Abstract
Alcohol activates reward systems through an unknown mechanism, in some cases leading to alcohol abuse and dependence. Herein, we utilized a two-choice Capillary Feeding assay to address the neural and molecular basis for ethanol self-administration in Drosophila melanogaster. Wild-type Drosophila demonstrates a significant preference for food containing between 5 and 15% ethanol. Preferred ethanol self-administration does not appear to be due to caloric advantage, nor due to perceptual biases, suggesting a hedonic bias for ethanol exists in Drosophila. Interestingly, rutabaga adenylyl cyclase expression within intrinsic mushroom body neurons is necessary for robust ethanol self-administration. The expression of rutabaga in mushroom bodies is also required for both appetitive and aversive olfactory associative memories, suggesting that reinforced behavior has an important role in the ethanol self-administration in Drosophila. However, rutabaga expression is required more broadly within the mushroom bodies for the preference for ethanol-containing food than for olfactory memories reinforced by sugar reward. Together these data implicate cAMP signaling and behavioral reinforcement for preferred ethanol self-administration in Drosophila melanogaster.
Keywords: Drosophila, Ethanol Self-Administration, Learning, cAMP, Mushroom Body
Introduction
Alcohol can act as a complex rewarding stimulus in humans, and positive reinforcement of alcohol consumption may ultimately lead to compulsive drinking and addiction. Since alcohol broadly affects neural activity, both presynaptically and postsynaptically, by interacting with several classes of receptors, the underlying neurobiology of continued alcohol self-administration has been difficult to unravel.
The rodent two-bottle choice paradigm has been widely used to model alcohol reward during alcohol self-administration (Belknap et al., 1993, Cicero, 1980). A similar two-choice assay has been developed for Drosophila melanogaster. In this paradigm, known as the Capillary Feeder (CAFE) assay, Drosophila are provided with a choice of drinking from two microcapillaries; one capillary tube contains liquid food and the other liquid food plus ethanol (Devineni & Heberlein, 2009, Ja et al., 2007). In the CAFE assay, Drosophila display significant preference for food containing up to 15% ethanol over the non-ethanol food. Ethanol self-administration is independent of an olfactory or gustatory basis for ethanol within the liquid food (Devineni & Heberlein, 2009). Moreover, altering the ratio of calories between the normal food, and the ethanol containing food in the CAFE assay did not significantly change the ethanol preference, suggesting ethanol self-administration is not due to the additional calories potentially provided by the ethanol (Devineni & Heberlein, 2009). Interestingly, Drosophila also will prefer alcohol even when mixed with the bitter tasting quinine, and will rapidly display strong preference after deprivation, suggesting relapse (Devineni & Heberlein, 2009). The continued preference despite the negative consequences of bitterness and the relapse after deprivation are features of addiction (Morse & Flavin, 1992, Rodd et al., 2004). The CAFE assay is hence a capable behavioral model to genetically dissect the molecular mechanisms involved in ethanol self-administration.
Herein, we have used a modified CAFE assay to uncover a molecular mechanism and neural center involved in the preference for ethanol containing food. We initially verified that the ethanol preference of wild-type flies is independent of a gustatory, olfactory, or caloric bias for the ethanol-containing food. We further found that a mutation in the learning gene rutabaga has significant reduction in preference for ethanol- containing food. The targeted rescue of this mutant phenotype has identified intrinsic mushroom body neurons as important in the display of an ethanol preference. These neurons are central to positively-reinforced olfactory conditioning with either sugar or ethanol as the rewarding stimulus. (Kaun et al., 2011, Schwaerzel et al., 2003, Thum et al., 2007). Hence, rutabaga dependent ethanol self-administration may function through alcohol’s positive reinforcement of feeding behavior.
Methods and Materials
1. Fly strains and husbandry
All flies were cultured on standard medium at 25°C, approximately 60% relative humidity and a 12 h light/dark cycle. With the exception of the Gal4 lines, mutations and transgenes used in this study were out-crossed into the Canton-S background for a minimum of six generations prior to behavioral analysis. All the Gal4 lines were out-crossed into w1118 (CS10) background for at least seven generations. The orco2 mutation is a loss-of-function disruption of the orco locus (Larsson et al., 2004). The lush1 mutation has a deletion of lush coding sequences (Laughlin et al., 2008). The rut2080 mutation is severe hypomorphic allele caused by a P{lArB} insertion within rutabaga (Levin et al., 1992). The UAS-rutabaga line is capable of rescuing rut2080 mutant phenotypes when expressed by Gal4 (Zars et al., 2000a). The elav-GeneSwitch transgene is an RU486 inducible UAS driver (Osterwalder et al., 2001). GH146-Gal4 drives expression within the projection neurons of the antennal lobe (Stocker et al., 1997). MB247-Gal4 is a Gal4 line driving expression in the α, β and γ lobes of mushroom body, and the c305a Gal4 transgene drives expression in α’/β’ lobes (Aso et al., 2009, Zars et al., 2000a). The c305a and MB247 Gal4 lines were combined in a single genotype to drive expression in all the classes of mushroom body neurons. OK107, c772 and 238y express Gal4 in all classes of intrinsic mushroom body neurons (Aso et al., 2009). The MB-specific Gal80 represses Gal4 expression in the mushroom body (Krashes et al., 2007). For the genetic rescue of rut2080, the experimental genotype was generated by crossing virgin females of rut2080; UAS-rut/CyO (or rut2080; +; UAS-rut) with males of the indicated Gal4 lines. All Gal4 insertions were autosomal. F1 male progeny were selected for analysis.
2. Ethanol preference assay
The ethanol preference feeding assay is as described with some modification (Devineni & Heberlein, 2009, Ja et al., 2007). The feeding apparatus is composed of two nested vials: an inner feeding vial and an outer humidity chamber. The feeding inner vial (1.5-cm diameter, 3.5 cm long) was plugged by a rubber stopper (standard 0#) with two holes inserted by two 200-µl pipette tips that were cut to fit microcapillary pipettes. One of the tips was labeled. The bottom of inner vial was pierced to allow entry of water vapor and air from the outer vial, which was standard 25 mm Drosophila vial filled with 5 ml of water. Calibrated glass 5 µl micropipettes (VWR, West Chester, PA) filled with liquid food by capillary action were inserted through the 200-µl pipette tips. A mineral oil (CAS: 8042-47-5; Sigma-Aldrich, St. Louis, MO) overlay was applied in micropipette to minimize evaporation.
Liquid food was prepared with the designated amount of sucrose (CAS: 57-50-1; Sigma-Aldrich, St. Louis, MO), yeast extract (Bacto yeast extract; BD Diagnostic Systems, Franklin Lakes, NJ), and ethanol (CAS: 64-17-5; AAPER Alcohol and Chemical CO., Shelbyville, KT). All the solutions were made fresh each week and stored at 4°C.
Flies of the desired genotypes were collected at 0–1 day old. After 2 days, flies were anaesthetized by CO2, and males were placed individually into the feeding chamber. They were habituated in the CAFE apparatus for 24 h, with ad libitum medium. After 24 h, the micropipettes which contain liquid food and liquid food plus ethanol were added to each feeding chamber. As a control, micropipettes were also applied in three empty vials, to measure the vaporized volume. The micropipettes were changed daily, while also altering the position of the ethanol containing microcapillary pipette. The testing conditions were four days at 25°C, 60%–70% humidity, with 12L:12D cycle.
To measure the liquid consumed, first the difference between the liquid surface present at the beginning and end of each 24 h was determined. The amount of liquid consumed was then determined as this difference minus the average evaporated liquid. Food consumption was defined as food with ethanol consumption + food without ethanol consumption. A Preference Index was defined as (food with ethanol consumption-food without ethanol consumption)/food consumption. The caloric content of the medium was calculated on the basis of the following values: 4 kcal/g (sucrose), 1.58 kcal/g (yeast extract), and 7 kcal/g (ethanol) (Ja et al., 2007). Energy intake was defined as energy from ethanol containing food + energy from non-ethanol food.
3. Proboscis extension response assay
Our proboscis extension response (PER) assay was developed from a preceding assay (Kimura et al., 1986). Three-day-old flies were starved for 16 to 20 hours to increase their gustatory sensitivity. A vial containing the flies was placed on ice for 2 minutes to briefly immobilize the flies. The flies were then transferred to a Petri dish. Small drops, each approximately 0.7 µl of Loctite 404 (Henckel CO., Lewisville, TX), were placed on a glass slide lined with labeling tape. The flies were then glued to the glass slide on their dorsal side with their legs free. Then the flies were covered with moist Kimwipes and allowed 15 to 30 minutes to recover from the cold stress. After the recovery period, a drop of water was applied to each fly’s foreleg in order to habituate the PER to water. To measure the gustatory sensitivity, a drop of solution was briefly introduced to each fly’s foreleg. The proboscis extension of the fly was recorded as an all-or-none event. For each trial, 10 flies were tested first at 5% sucrose + 5% yeast, then at 5% sucrose + 5% yeast + 5% (or 10%, or 15%) ethanol. PER index was defined as the ratio between the flies showing proboscis extension and total flies tested.
4. RU486 feeding
As previously described (Roman & Davis, 2002, Roman et al., 2001), a 10 mM stock solution of RU486 (mifepristone; CAS: 84371-65-3; Sigma-Aldrich, St. Louis, MO) dissolved in 80% ethanol was diluted to 500 µM in 2% sucrose. Adult males were transferred to vials, each containing one Kimwipe wetted with 2 ml of diluted RU486 solution. Control flies were fed on 2% sucrose with 4% ethanol. Flies were kept in these vials at 25°C, 60%–70% relative humidity, with 12L:12D cycle for 24 h and then transferred to the CAFE apparatus for a 24 h habitation period before beginning the ethanol preference assay.
5. Survivorship assays
The survivorship assay was a modification from a previous protocol(Libert et al., 2007). In this assay, all flies were aged from synchronize embryos. Upon eclosion, they were collected for a 24-hour period and allowed to mate for 2–3 days. After this time, females were collected under light CO2 anesthesia and housed at 25 flies per group, and transferred to fresh food every other day. At ten days, the flies were transferred into vials containing 1% Agar, 1% Agar mixed with 0.7% ethanol, or 1% Agar mixed with 0.89% Sucrose. In the sucrose top experiments, the sucrose was layered on top of the agar to ensure the fly had full access to the sugar (no significance was seen in this treatment vs. mixing the sucrose with the agar). Deaths were recorded approximately every 3–5 hours during the period of highest mortality until 100% mortality was reached.
6. Immunohistochemistry
The methods for immunohistochemical detection of GFP were largely based on previously published techniques (Wu & Luo, 2006). Adult flies between 2 and 7 days after eclosion, containing the indicated Gal4 driver and the UAS-GFP responder, were used. The brains were dissected in phosphate-buffered saline (PBS) solution, fixed with 4% paraformaldehyde (CAS: 30525-89-4; Sigma-Aldrich, St. Louis, MO) in PBS for 20 minutes at room temperature and washed with PBS containing 0.4% Triton X-100 (PBT) (CAS: 9002-93-1; Sigma-Aldrich, St. Louis, MO). The fixed brains were subsequently rinsed with PBT for 20 minutes three times. After being blocked with PBT containing 5% normal goat serum (Sigma-Aldrich, St. Louis, MO) for 30 minutes at room temperature, the brains were incubated with the mouse polyclonal antibody against green fluorescent protein (GFP) (1:200; Roche, Germany) in PBT at 4 °C for 48 hrs. The brains were then washed with PBT for 20 minutes three times. The prepared labeled brains were next incubated with Alexa Fluo®488 conjugated goat anti-mouse (1:500; Invitrogen, U.S.A) in blocking solution for 48 hr at 4 °C. Finally, the brains were rinsed with PBT (3×20 minutes) and mounted in mounting medium (Vectashield; Vector Laboratories, U.S.A). The prepared brains were imaged with confocal microscopy on a Fluoview FV1000 microscope (Olympus, Japan).
7. Statistical analysis
All statistical analysis was performed using Statview software (SAS Institute, Inc., USA). In the CAFE assay, energy intake, food consumption or preference index was analyzed using Student’s t-test, or one-way ANOVA, after tested by Kolmogorov-Smirnov normality test. Bonferroni correction was performed when more than two groups were compared. As described before (Devineni & Heberlein, 2009, Masek & Scott), PER score were analyzed using one-way ANOVA or Student’s t-test. In the survivorship assay, Cox regression and log-rank tests were used to identify statistically significant differences in survival between treatments.
Results
Wild-type Drosophila prefer food containing ethanol
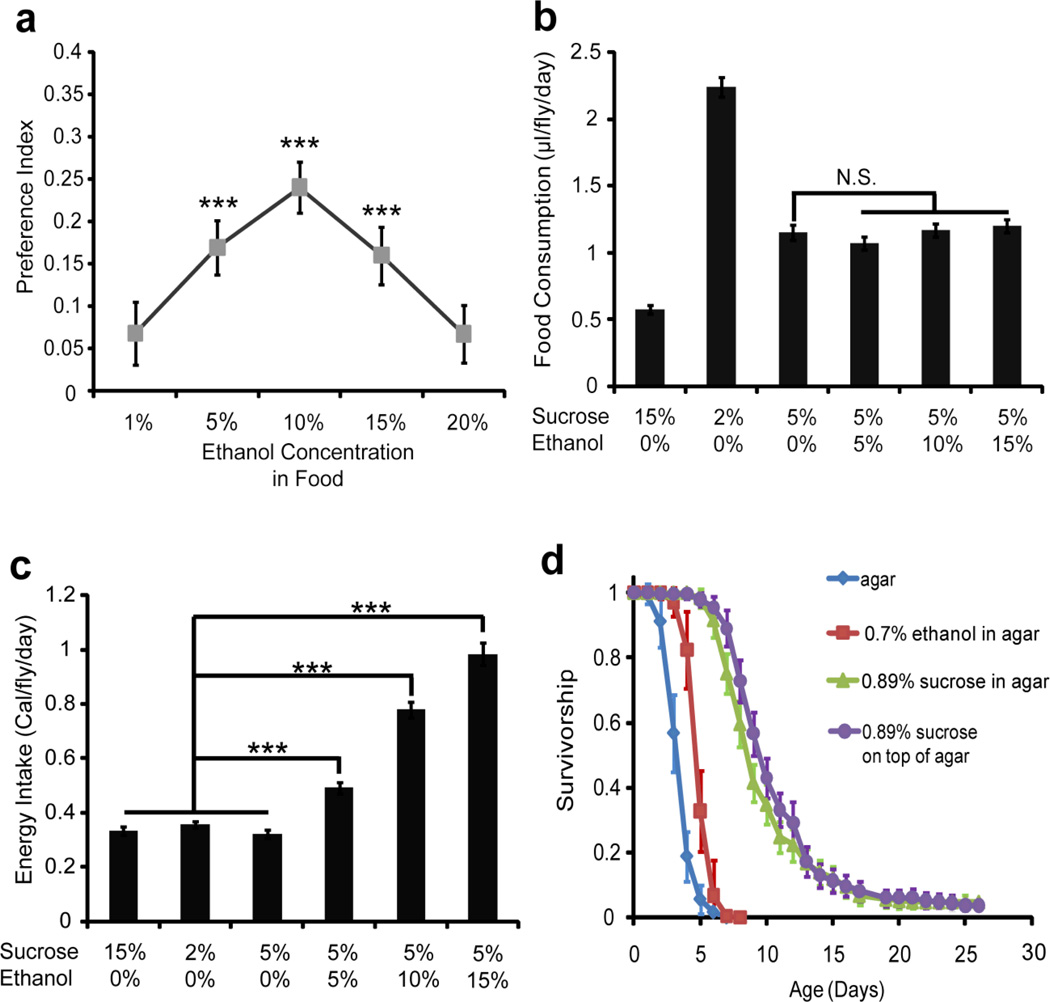
To measure ethanol preference in Drosophila, we modified the Capillary Feeder (CAFE) assay (Devineni & Heberlein, 2009, Ja et al., 2007). The CAFE assays are conceptually similar to the two-bottle choice assays commonly used to assess ethanol preference in rodents (Belknap et al., 1993, Cicero, 1980). In our CAFE assay, single males were placed in a small humidified chamber and allowed to feed from two microcapillary tubes containing liquid food (5% yeast extract, 5% sucrose) or liquid food plus ethanol for four days (Sup. Fig. 1). The preference index was calculated as previously described to measure the daily surplus of food containing ethanol consumed (Devineni & Heberlein, 2009). Initially, we examined the preference index for food containing different concentrations of ethanol during a four day period. In this experiment, wild-type Canton-S (CS) male flies significantly preferred 5%, 10% and 15% ethanol (Fig. 1a; for 5% ethanol, t(30)=5.193, P<0.0001, n=30 flies; for 10% ethanol, t(31)=5.756, P<0.0001, n=31 flies; for 15% ethanol, t(28)=5.537, P<0.0001, n=28 flies), but did not display a preference for 1% and 20% ethanol (Fig. 1a; Fig. 1a; for 1% ethanol, t(33)=1.997, P=0.0545, n=33 flies; for 20% ethanol, t(34)=1.762, P=0.0873, n=34 flies). Ethanol preference was stable over the four days examined (data not shown). The preferred range of ethanol concentrations is similar to that found in C57BL, the most widely used mouse strain in alcohol self-administration assay (Belknap et al., 1993), but appears significantly lower than that found for w− Berlin strains of Drosophila melanogaster (Devineni & Heberlein, 2009).
Figure 1. The preference for ethanol containing food is independent of nutrient value.
(a) Canton-S (CS) flies were analyzed in the CAFE assay for a preference for liquid food containing ethanol at the indicated percentages. CS flies display a significant preference for food containing 5%, 10% and 15% ethanol. (b) The daily food consumption of CS flies over a four day period was determined for the indicated liquid foods that also contained 5% yeast extract. In this experiment, the flies were allowed access to only one kind of food if without ethanol in food, or were tested in the standard two choices CAFE assay. Although CS modulates the amount of food consumed per day based on the sucrose content, they did not alter food consumption based on ethanol concentration. (c) The caloric value consumed by each fly daily for each food substrate was determined. In this assay, CS flies balanced the energy intake from sugar, but failed to make a similar adjustment to calories provided by ethanol, because the daily energy intake from each kind of ethanol-containing food was more than these of three kinds of non-ethanol food. (d) The ability of CS flies to utilize ethanol as an energy source was examined through a survivorship assay. CS flies fed with 0.7% ethanol survive significantly longer than completely starved flies, but significantly shorter than flies fed with 0.89% sucrose. Since 0.7% ethanol contains similar caloric content as 0.89% sucrose, the shorter survival time on ethanol as a food source compared to sucrose suggests that Drosophila cannot use a majority of the calories present in ethanol for sustenance. Data are means ± S.E.M. “N.S.” means no significance. ***P<0.001.
For a useful model of ethanol self-administration, the drug should be consumed for its pharmacological effects, rather than for calories, taste or smell (Cicero, 1980). Interestingly, flies can associate the nutritional strength of food with an odor to form a robust memory, which may reinforce the preference in CAFE assay (Burke & Waddell, 2011, Fujita & Tanimura, 2011). Yet, a previous study suggested that ethanol preference was unlikely to be a byproduct of caloric attraction (Devineni & Heberlein, 2009). To further confirm this result, we compared the daily food consumption and energy intake of CS males for which we varied either the sucrose content or the ethanol content in the food. When the concentration of sugar is varied in the CAFE assay, the flies will consume differing amounts of food, so they balance their daily caloric intake to approximately 0.33 calorie/fly/day (Fig. 1c; F5,113=132.111, P<0.0001, each n=18–20 flies; no significance for 5% sucrose group vs. 2% sucrose or 15% sucrose group). However, when the ethanol concentration within the food is varied, the flies consume the same total volume as the comparable non-ethanol containing food (Fig. 1b; F5,113=88.551, P<0.0001, n=18–20 flies; no significance between the 4 groups in which sucrose concentration is 5%), and do not balance food intake to compensate for any changes in caloric content added by the ethanol (Fig. 1c; F5,113=132.111, P<0.0001, each n=18–20 flies; P<0.001 for the 5% ethanol group vs. the other 3 non-ethanol groups, P<0.001 for 10% ethanol group vs. other 3 non-ethanol groups, P<0.001 for 15% ethanol group vs. other 3 non-ethanol groups). To confirm that Drosophila can utilize ethanol for metabolic energy, we examined the survivorship of CS and w1118 wild-type strains on agar alone, agar mixed with 0.7% ethanol, or agar mixed with 0.89% sucrose. Interestingly, ethanol prolongs survival without other energy source(Sup. Fig. 2; P<0.05, log-rank test, n=10 groups). However, Canton-S can survive much longer on the same amount of sugar calories than on ethanol calories (Fig. 1d; P<0.001, log-rank test, n=10 groups). In these experiments the ethanol was mixed into the food, which substantially reduces the evaporation (Gibson, 1981). Moreover, the ethanol vial was exchanged after 4 days, allowing for the flies access to ethanol as a food source during this experiment. Hence, ethanol contains significantly less nutrition than sucrose. Since Drosophila do not alter the total food consumed based on ethanol content, the few calories received from ethanol are likely ignored and an energy bias is not a source of innate ethanol preference in the CAFE assay.
A second explanation for ethanol preference is a sensory bias; the food containing ethanol may either smell or taste better than the normal food. To w− flies in a Berlin background, high ethanol concentrations suppressed the proboscis extension reflex (PER) to sucrose, indicating a gustatory aversion to ethanol. However, the PER response was not suppressed much by low ethanol concentrations (Devineni & Heberlein, 2009). We also found that food containing 5% to 15% ethanol did not inhibit PER significantly, relative to food without ethanol, while low ethanol concentrations are at best neutral (Sup. Fig. 3a; F3,52=1.948, P>0.05, n=12–14 groups). Previously, the w− Berlin flies were found to have an olfactory bias towards ethanol odor, but they preferred ethanol even when olfactory ability was removed, indicating that olfaction was not necessary for ethanol preference (Devineni & Heberlein, 2009). Interestingly, in an olfactory trap assay Canton-S flies in a w− background were attracted more by yeast than by yeast plus 25% ethanol odorants (Kim & Smith, 2001), which suggests that these flies have an olfactory bias for non-ethanol food over ethanol-containing food. To further explore possible roles for olfactory plasticity in ethanol preference, we examined w+; orco2 and w+; lush1 mutants in the CAFE assay. The w+; orco2 mutants are broadly anosmic, display increased stress resistance and altered metabolism, and are long- lived (Larsson et al., 2004, Libert et al., 2007). The w+; orco2 flies did not display any significant defects in ethanol preference (Sup. Fig. 3b; for 5% ethanol, t(44)=0.212, P>0.05, n=23 each; for 10% ethanol, t(74)=0.959, P>0.05, n=38 each; for 15% ethanol, t(48)=0.542, P>0.05, n=25 each). The lush gene encodes an odorant binding protein required for detecting a pheromone, 11-cis vaccenyl acetate (Laughlin et al., 2008). The lush1 mutants are more attracted to high ethanol concentrations in olfactory trap assays than wild-type flies (Kim et al., 1998). However, the lush1 mutants did not demonstrate a preference phenotype within our CAFE assay (Sup. Fig. 3c; for 5% ethanol, t(36)=0.421, P>0.05, n=19 each; for 10% ethanol, t(56)=1.303, P>0.05, n=29 each; for 15% ethanol, t(38)=0.328, P>0.05, n=20 each). The absence of a phenotype in the anosmic w+; orco2 and the ethanol preferring lush1 mutants support an absence of a significant role for an olfactory biasin the ethanol self-administration (Devineni & Heberlein, 2009).
The rutabaga type I adenylyl cyclase in mushroom body is essential for the expression of an ethanol preference
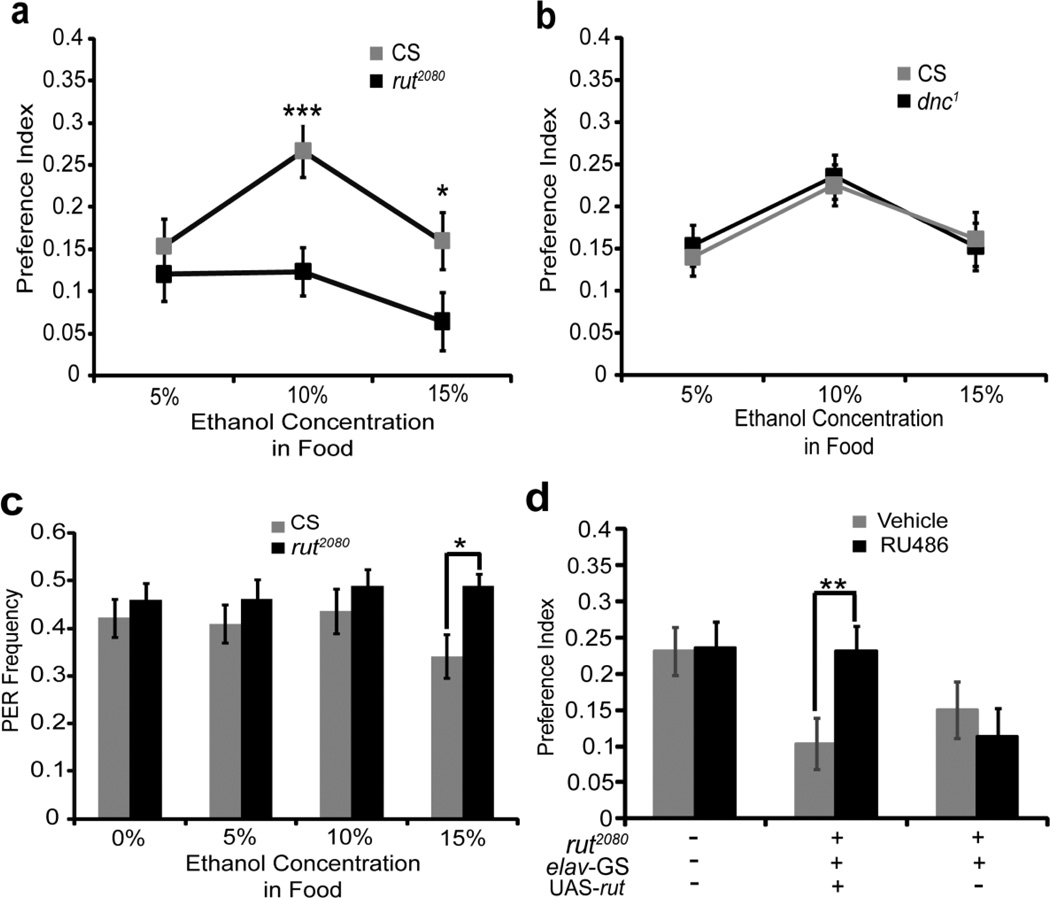
The rutabaga (rut) gene encodes a type I calcium-activated adenylyl cyclase and is essential for both associative learning and for normal responses to the sedative effects of ethanol vapor (Levin et al., 1992, Moore et al., 1998, Tempel et al., 1983). We therefore examined rut2080 reduction-of-function mutants in our CAFE assay. The rut2080 mutants displayed a normal preference to 5% ethanol; however, the preference for 10% and 15% ethanol was significantly less than that of Canton-S flies (Fig. 2a; for 5% ethanol, t(46)=0.951, P>0.05, n=24 each; for 10% ethanol, t(69)=3.545, P=0.0007, n=36 each; for 15% ethanol, t(52)=2.329, P=0.025, n=27 each). These data suggest a role for cAMP signaling in the expression of an ethanol preference. The rut2080 mutants also displayed decreased food consumption in the CAFE assay, raising the possibility that deceased total food consumption may shape the expressed ethanol preference (Sup. Fig. 4a; for 5% ethanol, t(46)=2.989, P=0.0045, n=24 flies; for 10% ethanol, t(69)=2.722, P=0.0082, n=35–36 flies; for 15% ethanol, t(52)=2.550, P=0.014, n=27 flies).
Figure 2. The rutabaga adenylyl cyclase is acutely required for an ethanol preference.
(a) rut2080 mutants and control CS flies were examined in the CAFE assay. The rut2080 mutants display a significantly reduced preference for 10% and 15% ethanol compared to wild-type CS. (b) Mutants in the dunce phosphodiesterase were also examined in the CAFE assay. The dnc1 loss-of-function mutants exhibited a normal preference for 5%, 10% and 15% ethanol. (c) In the PER assay, rut2080 did not show a significant difference with CS, to liquid food without or with ethanol, except for 15% ethanol, indicating that the rut2080 reduced preference is not due to altered gustatory responses to ethanol. (d) The reduced ethanol preference of rut2080 can be rescued by the induced expression of a rut cDNA in the nervous system using the elav-Geneswitch driver. In this experiment rutabaga expression was induced by RU486, resulting in significantly greater preference for 10% ethanol than that within the same genotype but vehicle-fed flies, and also more than the genotype rut2080;+;elav-GS/+ induced by RU486. The same treatment did not influence the ethanol preference in CS flies and the negative control flies, rut2080;+;elav-GS/+. Data are means ± S.E.M. *P<0.05, **P<0.01, and ***P<0.001.
The dunce gene (dnc) encodes a cAMP phosphodiesterase that is required during negatively-reinforced associative conditioning, but is not required in a positively-reinforced olfactory learning assay (Tempel et al., 1983, Tully & Quinn, 1985). The dnc gene product is also the primary and perhaps only phosphodiesterase active in adult mushroom body neurons (Gervasi et al., 2010). Interestingly, the dnc1 mutants displayed a normal preference for 5%, 10% and 15% ethanol (Fig. 2b; for 5% ethanol, t(57)=0.422, P>0.05, n=29 flies; for 10% ethanol, t(58)=0.450, P>0.05, n=30 flies; for 15% ethanol, t(38)=0.105, P>0.05, n=20 flies). These differences between dnc1 and rut2080 in the CAFE assay also mirrors that of an ethanol sedation assay, where the rut2080 mutants are hypersensitive, and the dnc1 mutants lack an obvious phenotype (Moore et al., 1998).
To eliminate trivial causes for the rut2080 ethanol preference phenotype, we examined the gustatory response of this mutant to ethanol-containing food. The PER elicited by 5% or 10% ethanol containing food by rut2080 mutants was not significantly different from the wild-type controls (Fig. 2c; for 5% ethanol, t(18)=0.924, P>0.05, n=9–11 groups; for 10% ethanol, t(17)=0.907, P>0.05, n=8–11 groups). In contrast, the PER index of rut2080 flies to food containing 15% ethanol was significantly greater than that of Canton-S flies (Fig. 2c; t(17)=2.64, P=0.017, n=9–10 groups), which suggested that 15% ethanol was less aversive to rut2080 than to Canton-S controls. The expectation from the reduced aversive gustatory response of rut2080 flies to 15% ethanol-containing food is that they would drink more ethanol than Canton-S. Thus, the reduced ethanol preference of the rut2080 mutants is unlikely to be due to reduced aversive taste of ethanol. Furthermore, it was previously reported that: 1) the olfactory response of rut2080 flies was not different than that of wild-type flies and 2) the absorption and metabolism of ethanol is not altered between rut2080 and wild-type flies (Moore et al., 1998, Tully & Quinn, 1985).
Loss of rut activity may lead to developmental defects in neural function (Zhong & Wu, 1993). To examine the possibility of defects in developmental underlying the rut function in ethanol self-administration we determined whether the deficit in ethanol preference of rut2080 flies could be rescued by rut expression post-developmentally. This was accomplished using the RU486-inducible elav-GeneSwitch driver (Osterwalder et al., 2001, Roman, 2004). One to two day old Canton-S, rut2080;+; elav-GeneSwitch/+ and rut2080; UAS-rut/+; elav-GeneSwitch/+ flies were fed either RU486 or vehicle for 24 h, followed by a second 24 h period of habituation to the CAFE chamber. The RU486 did not alter the ethanol preference behavior of Canton-S and rut2080; +; elav-GeneSwitch/+ flies (Fig. 2d). However, the RU486-induced expression of the rut cDNA within the nervous tissue completely rescued the rut2080 ethanol preference phenotype (Fig.2d; t(71)=3.08, P=0.003 for induced rut2080;UAS-rut/+;GS/+ vs. un-induced rut2080;UAS-rut/+;GS/+ planned comparison, n=36–37 each). Also, this post-developmental expression of the rut cDNA did not significantly increase the total food consumption phenotype (Sup. Fig. 4b; planned comparison for rut2080;UAS-rut/+;GS/+ with un-induced rut2080;UAS-rut/+;GS/+, t(71)=0.500, P>0.05, n=35 flies), genetically distinguishing these two phenotypes. Moreover, the OK107 or c772 driven rutabaga expression rescued the abnormal ethanol preference in rut2080 (see below), but did not rescue the total food consumption phenotype (Sup. Fig. 4c; F(3.150)=9.891, P<0.0001, in post hoc comparisons, P>0.05 for rut2080;c772/+;UAS-rut/+ vs. rut2080;c772/+, n=37–39 each; Sup. Fig. 4d; F(3.140)=17.650, P<0.0001; in post hoc comparisons, P>0.05 for rut2080;;UAS-rut/+;OK107/+ vs. rut2080;;;OK107/+. n=27 for the rut2080;;;OK107/+ group and n=39 for the rut2080;;UAS-rut/+;OK107/+ group). These data suggested that total food consumption is not measurably affecting the preference for ethanol in our CAFE assay. Thus, the ethanol preference phenotype is due to the acute lack of rut expression within the nervous system, and not due to the absence of rut during development.
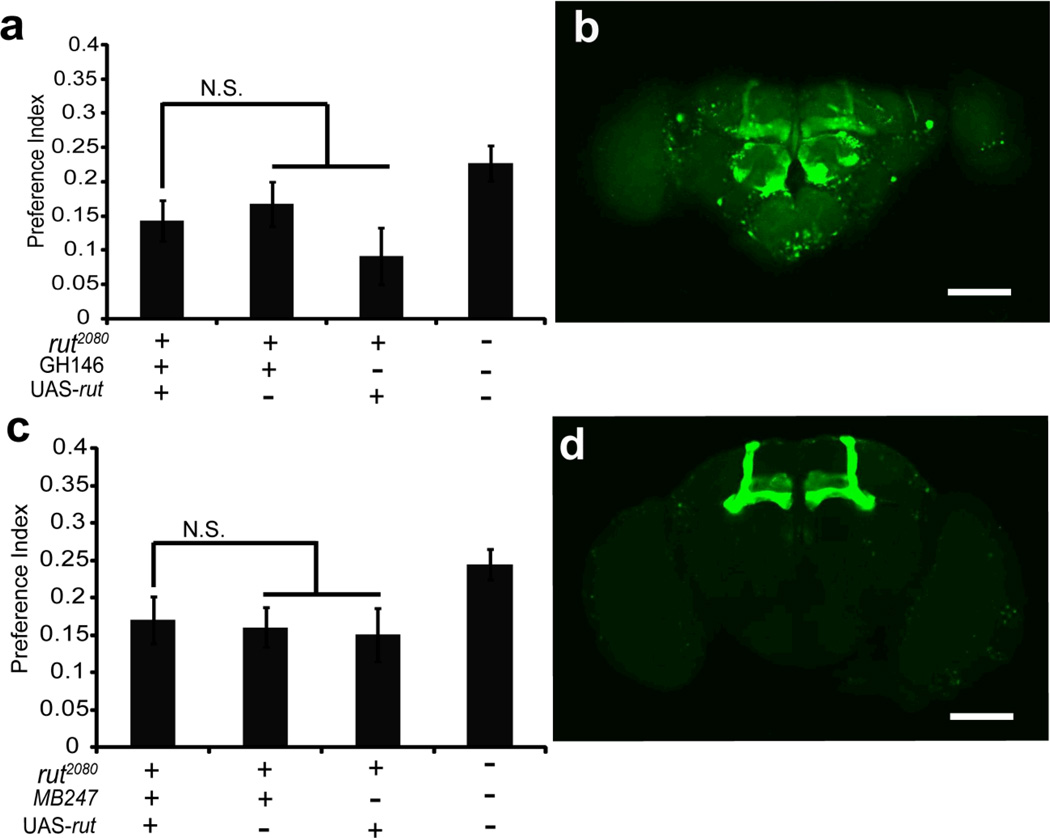
The identification of the neural foci for the rut dependent function in ethanol self-administration will elucidate neurons that are required for this behavior and may also provide insight into the underlying behavioral mechanisms. The rut gene is broadly expressed in the adult nervous system, and is enriched within the mushroom body neurons of the central brain (Levin et al., 1992). These mushroom body neurons are critical sites for both positively-reinforced and negatively-reinforced olfactory learning and memory formation (reviewed by Zars, 2011). The decreased ethanol self-administration of rut2080 mutants could be due to the role of this gene in forming positively reinforced associations. Ethanol can act as both a long term positive-reinforcer and shorter term negative-reinforcer in an olfactory learning paradigm in Drosophila (Kaun et al., 2011), but it is not known whether rut has a role in acquiring these memories. The activity of rut is however required for wild-type levels of sugar-reinforced olfactory memories in Drosophila (Schwaerzel et al., 2003, Tempel et al., 1983). We next considered the possibility that the rut-dependent component of the ethanol preference behavior in the CAFE assay is due to similar appetitive olfactory learning. The sugar-reinforced olfactory memory deficit of rut2080 can be fully rescued through the expression of a wild-type rut cDNA within the antennal lobe projection neurons defined by the GH146-Gal4 driver, or the mushroom body neurons defined by the MB247-Gal4 driver (Thum et al., 2007). The expression of rut driven by either GH146 or MB247 failed to rescue the rut2080 ethanol preference phenotype (Fig. 3; F(3.123)=2.800, P=0.042, no significance for rut2080;GH146/+;UAS-rut/+ vs. rut2080;GH146/+ or rut2080;;UAS-rut/+, n=30–32 each; Fig. 3b; F(3.190)=2.047, P=0.109 n=43–46 each). This result genetically separates the role of rut in sugar-reinforced olfactory memory formation from ethanol self-administration.
Figure 3. The ethanol preference phenotype of rut2080 mutants is separable from simple appetitive olfactory learning.
Both GH146 and MB247 rescue the olfactory appetitive learning defect in rut2080. However, they cannot rescue ethanol preference defect in rut2080. (a) The expression of rut driven by the projection neuron driver GH146-Gal4 failed to rescue the rut2080 ethanol preference phenotype, since the ethanol preference of the genotype rut2080; GH146/+; UAS-rut/+ was not significantly difference from both negative controls. (b) GH146-Gal4 drives GFP expression in antennal lobe, olfactory projection neurons and partial mushroom body. (c) Likewise, the expression of rut driven by the MB247-Mushroom body Gal4 line also failed to rescue the rut2080 ethanol preference phenotype, since the preference index of the genotype rut2080;;UAS-rut/MB247 was not significantly difference from both negative controls. (d) MB247-Gal4 drives GFP expression in α/β and γ lobe. Data are mean ± S.E.M. “N.S.” means no significance.
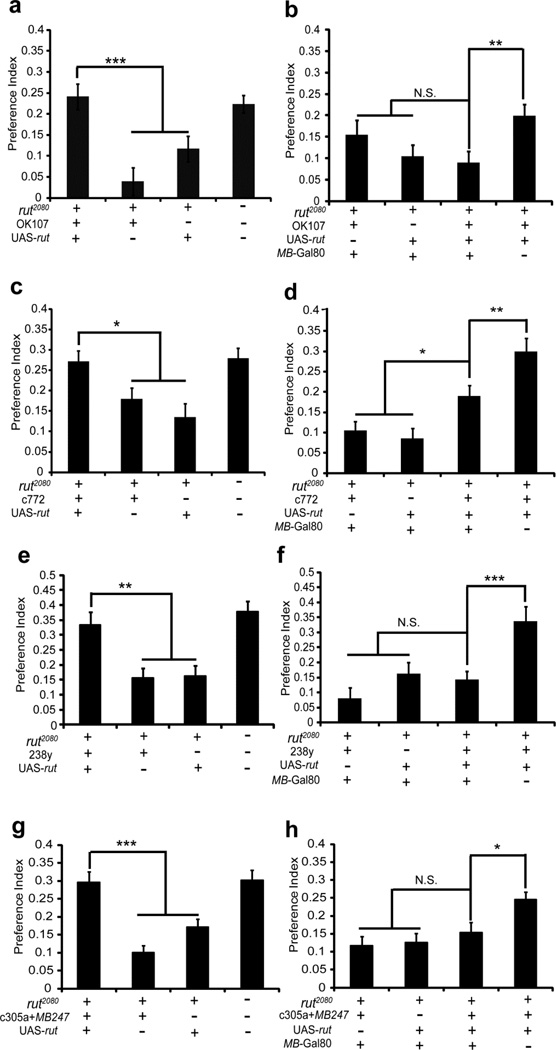
In contrast, the expression of rut driven by either the OK107, c772, 238y or c305a+MB247 Gal4 mushroom body drivers rescued the ethanol preference phenotype of rut2080 (In Fig. 4a, F(3.140)=10.398, P<0.0001, in post hoc comparisons, P<0.001 for both rut2080;;UAS-rut/+;OK107/+ vs. rut2080;;;OK107/+ or rut2080;;UAS-rut/+, in rut2080;;;OK107/+ group, n=27 flies and each n=39 in other groups. In Fig. 4c, F(3.150)=6.638, P=0.0003, in post hoc comparisons, P=0.023 for rut2080;c772/+;UAS-rut/+ vs. rut2080;c772/+, and P=0.0005 for rut2080;c772/+;UAS-rut/+ vs. rut2080;;UAS-rut/+.n=38–40 each. In Fig. 4e, F(3,115)=9.623, P<0.0001, in post hoc comparisons, P<0.01 for both rut2080;;UAS-rut/238y vs. rut2080;;238y/+ or rut2080;;UAS-rut/+, n=28–30 each. In Fig. 4g, F(3.175)=14.238, P<0.001, in post hoc comparisons, P<0.001 for rut2080;c305a/+;UAS-rut/MB247 vs. rut2080;c305a/+;UAS-rut/MB247 or rut2080;;UAS-rut/+, n=44–45 each). These four Gal4 lines drive expression within the whole mushroom bodies, and also provide weak expression in antennal lobe, optic lobe, pars intercerebralis, antennal nerve, tritocerebrum, and subesophageal ganglion (Fig. 5)(Aso et al., 2009). The rut foci for ethanol preference were further narrowed down by inhibiting Gal4 with the MB-Gal80 transgene (Krashes et al., 2007). In the MB-Gal80 transposon, Gal80 is driven by the dmef2 mushroom body enhancer, which is as also used in the MB247-Gal4 driver (Zars et al., 2000a). However, this enhancer drives the Gal80 expression in the whole mushroom body, rather than in the α/β and γ lobes in MB247-Gal4 (Krashes et al., 2007). In the presence of MB-Gal80, the GFP expression in mushroom bodies driven by the OK107, c772, 238y Gal4 lines or the c305a + MB247 combination was significantly reduced or eliminated (Fig. 5). Moreover, the four Gal4 drivers were no longer capable of rescuing the rut2080 ethanol preference phenotype in the presence of MB-Gal80 (In Fig. 4b, F(3.114)=3.011, P=0.033, no significance for rut2080; MB-Gal80/+;UAS-rut/+;OK107/+ vs. rut2080; MB-Gal80/+;;OK107/+ or rut2080; MB-Gal80/+;UAS-rut/+, however, P=0.0074 for rut2080; MB-Gal80/+;UAS-rut/+;OK107/+ vs. rut2080;;UAS-rut/+;OK107/+.In Fig. 4d, F(3.110)=13.898, P<0.0001, in post hoc comparisons, P=0.018 for rut2080;MB-Gal80/c772;UAS-rut/+ vs. rut2080;MB-Gal80/c772, however, P=0.0013 for rut2080; MB-Gal80/c772;UAS-rut/+ vs. rut2080;c772/+;UAS-rut/+,n=28–30 each. In Fig. 4f, F(3.80)=7.73, P=0.0001, no significance for rut2080; MB-Gal80/+;UAS-rut/238y vs. rut2080; MB-Gal80/+;238y/+ or rut2080; MB-Gal80/+;UAS-rut/+, however, P=0.0008 for rut2080; MB-Gal80/+;UAS-rut/238y vs. rut2080;;UAS-rut/238y, n=20–22 each. In Fig. 4h, F(3.165)=4.938, P=0.0026, no significance for rut2080; MB-Gal80/c305a;UAS-rut/MB247 vs. rut2080; MB-Gal80/c305a;MB247/+ or rut2080; MB-Gal80/+;UAS-rut/+, however, P=0.0209 for rut2080; MB-Gal80/c305a;UAS-rut/MB247 vs. rut2080;c305a/+;UAS-rut/MB247, n=41–42 each). The expression of rut in the α/β, α’/β’ and γ lobe neurons is capable of fully rescuing the rut2080 ethanol self-administration phenotype, whereas rut expression in the α/β and γ lobe neurons driven by MB247 alone is insufficient for rescue. This difference suggests that the expression of rutabaga in the α’/β’ neurons may be sufficient to rescue the rut2080 ethanol self-administration phenotype. We examined the isolated c305a α’/β’ Gal4 driver for an ability to rescue the rut2080 self-administration phenotype. The resulting phenotype lies between the Canton-S positive control and the rut2080; c305/+ genotype control, and is not significantly different from either (Sup. Fig. 5; F(3.147)=5.767, P=0.0009. In post hoc comparisons, P=0.487 for rut2080;c305a/+;UAS-rut/+ vs. CS, P=0.137 for rut2080;c305a/+;UAS-rut/+ vs. rut2080; c305a/+, and P=0.002 for rut2080;c305a/+;UAS-rut/+ vs. rut2080;; UAS-rut/+. n=36–38 each). Hence, it remains possible that rut expression within the α’/β’ lobe neurons is critical for the expression of a normal ethanol self-administration. Together, these data indicate that rut expression in the mushroom body is essential for ethanol preference in Drosophila.
Figure 4. The rutabaga requirement for normal ethanol preference maps to the mushroom body neurons.
The indicated genotypes were examined in the CAFE assay for an ethanol preference. (a) In the presence of both the UAS-rut and the OK107 mushroom body Gal4 driver, the rut2080 ethanol preference phenotype is not significantly different than CS, and is significantly higher than the control rut2080 genotypes. (b) The MB-Gal80 transgene inhibits the expression of Gal4 in specifically mushroom body neurons (Krashes et al., 2007). In the presence of MB-Gal80, UAS-rut and OK107 did not display a significantly rescued ethanol presence in comparison to the rut2080; UAS-rut/+;; OK107/+ genotype, and also no significance in comparison to rut2080; MB-Gal80/+;; OK107/+ and rut2080; UAS-rut/MB-Gal80 genotype. (c) Similarly, the rut2080 ethanol preference phenotype is only rescued in the presence of both the UAS-rut and c772 mushroom body Gal4 driver. (d) The MB-Gal80 transgene similarly reduced the ethanol preference of rut2080 flies carrying the UAS-rut and c772 transgenes. However, even in the presence of MB-Gal80, UAS-rut and c772 Gal4 displayed a significantly increased ethanol preference, compared with the two negative control. (e) In the presence of both the UAS-rut and the 238y Gal4 driver, the rut2080 ethanol preference phenotype is significantly higher than the control rut2080 genotypes. In all genotypes, the ry506 allele was also present. (f) In the presence of MB-Gal80, UAS-rut and 238y-Gal4 display a significantly rescued ethanol presence in comparison to the rut2080; UAS-rut/+; 238y/+ genotype, and also no significance in comparison to rut2080; MB-Gal80/+;238y/+ and rut2080; UAS-rut/MB-Gal80 genotype. In all genotypes, a ry506 allele was taken in third chromosome. (g) In the presence of the UAS-rut and the combination of c305a + MB247 Gal4 drivers, the rut2080 ethanol preference phenotype is significantly higher than the control rut2080 genotypes. (h) The MB-Gal80 transgene similarly reduced the ethanol preference of rut2080 flies carrying the UAS-rut and c305a+MB247 transgenes, and also displayed no significance in comparison to rut2080; MB-Gal80/c305a; MB247/+ and rut2080; UAS-rut/MB-Gal80 genotype. Data are means ± S.E.M. “N.S.” means no significance. *P<0.05, **P<0.01 and ***P<0.001.
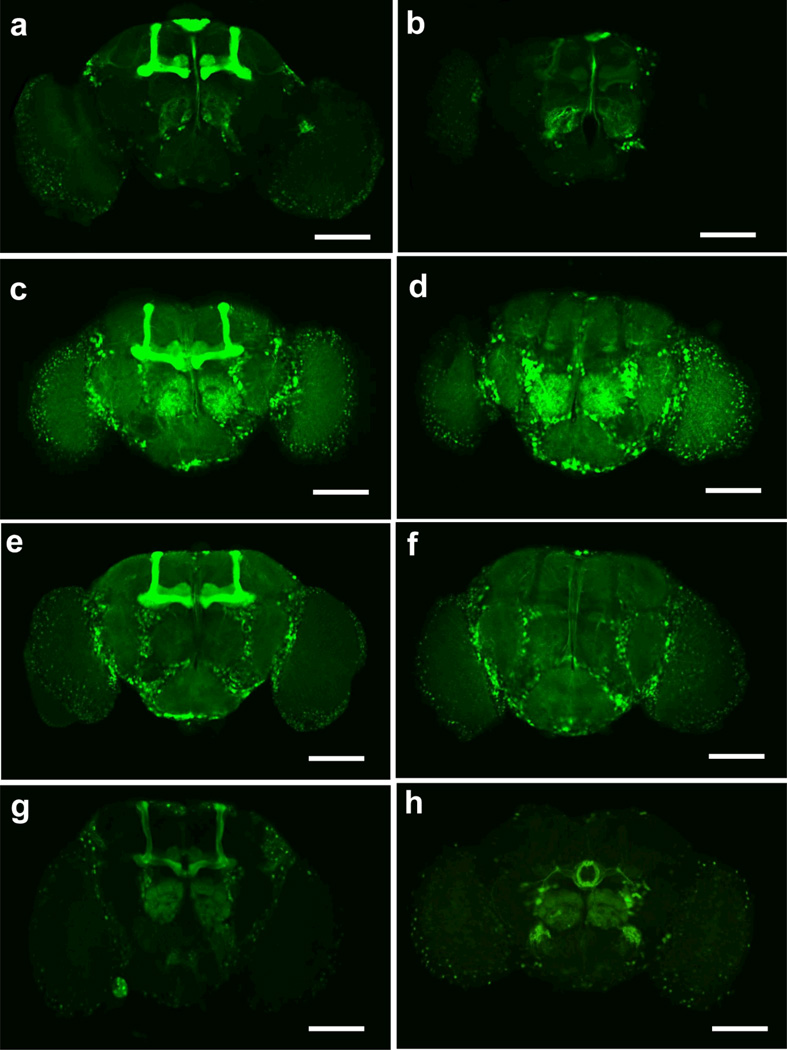
Figure 5. MBGal80 inhibits Gal4 activity within all the mushroom body neurons.
GFP was driven by four Gal4 drivers (a, OK107; c, c772; e, 238y; g, c305a+MB247), and detected by immunohistochemistry. The genotypes in panels b, d, f and h contain the MB-Gal80 transgene which effectively removes the expression of GFP from the mushroom bodies. Scale bar, 100µm.
Discussion
Why alcohol is consumed habitually and compulsively remains a fundamental unanswered question in addiction research. The use of diverse model systems and approaches appears necessary to unravel this question. We have used the CAFE self-administration assay to address the behavioral mechanism, molecular mechanism, and neural circuitry of ethanol feeding preference in Drosophila melanogaster. In our assay, wild-type Canton-S flies exhibit a significant preference for ethanol concentrations from 5% to 15%. This preference does not depend on the recognition of extra calories gained from the ethanol, nor does it depend on olfactory or gustatory biases for ethanol. The preference does, however, require the activity of the rut type I Ca+2-stimulated adenylyl cyclase within the mushroom body neurons, indicating a role for cAMP signaling in ethanol consumption behavior. This result is very significant as rut activity within the mushroom bodies is also involved in both reward and punishment learning. Since ethanol can act as a positive unconditioned stimulus in olfactory learning, and mushroom bodies are required to learn the association of odorants with alcohol (Kaun et al., 2011), there is a strong likelihood that the ethanol preference found in our assay is due to the rewarding properties of this drug.
The rut2080 mutants are defective in ethanol preference. This mutant phenotype was rescued acutely through the induced expression of a wild-type rut cDNA within the nervous system, indicating a physiological need for this cyclase for ethanol self-administration behavior. Knock-out mutants of the mouse calcium-activated type VIII adenylyl cyclase (AC8), but not knock-outs of the type I adenylyl cyclase, also display reduced ethanol self-administration (Maas et al., 2005). In mouse cortical membrane preparations, ethanol does not increase Ca2+-stimulated adenylyl cyclase activity, but in the AC8 knock-out mutants there is a reduction of ethanol-induced PKA phosphorylation events (Maas et al., 2005). Moreover, the reduction of G(s)α activity within mice results in reduced adenylyl cyclase activity and ethanol self-administration (Wand et al., 2001). A knock-out mutation of Protein kinase A Regulatory subunit IIβ leads to a decrease in basal cAMP stimulated PKA activity and an increase in ethanol self-administration, indicating additional complexities in cAMP pathway modulation of ethanol self-administration (Thiele et al., 2000). These findings suggest a phylogenically conserved role for cAMP production in modulating ethanol consumption, however in vertebrates the role for this pathway during development and the critical neural foci are currently unknown.
The spatial rescue of the rut self-administration phenotype suggests that cAMP signaling within the mushroom bodies is critical for this behavior. Recent imaging data has demonstrated that both dopamine and octopamine induce rut-dependent increases in cAMP and PKA activation within mushroom body neurons (Gervasi et al., 2010, Tomchik & Davis, 2009). Interestingly, the dnc1 phosphodiesterase mutants exhibited a normal ethanol preference even though the cAMP concentration within the heads is almost two-fold higher than in wild-type Canton-S (Byers et al., 1981), and the foskolin-induced PKA activity within mushroom body axons is increased by more than 20% (Gervasi et al., 2010). Within the mushroom bodies of dnc1 mutants, the spatial dynamics of PKA activation induced by dopamine but not octopamine is altered. In wild-type flies, the application of dopamine leads to significant PKA activation in the vertical α lobe, but not the horizontal β and γ lobes. In the dnc1 mutants, this PKA restriction is removed and PKA is significantly increased within both horizontal lobes (Gervasi et al., 2010). The compartmentalization of PKA activation by octopamine is unchanged in the dnc1 mutants (Gervasi et al., 2010). The absence of an ethanol self-administration phenotype in the dnc1 mutants suggests that the compartmentalization of dopamine induced PKA activity does not have a major role in this behavior. Octopamine may be responsible for rut activation during ethanol feeding, but additional neuromodulators are also possible (Feany & Quinn, 1995).
The expression of rut driven by either MB247 or GH146 Gal4 cannot rescue the defective ethanol preference. This result differentiates self-administration from sugar-reinforced olfactory appetitive learning (Thum et al., 2007). Yet, ethanol as a rewarding unconditioned stimulus likely requires different neural populations for reinforcement. In a recent adaptation of the olfactory learning paradigm using ethanol vapor as the unconditioned stimulus, learning ethanol reward required synaptic release from the γ lobe neurons during training, the α’/β’ lobes during consolidation, and the α/β lobe neurons during retrieval (Kaun et al., 2011). In negatively-reinforced olfactory memory, rut activity is required in different subsets of neurons for short-term and long-term memory formation, indicating both spatially and temporally distinct roles for rut within mushroom bodies (Blum et al., 2009). Thus, for the acquisition, consolidation, and retrieval of an appetitive memory formed during the CAFE assay, rut activity in multiple lobes of the mushroom bodies may be required.
If the rut-dependent ethanol preference found in the CAFE assay originates from a positively reinforced behavior, the actions or stimuli that may be specifically conditioned are currently unknown. Devineni and Heberlein (2009) showed that flies drink ethanol-containing food in shorter duration bouts than non-ethanol food and that flies visit ethanol-containing food more frequently. Since the ethanol preference is expressed by repeatedly going back to the ethanol containing capillary rather than continuously feeding, and the capillaries used in the CAFE assay are identical, a conditioning of feeding behavior per se would not produce the ethanol preference. In our experiments, we exchanged the locations and capillary tubes of regular food and ethanol-containing food daily to prevent a long-term place preference memory. The expression of rut in mushroom body neurons is not required in independent spatial or visual operant learning paradigms (Liu et al., 2006, Zars et al., 2000b). In another negatively reinforced place preference paradigm, the mushroom bodies have no appreciable role (Ofstad et al., 2011). Nevertheless, we cannot currently exclude the possibility that a shorter-term place preference for the ethanol containing capillary may have taken place during each 24 h period and this spatial memory is dependent on rut activity within the mushroom bodies. Alternatively, appetitive associations of ethanol reinforcement to specific ethanol sensory cues such as the taste of ethanol may be responsible for this rut-dependent component of the ethanol preference behavior. The brief and frequent drinking bout structure may be due to the complex negative and positive reinforcing properties of ethanol (Kaun et al., 2011); after consuming alcohol the drinking behavior is temporally inhibited by the early and temporarily aversive properties of this drug, but subsequently, the positive reinforcement brings the flies back to drink more.
The rut-dependent preference for ethanol-containing food may also result from a more general role for mushroom body plasticity in modulating motivation. Intrinsic mushroom body neurons are also critical for ethanol-induced locomotion hyperactivity (King et al., 2011), suggesting these neurons are important for the stimulating effects of ethanol (Wolf et al., 2002). The knock-down of the tequila neurotrypsin-like gene with the inducible mushroom body Gene-Switch driver displayed a reduced response to sugar after starvation (Colomb et al., 2009). If this requirement for teq in a motivated response to starvation lies within the mushroom body neurons, a similar role for a rut-dependent cAMP signaling within mushroom bodies for a motivated response toward alcohol is also plausible.
In conclusion, our data support previous findings that Drosophila displays an innate preference for ethanol containing food (Devineni & Heberlein, 2009, Ja et al., 2007). This preference is largely independent of an innate olfactory or gustatory bias for ethanol, and is not due to the nutrition in the ethanol containing food. We further found that the rut type I adenylyl cyclase is required acutely and broadly within the mushroom bodies for the expression of an ethanol preference. Hence, cAMP signaling within these neurons is likely required for continued ethanol self-administration. We propose that a major component of the preference for ethanol containing food comes from the hedonic reward of feeding behavior by ethanol and that this is regulated by a rut-induced cAMP signaling cascade in mushroom body neurons.
Supplementary Material
Acknowledgements
We thank Louise Parker for the MB-Gal80 line, Adriana Alcantara for helpful advice and discussion, Brigitte Dauwalder and Yanting Qi for technical support in immunohistochemistry; Tony Chen, Martha Espinosa, Amanda Coonce and Stacey Edwards for technical assistance. Funding for this research was provided by NIH grant AA015598 (G. Roman), a Grant in Aid from the ABMRF/The Foundation for Alcohol Research (G. Roman) and a GEAR Grant from the University of Houston (G. Roman). The research was also supported by US National Institutes of Health (R01AG030593 and R01AG023166) and the Ellison Medical Foundation to S. Pletcher, as well as by US National Institute of Health (F31AG33981 and T32AG18316) to T. Chan. This work utilized the Drosophila Aging Core of the Nathan Shock Center of Excellence in the Biology of Aging funded by the NIA (P30-AG-013283).
Footnotes
The authors report no conflict of interests.
References
- Aso Y, Grubel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 2009;23:156–172. doi: 10.1080/01677060802471718. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Blum AL, Li W, Cressy M, Dubnau J. Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr Biol. 2009;19:1341–1350. doi: 10.1016/j.cub.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CJ, Waddell S. Remembering nutrient quality of sugar in Drosophila. Curr Biol. 2011;21:746–750. doi: 10.1016/j.cub.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers D, Davis RL, Kiger JA., Jr Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature. 1981;289:79–81. doi: 10.1038/289079a0. [DOI] [PubMed] [Google Scholar]
- Cicero T. Alcohol self-administration, tolerance, and withdrawl in humans and animals: theoretical and methodological issues. In: Rigter H, Crabbe J, editors. Alcohol Tolerance and Dependence. Amsterdam: Elsvier/North Holland Biomedical Press; 1980. pp. 1–51. [Google Scholar]
- Colomb J, Kaiser L, Chabaud MA, Preat T. Parametric and genetic analysis of Drosophila appetitive long-term memory and sugar motivation. Genes Brain Behav. 2009;8:407–415. doi: 10.1111/j.1601-183X.2009.00482.x. [DOI] [PubMed] [Google Scholar]
- Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- Fujita M, Tanimura T. Drosophila evaluates and learns the nutritional value of sugars. Curr Biol. 2011;21:751–755. doi: 10.1016/j.cub.2011.03.058. [DOI] [PubMed] [Google Scholar]
- Gervasi N, Tchenio P, Preat T. PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron. 2010;65:516–529. doi: 10.1016/j.neuron.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Gibson JB, May TW, Wilks AV. Genetic Variation at the Alcohol Dehydrogenase Locus in Drosophila melanogaster in Relation to Enviornmental Variation: Ethanol Levels in Breeding Sites and Allozyme Frequencies. Oecologia. 1981;51:191–198. doi: 10.1007/BF00540600. [DOI] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14:612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Smith DP. The invertebrate odorant-binding protein LUSH is required for normal olfactory behavior in Drosophila. Chem Senses. 2001;26:195–199. doi: 10.1093/chemse/26.2.195. [DOI] [PubMed] [Google Scholar]
- Kimura K, Shimoawa T, Tanimura T. Muscle degeneration in the posteclosional development of a Drosophila mutant, abnormal proboscis extension reflex C (aperC) Dev Biol. 1986;117:194–203. doi: 10.1016/0012-1606(86)90361-1. [DOI] [PubMed] [Google Scholar]
- King I, Tsai LT, Pflanz R, Voigt A, Lee S, Jackle H, Lu B, Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, Reed RR. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Liu G, Seiler H, Wen A, Zars T, Ito K, Wolf R, Heisenberg M, Liu L. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- Maas JW, Jr, Vogt SK, Chan GC, Pineda VV, Storm DR, Muglia LJ. Calcium-stimulated adenylyl cyclases are critical modulators of neuronal ethanol sensitivity. J Neurosci. 2005;25:4118–4126. doi: 10.1523/JNEUROSCI.4273-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Scott K. Limited taste discrimination in Drosophila. Proc Natl Acad Sci U S A. 107:14833–14838. doi: 10.1073/pnas.1009318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Morse RM, Flavin DK. The definition of alcoholism. The Joint Committee of the National Council on Alcoholism and Drug Dependence and the American Society of Addiction Medicine to Study the Definition and Criteria for the Diagnosis of Alcoholism. JAMA. 1992;268:1012–1014. doi: 10.1001/jama.268.8.1012. [DOI] [PubMed] [Google Scholar]
- Ofstad TA, Zuker CS, Reiser MB. Visual place learning in Drosophila melanogaster. Nature. 2011;474:204–207. doi: 10.1038/nature10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Sable HJ, Murphy JM, McBride WJ. Recent advances in animal models of alcohol craving and relapse. Pharmacol Biochem Behav. 2004;79:439–450. doi: 10.1016/j.pbb.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Roman G. The genetics of Drosophila transgenics. Bioessays. 2004;26:1243–1253. doi: 10.1002/bies.20120. [DOI] [PubMed] [Google Scholar]
- Roman G, Davis RL. Conditional expression of UAS-transgenes in the adult eye with a new gene-switch vector system. Genesis. 2002;34:127–131. doi: 10.1002/gene.10133. [DOI] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Tempel BL, Bonini N, Dawson DR, Quinn WG. Reward learning in normal and mutant Drosophila. Proc Natl Acad Sci U S A. 1983;80:1482–1486. doi: 10.1073/pnas.80.5.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele TE, Willis B, Stadler J, Reynolds JG, Bernstein IL, McKnight GS. High ethanol consumption and low sensitivity to ethanol-induced sedation in protein kinase A-mutant mice. J Neurosci. 2000;20:RC75. doi: 10.1523/JNEUROSCI.20-10-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum AS, Jenett A, Ito K, Heisenberg M, Tanimoto H. Multiple memory traces for olfactory reward learning in Drosophila. J Neurosci. 2007;27:11132–11138. doi: 10.1523/JNEUROSCI.2712-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, Davis RL. Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron. 2009;64:510–521. doi: 10.1016/j.neuron.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol [A] 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Wand G, Levine M, Zweifel L, Schwindinger W, Abel T. The cAMP-protein kinase A signal transduction pathway modulates ethanol consumption and sedative effects of ethanol. J Neurosci. 2001;21:5297–5303. doi: 10.1523/JNEUROSCI.21-14-05297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Zars T. Short-term memories in Drosophila are governed by general and specific genetic systems. Learn Mem. 2011;17:246–251. doi: 10.1101/lm.1706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000a;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- Zars T, Wolf R, Davis R, Heisenberg M. Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: in search of the engram. Learn Mem. 2000b;7:18–31. doi: 10.1101/lm.7.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wu CF. Differential modulation of potassium currents by cAMP and its long-term and short-term effects: dunce and rutabaga mutants of Drosophila. J Neurogenet. 1993;9:15–27. doi: 10.3109/01677069309167273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.