Abstract
The fatty acid desaturase genes (FADS1 and FADS2) code for enzymes required for synthesis of omega-3 and omega-6 long-chain polyunsaturated fatty acids (LCPUFA) important in the central nervous system, inflammatory response, and cardiovascular health. SNPs in these genes are associated with numerous health outcomes, but it is unclear how genetic variation affects enzyme function. Here, lymphoblasts obtained from Japanese participants in the International HapMap Project were evaluated for association of expression microarray results with SNPs in the FADS gene cluster. Six SNPs in the first intron of the FADS2 gene were associated with FADS1 expression. A 10-SNP haplotype in FADS2 (rs2727270 to rs2851682) present in 24% of the population was associated with lower expression of FADS1. A highly conserved region coinciding with the most significant SNPs contained predicted binding sites for SREBP and PPARγ. Lymphoblasts homozygous for either the major or minor haplotype were treated with agonists for these transcription factors and expression of FADS1 and FADS2 determined. Simvastatin and the LXR agonist GW3965 both upregulated expression of FADS1 and FADS2; no response was found for PPARγ agonist rosiglitazone. The minor haplotype homozygotes had 20–40% higher induction of FADS1 and FADS2 after simvastatin or GW3965 treatment. A 22 bp polymorphic insertion-deletion (INDEL) was found 137 bp downstream from the putative sterol response element, as well as a 3 or 1 bp INDEL 81–83 bp downstream. All carriers of the minor haplotype had deletions while all carriers of the major haplotype had insertions. Individuals carrying the minor haplotype may be vulnerable to alterations in diet that reduce LCPUFA intake, and especially responsive to statin or marine oil therapy.
Keywords: FADS2, FADS1, Insertion-deletion, Intron, LCPUFA, LXR, statin, SREBP-1c
Introduction
The delta-5 and delta-6 desaturases (encoded by FADS1 and FADS2, respectively) are essential for biosynthesis of long-chain omega-3 and omega-6 fatty acids. These long-chain polyunsaturated fatty acids (LCPUFA), such as docosahexaenoic acid (DHA) and arachidonic acid (ARA), are major components of cell membranes in the central nervous system, act as precursors for signaling molecules such as eicosanoids and docosanoids, and directly affect gene expression to influence important physiological functions such as inflammation and blood clotting [1–3]. Although both ARA and DHA can be obtained pre-formed through diet, most populations do not consume enough to completely remove any dependence on biosynthesis. Indeed, genetic studies have underscored the importance of FADS1 and FADS2 by linking single nucleotide polymorphisms (SNPs) in these genes with numerous health outcomes, including coronary artery disease, total cholesterol, LDL, C-reactive protein levels, allergy and atopic eczema, as well as cognitive outcomes such as attention-deficit hyperactivity disorder and IQ in children [4–9].
However, mechanisms describing how gene variants affect FADS gene function are lacking, so it is difficult to translate SNP associations into disease prevention or treatment. Despite numerous SNPs in FADS2 associated with health outcomes, there are no validated non-synonymous SNPs in the FADS2 gene. This suggests that mechanisms are likely to be regulatory rather than through protein structural changes for any causal SNPs in FADS2. A causal SNP in the promoter region of FADS2 has been reported [10], but this SNP is not polymorphic in the Han Chinese, Japanese, and Yoruba International HapMap project populations (HapMap Genome browser release #28). Thus, this one SNP does not explain all associations found in other world populations. In addition, this SNP only affects FADS2 expression, so it does not explain results in European or majority European-descendent populations suggesting lower 35-desaturase activity associated with some SNPs [11–13].
Some key molecules involved in regulation of FADS1 and FADS2 gene expression have been identified, but regulatory regions and functional binding sites within these genes have not been fully elucidated. The FADS genes reside as a cluster on chromosome 11 consisting of FADS1, FADS2 and a third putative desaturase designated FADS3. All reported mechanisms for regulation, such as dietary fatty acid or hormonal responses, affect both FADS1 and FADS2 in concert. For example, the end-product DHA is known to down-regulate expression of both FADS1 and FADS2 by lowering activity of the sterol response element binding protein 1c (SREBP-1c) [14]. Other transcription factors that have been implicated in regulation of FADS genes include the peroxisome proliferator-activated receptors (PPARs) [15]. However, no information is available to link specific SNPs and phenotypes with response to specific transcription factors.
Here, we have taken advantage of dense genotyping conducted for the International HapMap Project to achieve fine mapping of an expression quantitative trait locus (eQTL) in the FADS gene cluster. Because previous studies have focused mostly on European-derived populations, the Japanese in Tokyo (JPT) population was chosen for study here. The JPT population has a different linkage disequilibrium (LD) block structure from Europeans, and SNPs have different minor allele frequencies, so that results are likely to provide new information complementary to existing studies in Europeans. We evaluated gene expression in lymphoblast cell culture, which controls for environmental and hormonal influences that would otherwise reduce power to detect effects; this approach is ideal for a Japanese population, where exposure to pre-formed DHA from high seafood consumption in the traditional diet would otherwise confound any study of FADS gene expression. Single SNPs and haplotypes were evaluated for association with FADS gene expression, and the region of highest association was searched for putative transcription factor binding sites. Cells were treated with transcription factor agonists to test hypotheses, and a novel eQTL associated with SREBP-1c response was identified. Finally, sequencing of the region flanking the putative SREBP-1c binding site revealed two nearby insertion-deletion (INDEL) polymorphic variants specific to minor haplotype carriers.
Materials and Methods
Single SNP association analysis
SNP associations were carried out for SNPs in the FADS gene cluster using publicly available data from 46 International HapMap lymphoblast cell lines for the Japanese in Tokyo (JPT) population. Normalized expression data from the Illumina Sentrix Human-6 Expression BeadChip Microarray were obtained from the Gene Expression Omnibus (Series GSE6536) [16, 17]. Genotype information for lymphoblast cell lines was obtained from the Coriell Institute for Medical Research SNP Browser. All genotyped SNPs were used, excluding SNPs that are non-polymorphic in the JPT population. All 41 SNPs used in the analysis had minor allele frequencies > 1% and did not deviate from Hardy-Weinberg equilibrium. Single SNP associations and multiple test correction were carried out in PLINK v1.07 [18] using linear regression.
Haplotype association analysis
Phase was imputed and haplotypes identified in the Japanese and European (CEU) HapMap populations using Haploview v4.2 [19]. Linkage disequilibrium blocks were defined by the confidence interval-based algorithm of Gabriel et al. [20]. Identical parameters were used to calculate blocks in both populations. Haplotype associations were carried out in PLINK v1.07 by linear regression with an additive model, using the max(T) permutation procedure with 10,000 permutations to correct for multiple testing. Exclusion criteria were the same as for individual SNP associations, except that SNPs or individuals missing more than 10% of genotypes were excluded. Haplotype allele frequencies lower than 5% were also excluded from analysis.
Conserved regions and transcription factor binding predictions
Genomic sequence alignment calculating percent identity of multiple species compared with the human sequence was carried out with mVISTA <http://genome.lbl.gov/vista/mvista/about.shtml> [21]. Predicted transcription factor binding sites were identified by TRANSFAC Professional 9.2 database search combined with comparative sequence analysis, using rVISTA (regulatory VISTA, <http://genome.lbl.gov/vista/rvista/about.shtml> [22].
Lymphoblast cell culture, treatments, and RNA extraction
Immortalized B-lymphocyte (lymphoblast) cell lines from Japanese HapMap participants were obtained from the Coriell Institute for Medical Research, and used within 10 passages of receipt from the repository. Cells were grown in RPMI 1640 with 2 mM L-alanyl-glutamine (Sigma) and 15% fetal bovine serum (media and serum obtained from HyClone) in a humidified environment at 37°C with 5% CO2. All experiments were conducted on cells grown and treated in parallel in identical media and growth conditions. Cells were treated with 5 μM simvastatin (Sigma), 1 μM GW3965 (Sigma), or 20 μM rosiglitazone (Cayman Chemicals), or vehicle for 24 hours before collecting lysates. RNA was extracted using the RNeasy kit (Qiagen), and RNA quality was checked by agarose gel electrophoresis to verify RNA integrity and by 260/280 nm ratios on a NanoDrop 2000 (Thermo Scientific). cDNA was prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol.
Quantitative real-time PCR
Quantitative real-time PCR was carried out using SYBR Green Master Mix (Roche) on a LightCycler 480 instrument (Roche). PCR primers were obtained from Integrated DNA Technologies (sequences available upon request), except for 18S, which was obtained from Qiagen as a QuantiTect Primer Assay. PCR reaction efficiency was calculated from standard curves, and reactions were assessed by both melting curves and by running on agarose gels to verify reaction products and the absence of primer-dimers. Quantitative cycle (Cq) values were determined using LightCycler 480 SW1.5.0SP3 software, version 1.5.0.39 (Roche). Relative quantification was carried out using the Pfaffl method [23], taking into account reaction efficiency and using multiple reference genes for greater accuracy (β-actin, GAPDH, and 18S). Before proceeding with analysis, basal expression was shown to be invariant by vehicle-only treatment for all genes studied. Statistical significance of differences in fold changes between genotypes in response to cell treatments (normalized to vehicle treatment for each genotype) was assessed by the Mann-Whitney U test. Bootstrapping and randomization techniques were used in REST 2009 software (Qiagen) to calculate significance of fold changes normalizing both genotypes to basal expression in major allele homozygotes.
Sequencing of candidate sterol response element regions
A total of about 5 million cells from lymphoblast cultures were harvested for DNA extraction. DNA extraction was performed using DNeasy Blood & Tissue Kit (Qiagen). A 629 base pair portion (bases 6908015-6908643; GenBank Accession Number NT_167190.1) of FADS2 intron 1 and a 291 base pair portion of FADS1 intron 5 (bases 6882783-6882505; GenBank Accession Number NT_167190.1) flanking the sterol regulatory element (SRE) DNA sequence were amplified using the following primer pairs: FADS2 forward primer 5′ TTTCTCAAAGGCCGTGGTGT 3′, FADS2 reverse primer 5′ AGTGCTAACCACTCCTGGAA 3′ and FADS1 forward primer 5′ ACAGAGAAT GAAGGGACGCA 3′, FADS1 reverse primer 5′ ACCCGAAGGAGGCCATATCT 3′. The PCR reactions were carried out using GeneAmp High Fidelity PCR System (Invitrogen). Thermal cycling conditions were: initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 45 s and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products were separated on 2% agarose gels, and the DNA bands were gel eluted and purified using PureLink Quick Gel Extraction Kit (Invitrogen, USA). The purified products were sequenced at the Cornell University Life Sciences Core Laboratories Center.
Results
Identification of eQTL associated with FADS1 expression
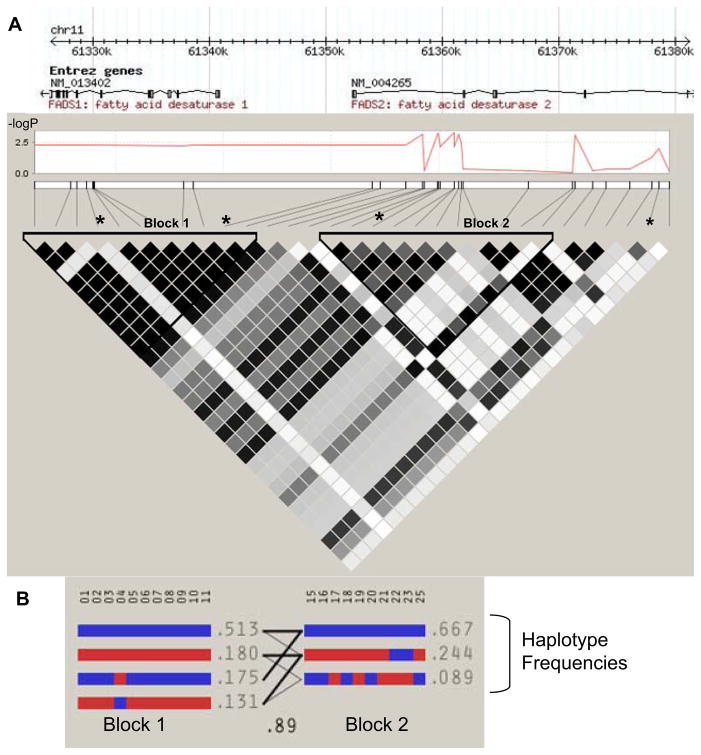
Lymphoblast cell cultures derived from all 46 Japanese participants in the International HapMap Project were used to search for genetic variants associated with FADS gene expression. Archived Illumina expression microarray data was obtained from the Gene Expression Omnibus database repository [16, 17]. Analysis was carried out on 41 SNPs covering the FADS gene cluster. The Manhattan plot shown in Figure 1A demonstrated a highly significant region located in FADS2 intron 1 that was associated with lower FADS1 expression for the minor allele. Within this region, six SNPs passed Bonferroni correction for multiple testing, as shown in Table 1. There were a few nominally significant SNPs associated with FADS2 or FADS3 expression, but none of these passed correction for multiple testing (not shown). Analysis of the linkage disequilibrium (LD) block structure for the Japanese HapMap population revealed two LD blocks in the region of interest, with the most highly significant SNPs primarily in the second block. Three haplotype alleles were observed within Block 2, as shown in Figure 1B. SNPs in this block were: rs2727270, rs2727271, rs174576, rs2524299, rs174577, rs2072114, rs174578, rs174579, rs174585, and rs2851682. The minor haplotype present in about one quarter of the population was significantly associated with lower FADS1 expression, as summarized in Table 2.
Figure 1. Expression quantitative trait locus in FADS2 intron 1 and associated haplotype blocks in the Japanese HapMap population.
(A) A Manhattan plot of -log(p-value) vs. distance in Kb is shown for the association of individual SNPs with FADS1 expression, with gene annotations above. Two LD blocks for this population are shown, with darkness of shaded cells representing the degree of correlation (R2) between pairs of SNPs. * Marked SNPs were associated with the ratio of arachidonic to linoleic acid (a measure of apparent total desaturase activity) in other Asian populations [11, 39]. (B) Haplotypes for each LD block are shown, with major alleles for each SNP in blue, and minor alleles in red. Recombination between blocks is depicted by the density of lines connecting individual haplotypes, and the multiallelic D′ (0.89).
Table 1.
Single SNPs in FADS2 intron 1 significantly associated with FADS1 expression.
| SNP | Base positiona | p-value | Betab | Bonferronic | FDRd |
|---|---|---|---|---|---|
| rs2845573 | 61358484 | 0.00073 | −0.21 | 0.0299 | 0.00597 |
| rs2727270 | 61359813 | 0.00073 | −0.21 | 0.0299 | 0.00597 |
| rs2727271 | 61359934 | 0.00065 | −0.21 | 0.0265 | 0.00597 |
| rs2524299 | 61361358 | 0.00053 | −0.23 | 0.0216 | 0.00597 |
| rs2072114 | 61361791 | 0.00073 | −0.21 | 0.0299 | 0.00597 |
| rs2851682 | 61372588 | 0.00091 | −0.21 | 0.0374 | 0.00623 |
Base positions from NCBI Build 36
Linear regression coefficient
Bonferroni adjusted p-value
False discovery rate (Benjamini-Hochberg method)
Table 2.
Haplotype testing for association with FADS1 expression.
| Haplotype | Frequency | Betaa | p-value | Empirical p-valueb |
|---|---|---|---|---|
| CACACATCGA | 0.67 | 0.090 | 0.061 | 0.15 |
| TTATAGACGG | 0.24 | −0.12 | 0.010 | 0.028 |
| CAAAAAATAA | 0.089 | 0.068 | 0.36 | 0.64 |
Linear regression coefficient
Corrected empirical p-value from max(T) permutation testing, controlling familywise error rate
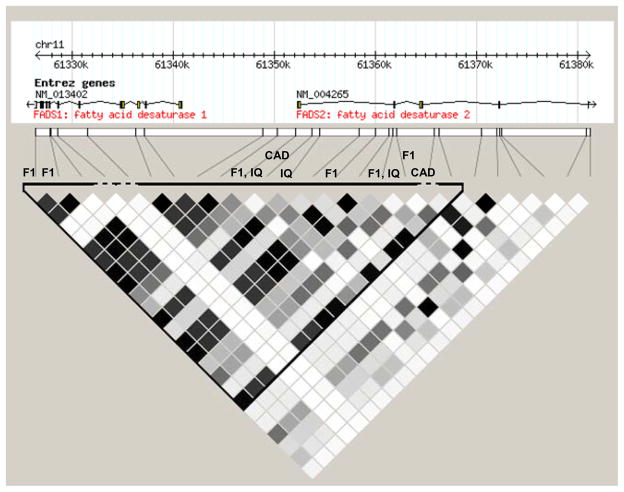
To estimate relevance of these results to studies in other populations, the LD block structure of the European (CEU) HapMap population is visualized in Figure 2. Previous large-scale studies, with associated SNPs marked in Figure 2, found a variety of outcomes associated with the first intron of the FADS2 gene in European-derived populations. In particular, fatty acid changes suggestive of lower FADS1 enzyme activity were found for SNPs in close proximity and in LD with the most highly significant region in the Japanese population.
Figure 2. Haplotype block in European CEU HapMap population.
FADS2 intron 1 is contained within a large single LD block in the European HapMap population. SNPs associated with apparent FADS1 activity are in close proximity and in LD with the region highly associated with FADS1 expression in the Japanese population. Abbreviations: F1 = SNPs associated with apparent FADS1 activity, inferred from fatty acid product/substrate ratios [11–13]; IQ = SNPs associated with IQ in breastfed children [9, 40]; CAD = SNPs associated with coronary artery disease and c-reactive protein levels [7].
Predicted transcription factor binding sites
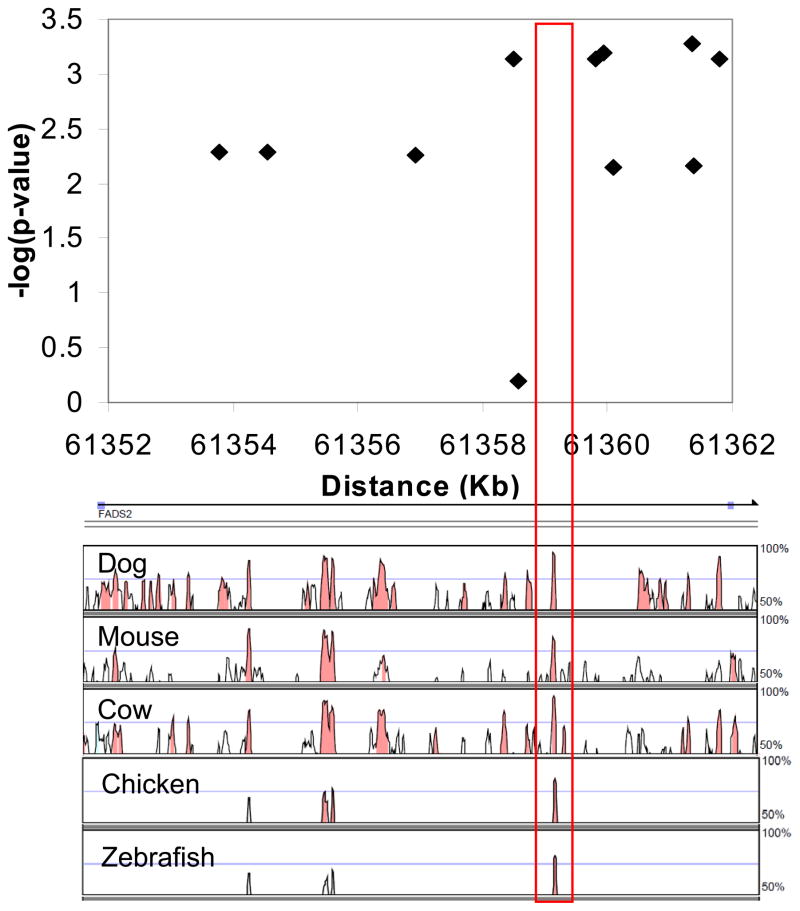
To investigate possible causal mechanisms for the lower FADS1 expression associated with this minor haplotype, the area immediately surrounding the most strongly associated region, and thus most likely to contain the actual causal locus, was examined for predicted regulatory elements. A region highly conserved from zebrafish to humans was identified that overlapped the area containing the most highly significant associated SNPs, as shown in Figure 3. TRANSFAC database search using the bioinformatics tool rVISTA revealed predicted consensus binding sites in the conserved region for two transcription factors: peroxisome proliferator-activated receptor gamma (PPARγ) and sterol response element binding protein (SREBP). We hypothesized that the significant SNPs identified above could be associated with a genetic variant within one of these binding sites, resulting in altered binding of a transcription factor responsible for regulating FADS1 expression.
Figure 3. Conserved region overlapping with significant SNPs in FADS2 intron 1.
Zoomed-in detail of FADS2 intron 1 in the Manhattan plot of -log(p-value) vs. base position depicts association of SNPs in the Japanese HapMap population with FADS1 expression, with conserved regions depicted below as percent identity with the human sequence. The red box outlines a region overlapping with the area of highest significance that is conserved from zebrafish to humans, and contains predicted binding sites for SREBP and PPARγ.
Lymphoblast FADS1 and FADS2 expression and transcription factor agonists
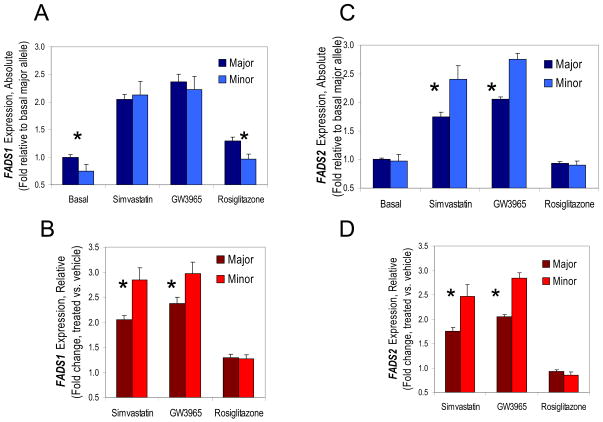
Follow-up experiments were carried out in Japanese HapMap lymphoblast cell lines homozygous for the minor haplotype associated with FADS1 expression, or homozygous for the major haplotype. All available minor haplotype homozygote cell lines were used (n = 5), as well as randomly chosen major allele homozygote cell lines (n =11). FADS1 expression was examined by quantitative real-time PCR, confirming the initial microarray results showing lower FADS1 expression for minor haplotype homozygotes (Figure 4A).
Figure 4. Basal FADS1 and FADS2 expression and drug response in lymphoblasts homozygous for major or minor haplotypes.
(A) FADS1 expression normalized to the major haplotype cells grown under basal conditions (ordinary growth media = 1). Under basal conditions, the minor haplotype homozygotes had significantly lower basal FADS1 expression than the major haplotype homozygotes, and this pattern persisted with rosiglitazone treatment. In contrast, simvastatin or GW3965 treatment upregulated expression in both genotypes such that they were no longer significantly different. (B) The same data as in (A) expressed as the change in FADS1 expression relative to drug treatment, normalized to vehicle treatment (not shown) for each genotype. Minor haplotype homozygotes confirm the findings in (A), showing significantly greater fold increase in FADS1 in response to simvastatin and GW3965 and no difference from major haplotype homozygotes in response to rosiglitazone. (C) FADS2 expression normalized to basal condition for the major haplotype is equivalent for the two haplotypes under basal conditions and for rosiglitazone. Simvastatin and GW3965 differentially upregulate haplotype FADS2 expression so that the minor haplotype is greater than the major haplotype. (D) The change in FADS2 expression relative to each genotype’s vehicle control is significant for simvastatin and GW3965. *p<0.05
To investigate the possibility of a causal genetic variation in a transcription factor binding site, cells were treated with drugs acting directly or indirectly on PPARγ or SREBP-1c. The PPARγ agonist rosiglitazone produced identical modest FADS1 upregulation responses in major and minor allele homozygotes, so the basal difference in FADS1 expression was maintained even after rosiglitazone treatment, as shown in Figure 4. Next, two drugs of different classes that alter SREBP-1c by distinct mechanisms were examined: the statin drug simvastatin, and the LXR agonist GW3965. Both treatments upregulated FADS1 by about two-fold in major haplotype homozygotes. Unexpectedly, minor haplotype homozygotes responded even more strongly to modulators of SREBP-1c; upregulation of FADS1 was 40% higher in response to simvastatin, and 25% higher in response to GW3965 for the minor haplotype (Figure 4B). These strong responses to SREBP-1c eliminated the difference in FADS1 expression observed in untreated cells, so that the two genotypes did not significantly differ in final FADS1 expression after treatment with simvastatin or GW3965 (Figure 4A).
Although no difference in basal FADS2 expression for each genotype was observed in the archived microarray data (not shown), both PPARγ and SREBP-1c have previously been implicated as regulators of FADS2 expression. Thus, FADS2 expression with and without the same drug treatments was evaluated. As shown in Figure 4C, the qRT-PCR results confirmed the microarray results for basal FADS2, with no difference between genotypes, and neither genotype showed any response to rosiglitazone treatment. As with FADS1, SREBP-1c modulators produced about a two-fold increase in FADS2 levels for major haplotype homozygotes, and minor haplotype homozygotes increased FADS2 expression even more in response to simvastatin or GW3965 (Figure 4D). Because there was no difference in basal FADS2 levels by genotype, final levels of FADS2 were at least 20% higher after statin or LXR agonist treatment in minor haplotype homozygotes compared to the major haplotype group (Figure 4C). The pattern of response for FADS1 and FADS2 is remarkably consistent (Figures 4B and 4D) and the differences in the pattern of total expression (Figures 4A and 4C) are due to differences in basal expression.
Sequence differences near the putative SREBP-1c binding site in FADS2 intron 1
Based on the difference in response to SREBP-1c, we hypothesized that a sequence variation between the major and minor haplotypes might exist within or in close proximity to a putative sterol response element (SRE) binding site for SREBP-1c. Analysis of FADS1 and FADS2 genes with the rVISTA program revealed two decamer SREs, one in FADS2 intron 1 (5′-ATCACCCCAC-3′), and another in FADS1 intron 5 (5′-ATCACGCCAC-3′). The sequencing of a 291 base pair (bp) fragment flanking the SRE DNA sequence of FADS1 showed no differences between the major and minor haplotypes. However, sequencing of a 629 bp fragment flanking the SRE DNA sequence of FADS2 showed two INDEL polymorphic variations exclusively in the minor haplotype. As shown in Table 3, a 22 bp homozygous deletion (rs66698963) was identified by sequencing in all five minor haplotype homozygotes within a minisatellite sequence located 137 bases downstream of the putative SRE. In addition, a 3 bp deletion (-CCA, rs138766446) located 81 bases downstream of the putative SRE was observed in all but one of the minor haplotype homozygotes, with the remaining one individual missing only one of the 3 bp (-A, rs149597144). None of these deletions were observed in any of the eleven major haplotype homozygotes.
Table 3.
INDEL Mutations near the putative SRE in FADS2 intron 1.
| ID number | Sequence | Distance to SRE (bp) | Minor haplotype |
|---|---|---|---|
| rs66698963 | -/ACTTCTCCCTGCCTCCCCAGGG - / CCA or - / A | 137 | Deletion |
| rs138766446 or rs149597144 | 81 or 83 | Deletion |
Discussion
Here, single SNPs and a haplotype in FADS2 intron 1 were found to be significantly associated with FADS1 expression, measured by both microarray and independently by qRT-PCR. The FADS2 gene is positioned immediately adjacent at a distance of 11.3 kb to FADS1 in a head-to-head orientation [24], so that FADS2 intron 1 is upstream of the transcription start site for FADS1. Thus, the existence of an important regulatory region for FADS1 transcription in intron 1 of the FADS2 gene is quite plausible. It has been shown that CYP1A1 and CYP1A2 genes that are adjacent to each other in head-to-head orientation on human chromosome 15 share common bidirectional regulatory regions [25]. The region most highly associated with FADS1 expression overlapped with a conserved region containing predicted binding sites for PPARγ and SREBP. In follow-up experiments, no difference was observed in response to a PPARγ agonist, rosiglitazone, but homozygotes for a minor haplotype were significantly more sensitive to expression regulation by SREBP-1c modulation. The enhanced response to SREBP-1c was consistently observed for drugs activating SREBP-1c by two different mechanisms: the statin simvastatin upregulates SREBP-1c levels as part of its pleiotropic effects [26], and the LXR agonist GW3965 stimulates the LXR/RXR heterodimer to activate SREBP-1c [27]. Both genotypes upregulated both FADS1 and FADS2 in response to the drugs, but homozygotes for the minor haplotype exhibited a significantly greater increase in expression of both genes after drug treatment. Minor haplotype homozygotes had final FADS1 levels equivalent to major haplotype homozygotes, and 20% higher FADS2 levels. The minor haplotype was thus associated with two paradoxical states: lower FADS1 expression in the basal state, and stronger upregulation of FADS1 and FADS2 in response to SREBP-1c. The results suggest that the minor haplotype is associated with enhanced response to SREBP-1c in a binding site with shared regulatory activity for both FADS1 and FADS2. Our hypothesized model for bidirectional regulation through this locus is shown in Figure 5. These findings do not explain the reason for lower FADS1 expression in the basal state, but it is possible that a mutation that enhances SREBP-1c binding may be mutually exclusive to binding of another transcription factor in a shared binding region. Sequencing of the candidate region in FADS2 led to the identification of two INDEL variants present only in the minor haplotype homozygotes, located 81 and 137 bases downstream of the putative SREBP-1c binding site. A recent study using DNase-Seq to map chromatin structure resulted in the estimate that 55% of eQTL SNPs cause regulatory changes by altering chromatin accessibility to transcription factors [28], suggesting a possible mechanism by which SREBP-1c binding could be modified by sequences near the binding site. Although the close proximity to the SRE is highly suggestive of a causal role, further in vitro experimentation is needed to determine whether these INDEL variants directly affect SREBP-1c binding and activity. Such a mechanism would be analogous to previous observations. For instance, an INDEL variant within intron 16 of angiotensin-converting enzyme (ACE) accounted for 47% of variance in serum ACE activity [29] and a 14 bp INDEL variant within intron 2 of NCX1 in an East European population was associated with increased risk for coronary artery disease [30].
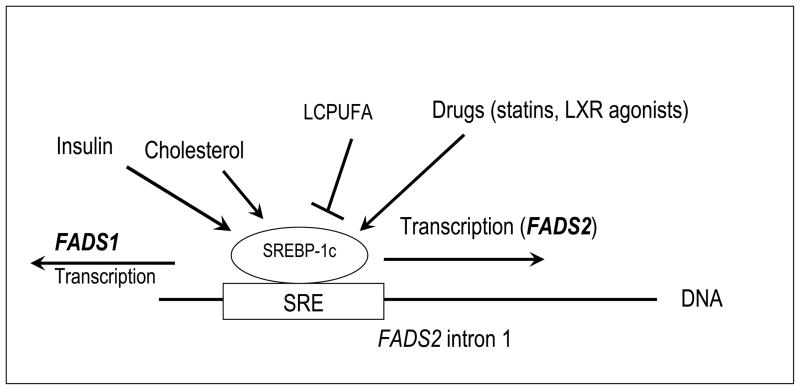
Figure 5. Hypothesized model for bidirectional regulation of FADS1 and FADS2 through a sterol response element in FADS2 intron 1.
Genetic variants near a putative SRE in the first intron of FADS2 are associated with altered response to SREBP-1c agonists. Known environmental and physiological factors modulating SREBP-1c activity are shown.
LCPUFA biosynthesis via FADS-encoded desaturase activity is redundant with direct dietary intake of preformed LCPUFA, specifically 20:4n-6, 20:5n-3 and 22:6n-3. Fish intake is recommended in part for its high content of LCPUFA. The traditional Japanese diet delivers on average more than one gram of LCPUFA daily through regular fish consumption [31]. At these levels, only minimal biosynthesis of LCPUFA from linoleic and linolenic acids is metabolically necessary at any life stage, and any differences in desaturase expression and consequent LCPUFA biosynthetic activity between major and minor haplotypes would be masked and presumably of little health consequence. In contrast, typical North American diets provide, on average, less than 300 mg LCPUFA total and thus biosynthesis may be much more important.
Our in vitro results on human lymphoblasts appear to be relevant to free living humans. Previous studies in Asian populations have linked SNPs in this region with serum fatty acid changes consistent with lower total desaturase activity (noted in Figure 1), suggesting that the genetic associations observed here can be replicated in human populations. These previous studies suggest that the usual state for individuals carrying these genetic variants is to have basal FADS1 levels so low that they become rate-limiting, reducing overall synthesis of fatty acid end-products. If this is the case, individuals with the minor haplotype may particularly benefit from diets incorporating higher levels of preformed LCPUFA from fatty fish or marine oil supplements to augment biosynthesis. Thus, although the minor haplotype is common (present in about one quarter of the Japanese HapMap population), any detrimental effects of the minor genotype are likely to be masked by the large amounts of fatty fish in the traditional Japanese diet. Importantly, the prevalence of this genetic variant may represent an additional risk to adopting a western diet for many individuals of Japanese descent. Put another way, individuals with the minor haplotype are predicted to be particularly vulnerable to ill-health when adopting diets that severely reduce preformed LCPUFA intake.
Further studies are needed to determine whether the expression results extend to other populations. Our data suggest the hypothesis that in populations with low LCPUFA intake, possibly due to low fish consumption, individuals with these genetic variants may have a conditional requirement for LCPUFA. At least some of these populations, such as Americans, derive cardiovascular benefits from statins. A number of previous studies have associated SNPs in FADS2 intron 1 with apparent FADS1 enzyme activity in large studies of European-derived populations (Figure 2), suggesting that the same causal locus may exist in Europeans as well as Japanese. Figure 2 also shows that the same region has been associated with IQ in breastfed children, but several studies in various European populations have produced conflicting and sometimes paradoxical results that were not accounted for by differences in fish consumption between populations [6, 9]. However, these studies did not take into account cholesterol, an activator of SREBP-1c [27] present in breastmilk but not in infant formula; population differences in maternal dietary fatty acids and phytosterols may affect breastmilk and infant plasma cholesterol and phytosterol levels [32–34]. In addition, individuals with the minor haplotype might have lower FADS1 expression in the absence of SREBP-1c activators (such as with infant formula, which does not contain cholesterol), but higher total FADS gene expression in the presence of cholesterol (breastmilk); a similar trend of significantly lower IQ in formula-fed, and almost-significant higher IQ in breastfed children, was observed in one IQ study for several SNPs in FADS2 intron 1 [9].
SNPs nearby and in LD with the SNPs most strongly associated with FADS1 expression here have also been associated with coronary artery disease in European studies (Figure 2). Thus, the finding of enhanced responsiveness to simvastatin for the minor haplotype may be of special interest. Statins have long been known to increase LCPUFA levels, and there is some previous evidence of changes in apparent desaturase activity in response to statin treatment [35, 36]. Although statins primarily are known for lowering cholesterol by inhibiting HMG-CoA reductase, they also have numerous pleiotropic effects that contribute to their medical benefits, including improved endothelial function and reduced inflammation and thrombosis. It has been theorized that statin pleiotropic effects may be primarily due to alterations in LCPUFA levels [36]. Interestingly, diets high in omega-3 LCPUFA were found to reduce overall and cardiac mortality by the same amount as statin treatment [37]. Here, we show that simvastatin upregulates both FADS1 and FADS2. The minor haplotype identified here was associated with an especially strong response to simvastatin. These results were obtained in B lymphocyte-derived cells, and may not be reproducible in all tissues. However, SREBP-1c is known to be a primary regulator of FADS1 and 2 expression [14], so differential responsiveness to SREBP-1c is likely to be important beyond lymphocytes. Moreover, B lymphocyte activity is important in the inflammatory component of atherosclerosis [38], which is likely to be modulated by desaturase expression through eicosanoids and docosanoids derived from LCPUFA. Assuming living humans have similar responses as observed in lymphoblasts, individuals with the minor haplotype may have lower basal FADS1 expression, leading to lower LCPUFA production. However, our results predict that minor haplotype carriers would have especially high LCPUFA synthesis after simvastatin treatment, and thus would especially benefit from statin pleiotropic effects attributed to LCPUFA production.
Here, we identify a minor haplotype associated with either lower, or higher, desaturase expression depending on levels of SREBP-1c or LXR agonists. Environmental factors undoubtedly interact with genotype to alter desaturase expression: for example, diet modulates SREBP-1c or LXR activity via natural ligands, or and sufficient pre-formed dietary LCPUFA minimizes the need for biosynthesis. Moreover, carriers of this minor haplotype who are candidates for statin treatment may especially benefit from these drugs, as well as possibly from future LXR agonist drugs currently under development. Future studies in large cohorts should incorporate screening of these polymorphisms to clarify their role in PUFA regulation.
Acknowledgments
This work was supported by the Cornell Division of Nutritional Sciences internal seed funding and the National Institutes of Health. HTR was supported by a Ruth L. Kirchstein-NRSA predoctoral training fellowship in reproductive sciences and genomics (Grant Number T32HD052471) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NICHD or the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schmitz G, Ecker J. The opposing effects of n-3 and n-6 fatty acids. Prog Lipid Res. 2008;47(2):147–55. doi: 10.1016/j.plipres.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Brenna JT, Diau GY. The influence of dietary docosahexaenoic acid and arachidonic acid on central nervous system polyunsaturated fatty acid composition. Prostaglandins Leukot Essent Fatty Acids. 2007;77(5–6):247–50. doi: 10.1016/j.plefa.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calder PC. Polyunsaturated fatty acids and inflammatory processes: New twists in an old tale. Biochimie. 2009;91(6):791–5. doi: 10.1016/j.biochi.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Brookes KJ, et al. Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2006;60(10):1053–61. doi: 10.1016/j.biopsych.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Schaeffer L, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15(11):1745–56. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 6.Caspi A, et al. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc Natl Acad Sci U S A. 2007;104(47):18860–5. doi: 10.1073/pnas.0704292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinelli N, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88(4):941–9. doi: 10.1093/ajcn/88.4.941. [DOI] [PubMed] [Google Scholar]
- 8.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steer CD, et al. FADS2 polymorphisms modify the effect of breastfeeding on child IQ. PLoS One. 2010;5(7):e11570. doi: 10.1371/journal.pone.0011570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lattka E, et al. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 2010;51(1):182–91. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merino DM, et al. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol Genet Metab. 2011;103(2):171–178. doi: 10.1016/j.ymgme.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Bokor S, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51(8):2325–33. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawar A, et al. The role of liver X receptor-alpha in the fatty acid regulation of hepatic gene expression. J Biol Chem. 2003;278(42):40736–43. doi: 10.1074/jbc.M307973200. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaka T, et al. Dual regulation of mouse Delta(5)- and Delta(6)-desaturase gene expression by SREBP-1 and PPARalpha. J Lipid Res. 2002;43(1):107–14. [PubMed] [Google Scholar]
- 16.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Frazer KA, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loots GG, et al. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;(5):832–9. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquardt A, et al. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66(2):175–83. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 25.Ueda R, et al. A common regulatory region functions bidirectionally in transcriptional activation of the human CYP1A1 and CYP1A2 genes. Mol Pharmacol. 2006;69(6):1924–30. doi: 10.1124/mol.105.021220. [DOI] [PubMed] [Google Scholar]
- 26.Rise P, et al. Delta5 desaturase mRNA levels are increased by simvastatin via SREBP-1 at early stages, not via PPARalpha, in THP-1 cells. Eur J Pharmacol. 2007;571(2–3):97–105. doi: 10.1016/j.ejphar.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14(22):2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482(7385):390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigat B, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kepp K, et al. Hypervariable intronic region in NCX1 is enriched in short insertion-deletion polymorphisms and showed association with cardiovascular traits. BMC Med Genet. 2010;11:15. doi: 10.1186/1471-2350-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poudel-Tandukar K, et al. Long chain n-3 fatty acids intake, fish consumption and suicide in a cohort of Japanese men and women--the Japan Public Health Center-based (JPHC) prospective study. J Affect Disord. 2011;129(1–3):282–8. doi: 10.1016/j.jad.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Hibberd CM, et al. Variation in the composition of breast milk during the first 5 weeks of lactation: implications for the feeding of preterm infants. Arch Dis Child. 1982;57(9):658–62. doi: 10.1136/adc.57.9.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellies MJ, et al. Effects of varying maternal dietary cholesterol and phytosterol in lactating women and their infants. Am J Clin Nutr. 1978;31(8):1347–54. doi: 10.1093/ajcn/31.8.1347. [DOI] [PubMed] [Google Scholar]
- 34.Potter JM, Nestel PJ. The effects of dietary fatty acids and cholesterol on the milk lipids of lactating women and the plasma cholesterol of breast-fed infants. Am J Clin Nutr. 1976;29(1):54–60. doi: 10.1093/ajcn/29.1.54. [DOI] [PubMed] [Google Scholar]
- 35.Harris JI, et al. Statin treatment alters serum n-3 and n-6 fatty acids in hypercholesterolemic patients. Prostaglandins Leukot Essent Fatty Acids. 2004;71(4):263–9. doi: 10.1016/j.plefa.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Jula A, et al. Effects of diet and simvastatin on fatty acid composition in hypercholesterolemic men: a randomized controlled trial. Arterioscler Thromb Vasc Biol. 2005;25(9):1952–9. doi: 10.1161/01.ATV.0000177812.84927.fa. [DOI] [PubMed] [Google Scholar]
- 37.Studer M, et al. Effect of different antilipidemic agents and diets on mortality: a systematic review. Arch Intern Med. 2005;165(7):725–30. doi: 10.1001/archinte.165.7.725. [DOI] [PubMed] [Google Scholar]
- 38.Major AS, Fazio S, Linton M. B-lymphocyte deficiency increases atherosclerosis in LDL receptor-null mice. Arterioscler Thromb Vasc Biol. 2002;22(11):1892–8. doi: 10.1161/01.atv.0000039169.47943.ee. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama K, et al. A single nucleotide polymorphism in the FADS1/FADS2 gene is associated with plasma lipid profiles in two genetically similar Asian ethnic groups with distinctive differences in lifestyle. Hum Genet. 2010;127(6):685–90. doi: 10.1007/s00439-010-0815-6. [DOI] [PubMed] [Google Scholar]
- 40.Morales E, et al. Genetic Variants of the FADS Gene Cluster and ELOVL Gene Family, Colostrums LC-PUFA Levels, Breastfeeding, and Child Cognition. PLoS One. 2011;6(2):e17181. doi: 10.1371/journal.pone.0017181. [DOI] [PMC free article] [PubMed] [Google Scholar]