Abstract
The rapid proliferation of progenitors during neurogenesis requires a stringent genomic maintenance program to ensure transmission of genetic fidelity. However the essential factors that govern neural progenitor genome integrity are unknown. Here we report that conditional inactivation of mouse TopBP1, a protein linked to DNA replication, and a key activator of the DNA damage response kinase ATR (ataxia telangiectasia and rad3 related) is critical for maintenance of early-born neural progenitors. During cortical development TopBP1 prevented replication-associated DNA damage in Emx1-progenitors which otherwise resulted in profound tissue ablation. Importantly, disrupted neurogenesis in TopBP1-depleted tissues was substantially rescued by p53- but not ATM-inactivation. Our data establish that TopBP1 is essential for preventing replication-associated DNA strand breaks, but is not essential per se for DNA replication. Thus, TopBP1 is crucial for maintaining genome integrity in the early progenitors that drive neurogenesis.
Maintaining genome stability is critical for mammalian development, and is achieved by the functional integration of DNA replication and DNA damage signaling1–4. This is particularly important during neural development, where defective DNA damage signaling can result in a wide spectrum of human diseases that are characterized by neurodegeneration, neurodevelopmental disease or brain tumors5. However, little is known about specific factors that modulate and maintain stem and progenitor genome integrity in vivo during neurogenesis.
Topoisomerase II binding protein 1 (TopBP1) is important for DNA replication initiation and cell cycle checkpoint activation6–10. Mammalian TopBP1 contains eight BRCA1 C-terminal (BRCT) domains that facilitate protein-protein interactions between key replication and checkpoint factors11–14. TopBP1 has been linked to the pre-replication complex because, together with factors such as TRESLIN/Ticrr and GEMC1, it is required for loading essential replication initiation components including Cdc456, 9, 10,15.
DNA damage arises during replication via frequent stalled replication forks or when DNA breaks occur by free radicals generated during cellular metabolism1, 15, 16. When replication forks stall, TopBP1 is important for initiating a cellular checkpoint. In this capacity, a key function appears to be activation of the ataxia telangiectasia and rad3-related (ATR) kinase to facilitate establishment of a checkpoint by phosphorylation of Chk18, 17–20. The TopBP1-ATR-Chk1 signaling axis is key to DNA damage resolution during replication. When any of these factors are inactivated in the mammalian germline, replication failure occurs resulting in embryonic lethality13, 21–23. Many essential cellular components interface with TopBP1 to establish ATR-dependent checkpoint activation. These include the Rad9-Rad1-Hus1 (9-1-1) complex; a DNA binding clamp that is loaded onto DNA by Rad17 in response to DNA damage24–26. TopBP1 then interacts with this complex via Rad9 in a damage-dependent manner to facilitate ATR activation; this occurs through TopBP1 binding to ATRIP, the obligate ATR binding partner19, 25, 27,28. This is a key event in the response to DNA damage as ATR is inactive until stimulated by TopBP117. Other factors are also important in TopBP1 function during establishment of the replication checkpoint. These include the BACH1/FANCJ helicase and RHINO, which can bind TopBP1 to modulate ATR activation29–31.
While in vitro data indicate TopBP1 is critical for DNA replication, the physiological settings requiring this function are unknown. Here we show that there is a broad requirement for TopBP1 throughout neurogenesis where it functions to prevent DNA damage accumulation in progenitor cells. Notably, unlike ATR inactivation32, 33, DNA damage signaling after TopBP1 loss is p53-dependent. Finally, our data show that TopBP1 is not essential per se for replication; rather, the critical biological function of this protein during neurogenesis is most likely the activation of cellular checkpoints after replication-associated DNA damage.
Results
Neural inactivation of TopBP1
To understand the biological function of TopBP1 we determined its role in the nervous system, as this tissue is often primarily affected in human syndromes associated with DNA repair deficiency5. We generated a TopBP1 allele in which LoxP sites flank exons 3–6, which resulted in an out-of-frame mutation when cre recombinase is used to drive gene deletion (Fig. 1a). We used Nestin-cre (Nes-cre) to generate TopBP1Nes-cre mice in which TopBP1 was inactivated throughout the nervous system. In TopBP1Nes-cre mice, TopBP1 was efficiently inactivated in neural tissues, while in non-neural tissues, such as the thymus and spleen, normal protein levels were present (Fig. 1b). TopBP1Nes-cre mice were viable and were similar in size to wild-type littermates at birth, although they became runted within a week (Fig. 1c) and generally did not survive beyond two weeks of age.
Figure 1. TopBP1 deletion in the nervous system.
(a) TopBP1 contains eight BRCA1 C-terminal (BRCT) domains. These encompass an amino terminal TRESLIN/Ticrr interacting domain, a C-terminal ATR-activating, a BACH1-binding region and an ATM phosphorylation site in BRCT domain 5. A conditional TopBP1 allele has LoxP sites flanking exons 3 and 6 and generates an out-of-frame mutation after cre-mediated recombination. (b) TopBP1 protein is absent in P0 TopBP1Nes-cre brain tissues but is present at normal levels in the thymus and spleen, while Nbs1 and actin levels are similar in all mutant tissues. (c) TopBP1Nes-cre neonates (asterisks) are initially similar to littermate controls, but become runted by P4. (d) The TopBP1Nes-cre cerebellum lacks granule neurons resulting in a disorganized Purkinje cell placement as shown at P0 using calbindin staining and at P12 using the neuronal differentiation marker βtubulin III (Tuj1). (e) Quantitative real-time PCR shows TopBP1 deletion occurs by E12.5 in the cortex but not the liver. (f) Western blot analysis of E14.5 TopBP1Nes-cre tissues shows that TopBP1 is deleted in the cortex but not the liver; p53pSer18 expression is also elevated in TopBP1Nes-cre tissue. Nbs1 and Ponceau staining are loading and blotting controls, respectively. (g) Apoptosis (arrows) occurs in proliferating cells in the developing cortex at E13.5.
TopBP1Nes-cre mice were characterized by neurodevelopmental abnormalities (Fig. 1d and Suppl. Fig. 1). We found a general reduction in cellularity in the TopBP1Nes-cre brain and a pronounced defect in cerebellar development. Analysis of the postnatal day 0 (P0) cerebellum showed disorganized Purkinje cells and an absence of an external granule layer (EGL), which resulted from apoptosis in the rhombic lip during development (Fig. 1d and see later). Elsewhere, less striking effects were observed, although the hippocampal region was also markedly affected (Suppl. Fig. 1). Additionally, developmental gliogenesis was similar in TopBP1Nes-cre tissue to controls (Suppl. Fig 2).
TopBP1 loss affects cortical development
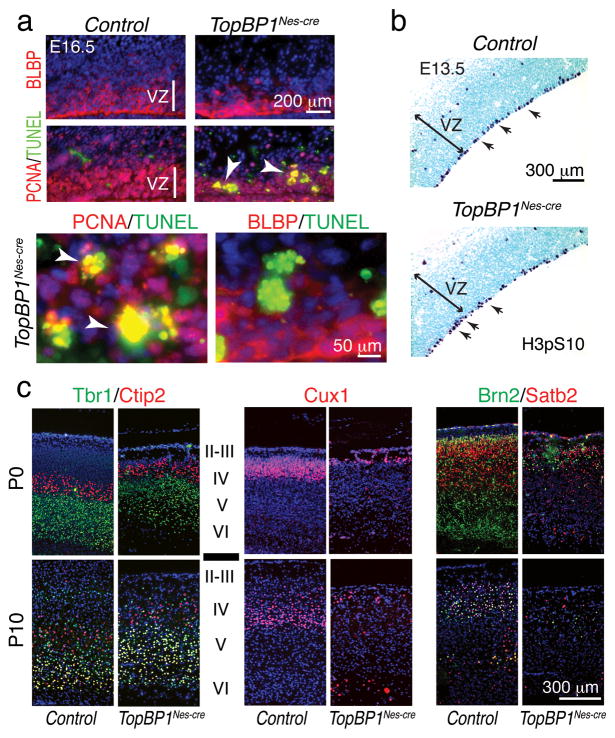
To further determine the role of TopBP1 during neural development, we focused on cortical development, which commences from embryonic day 10.5 (E10.5)34, 35. We initially determined TopBP1 gene deletion levels and protein loss in the TopBP1Nes-cre developing cortex. Quantitative analysis of isolated neopallium using real-time PCR indicated that TopBP1 deletion was maximal between E12.5 and E13.5 (Fig. 1e), and Western blot analysis confirmed TopBP1 protein was absent at E14.5 (Fig. 1f). Of note, TopBP1 loss led to p53 phosphorylation, consistent with DNA damage-induced activation of p53 (Fig. 1f). TopBP1 deletion resulted in widespread apoptosis by E13.5 as shown by terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) positive cells in the ventricular zone (VZ) of the developing cortex (Fig. 1g). These proliferating areas also accumulated DNA damage as revealed by immunostaining for phosphorylated H2AX (γH2AX), which occurred early and was widespread between E11.5 and E12.5, and preceded apoptosis (Suppl. Fig. 3a,b). Analysis of cell types undergoing apoptosis indicated that BLBP-positive radial glial progenitors were largely intact, although they contained γH2AX (data not shown), indicating that TopBP1 loss was associated with apoptosis of newborn progenitors as abundant overlap between TUNEL- and PCNA-positive cells occurred at E16.5 (Fig. 2a). Moreover, apical progenitors identified by phospho-histone H3 immunostaining were intact and largely unperturbed in TopBP1Nes-cre tissue at E13.5.
Figure 2. Apoptosis and cortical layering disruption in the TopBP1Nes-cre brain.
(a) Apoptosis in the TopBP1Nes-cre E16.5 cortex is associated with PCNA+ proliferating cells but not BLBP+ radial glia progenitors. (b) Apical progenitors (arrows) were identified using H3pSer10 immunostaining in E13.5 TopBP1Nes-cre and control neocortex. (c) Analysis of cortical development in P0 and P10 TopBP1Nes-cre and WT tissue using Tbr1 and Ctip2 to mark layers VI-IV, Cux1 to identify layers IV-II and Brn2/Satb2 to identify layers V-II reveals disrupted layer development in the TopBP1Nes-cre cortex. VZ is ventricular zone. Capital Roman numerals indicate the cortical layers.
Apoptosis in the TopBP1Nes-cre neocortex resulted in an overall reduction in cortical size (Suppl. Fig. 1b). The cortex comprises six layers derived from inside-out patterning, with early cortical progenitors fated to produce deep-layer (layers VI–V) neurons34, 35. To determine how TopBP1 deletion during progenitor proliferation and differentiation affected layering in the TopBP1Nes-cre cortex we used layer-specific markers. Analysis at P0 and P10 using Tbr1 and Ctip2 showed that the deep layers (Layers VI–V) formed to a large extent in the TopBP1Nes-cre cortex (Fig. 2c). In contrast, upper cortical layers identified by Cux1, Brn2 and Satb2 immunostaining, were reduced in size, suggesting persistent DNA damage in longer-cycling progenitors36 eventually compromised upper cortical layers (Fig. 2c).
Expansion of Emx1 cortical progenitors requires TopBP1
Our data indicate that despite efficient deletion of TopBP1 during neurogenesis substantial cortical development occurs in the TopBP1Nes-cre brain. Given the in vitro studies identifying a crucial role for TopBP1 function during replication9, 13, the large amount of cell proliferation associated with neurogenesis in the TopBP1Nes-cre tissue was unexpected.
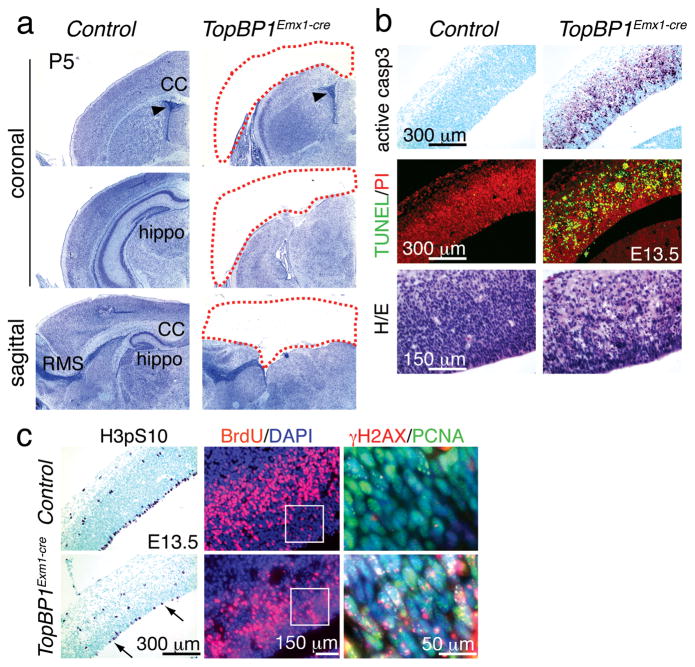
To understand these findings we considered if the timing of gene deletion influences the outcome of TopBP1 inactivation. We assessed this using Emx1-cre to drive TopBP1 deletion in early cortical progenitors. Emx1-cre expresses in the dorsal telencephalon earlier than Nestin-cre (i.e. ~E10 vs. E11)37, 38. Emx1-cre expression occurs in cells that give rise to the hippocampus and parts of the motor, somatosensory and visual cortex, but does not express in the ventral telencephalon37, 38. In striking contrast to TopBP1Nes-cre, there was a loss of all cortical structures derived from Emx1-expressing progenitors in TopBP1Emx1-cre mice (Fig. 3a and Suppl. Fig. 4a). To ascertain the cause of this phenotype, we examined the embryonic neocortex at E13.5, when Emx1-expressing progenitors are rapidly expanding37. In TopBP1Emx1-cre, but not control tissue, we found abundant apoptosis indicated by activated caspase-3 immunostaining, TUNEL and resultant reduced cellularity after hematoxylin and eosin staining (Fig. 3b and Suppl. Fig. 4b). Cell loss was also evident by reduced BrdU incorporation and fewer H3pS10 positive G2/M phase cells at the ventricular/apical surface (Fig. 3c, arrows). This extensive cell loss is sufficient to account for the absence of the cortices at birth. We also found that TopBP1 inactivation led to widespread γH2AX immunostaining by E11.5 that co-localized with proliferating PCNA-positive cells in the dorsal telencephalon (Suppl. Fig. 4c). While the dorsal telencephalon was affected in the TopBP1Emx1-cre brain, other regions including the ventral telencephalon and the cerebellum showed no developmental abnormalities, underscoring the specificity of Emx1-cre expression (Suppl. Fig. 4d).
Figure 3. Dorsal telencephalon progenitors are lost in TopBP1Emx1-cre mice.
(a) Cortices and other structures developing from the dorsal telencephalon are missing in the TopBP1Emx1-cre brain; shown by Nissl staining at P5. CC is the corpus callosum, hippo is the hippocampus and RMS is the rostral migratory stream. (b) Abundant apoptosis is observed in the E13.5 TopBP1Emx1-cre neopallial cortex shown using TUNEL and activated caspase-3 staining. Differences in the numbers of apoptotic cells between genotypes calculated using a student’s t-test were significantly different; active caspase 3 positive cells in 1 mm2 (± SEM) were; control = 0; TopBP1Emx1-cre = 734.5 (± 30.58), and TUNEL positive cells in 1 mm2 (± SEM) were; control = 7 (± 0.81) and TopBP1Emx1-cre = 987.5 (± 27.381). (c) Fewer proliferating cells indicated by histone H3pSer10 or BrdU incorporation staining are seen in the E13.5 TopBP1Emx1-cre neopallial cortex. Differences in the numbers of proliferating cells between genotypes calculated using a t-test were significantly different; BrdU positive cells in 0.27 mm2 (± SEM) were; control = 438.4 (±26.48) TopBP1Emx1-cre = 205.6 (± 10.31) and H3pS10 positive cells in 1 mm2 (± SEM) were; control = 91 (± 4.589), TopBP1Emx1-cre = 27.69 (± 1.707). Widespread DNA damage shown by γH2AX and PCNA immunostaining in regions indicated in white boxes in adjacent panels.
Emx1 progenitors proliferate after Lig4 or Xrcc1 loss
To further gauge the essential requirement for TopBP1 in the cortical progenitors, we determined the comparative importance of the key DNA repair factors DNA ligase IV (Lig4) and Xrcc1. These factors are central for DNA strand break repair via non-homologous end-joining and base excision repair respectively; germ line deletion of either is embryonic lethal and deletion throughout the nervous system using Nestin-cre results in microcephaly and DNA damage accumulation39, 40.
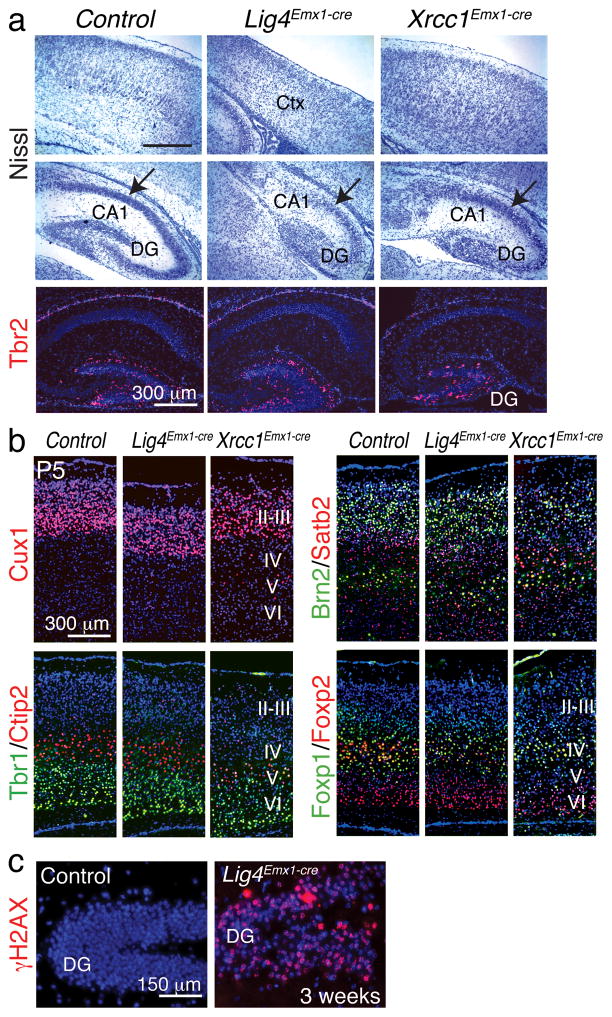
We therefore compared cortical development in Lig4Emx1-cre and Xrcc1Emx1-cre to TopBP1Emx1-cre mice. Loss of Lig4 or Xrcc1 resulted in cortices with milder developmental perturbations, although a marked effect upon the hippocampus was apparent (Fig. 4a). Accordingly, cortical layering was relatively intact in both the Lig4Emx1-cre and Xrcc1Emx1-cre mice with each of the six layers present (Fig. 4b). Nonetheless, substantial levels of DNA damage accumulated, as γH2AX was abundant in Lig4Emx1-cre tissue, consistent with Lig4 gene deletion throughout the dorsal cortex leading to persistent unrepaired DNA (Fig. 4c). These data indicate that despite the essential neural repair functions of Lig4 and Xrcc1, TopBP1 fulfills a unique role in maintaining genome stability of progenitors.
Figure 4. Analysis of cortical development in Lig4Emx1-cre and Xrcc1Emx1-cre brain.
(a) Nissl staining identifies the Lig4Emx1-cre and Xrcc1Emx1-cre cortex (Ctx) and hippocampus (DG and CA1) and shows the relative levels of defective development after Lig4 or Xrcc1 loss. Relative gene deletion (± SEM) was determined in genomic DNA isolated from the cortex using realtime PCR. For Xrcc1; WT = 1.22 (± 0.033), Xrcc1Emx1-cre = 0.46 (± 0.006). For Lig4; WT = 0.86 (± 0.038), Lig4Emx1-cre = 0.19 (± 0.004). Tbr2 marks the dentate gyrus (DG). (b) Cortical markers were used to assess development in the Lig4Emx1-cre and Xrcc1Emx1-cre cortex. Roman numerals indicate the cortical layers. (c) DNA damage (γH2AX) accumulates in the Lig4Emx1-cre cortex; DG is the dentate gryus.
Progenitor loss after TopBP1 inactivation requires p53
In the nervous system, DNA damage-induced apoptosis is often p53- or Atm-dependent41. To determine if this signaling pathway contributed to the TopBP1-deleted phenotype, we generated compound TopBP1 mutants with either coincident p53 or Atm deletion.
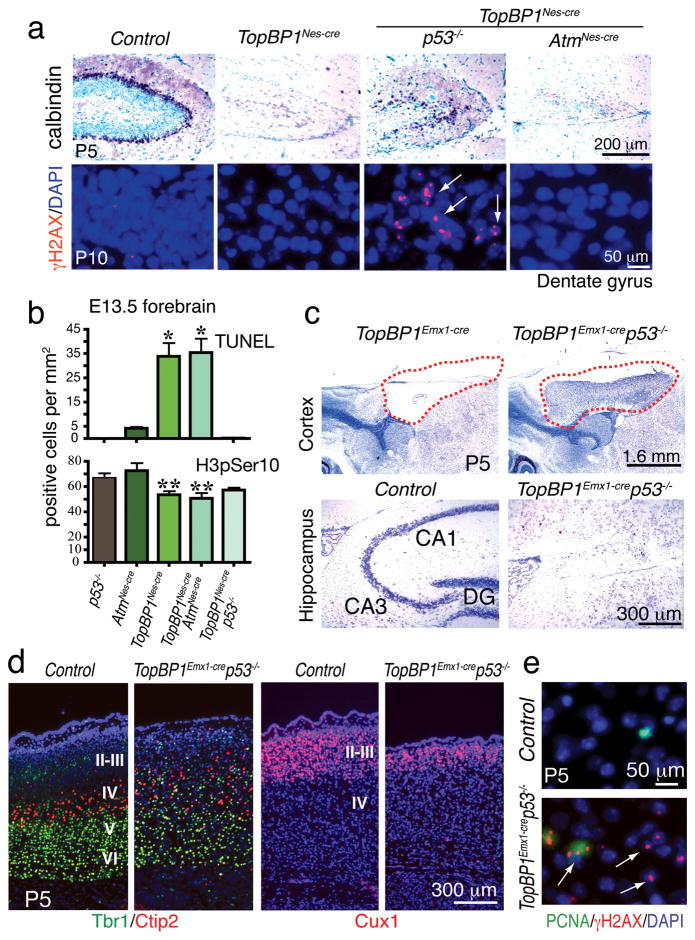
Loss of p53 resulted in an obvious amelioration of the TopBP1Nes-cre cortical defects that were particularly noticeable in the case of hippocampal development, although this rescue was only partial (Fig. 5a and Suppl. Fig. 5a). Notably, widespread DNA damage was observed in the rescued TopBP1Nes-cre;p53−/− dentate gyrus, indicating that chronic genome instability was a feature of this tissue (Fig. 5a and Suppl. Fig. 6a,b). Consistent with this developmental rescue, apoptosis resulting from TopBP1 loss was also abrogated in TopBP1Nes-cre;p53−/− tissue and cell proliferation was recovered as determined by histone H3 phosphorylation (Fig. 5b). However, in contrast to p53 loss, inactivation of Atm did not influence the TopBP1-null phenotype (Fig. 5a,b). The rescue of the TopBP1Nes-cre phenotype by p53 also occurred in other TopBP1Nes-cre brain regions. The E13.5 TopBP1Nes-cre embryonic cerebellum showed apoptosis in proliferating cells of the ventricular zone, which was blocked after loss of p53, but not Atm (Suppl. Fig. 7a). Quantification of both proliferation and apoptosis showed that inactivation of p53 blocked apoptosis and restored proliferation to WT levels in the TopBP1Nes-cre;p53−/−cerebellum and ganglionic eminence, while Atm loss worsened the phenotype somewhat (Suppl. Fig. 7b).
Figure 5. Defective neurogenesis after TopBP1 inactivation requires p53, but not Atm signaling.
(a) The hippocampal defects in TopBP1Nes-cre brain involve p53 signaling as TopBP1Nes-cre;p53−/− mice show partial rescue of hippocampal dentate gyrus development as shown using calbindin immunostaining to assess histology at postnatal day 5 (P5). In contrast to p53, Atm loss failed to rescue defective TopBP1Nes-cre hippocampal development. Despite recovered development, TopBP1 loss nonetheless resulted in persistent DNA damage as demonstrated by γH2AX foci. (b) Analysis of apoptosis using TUNEL or proliferation using H3pSer10 indicates that p53, but not Atm signaling is involved in the TopBP1Nes-cre phenotype. (c) Cortices that fail to develop in TopBP1Emx1-cre mice are rescued by coincident loss of p53 (red-hatched outlines). However, the hippocampal formation is not rescued in the TopBP1Emx1-cre;p53−/− brain. (d) Although substantial cortical rescue occurs in TopBP1Emx1-cre;p53−/− mice, cortical layering shows disorganization as indicated using Cux1 or Tbr1 and Ctip2 co-staining. (e) DNA damage is present throughout the P5 TopBP1Emx1-cre;p53−/− cortices (arrows) and γH2AX foci are frequently observed in PCNA-positive proliferating cells (arrowhead); P5 is postnatal day 5. DG is dentate gyrus.
Although rescue of the TopBP1Nes-cre phenotype by p53-loss was extensive, it only extended the lifespan of the TopBP1Nes-cre mice by around 10 days (Suppl. Fig. 5b). This likely reflects the profound functional consequence of persistent DNA damage after TopBP1 loss in replicating cells throughout the brain. These data indicate that DNA damage-associated apoptosis after TopBP1 loss is p53-dependent and that cells can proliferate in the absence of TopBP1, but at the expense of DNA damage accumulation.
Given the level of development in TopBP1Nes-cre;p53−/− neural tissue, we also determined how p53 inactivation impacted tissue loss in the TopBP1Emx1-cre brain and found a remarkable degree of cortical development in the TopBP1Emx1-cre;p53−/− mice (Fig. 5c; red dashed lines). This rescue resulted in establishment of cortical layering, albeit less organized compared to WT tissue (Fig. 5d). Most notably, the upper cortical layers were more affected, probably as a result of chronic DNA damage during neurogenesis. Recovered development in TopBP1Emx1-cre;p53−/− tissue showed perturbation of homeostasis as these regions were marked by gliosis (Suppl. Fig. 6c, d). One exception to the rescue in the TopBP1Emx1-cre;p53−/− tissue was the hippocampus, as this structure was absent from the TopBP1Emx1-cre;p53−/− brain (Fig. 5c). This situation contrasts the partially recovered hippocampal development in the TopBP1Nes-cre;p53−/− brain (Fig. 5a). The different hippocampal phenotypes between the TopBP1Emx1-cre;p53−/− and TopBP1Nes-cre;p53−/− likely reflect the enhanced sensitivity of early hippocampal progenitors towards genotoxic stress in the absence of TopBP1. The TopBP1Emx1-cre;p53−/− cortex was also associated with γH2AX immunopositive cells that co-localized with PCNA immuno-positive cells, indicating that TopBP1-null cells have survived and are replication competent (Fig. 5e).
DNA strand breaks accumulate in TopBP1-null progenitors
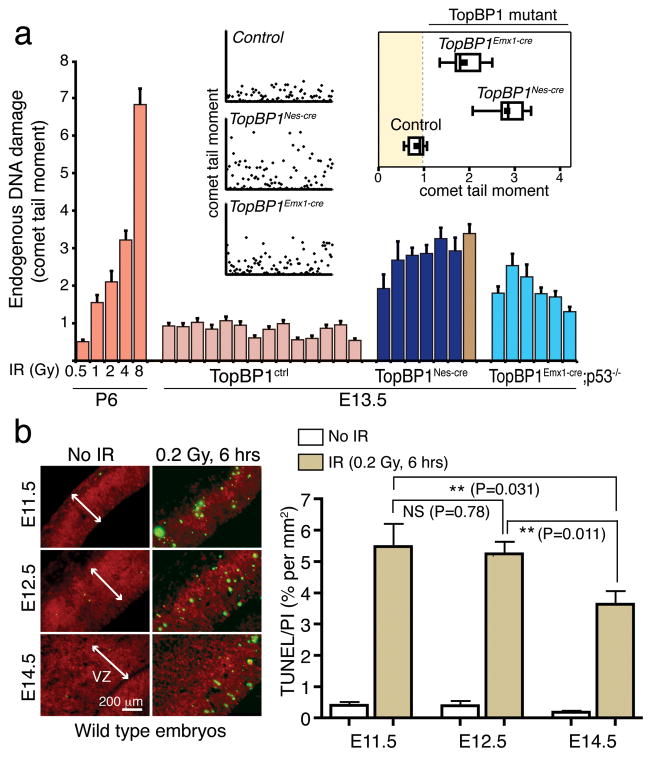
Despite the rescue of developmental defects by p53 deficiency after TopBP1 inactivation, DNA damage was associated with TopBP1 loss and was frequently co-localized with proliferating cells (Fig. 5e and Suppl. Fig. 3 and 6a). Although this reinforces the notion that TopBP1 is critical for maintaining genome stability but is dispensable for DNA replication, it raises the question of the physical nature of the DNA damage. As γH2AX is only a surrogate marker for DNA damage, we directly addressed DNA integrity in cortical progenitors in situ using the alkaline comet assay (ACA) to measure DNA strand breaks. We isolated progenitors from E13.5 neocortex from both TopBP1Nes-cre and TopBP1Emx1-cre;p53−/− and control embryos and then immediately subjected these to ACA. In this way we were able to directly assess DNA integrity in vivo at a specific developmental stage (Fig. 6a). To determine the amount of strand breaks, we included an irradiated series of acutely isolated WT cerebellar granule neuron precursors (GNP) as a comparison to the cortical progenitors. We found that all individual cortical isolates from TopBP1 mutant tissue had markedly increased DNA strand breaks. The levels of DNA damage were similar to ~2Gy of radiation in GNPs (Fig. 6a), which is estimated to be about 80 double-strand breaks/nucleus42, although the damage measured by the ACA also includes single-strand breaks. Notably, the levels of in situ DNA breaks were similar between cortical progenitors from TopBP1Nes-cre and TopBP1Emx1-cre progenitors indicating that both cre drivers are effective at TopBP1 deletion despite the more profound defects of cortical development in TopBP1Emx1-cre tissue.
Figure 6. TopBP1-deficiency during neurogenesis results in DNA strand break accumulation in cortical progenitors.
(a) Embryonal neural cells isolated from TopBP1Nes-cre and TopBP1Emx1-cre accumulate significant DNA damage compared to wild type controls. Comet analysis of acutely isolated cells from E13.5 TopBP1Nes-Cre (dark blue; six individual TopBP1Nes-cre embryo isolates and brown is data from a TopBP1Nes-cre;p53−/− embryo) or TopBP1Emx1-cre;p53−/− (light blue; six individual embryos) dorsal telencephalon show accumulated DNA damage in situ compared to controls (mauve; 14 individual embryo isolates); a minimum of 300 comet-tail moments were measured for each sample and mean comet-tail moments for each replicate are shown. Mean comet tail moments of irradiated P6 cerebellar granule neurons at increasing doses (orange) are shown in comparison as a standard to quantify DNA strand breaks. Inset scatterplots indicate actual comet tail moment values of 100 representative cells analyzed from the respective genotypes. The inset boxplot indicates the mean comet tail moment of all replicates within each genotype. (b) Cortical progenitors at E11.5 and E12.5 are significantly more susceptible to DNA damage-induced apoptosis than at those at E14.5. Quantitation of TUNEL positive cells was determined in proportion to propidium iodide-stained cells in the neocortex after 0.2Gy of radiation. VZ is ventricular zone.
Because TopBP1 deletion by either Nes-cre or Emx1-cre resulted in similar amounts of DNA damage in progenitors at E13.5, we determined the relative sensitivity of progenitor populations to DNA damage at different early embryonic developmental times. Using very low doses (0.2Gy) of ionizing radiation (IR) that generate ~8 breaks/G1-genome42 we quantified the levels of apoptosis by determining the ratio of TUNEL+ cells to total cells in a defined area in the ventricular zone of E11.5, E12.5 and E14.5 embryos. We found that there was a significantly enhanced susceptibility to DNA damage-induced apoptosis in early embryos compared with later-stage embryos, indicating a lower threshold for apoptosis in earlier progenitors compared with those generated later (Fig. 6b).
Collectively these data indicate that TopBP1 deletion results in the accumulation and persistence of DNA strand breaks in cortical progenitors, and also that the relative levels of DNA damage are similar in TopBP1Emx1-cre and TopBP1Nes-cre progenitors. Thus the striking difference in phenotype after deletion via the different cre-drivers is not due to different amounts of DNA strand breaks, rather the enhanced susceptibility of earlier-born cortical progenitors to DNA damage after TopBP1 loss.
Defective DNA damage signaling in TopBP1 deficient cells
As TopBP1 loss resulted in elevated DNA damage, we determined the competency of TopBP1 deficient cells to respond to and repair DNA damage. We initially attempted to use TopBP1LoxP/LoxP mouse embryonic fibroblasts (MEFs) and retroviral transduction with MSCV-cre to delete TopBP1. However, retroviral introduction of cre caused widespread apoptosis of the TopBP1LoxP/LoxP cells after 48hrs, while TopBP1+/LoxP used as a control were unaffected by MSCV-cre. This likely reflects hypersensitivity of the TopBP1-deleted cells to DNA breaks associated with retroviral integration.
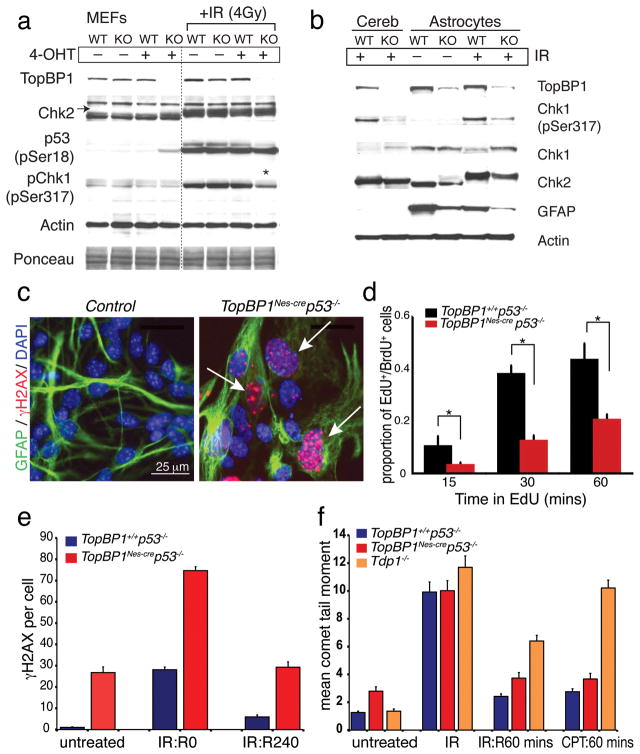
Therefore, we established TopBP1creTM cells in which TopBP1 can be inactivated after tamoxifen administration (creTM is a tamoxifen-inducible version of cre that uses the actin promoter and CMV enhancer). We found that efficient TopBP1 deletion occurred in TopBP1creTM MEFs after 4-hydroxytamoxifen (4-OHT) administration and that loss of TopBP1 resulted in a modest upregulation of p53, probably from replication-associated DNA damage (Fig. 7a). After 4-OHT-mediated deletion (48hr treatment) TopBP1creTM MEFs ceased proliferation, although they remained viable. Using these, we found that loss of TopBP1 did not alter Chk2 activation or p53ser18 phosphorylation after DNA damage by IR. However, consistent with ATR activation requiring TopBP1, Chk1ser317 phosphorylation was reduced after radiation (Fig. 7a, asterisk). As a comparison to the in vitro data, we examined the TopBP1Nes-cre cerebellum after radiation and found a similar defect in Chk1 phosphorylation to that in MEFs (Fig. 7b). Overall, these data suggest that TopBP1 is not required for DNA double strand break (DSB) signaling, but rather is central to maintaining DNA integrity and ATR-dependent checkpoint activation during replication.
Figure 7. Analysis of DNA damage responses in TopBP1-depleted cells.
(a) The radiation-induced DNA damage response was investigated using tamoxifen (4-OHT)-induced deletion of TopBP1 in mouse embryonic fibroblasts (MEFs): WT are TopBP1+/+;CreTM and KO are TopBP1LoxP/LoxP;CreTM. TopBP1, Chk2, p53(p-ser18), Chk1(p-ser317) and actin indicate the result of probing with the respective antibodies. Activation of Chk2 is indicated by mobility shift (arrow). Ponceau staining and actin immunostaining indicates protein loading. Asterisk indicates decreased Chk1(p-ser317) immunostaining after TopBP1 inactivation. (b) Cerebellar extracts or astrocytes were from wild type or TopBP1Nes-cre mice and were immunoblotted as described above for MEFs, but also included antibodies recognizing total Chk1 and glial fibrillary acidic protein (GFAP); longer exposure reveals clear GFAP staining in the cerebellum (not shown). Trace TopBP1 signal in replication-defective null-astrocytes is from meningeal fibroblast contamination. Radiation was 4Gy with 2h recovery. (c) TopBP1Nes-cre;p53−/− astrocytes show elevated levels of endogenous DNA damage indicated by γH2AX immunostaining (arrows). Astrocyte purity is shown by GFAP immunostaining. (d) DNA replication in control and TopBP1Nes-cre:p53−/− astrocytes was determined using EdU pulsing after BrdU pre-labeling. TopBP1 deficiency markedly compromised DNA replication as shown by a significant reduction of EdU uptake; *= p<0.001. (e) DNA repair in TopBP1Nes-cre;p53−/− astrocytes determined by quantifying γH2AX foci removal after IR (n=50 cells); R0 and R240 represent recovery after 0 and 240 minutes. (f) DNA repair was assessed after IR (10 Gy) and camptothecin (14μM) using the alkaline comet assay; Tdp1−/− primary astrocytes were established from tyrosyl-DNA phosphodiesterase- 1 null mice. R60 represents recovery after 60 minutes.
DNA repair is near normal in TopBP1 deficient cells
We also determined DNA repair competency in TopBP1-depleted cells using primary TopBP1Nes-cre astrocytes challenged with IR or camptothecin. Although we were unable to establish TopBP1Nes-cre astrocytes, probably as a result of DNA damage induced cell cycle arrest, we were able to generate TopBP1Nes-cre;p53−/− astrocytes (that presumably overcame DNA damage-induced p53-dependent cell cycle arrest). However, compared with WT cultures, TopBP1Nes-cre;p53−/− astrocyte cultures only reached ~30 % confluency and were characterized by chronic DNA damage accumulation indicated by widespread γH2AX foci (Fig. 7c). This DNA damage accumulation was associated with replication abnormalities, as BrdU/EdU incorporation in TopBP1 astrocytes showed a marked reduction compared with controls (Fig. 7d, Suppl. Fig. 8).
Despite a normal IR-induced DNA damage response, the high endogenous levels of γH2AX indicated extensive DNA damage; most likely strand breaks associated with compromised replication. Therefore, we assessed DNA strand break repair after IR, by measuring resolution of the resultant γH2AX foci in TopBP1Nes-cre;p53−/− cells. We found that IR-induced γH2AX foci were resolved in TopBP1Nes-cre;p53−/− astrocytes suggesting that this type of exogenously induced DNA damage could be repaired effectively after TopBP1 loss (Fig. 7e). Additionally, we measured DNA repair after genotoxic stress using the ACA. We found that while TopBP1Nes-cre cells had a higher basal level of DNA damage, these cells repaired DNA breaks at a rate similar to control cells after either IR or camptothecin. As a comparison to TopBP1Nes-cre we included DNA repair deficient tyrosyl-DNA phosphodiesterase 1 (Tdp1−/−) cells43 and confirmed that these were defective after both genotoxic agents (Fig. 7f). Together, these data indicate that TopBP1 is not essential for repair of DNA strand breaks induced by agents that require BER and NHEJ/HR pathways. Thus, DNA strand break pathways are competent in TopBP1-null tissue, and the persistence of DNA breaks likely reflects excessive DNA damage accumulation and perturbed steady-state repair.
Discussion
Coordinated integration of DNA replication and damage signaling is necessary for unperturbed mammalian development. While our understanding of the biochemistry of DNA replication and repair has rapidly advanced during the last few years, it nonetheless remains a challenge to transition this knowledge to physiological systems. The nervous system is particularly relevant in this regard as it is often a primary tissue affected in human diseases associated with defects in the response to DNA damage5. In the present study we show that the ATR activator, TopBP1, is critical for the prevention of DNA damage accumulation and apoptosis in early-born neural progenitors, thereby maintaining homeostasis at a critical stage of nervous system development (Suppl. Fig. 9).
We found that TopBP1 inactivation by Emx1-cre in dorsal telencephalon progenitors during the very early stages of cortical neurogenesis (from ~E10) resulted in cortical ablation. In contrast, TopBP1 inactivation at later stages by Nes-cre resulted in less substantial defects in cortical development. The main reason for these different outcomes is the timing of cre-mediated deletion; Emx1-cre targets specific progenitors at least one day earlier than Nes-cre. Although Emx1-cre targets earlier-born cortical progenitors we found similar levels of DNA strand-breaks in dorsal telencephalon progenitors at E13.5 after TopBP1 inactivation by either cre-driver, indicating equivalent levels of damage. This raises the likelihood that there is a different threshold to DNA damage in the early-born progenitors, perhaps reflective of an increased need for vigilance of genome integrity in stem/early-progenitor populations. This assertion is supported by our finding of an increased sensitivity of cortical progenitors to DNA damage at E11 compared to E14. Indeed, although they cycle rapidly, early-born cortical progenitors have an extended S-phase that would allow enhanced genome surveillance and integrity. When switching to neuron production, S-phase shortens, suggesting genome maintenance is an important feature of uncommitted progenitors44. Additionally, as cortical expansion occurs G1 also lengthens in progenitors undergoing differentiation, highlighting the dynamic events associated with cell cycle regulation during neurogenesis36, 44, 45.
Altered transcriptional programming in cortical progenitors may also influence the outcome of genotoxic stress. For example, by the time they are targeted using Nes-cre the Emx1-expressing progenitor cells in the dorsal telencephalon have already undergone substantial expansion and cell fate determination by Lhx2 to commit to cortical genesis38. Therefore, progression from stem/uncommitted progenitor can result in altered cell cycle dynamics, intrinsic replication differences and differential sensitivity to DNA damage. In contrast to TopBP1, inactivation of the key DNA repair factors Xrcc1 and DNA Ligase IV by Emx1-cre failed to fully disrupt cortical genesis. This was also the case for Brca1 inactivation by Emx1-cre, although in that scenario early-born but not late-born progenitors were lost by apoptosis46. Taken together, one interpretation of our data is that the outcome of DNA damage is regulated differently as progenitors mature, and that TopBP1 contributes uniquely to genome stability in early-born progenitors. Alternatively, and despite equal levels of DNA strand breaks in both TopBP1Emx1-cre and TopBP1Nes-cre progenitors at E13.5, the greater cell loss in TopBP1Emx1-cre may reflect the more protracted period of proliferation prior to Nes-cre expression.
The consequences of TopBP1 loss towards newborn progenitors in the cerebellum was similar to the cortex. TopBP1Nes-cre mice showed pronounced cerebellar agenesis, as the EGL failed to expand from the Rhombic lip. The cerebellar ventricular zone progenitors were also markedly affected, and the resultant lack of development lead to a cerebellum consisting of a disorganized collection of cells such as Purkinje cells that arise prior to Nes-cre activation.
While our data highlight the importance of TopBP1 for preventing replication-associated DNA damage, most prominently they show that it is not required per se for replication. This is because neural development defects in TopBP1Nes-cre or TopBP1Emx1-cre can be substantially rescued by concomitant p53 loss, leading to continued tissue development. These data contrast other studies that show TopBP1 is essential for the assembly of DNA replication pre-initiation complexes, a function tightly linked to factors such as Treslin/Ticrr and GEMC16, 9, 10. The in vivo nature of our study maintains the physiological parameters that support development and cell proliferation, unlike other studies that utilize cell-free extracts or in vitro cell culture systems. However, consistent with those other studies, we find severely compromised proliferation of TopBP1-null cells in culture, and coincident inactivation of p53 is required to extend their viability. Nonetheless, TopBP1Nes-cre;p53−/− astrocytes can only undergo a few rounds of division, during which they accumulate abundant DNA damage.
While a key function of TopBP1 is activation of ATR, the more severe neurogenesis defects after TopBP1-loss than those of ATR inactivation47 argue for a broader role in genome maintenance. Furthermore, TopBP1-loss activated a DNA damage response resulting in p53-mediated apoptosis. This contrasts the different outcomes of Atr and p53 inactivation32, 33, 47, suggesting loss of TopBP1 in vivo results in DNA lesions and/or DNA damage signaling that are qualitatively different to those in Atr-deficient tissues. Although Atm can also be important in modulating p53-dependent DNA damage-induced apoptosis in the nervous system48, Atm loss failed to rescue defects in TopBP1Nes-cre mice. This is reflective of other scenarios in the nervous system whereby DNA damage leads to Atm-dependent apoptosis in immature post-mitotic neural cells, but not in proliferative cells49, 50.
Taken together, our data are compatible with a model in which TopBP1 maintains replication fidelity via checkpoint activation, rather than an essential role in replication pre-initiation complex assembly. In its absence replication proceeds, but DNA fidelity is compromised via the accumulation of stalled replication forks and other DNA damage during S-phase. This damage would normally signal ATR and other factors such as the BACH1 helicase30 for checkpoint activation and homologous recombination repair17. Thus, our study reveals the indispensible requirement for TopBP1 to prevent DNA damage accumulation in early-born progenitors, where maintenance of genome integrity is of paramount importance for neurogenesis.
Methods
TopBP1LoxP mice
A targeting vector was constructed using recombineering to generate a TopBP1 allele with exons 3–6 flanked by LoxP sites. To do this a single LoxP site was inserted between exons 2 and 3 and a neomycin-thymidine kinase selection cassette flanked by Frt sites with a single LoxP site was inserted between exons 6 and 7. The targeting construct was transfected into embryonic stem (ES) cells via electroporation and after selection with G418, homologous recombinants were screened by Southern blotting using both 3′ and 5′ probes outside the targeted region. Cells transfected with Flp recombinase were selected using FIAU to remove cells retaining the thymidine kinase and further screened by PCR using primers flanking the LoxP site or the LoxP/Frt remaining after recombination. Positive clones were blastocyst-injected to generate chimeric mice. The male chimeras were bred with C57BL/6 females to establish mice carrying the conditional TopBP1 allele. Further breeding with cre mice under the control of the Nestin promoter (JAX #003771: B6.Cg-Tg(Nes-cre)1Kln/J) resulted in deletion throughout the nervous system (http://cre.jax.org/Nes/Nes-Cre.html), while breeding with the Emx1-cre mice (JAX #005628: B6.129S2-Emx1tm1(cre)Krj/J) deleted TopBP1 in Emx1 expressing neural progenitors (http://cre.jax.org/Emx1/Emx1_cre.html). CreTM mice (JAX #004682: B6.Cg-Tg(CAG-cre/Esr1)5Amc/J) provided tamoxifen inducible cre expression driven from the actin promoter and were used to obtain primary cell cultures. Other mice used were p53−/−, Lig4LoxP/LoxP and Xrcc1LoxP/LoxP have been described previously39, 40, 48. The Atm conditional allele was engineered to have exon 58 flanked by LoxP sites47. All animal experiments were carried out according to NIH regulations and were approved by the SJCRH animal care and use committee.
Histology
Mice were perfused transcardially or embryos were drop-fixed in 4 % buffered paraformaldehyde at several different time points, and cryoprotected in buffered 25% sucrose solution. Brains and embryos were sectioned at 10 μm sagittally or coronally using a HM500M cryostat (Microm). Nissl staining was carried out with 1 % (w/v) thionin, and hematoxylin/eosin staining was done according to standard procedures. Immunohistochemical and immunocytochemical staining of tissues and cells were carried out with the antibodies listed below. For colorimetric visualization of positive signals, sections were incubated with antibodies overnight at room temperature after quenching endogenous peroxidase using 0.6 % H2O2 in methanol. Slides were washed with phosphate-buffered saline (PBS) three times, followed by incubation with biotinylated secondary antibody and avidin-biotin complex (Vectastain Elite kit, Vector Labs). Antibodies were applied after citrate buffer–based antigen retrieval. Immunoreactivity was visualized with the VIP substrate kit (Vector Labs) using the manufacturer’s protocol. After staining, sections were counterstained with 0.1 % methyl green, dehydrated, and mounted in DPX (Sigma). For immunofluorescence, FITC- or Cy3-conjugated secondary antibodies (Jackson Immunologicals) were used and counterstained with 4′6-diamidino-2-phenylindole (DAPI) or propidium iodide (Vector Laboratories). For immunohistochemistry and immunocytochemistry, we used the following antibodies at the indicated dilutions: anti-Calbindin (mouse, 1:2,000, Sigma), anti-PCNA (mouse, 1:500, Santa Cruz Biotechnology), anti-Pax2 (rabbit, 1:500, Zymed), anti-Ki67 (rabbit, 1:250, Vector Laboratories), anti-BrdU (Rat, 1:500, Oxford Biotechnology), anti-active Caspase-3 (rabbit, 1:100, BD Biosciences), anti-p53ser15 (rabbit, 1:100, Cell signaling; which recognizes mouse p53ser18), anti-pH2AXser139 (rabbit 1:500, Abcam), anti-pH3ser10 (rabbit, 1:500, Cell Signaling), anti-β Tubulin III (Tuj1, mouse, 1:500, BabCo), BLBP (brain lipid binding protein, rabbit, 1:300, Millipore), GFAP (Glial fibrillary acidic protein, mouse, 1:500, Sigma). Double immunohistochemistry of BLBP and γH2AX was differentially stained using donkey-FITC and goat-Cy3 secondary antibodies.
Layer markers for the cerebral cortex were: anti-Cux1 (CDP, rabbit, 1:100, Santa Cruz Biotechnology), anti-Brn2 (rabbit, 1:200, GeneTex), anti-Satb2 (mouse, 1:50, Abcam), anti-Tbr1 (rabbit, 1:100, Abcam), anti-Ctip2 (rat, 1:100, Abcam), anti-Tbr2 (rabbit, 1:200, Abcam), anti-Foxp1 (mouse, 1:50, Thermo Fisher Scientific), and anti-Foxp2 (rabbit, 1:50, Abnova). All antibodies except Cux1, Foxp1 and Foxp2 underwent the citrate buffer antigen-retrieval prior to the immunohistology procedure. Actin filaments in astrocytes were stained with phalloidin conjugated with Alexa Fluro 488 (Molecular Probes). For in vivo proliferation assays, newborn mice or pregnant females were intraperitoneal-injected with 5-bromo-2′-deoxyuridine (BrdU, 50μg/g body weight, Sigma-Aldrich). Embryos or brains were removed 2 h after BrdU injection and fixed in 4 % PBS-buffered paraformaldehyde. TUNEL analysis was performed using cryosections with the ApopTag® fluorescein In Situ Apoptosis Detection Kit (CHEMICON) according the manufacturer’s directions. Double-labeling of TUNEL with PCNA or BLBP was as described previously50.
Western blot analysis
Western blot analysis was done using astrocyte cell lines (genotypes as indicated in the main text and figures) at passage 1 or 2, or tissues (cortex, cerebellum, liver, thymus and spleen) from both P4/P5 control mice (TopBP1LoxP/+;Nes-cre) and conditional knockout mice (TopBP1Nes-cre). Protein extracts were prepared by using lysis buffer (50 mM Tris-HCl, 200 mM NaCl, 0.2 % NP-40, 1 % Tween-20, 1 mM NaF, 1 mM sodium vanadate, 50 mM β-glycerophosphate, 2 mM PMSF, and protease inhibitor cocktail (Roche)) and quantified using the Bradford assay (Bio-Rad). Proteins (50 μg per lane) were separated through a 4–12 % Bis-Tris SDS polyacrylamide gel (Invitrogen) and transferred onto nitrocellulose membrane (Bio-Rad). Blots were sequentially immunostained with mouse anti-TopBP1 antibody (rabbit, 1:1,000, Millipore/Chemicon) followed by horseradish peroxidase–conjugated secondary antibody (1:1,000, GE Healthcare) and detected using ECL Plus chemiluminescence (GE Healthcare). The blots were also probed with with anti-Nbs1 antibody (rabbit, 1:500, Cell Signaling), anti-Chk2 (mouse, 1:1000, Millipore), anti-p53ser15 (rabbit, 1:1,000 Bio-labs), anti-Chk1 (rabbit, 1:500, Cell Signaling), anti-pChk1ser317 (rabbit, 1:800, Bethyl) and processed as described above. Anti-actin (goat, 1:500, Santa Cruz Biotech) Western blots or Ponceau-stained transfer membrane were used as protein-loading controls.
Quantitative Real-Time PCR
To confirm deletion of the TopBP1 gene using cre lines (both Emx1- and Nestin-cre), genomic DNA was isolated at several different time points. For Exm1-Cre, the dorsal portion of the cerebral cortex was separated from the rest of brain. Similarly, the entire cerebral cortex was removed from the embryonic brain for Nestin-cre. Quantitative Real-time PCR using SYBR Green was performed according to the manufacturer’s guideline (iQ5 multicolor real-time PCR detection system, Bio-Rad). Following primer sets were used to measure the quantity of exon 4 (targeted exon) and exon 11 of the TopBP1 gene. The deletion efficiency was calculated as a ratio between exon 4 and exon 11. Each groups contained 2 to 4 embryos. The primers are; Exon 4 forward: 5′-GCGCCACCAGCAATGTG, Exon 4 reverse: 5′-TGTACAGGATACAGTTACGTCAGACATTA. Exon 11 forward: 5′-GCATGCTGGCAAGATTGTGT, Exon 11 reverse: 5′-CTTCCCAGCAGAATTGTTGCTGACTATGC. Real-time PCR for Xrcc1 and Lig4 were as described previously39, 40.
In situ cell counts
To determine cell cycle indices and apoptosis in TopBP1Nes-cre tissue during brain development, E13.5 embryos were subjected to quantitative analysis. Three embryos for each genotype were analyzed. Immunopositive signals for BrdU, PCNA, pH3ser10, TUNEL or active caspase-3 were measured within 1 or 0.27 mm2 from at least three representative sections of the neopallial cortex, cerebellum and ganglionic eminence per embryo. Similar quantification methods were applied to IR-induced apoptosis (TUNEL) in embryos collected 6 hs after 0.2 Gy IR at E11.5, E12.5 and E14.5. In this case, the ratio of TUNEL positive signals to propidium iodide (PI) positive neurons was calculated using ImageJ (1.46b, NIH), and analyzed using Prism v4 software (Graphpad)..
Isolation of primary astrocytes and embryonic fibroblasts
Primary astrocytes were prepared from P1-P2 mouse brains. Cortices were dissociated by passage through a 5 ml pipette and cells were suspended in Dulbecco’s modified Eagle’s medium and Ham’s nutrient mixture F-12 (1:1 DMEM/F12, Gibco-BRL) supplemented with 10 % fetal bovine serum (v/v), 100 U/ml penicillin, 100μg/ml streptomycin and 20 ng/mlepidermal growth factor (Millipore). Primary astrocytes were established in Primeria T-25 tissue culture flasks (Falcon) at 37° C in a humidified CO2-regulated (5 %) incubator. Mouse embryonic fibroblasts (MEFs) were isolated from E13.5 embryos and grown at 37° C in DMEM/10 % FCS supplemented with β-mercaptoethanol in a humidified CO2-regulated (5%) incubator.
For DNA replication analysis, astrocytes were cultured in the presence of 10 μm BrdU (Sigma). Five days later cells were trypsinize, resuspended in media containing BrdU and seeded onto 1 mm glass coverslips (Fisher) at ~20 % confluency. Twenty hours later, the BrdU-enriched media was removed and cells were incubated with pre-warmed media alone (0 min) or media containing 10 μm EdU (Invitrogen) for 15, 30 or 60 mins. Cells were then washed, fixed in 4 % PBS-buffered paraformaldehyde and permeabilized with 0.5 % PBS-buffered Triton X-100. Labeling of EdU was performed using the Click-It EdU Alexa 488-azide detection kit (Invitrogen) using the manufacturer’s suggested protocol. Cells were then treated with 2 N HCl for 30 mins at 37° C and then immunostained with Alexa 555-conjugated anti-BrdU antibody (Invitrogen, 1:500, clone MoBU-1). Cells were counterstained with 4′6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Cells counts were analyzed using a student t-test.
Comet Assays
Quiescent primary astrocytes were treated with either 14μM camptothecin for 60 mins at 37° C or γ irradiation (20Gy, Cs137). Following treatments, cells were incubated for various times in drug-free medium at 37° C to assess recovery from damage. Cells were trypsinized and suspended in pre-chilled PBS, mixed with an equal volume of 1.2 % low-melting point agarose (Invitrogen), maintained at 42° C and immediately layered onto frosted glass slides (Fisher) pre-coated with 0.6 % agarose and maintained in the dark at 4°C for all subsequent steps. Slides were immersed in pre-chilled lysis buffer (2.5 M NaCl, 10 mM Tris-HCl, 100 mM EDTA (pH 8.0), 1 % (v/v) Triton X-100, 3 % (v/v) DMSO, pH10) for 1h, washed with pre-chilled distilled water (twice for 10 min each) and placed into pre-chilled alkaline electrophoresis buffer (50 mM NaOH, 1 mM EDTA, 1 % DMSO) for 45 min. Electrophoresis was carried out at 95 mA for 25 min, followed by neutralization in 0.4 M Tris-HCl (pH 7.0). Comets were stained with SYBR Green (1:10,000 in PBS, Sigma) for 10 min. Experiments were performed in triplicate and a minimum of 100 comet tail moments were measured using the Comet Assay IV system (Perceptive Instruments) coupled to an Axioskop2-plus microscope (Zeiss) at 200x magnification.
For in vivo comet analysis, tissue from E13.5 embryonal dorsal telencephalon was triturated into a single-cell suspension in pre-chilled PBS-buffered 2 % glucose. Cells were embedded into LMP agarose and underwent alkaline comet analysis as outlined above. Samples were prepared and analyzed from every embryo collected and genotyping was done post-experimentally ensuring a blinded analysis of strand break levels. Cerebellar granule neurons were prepared from P6 WT mice via a percoll gradient as previously described43; neurons were irradiated on ice at the specified dose rate and immediately embedded into agarose for alkaline comet analysis. At least 300 comet tail moments were measured from each sample.
Statistical analysis
Cell counts were determined using a student t-test and were calculated using Prism V4 software (Graphpad); P<0.05 was considered significant. Box plots representing comet data were calculated using Deltagraph 6 (Redrock software) from tail moment data generated using the Comet Assay IV system (Perceptive Instruments).
Supplementary Material
Acknowledgments
We thank the Hartwell Center for biotech support, the Transgenic core facility for blastocyst injections and chimera production and the ARC for animal husbandry. PJM is supported by the NIH (NS-37956, CA-21765), the CCSG (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital. S.K. is a Neoma Boadway AP Endowed Fellow.
Footnotes
Competing financial Interests: The authors declare no competing financial interests.
Nine supplemental figures are associated with this manuscript.
References
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 4.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 5.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol. 2010;12:484–491. doi: 10.1038/ncb2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Kumagai A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sansam CL, et al. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makiniemi M, et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- 12.Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem. 1997;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- 13.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 15.Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, et al. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navadgi-Patil VM, Burgers PM. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 2009;8:996–1003. doi: 10.1016/j.dnarep.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeon Y, et al. TopBP1 deficiency causes an early embryonic lethality and induces cellular senescence in primary cells. The Journal of biological chemistry. 2011;286:5414–5422. doi: 10.1074/jbc.M110.189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 23.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 24.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 28.Greer DA, Besley BD, Kennedy KB, Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 2003;63:4829–4835. [PubMed] [Google Scholar]
- 29.Cotta-Ramusino C, et al. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung CC, Gong Z, Chen J, Glover JN. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. The Journal of biological chemistry. 2011;286:4292–4301. doi: 10.1074/jbc.M110.189555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murga M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruzankina Y, et al. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–1149. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gotz M, Huttner WB. The cell biology of neurogenesis. Nature reviews Molecular cell biology. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 35.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 36.Caviness VS, Jr, Nowakowski RS, Bhide PG. Neocortical neurogenesis: morphogenetic gradients beyond. Trends Neurosci. 2009;32:443–450. doi: 10.1016/j.tins.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou SJ, Perez-Garcia CG, Kroll TT, O’Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shull ER, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y, Chong MJ, McKinnon PJ. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J Neurosci. 2001;21:6687–6693. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katyal S, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. Embo J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arai Y, et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 46.Pulvers JN, Huttner WB. Brca1 is required for embryonic development of the mouse cerebral cortex to normal size by preventing apoptosis of early neural progenitors. Development. 2009;136:1859–1868. doi: 10.1242/dev.033498. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y, et al. ATR maintains select progenitors during nervous system development. Embo J. 2012;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 49.Lee Y, Barnes DE, Lindahl T, McKinnon PJ. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm. Genes Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci U S A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.