for clarity. Neighboring residues to ARG212 are shown only as ribbon for the back bone and atoms hidden also for clarity. Image generated (A) with PYMOL, and (B) with UCSF CHIMERA.
Analysis of the representative structures used here identifies that different regions of the CYP3A4 active site are perturbed by the open or closed state of ingress and egress channels to different degrees. These more malleable regions appear to have multiple states available to them. Thus, we recommend future docking studies of CYP3A4 with larger substrates, such as 4OHT, to include multiple structures with multiple combinations of open channels. In fact, using a number of CYP3A4 structures with different combinations of open channels may prove to be an important metric of achieving adequate sampling for identifying modes supportive of different SOM and P450-catalyzed reaction mechanisms.
Incubations and liquid-chromatography mass-spectroscopy analysis (LC-MS)
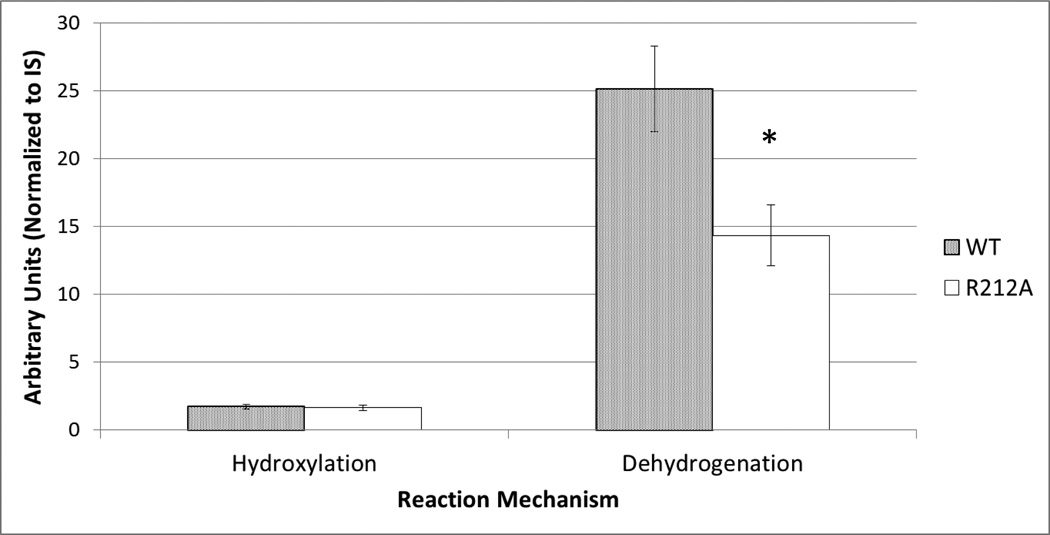
LC-MS analysis of incubations with CYP3A4 and the CYP3A4-R212A mutant showed a dramatically altered rate of dehydrogenation versus oxygenation, adding potent confirmation of the results from the modeling studies. LC-MS semiquantitative analysis of incubations of 4OHT with CYP3A4 wild-type and CYP3A4-R212A produced both hydroxylated and dehydrogenated metabolites. However, CYP3A4-R212A produced significantly less (~50%) dehydrogenated product (p=0.016), while the amount of hydroxylated metabolite was essentially unalterered (p=0.677), (Figure 9).
Figure 9.
4OHT relative dehydrogenation and hydroxylation 4OHT products of Wild-Type (WT) versus Mutant (R212A) CYP3A4. Statistical significance indicated with (*).
Conclusions
The dH2 binding mode correctly predicted the potential role of Arg212 in the steric hindrance required to inhibit the rebound process of the dehydrogenation of 4OHT. Arg212 could also possibly contribute an electrostatic repulsion of the cationic intermediate formed during the dehydrogenation reaction coordinate. Our docking results show that population of the dH2 binding pose is sensitive to both inclusion of atomic partial charges and Compound I coordinates. Thus, Arg212 may provide both steric and electrostatic hindrance of the hydroxyl rebound process. However, the possible contributions from the dH1 mode, with inhibition of the rebound process due to thermodynamic fluctuations of the active site architecture as a whole, cannot be ruled out. Nor can we rule out the possibility that other biologically relevant binding modes may exist that we have yet not identified.
Interestingly, the key residue involved in the dehydrogenation of 4OHT is different than that identified in previous work with RALX, suggesting that these two selective estrogen receptor modulators have diverse conformational landscapes in the CYP3A4 active-site. Even though they are able to bind the ER in the same manner, their P450 metabolism requires very different binding modes.
QM-based energy minimizations identified multiple conformational minima for 4OHT that are likely thermally accessible at physiological temperatures. Therefore, performing MM-based studies requires sampling from all of these minima. The ab initio methods used here did not unambiguously identify the dehydrogenation SOM and a priori knowledge was required to do so. However, QM-based methods did consistently support the energetically favored sequence of hydrogen atom abstraction and provided a physical basis for the docking scoring scheme used here.
MD simulations of an x-ray structure, supportive of only one productive binding mode (PDB ID: 1TQN) produced models with ingress and egress channels observed to be present in other CYP3A4 experimental structures but in different combinations. These models, when used with computationally inexpensive docking tools, identified binding modes supportive of the entire metabolism of 4OHT by the most promiscuous and biomedically relevant hepatic P450, CYP3A4. One unique enzyme-substrate configuration was identified in a MD-based model that involved the potential interaction of a single residue, Arg212, and was supportive of the dehydrogenation reaction. Enzymatic incubations with a R212A mutant of CYP3A4 confirmed the role of this residue in the dehydrogenation of 4OHT. Use of these different models with QM-based heme parameters for the heme and charge states for 4OHT showed that for this hydrophobic substrate, the contribution of atomic partial charges was not as important as for RALX.
Comparison of these theoretical enzyme-substrate configurations have identified regions of the enzyme active site that differ in plasticity, and different binding modes supportive of the same reaction mechanisms. We postulate that the difficulties in identifying the SOM and PRM for CYP3A4 are related to the binding modes within the active-site. Depending on the size, rigidity and polarity of the substrate, multiple binding modes may be available and be supportive of the same PRM. This is further complicated by the fact that certain regions of the CYP3A4 active site appear to be more malleable than others. Thus, for some substrates, multiple binding modes may be available, and some of these may exist within these plastic regions which cannot be modeled using x-ray data alone. We speculate that these plastic regions, particularly those surrounding channel 4, which is formed with the long insertion in the F–G loop, unique to mammalian P450s [5], may play an important part in the ability of a number of P450 enzymes to accommodate chemically diverse substrates. Furthermore, the contribution of the cavities formed by the channels may play a more important role than that of individual residues for binding of more flexible and nonpolar substrates.
Based on our findings, we recommend that future computational CYP3A4 metabolism studies identify all the available minima for the substrate and verify that the interconversion between these minima is thermally possible so as to provide adequate sampling. We recommend that docking studies be performed with the penta- and hexa- coordinates, since inclusion of these parameters has been shown to help increase the observed number of productive binding modes for molecules with both rigid and flexible cores, albeit in different manners. Regarding models of the enzyme, it is recommended that in the refinement of existing experimental models, adequate sampling also be considered when attempting to identify different states of the enzyme in regards to the opening and closing of channels. This is especially important for larger, more flexible substrate molecules that can occupy these volumes adjacent to the active-site. To identify binding modes supportive of the entire metabolism of CYP3A4 substrates, including minor reaction pathways such as dehydrogenation, that produce unstable and reactive metabolites, it is important to adequately sample the entire conformational space of the enzyme.
Supplementary Material
Table 2.
Identification and classification of (i) ingress/egress channels and (ii) binding modes supportive of different reaction mechanisms for refined molecular dynamics based (m1–5-cpdi) and x-ray-based (1w0e-cpdi & 1tqn-cpdi) models of CYP3A4.
| (i) | |||||||
|---|---|---|---|---|---|---|---|
| model | # channel | 2a | 2b | 2e | 4 | 5 | Solvent |
| m1-cpdi | 2 | 5 | S | ||||
| m2-cpdi | 3 | 2b | 4 | S | |||
| m3-cpdi | 4 | 2b | 2e | 4 | S | ||
| m4-cpdi | 3 | 2b | 5 | S | |||
| m5-cpdi | 4 | 2a | 2b | 2e | 4 | ||
| 1w0e-cpdi | 4 | 2b | 2e | 5 | S | ||
| 1tqn-cpdi | 3 | 2a | 2e | S | |||
| (ii) | ||||||
|---|---|---|---|---|---|---|
| model | # productive modes | α-OH | dH1 | dH2 | NdM | Non |
| m1-cpdi | 2 | NdM | Non | |||
| m2-cpdi | 3 | α-OH | dH2 | NdM | Non | |
| m3-cpdi | 1 | Non | ||||
| m4-cpdi | 1 | NdM | Non | |||
| m5-cpdi | 2 | α-OH | dH1 | NdM | Non | |
| 1w0e-cpdi | 3 | α-OH | dH1 | NdM | Non | |
| 1tqn-cpdi | 1 | α-OH | Non | |||
Highlights.
MD-refined P450 structures identify the importance of channels in CYP3A4 docking
A unique configuration is identified involving Arg212 in the dehydrogenation of 4OHT
CYP3A4 and CYP3A4-R212A show a decreased rate of dehydrogenation versus oxygenation
Acknowledgment
Thomas E. Cheatham III acknowledges financial support from NIH GM079383 and computer time from the Center for High Performance Computing at Utah and from the NSF TeraGrid MCA01S027. Garold S. Yost acknowledges financial support from NIH GM0742249. We thank Drs. Chad D Moore and Chris Reilly for advice and help in the preparation of this manuscript.
Abbreviations
- P450
cytochrome P450 enzymes
- 4OHT
4-hydroxy-tamoxifen
- SOM
sites of metabolism
- RALX
raloxifene
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Furge LL, Guengerich FP. Cytochrome P450 enzymes in drug metabolism and chemical toxicology: An introduction. Biochemistry and Molecular Biology Education. 2006;34:66–74. doi: 10.1002/bmb.2006.49403402066. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz de Montellano P, Voss J. In: Substrate Oxidation by Cytochrome P450 Enzymes Cytochrome P450. Ortiz de Montellano PR, editor. Springer US: 2005. pp. 183–245. [Google Scholar]
- 3.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid Redox Signal. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halpert JR. Structure and function of cytochromes P450 2B: from mechanism-based inactivators to X-ray crystal structures and back. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2011;39:1113–1121. doi: 10.1124/dmd.111.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otyepka M, Skopalík J, Anzenbacherová E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Biochimica et Biophysica Acta (BBA) -General Subjects. 2007;1770:376–389. doi: 10.1016/j.bbagen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Hendrychová T, Anzenbacherová E, Hudeček J, Skopalík J, Lange R, Hildebrandt P, Otyepka M, Anzenbacher P. Flexibility of human cytochrome P450 enzymes: Molecular dynamics and spectroscopy reveal important function-related variations. Biochimica et Biophysica Acta (BBA) -Proteins & Proteomics. 2011;1814:58–68. doi: 10.1016/j.bbapap.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 7.DeVore NM, Meneely KM, Bart AG, Stephens ES, Battaile KP, Scott EE. Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine. FEBS Journal. 2011 doi: 10.1111/j.1742-4658.2011.08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzenbacher P, Otyepka M. Flexibility of Human Cytochromes P450: Molecular Dynamics Reveals Differences between CYPs 3A4, 2C9, and 2A6, which Correlate with Their Substrate Preferences. The Journal of Physical Chemistry B. 2008;112:8165–8173. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 10.Guengerich FP. Cytochrome P450 and Chemical Toxicology. Chemical research in toxicology. 2007;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 11.Afzelius L, Arnby CH, Broo A, Carlsson L, Isaksson C, Jurva U, Kjellander B, Kolmodin K, Nilsson K, Raubacher F, Weidolf L. State-of-the-art tools for computational site of metabolism predictions: comparative analysis, mechanistical insights, and future applications. Drug Metabolism Reviews. 2007;39:61–86. doi: 10.1080/03602530600969374. [DOI] [PubMed] [Google Scholar]
- 12.Sun H, Yost GS. Metabolic activation of a novel 3-substituted indole-containing TNF-alpha inhibitor: dehydrogenation and inactivation of CYP3A4. Chemical Research in Toxicology. 2008;21:374–385. doi: 10.1021/tx700294g. [DOI] [PubMed] [Google Scholar]
- 13.Smith DA, Obach RS. Metabolites in Safety Testing (MIST): Considerations of Mechanisms of Toxicity with Dose, Abundance, and Duration of Treatment. Chemical Research in Toxicology. 2009;22:267–279. doi: 10.1021/tx800415j. [DOI] [PubMed] [Google Scholar]
- 14.Baillie TA. Metabolism and Toxicity of Drugs. Two Decades of Progress in Industrial Drug Metabolism. Chemical Research in Toxicology. 2007;21:129–137. doi: 10.1021/tx7002273. [DOI] [PubMed] [Google Scholar]
- 15.Baillie TA, Cayen MN, Fouda H, Gerson RJ, Green JD, Grossman SJ, Klunk LJ, LeBlanc B, Perkins DG, Shipley LA. Drug Metabolites in Safety Testing. Toxicology and Applied Pharmacology. 2002;182:188–196. doi: 10.1006/taap.2002.9440. [DOI] [PubMed] [Google Scholar]
- 16.Yost GS. Bioactivation of toxicants by cytochrome p450-mediated dehydrogenation mechanisms. Adv Exp Med Biol. 2001;500:53–62. doi: 10.1007/978-1-4615-0667-6_6. [DOI] [PubMed] [Google Scholar]
- 17.Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 18.Guengerich FP, Macdonald TL. Chemical mechanisms of catalysis by cytochromes P-450: a unified view. Accounts of Chemical Research. 1984;17:9–16. [Google Scholar]
- 19.Ortiz de Montellano PR, De Voss JJ. In: Cytochrome P450, Structure, Mechanism and Biochemistry. Third Edition. Ortiz de Montellano Paul R., editor. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 185–245. [Google Scholar]
- 20.Sun H, Scott DO. Structure-based Drug Metabolism Predictions for Drug Design. Chemical Biology & Drug Design. 2010;75:3–17. doi: 10.1111/j.1747-0285.2009.00899.x. [DOI] [PubMed] [Google Scholar]
- 21.Davydov DR, Davydova NY, Tsalkova TN, Halpert JR. Effect of glutathione on homo-and heterotropic cooperativity in cytochrome P450 3A4. Archives of Biochemistry and Biophysics. 2008;471:134–145. doi: 10.1016/j.abb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott EE, Halpert JR. Structures of cytochrome P450 3A4. Trends in Biochemical Sciences. 2005;30:5–7. doi: 10.1016/j.tibs.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science (New York, N.Y. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 24.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. The Journal of Biological Chemistry. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 25.Sevrioukova IF, Poulos TL. Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18422–18427. doi: 10.1073/pnas.1010693107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz de Montellano PR. Hydrocarbon Hydroxylation by Cytochrome P450 Enzymes. Chemical Reviews. 2009;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar D, De Visser SP, Shaik S. Oxygen economy of cytochrome P450: what is the origin of the mixed functionality as a dehydrogenase-oxidase enzyme compared with its normal function? Journal of the American Chemical Society. 2004;126:5072–5073. doi: 10.1021/ja0318737. [DOI] [PubMed] [Google Scholar]
- 28.Moore CD, Shahrokh K, Sontum SF, Cheatham TE, 3rd, Yost GS. Improved cytochrome P450 3A4 molecular models accurately predict the Phe215 requirement for raloxifene dehydrogenation selectivity. Biochemistry. 2010;49:9011–9019. doi: 10.1021/bi101139q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahrokh K, Orendt A, Yost GS, Cheatham TE., 3rd Quantum mechanically derived AMBER-compatible heme parameters for various states of the cytochrome P450 catalytic cycle. Journal of Computational Chemistry. 2011 doi: 10.1002/jcc.21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. The Journal of Pharmacology and Experimental therapeutics. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 31.Fan PW, Zhang F, Bolton JL. 4-Hydroxylated metabolites of the antiestrogens tamoxifen and toremifene are metabolized to unusually stable quinone methides. Chemical Research in Toxicology. 2000;13:45–52. doi: 10.1021/tx990144v. [DOI] [PubMed] [Google Scholar]
- 32.Fan PW, Bolton JL. Bioactivation of tamoxifen to metabolite E quinone methide: reaction with glutathione and DNA. Drug Metabolism and Disposition: the Biological Fate of Chemicals. 2001;29:891–896. [PubMed] [Google Scholar]
- 33.Smigel K. Breast Cancer Prevention Trial shows major benefit, some risk. Journal of the National Cancer Institute. 1998;90:647–648. doi: 10.1093/jnci/90.9.647. [DOI] [PubMed] [Google Scholar]
- 34.MJT Frisch GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C.02. Vol. 2004. Wallingford CT: Gaussian, Inc.; 2004. [Google Scholar]
- 35.Dennington R, Keith T, Millam J. GaussView, Version 5. Semichem Inc., Shawnee Mission KS; 2009. [Google Scholar]
- 36.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. The Journal of Physical Chemistry. 1993;97:10269–10280. [Google Scholar]
- 37.Dupradeau FY, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys Chem Chem Phys. 2010;12:7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiser A, Šali A. Modeller: Generation and Refinement of Homology-Based Protein Structure Models. In: Carter Charles W, Jr, Robert MS., editors. Methods in Enzymology. vol. Volume 374. Academic Press; 2003. pp. 461–491. [DOI] [PubMed] [Google Scholar]
- 39.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith DE, Dang LX. Computer simulations of NaCl association in polarizable water. The Journal of Chemical Physics. 1994;100:3757–3766. [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 42.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 43.Hermans J, Berendsen HJC, Van Gunsteren WF, Postma JPM. A consistent empirical potential for water–protein interactions. Biopolymers. 1984;23:1513–1518. [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19:1639–1662. [Google Scholar]
- 46.Sanner MF. Python: A Programming Language for Software Integration and Development. J. Mol. Graphics Mod. 1998;17:57–61. [PubMed] [Google Scholar]
- 47.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLano W. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 49.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochimica et Biophysica Acta (BBA) - General Subjects. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 50.Ho B, Gruswitz F. HOLLOW: Generating Accurate Representations of Channel and Interior Surfaces in Molecular Structures. BMC Structural Biology. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun H, Moore C, Dansette PM, Kumar S, Halpert JR, Yost GS. Dehydrogenation of the indoline-containing drug 4-chloro-N-(2-methyl-1-indolinyl)-3-sulfamoylbenzamide (indapamide) by CYP3A4: correlation with in silico predictions. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2009;37:672–684. doi: 10.1124/dmd.108.022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu L, Liu H, Li W, Zhang F, Luckie C, van Breemen RB, Thatcher GRJ, Bolton JL. Oxidation of Raloxifene to Quinoids: Potential Toxic Pathways via a Diquinone Methide and o-Quinones. Chemical Research in Toxicology. 2004;17:879–888. doi: 10.1021/tx0342722. [DOI] [PubMed] [Google Scholar]
- 53.Fan PW, Zhang F, Bolton JL. 4-Hydroxylated Metabolites of the Antiestrogens Tamoxifen and Toremifene Are Metabolized to Unusually Stable Quinone Methides. Chemical Research in Toxicology. 1999;13:45–52. doi: 10.1021/tx990144v. [DOI] [PubMed] [Google Scholar]
- 54.Murphy CS, Parker CJ, McCague R, Jordan VC. Structure-activity relationships of nonisomerizable derivatives of tamoxifen: importance of hydroxyl group and side chain positioning for biological activity. Molecular Pharmacology. 1991;39:421–428. [PubMed] [Google Scholar]
- 55.Fang H, Tong W, Shi LM, Blair R, Perkins R, Branham W, Hass BS, Xie Q, Dial SL, Moland CL, Sheehan DM. Structure–Activity Relationships for a Large Diverse Set of Natural, Synthetic, and Environmental Estrogens. Chemical Research in Toxicology. 2001;14:280–294. doi: 10.1021/tx000208y. [DOI] [PubMed] [Google Scholar]
- 56.Philippe d M, Gilles F, Marc P. Multiple Targeting by the Antitumor Drug Tamoxifen: A Structure-Activity Study. Current Medicinal Chemistry -Anti-Cancer Agents. 2004;4:491–508. doi: 10.2174/1568011043352696. [DOI] [PubMed] [Google Scholar]
- 57.Hardcastle IR, Rowlands MG, Houghton J, Parr IB, Potter GA, Jarman M, Edwards KJ, Laughton CA, Trent JO, Neidle S. Rationally Designed Analogs of Tamoxifen with Improved Calmodulin Antagonism. Journal of Medicinal Chemistry. 1995;38:241–248. doi: 10.1021/jm00002a005. [DOI] [PubMed] [Google Scholar]
- 58.Gust R, Lubczyk V. Structure activity relationship studies on C2 side chain substituted 1,1-bis(4-methoxyphenyl)-2-phenylalkenes and 1,1,2-tris(4-methoxyphenyl)alkenes. The Journal of Steroid Biochemistry and Molecular Biology. 2003;87:75–83. doi: 10.1016/s0960-0760(03)00385-6. [DOI] [PubMed] [Google Scholar]
- 59.Menezes IRA, Leitão A, Montanari CA. Three-dimensional models of non-steroidal ligands: A comparative molecular field analysis. Steroids. 2006;71:417–428. doi: 10.1016/j.steroids.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Maximov PY, Myers CB, Curpan RF, Lewis-Wambi JS, Jordan VC. Structure–Function Relationships of Estrogenic Triphenylethylenes Related to Endoxifen and 4-Hydroxytamoxifen. Journal of Medicinal Chemistry. 2010;53:3273–3283. doi: 10.1021/jm901907u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuramochi H. Conformational Studies and Electronic Structures of Tamoxifen and Toremifene and Their Allylic Carbocations Proposed as Reactive Intermediates Leading to DNA Adduct Formation. Journal of Medicinal Chemistry. 1996;39:2877–2886. doi: 10.1021/jm960255g. [DOI] [PubMed] [Google Scholar]
- 62.Hossain MB, van der Helm D, Schmitz FJ, Pordesimo EO, Magarian RA, Meyer KL, Overacre LB, Day BW. Molecular Structures and Conformational Studies of Triarylcyclopropyl and Related Nonsteroidal Anti-Estrogens. Journal of Medicinal Chemistry. 1994;37:1670–1683. doi: 10.1021/jm00037a018. [DOI] [PubMed] [Google Scholar]
- 63.Huang M-J. Ab initio studies of tamoxifen and related compounds. International Journal of Quantum Chemistry. 2004;96:374–379. [Google Scholar]
- 64.Park S-J, Kufareva I, Abagyan R. Improved docking, screening and selectivity prediction for small molecule nuclear receptor modulators using conformational ensembles. Journal of Computer-Aided Molecular Design. 2010;24:459–471. doi: 10.1007/s10822-010-9362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knegtel RMA, Kuntz ID, Oshiro CM. Molecular docking to ensembles of protein structures. Journal of Molecular Biology. 1997;266:424–440. doi: 10.1006/jmbi.1996.0776. [DOI] [PubMed] [Google Scholar]
- 66.van Gunsteren WF, Bakowies D, Baron R, Chandrasekhar I, Christen M, Daura X, Gee P, Geerke DP, Glattli A, Hunenberger PH, Kastenholz MA, Oostenbrink C, Schenk M, Trzesniak D, van der Vegt NF, Yu HB. Biomolecular modeling: Goals, problems, perspectives. Angewandte Chemie (International ed. 2006;45:4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- 67.Simmerling C, Strockbine B, Roitberg AE. All-Atom Structure Prediction and Folding Simulations of a Stable Protein. Journal of the American Chemical Society. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 68.Steinbrecher T, Labahn A. Towards Accurate Free Energy Calculations in Ligand Protein-Binding Studies. Current Medicinal Chemistry. 2010;17:767–785. doi: 10.2174/092986710790514453. [DOI] [PubMed] [Google Scholar]
- 69.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nature Reviews. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.