1. Introduction
Biological systems are arguably the most complex ones studied by natural sciences. An average animal cell with the diameter of ~10 μm weighing ~1 ng contains >2 billion molecules of hundreds, if not thousands, different types of proteins, >80 billion molecules of various lipids, plus amino acids, nucleotides, metabolites, ions, etc. Thus, even if we don’t count ~20 trillion water molecules, the total number of biomolecules in an average cell exceeds the number of humans that ever lived since our species emerged. According to the laws of thermodynamics, the system of this complexity cannot be static. Indeed, each cell constantly receives and interprets hundreds of various stimuli, adjusting every aspect of its behaviour accordingly. Networks of signalling proteins that integrate inputs and coordinate responses govern these changes. Thus, in order to tell the cell what to do, we need to send our message via signalling proteins in a language the cell understands.
Elucidation of the fine molecular mechanisms of cell signalling is one of the greatest challenges of modern biology. The ability to produce expected outcome in a living cell by targeted manipulation of its signalling pathways is the ultimate test of our understanding of the mechanisms governing the cell behavior [1, 2]. Such ability may also be the ultimate therapeutic tool enabling us to restore normal behavior in cells where signalling is perturbed by a disease. We should not be embarrassed to acknowledge that we cannot build a living cell with desired functional characteristics from scratch: after all, this achievement took evolution more than a billion years of rigorous experimentation. Nor is it particularly necessary. However, if it were possible to reprogram a malfunctioning cell, thus restoring its normal behavior, that would certainly have a therapeutic value. Small molecule drugs are aiming at achieving just that but as tools they have inherent limitations, primarily because they are not a part of the cellular signalling network and thus are not responsive to regulatory feedbacks. In many cases, drugs offer relief but nor a cure. This is particularly obvious in case of neurological and psychiatric diseases, when even in the best case scenario a patient is maintained in a reasonably functional state by drugs, but that means taking the drugs for years without any hope of ever becoming disease-free. Another limitation of drugs is that not all protein functions involved in disease pathogenesis are amenable to regulation by small molecules. Receptors and enzymes are targeted by drugs quite successfully, whereas interfering with protein-protein interactions is much more complicated, particularly when the task is to enhance that interaction rather than disrupt it. Regulating by drugs of subcellular distribution, folding, or disposal of proteins involved in the disease process is also not easily accomplished.
An alternative to using small molecules to regulate cellular signalling is to employ signalling proteins as experimental and, ultimately, therapeutic tools. The simplest approach is to regulate the expression level of an endogenous signalling protein by overexpression of a wild type protein or by knockdown with some sort of an RNAi construct. Multiple attempts have been made to employ both these approaches for therapeutic purposes [3–6]. Recent and future advances in viral and non-viral delivery methods will make these techniques a viable clinical option for many diseases. However, manipulating an endogenous signalling protein simply by reducing or increasing its availability would inevitably affect all of its functions, which may not be always desirable or even safe in some cases. Targeted manipulation of specific functions of a multifunctional protein while preserving all other functions intact may be a preferred approach. To this end, mutant proteins with specific functions disabled or enhanced by precisely targeted mutations have to be employed. Furthermore, signalling can be redirected using novel scaffolding proteins assembled from existing domains, or new cellular functions could be created by expression of additional proteins in cells that do not normally express them. The problem is that these approaches require a much more extensive knowledge of structure-functional properties of signalling proteins involved in diseases. First, we need to know which function needs to be manipulated and how; second, we need to be able to construct mutants with desired properties to serve as tools. However, in recent years, a remarkable progress has been made in several such directions with therapeutic potential, largely using strategies previously validated by evolution. Here we overview some of these approaches, with particular focus on reengineering scaffolds to selectively suppress or enhance individual functions, using arrestins as an example of multi-functional organizers of cell signaling.
2. Modulating cell signalling by existing proteins and their elements
Ectopic expression of wild type proteins in sites other than their native location in order to compensate for the loss of function associated with the degeneration of cells that normally bear these proteins is an exciting approach with an enormous therapeutic potential. Recently, its feasibility has been proven experimentally, although admittedly there is still a long road ahead before this technique becomes a viable therapeutic choice. Mutations in dozens of human genes cause various forms of retinal degeneration, with a devastating result of complete blindness [7]. Recent successful gene therapy trials demonstrated that early intervention in case of some loss-of-function mutations can be successful. Three clinical trials attempted to cure Leber congenital amaurosis, which is caused by the deficiency of retinal pigment epithelium 65 (RPE65) [6, 8]. This protein performs a key step in the so called visual cycle, the conversion of all-trans-retinal released by light-activated rhodopsin into 11-cis-retinal necessary for rhodopsin regeneration [9, 10]. In its absence, rhodopsin cannot be regernerated, which leads to complete loss of rod function. The expression of fully functional RPE65 in retinas that lack this protein dramatically improved photoreceptor function and survival [6, 8]. However, the situation was considered hopeless after complete loss of photoreceptor cells. A recent study demonstrated that this is not necessarily the case [11]. Retina consists of multiple types of neurons, light-sensitive photoreceptors being the most prominent. Photoreceptor cells are the most vulnerable, dying off in retinitis pigmentosa and other types of retinal degenerations. The demise of photoreceptors results in blindness, but the other neurons remain in their place, although they undergo extensive rewiring [12]. Restoring vision in cases where photoreceptors are lost is an unmet challenge. Light-activated ion channels were expressed in non-light-sensitive ON bipolar cells in the retina of blind mice that lost photoreceptors due to retinal degeneration [11]. An exciting finding was that this expression conferred sufficient light sensitivity to allow these animals to successfully perform vision-guided behavioral tasks [11]. Although the animals only became sensitive to relatively bright light, this was a vast improvement. This study shows that the expression of an additional protein can generate a new functional modality, such as light sensitivity of bipolar cells, and this “unnatural” signal can be transmitted via existing circuits and successfully used by the brain to guide behaviour.
Colour blindness is another genetic disorder that was considered incurable. Recent experiments showed that the expression of a third type of cone pigment in photoreceptors of dichromatic adult monkeys successfully provided trichromatic color vision in these animals [13]. It is worth noting that here a particular cone opsin was expressed in the cells that never had it before. It apparently successfully used existing signalling machinery to confer the ability to discern light with specific wavelength to animals that were dichromatic from birth. In both above cases, a single additional protein was expressed in existing cells, and the brain was able to correctly interpret this new functional modality and successfully use additional information provided by it.
In many ways, the retina is unique, because it represents a sensory organ and a self-contained highly organized circuitry dedicated to the detection and analysis of the visual signals. This makes it easier for the brain to learn to correctly interpret retinal signals even when they come from the “wrong” cells, because they are partially made sense of at the retinal level due to the built-in properties of the circuit. Such circuit “reprogramming” is likely to be more difficult in other areas of the brain, although it is feasible in cases of well-defined circuit malfunctions. One such example comes from the field of Parkinson’s disease (PD). The classic model of PD pathophysiology posits that selective loss of dopaminergic neurons providing dopamine to the striatum leads to reduced activity of the direct and enhanced activity of the indirect output striatal pathway, resulting in a net increase of the inhibitory striatal output to the thalamus and excessive inhibition of the thalamo-cortical network [14, 15]. One of the main contributors to such an outcome is believed to be the elevated abnormal activity of the excitatory subthalamic nucleus. Numerous clinical data with deep brain stimulation of the nucleus support the notion that reduction in the activity of the subthalamic nucleus yields improvement in parkinsonian symptoms [16]. Recent report of a successful gene therapy trial [17] based on initial preclinical findings [18] demonstrated that by expressing glutamic acid decarboxylase, a rate-limiting enzyme for GABA synthesis, it was possible to partially convert subthalamic neurons from excitatory to inhibitory, thereby reducing the overall activity of the nucleus and ameliorating the disease symptoms. A conceptually similar approach was employed in study designed to restore the dopamine supply to the striatum in parkinsonian rats and monkeys by expressing enzymes requires for the dopamine synthesis, tyrosine hydroxylase, aromatic L-amino acid decarboxylase, and guanosine 5′-triphosphate cyclohydrolase, in striatal neurons [19, 20] that are GABAergic and do not possess these proteins. Considering that dopaminergic neurons of the substantia nigra that normally do the job degenerate in PD, this strategy to reprogram striatal neurons to supply dopamine for their own use seems sensible, and it successfully restored striatal dopamine and improved movement control in parkinsonian animals.
Most of the time, gene therapy is envisioned as a compensation for a disease-causing defect in the expression (too much or too little) or function (activating or inactivating mutations) of a signalling protein. In such cases, all that may be required is to supply a functional protein to appropriate cells or to adjust the level of expression. However, signalling proteins can be used to correct specific signalling deficits known to contribute to the disease pathogenesis even in cases when by themselves they are not defective. Such proteins could be used alone or in combination with drugs. G protein-coupled receptors (GPCRs) mediate cellular response to a wide variety of stimuli and are targeted by almost half of clinically used drugs [21]. However, the effectiveness of many drugs diminishes over time, necessitating increased doses to achieve therapeutic benefits, which often lead to debilitating side effects. For example, dopamine precursor L-DOPA routinely used to ameliorate the symptoms of Parkinson’s disease tends to lose its efficacy in the long term and almost inevitably produces dyskinesia, apparently associated with super-sensitivity of dopamine receptors [22, 23]. Most GPCRs are regulated by the phosphorylation of active receptors by G protein-coupled receptor kinases (GRKs) [24, 25] with subsequent arrestin binding to active phosphorylated receptor [26], which precludes further G protein activation [27] and redirects signalling to alternative pathways [28, 29]. Expression levels of GRKs and arrestins determine the strength of GPCR signalling [27] and are tightly regulated in cells [30, 31], which makes these two families of proteins potential tools for the regulation of GPCR signaling. Dopamine receptors belong to GPCR superfamily, and are regulated by GRKs and arrestins. It is conceivable that dopamine receptor supersensitivity associated with the loss of dopamine in the striatum in Parkinsonian patients can be ameliorated by an increased supply of GRKs, which would facilitate receptor desensitization and normalize signaling. Indeed, it has been recently shown that increased expression of GRK6 in the striatum reduces dopamine receptor sensitivity and manifestations of dyskinesia in the rat and monkey models of PD [32]. Interestingly, although there was a reduction in the expression of GRK6 and other GRK isoforms in the motor striatum in the rat model of PD [33], dyskinetic monkeys did not show any reduction in the concentration of GRK6 [34]. Nevertheless, overexpression of GRK6 significantly alleviated dyskinesia in both rat and monkey models [32]. Furthermore, over-expression of another protein, RGS9, that blunts signaling at the G protein level, also reduces L-DOPA-induced diskinesia in both rodent and primate models, although in both models the level of RGS9 expression was perfectly normal [35]. These data suggest that a signaling protein can be employed as a tool to achieve desired signaling outcome even when there is no obvious disease-associated deficit in its expression or function.
Proteins usually consist of multiple separately evolved domains that are independent folding units [36]. Individual protein domains that can be expressed separately and retain only one function have been shown to act in cells as dominant-negative constructs. For example, arrestin C-terminus is released upon receptor binding [37, 38] and becomes freely accessible in the arrestin-receptor complex. The C-termini of both non-visual arrestins contain short sequences that bind clathrin [39] and clathrin adaptor AP2 [40]. It has been shown that this C-terminal peptide expressed separately in the cell effectively competes with receptor-associated arrestins, suppressing clathrin-mediated GPCR internalization [41]. Another example is the C-terminal domain of GRK2, that mediates its recruitment to the plasma membrane by binding G protein βγ-subunits [42]. The expression of this element effectively scavenges free βγ-subunits in the cell, suppressing βγ-mediated signalling, as well as the recruitment of full-length GRK2 to GPCRs [43]. The association of excessive desensitization of beta-adrenoreceptors with congestive heart failure was discovered long ago [44]. Increased GRK2 expression in the heart exacerbates the problem (reviewed in [45, 46]). The expression of GRK2 C-terminus was shown to suppress GRK2-mediated desensitization of β-adrenergic receptors, ameliorate cardiac dysfunction [47], and prolong the survival in several animal models [45, 46]. Thus, dominant-negative action of separated protein elements is quite effective in living cells and whole animals. In some cases, the use of dominant negative constructs would serve as a functional equivalent of the protein knockdown. For example, preventing GRK2 from being recruited to GPCRs would inhibit GRK2-mediated receptor phosphorylation, which is the main (although not the only [24]) function of GRK2. Assuming there is no alternative way of targeting GRK2 to receptors, this would yield the result similar to that of the knockdown. However, in dealing with multifunctional proteins, which probably describes most existing proteins, dominant negative constructs may offer advantages over knockdown of precisely targeting the “offending” function of the protein while leaving the others unaffected. The dominant-negative arrestin is the case in point. Removing clathrin- and AP2 binding sites on the arrestin C-tail inhibited arrestin-mediated receptor internalization leaving arrestin-dependent receptor desensitization unaffected. These mutations are also unlikely to alter arrestin interaction with most non-receptor binding partners and interfere with arrestin-dependent signaling pathways, unless they strictly require receptor internalization, which is usually not the case. Therefore, such approach allowing for a reasonably precise inhibition of a defined function of a multifunctional signaling protein may be advantageous when such an inhibition is required for therapeutic purposes.
3. Creating new proteins by mixing and matching existing domains
In many cases, it would be advantageous to stimulate a particular signaling circuit in disease-affected cells using an external signal that these cells receive but that is not naturally coupled to the desired signal. The most obvious examples are neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, or retinal degenerations of different etiology, where any tool that can “tell” dying neurons to live longer would be of high therapeutic value. For example, connecting most common neurotransmitter inputs, such as glutamate, to a pro-survival pathway, such as ERK1/2 activation, can be envisioned as neuroprotective therapy. Indeed, non-specific protection of photoreceptor cells in the retina of genetically defective “retinal degeneration slow” mice by a mutant erythropoietin was shown to preserve photoreceptors [48], prolonging the time window during which more specific gene therapy corrections could be performed. In this particular case, the presence of erythropoietin receptors and endogenous signaling pathway in relevant cells were exploited, but this is not an obligatory requirement for this type of pro-survival therapy. Connecting an existing signal to an alternative pathway requires specifically engineered proteins with a combination of functions that is not represented in the proteome. One can envision a G protein or arrestin-based chimera that specifically interacts with the active form of one of the mGluRs (metabotropic glutamate receptors that belong to GPCR superfamily) and activates Ras or one of the Raf isoforms upon receptor binding, thereby connecting glutamate input and ERK1/2 activation via Raf-MEK-ERK pathway. Such protein could be expressed in vulnerable cells elevating the activity of the pro-survival ERK pathway, thereby preventing cell degeneration. Similarly, other generally pro-survival pathways such as Akt and NFκB could be targeted. Although this might sound like science fiction, successful engineering of chimeras combining functions that do not go together in existing proteins has been reported.
It is fairly well established that multiple-domain proteins consist of autonomously structured modules reshuffled by evolution and chained in various combinations [36, 49]. Thus, we can follow the example of nature and assemble novel proteins from existing domains. This idea looks particularly attractive in case of multi-domain signaling and scaffolding proteins, where individual structural elements bind distinct partners [50, 51]. Recent creative reengineering of Src-family kinase Hck provides a good example of the potential of this approach [51]. All Src-like kinases have SH2, SH3, and tyrosine kinase domains, where the first two regulate the activity and target the kinase to particular substrates. The replacement of SH2 and SH3 with a PDZ domain from syntropin redirected Hck activity towards novel substrates known to interact with this protein [51]. The introduction of the C-terminal PDZ ligand sequence into other proteins made them substrates of the engineered PDZ domain-containing kinase [51]. These data illustrate how known protein-protein interaction elements can be used to design customized cell signaling circuits [52].
The construction of 66 chimeras out of domains derived from 11 proteins in yeast mating pathway provided an even more striking demonstration of the potential of domain reshuffling [53]. Domain recombination yielded greater diversity in response dynamics than simple duplication of genes, and led to changes in mating phenotype. Interestingly, some recombinants demonstrated higher mating efficiency than wild type yeast, indicating how evolution creates and perfects signaling networks to generate new phenotypes [53]. Due to large number of mitogen-activated protein kinases (MAPKs), their direct activators MAPK kinases, and especially upstream kinases that activate the latter [54], MAPK pathways heavily rely on scaffolding proteins that bring appropriate partners together to create productive signaling complexes [55] and direct the flow of information within the cell [56]. Engineered scaffolds were successfully used to change the functional parameters of MAPK signaling pathways [57]. Building artificial signaling networks was also shown to be a powerful tool to elucidate how complex biological systems function (see [2] for a comprehensive review).
Collectively, these studies demonstrate that it is feasible to build novel signaling pathways in the cell by linking proteins sensing a particular input with the effectors of the pathway that needs to be turned on. All we need is a good understanding of the molecular mechanisms of protein-protein interactions involved. This approach has been used so far largely in research, but it clearly has high therapeutic potential.
4. Targeted reengineering of signalling proteins, one function at a time
Virtually every protein is multi-functional and interacts with various partners (a few examples can be found in [24, 29, 58, 59]). In many cases to correct disease-associated signaling imbalances, it is advantageous to enhance or suppress one of those functions, rather than all of them at the same time, as can be done by an increase or reduction of the expression of WT protein. This calls for the use of precisely engineered mutant forms of these proteins, which retain most of the capabilities of the WT form, but have individual functions exaggerated or subdued. Sometimes even in replacement gene therapy WT protein works less effectively than a mutant. For example, Royal College of Surgeons rats demonstrate retinal degeneration due to lack of protein tyrosine kinase MER (encoded by MERTK gene in humans), which results in the disruption of the retinal pigment epithelium phagocytosis pathway and leads to autosomal recessive retinitis pigmentosa, a form of retinal degeneration. Gene replacement therapy with AAV-delivered gene encoding WT MER kinase was found to be much less effective than when cDNA encoding a mutant “pre-activated” form of this enzyme was used [60].
Most proteins do not have a separable domain or a structural element responsible for each interaction. Arrestins are typical examples of this arrangement: they interact with hundreds of GPCRs and dozens of diverse non-receptor partners [26, 61], but consist of only two domains that are virtually inseparable in evolution [59]. Nonetheless, each partner binds to an identifiable site on the protein surface, which in many cases can be modified by mutations that do not dramatically affect other functions. Here we describe how this approach works using arrestins as a convenient model. Our ability to construct proteins in which individual functions are enhanced or suppressed by targeted mutations, leaving the rest of their repertoire unchanged, greatly expands the range and power of tools that can be used for research and therapy.
4.1. Severing links with internalization machinery
Arrestins are elongated soluble proteins consisting of the N- and C-domains, with the C-tail making strong contacts with the N-domain, which maintain the basal conformation of the molecule (Fig 1A) [62–65]. Clathrin-binding site (Fig. 1B) was the first properly identified interaction element in arrestins, and its localization in the C-tail was demonstrated at the time of the discovery of arrestin-clathrin interaction [39]. The interaction site of another element of internalization machinery, clathrin adaptor AP2, was also mapped to the arrestin C-tail (Fig. 1B)[40]. Arrestin with both sites simultaneously destroyed by mutagenesis was shown to act as dominant-negative, effectively suppressing GPCR internalization [66]. Obviously, to act in this manner the mutant has to retain normal receptor binding to be able to compete with endogenous arrestins. This was the first arrestin mutant with a single function selectively eliminated by targeted mutations. These mutants help to determine whether receptor internalization is required for a particular outcome in cells, although their therapeutic potential is unclear.
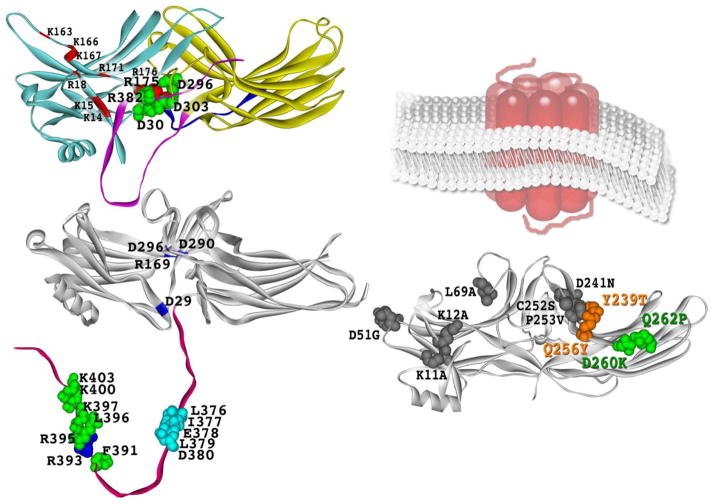
Fig. 1. Arrestin residues mediating the binding of GPCRs and trafficking proteins.
A. Crystal structure of arrestin-1 [62]. Phosphate-binding residues K14, K15, R18, K163, K166, K167, R171, R175, and K176 are indicated. Polar core residues D30, R175, D296, D303, and R382 are shown as CPK models. B. Crystal structure of arrestin-2 [63]. Polar core residues D29, R169, D290, D296, and R393 are highlighted. In the C-tail, polar core residue R393, key clathrin-binding residues L376, I377, E378, L379, D380, as well as AP2-binding residues F391, R395, L396, K397, K400, and K403 [66] are shown as CPK models. C. Crystal structure of arrestin-3 [120] showing the elements that determine its receptor specificity [105]. Schematic representation of membrane-imbedded GPCR is shown for comparison. The two lysines (K11, K12) substituted with alanines in receptor binding-deficient KNC mutant [85, 121] and mutations that change receptor preference of arrestin-3, D51G, L69A, Y239T, D241N, C252S, P253V, Q256Y, D260K, and Q262P [105], are shown as CPK models. Note that the combination of D260K and Q262P mutations greatly reduces arrestin binding to β2AR, but not M2 muscarinic or D1 and D2 dopamine receptors, increasing its selectivity for these receptors over β2AR >50-fold. The combination of Y239T and Q256Y mutations creates arrestin-3 with >5-fold preference for D1 over D2 dopamine receptor. The structures were rendered in ViewerPro; attached (panel A) and detached (panel B) C-tail that is not resolved in arrestin crystal structures was added in Adobe Photoshop.
4.2. Resetting the phosphate sensor
Mammals express hundreds of GPCR subtypes (http://sevens.cbrc.jp/), but only four different arrestins [59], two of which are largely restricted to photoreceptors [67]. This leaves the two non-visual subtypes, arrestin-2 and arrestin-31, to serve the great majority of GPCRs. Considering an amazing structural variety among GPCRs [68], it was not clear how just two arrestins can bind so many different receptors. The discovery of the mechanism of arrestin activation by receptor-attached phosphates [69] explained this mystery. All arrestins with known structure have five charged residues in the middle of the molecule, three aspartic acids and two arginines, that are essentially solvent-excluded and form a network of ionic bonds (Fig 1A) [62]. This arrangement, first discovered in visual arrestin-1 and termed the polar core [62], is rather unusual for a soluble protein, where charged residues are normally exposed to hydrophilic environment. Extensive mutagenesis demonstrated that the salt bridge between Arg175 and Asp296 in the polar core of arrestin-1 serves as the phosphate sensor: neutralization or reversal of either charge greatly increases the binding to unphosphorylated active rhodopsin [69–71]. This phenotype is consistent with the idea that the mutations turn the phosphate sensor “on”, thereby “tricking” the molecule into treating any active form of rhodopsin as phosphorylated [69]. Importantly, simultaneous reversal of both charges, which restores the salt bridge, also restores high arrestin-1 selectivity for active phosphorylated rhodopsin (P-Rh*) [69]. These data suggest that receptor-attached phosphates simply need to bind a positive charge and break the salt bridge, which signals to the rest of the molecule that the phosphates are in place, allowing its transition into high-affinity receptor-binding state [72]. This mechanism easily explains how very few arrestin subtypes can bind hundreds of structurally diverse GPCRs: arrestin activation only requires spatially concentrated negative charge on the receptor, making it insensitive to the sequence context of phosphorylated residues [67]. Indeed, crystal structures of other members of the arrestin family revealed the same polar core [63–65]. Moreover, its destabilization by charge reversal in non-visual arrestin-2 and -3 yielded phenotypically similar phosphorylation-independent mutants that bind with high affinity to any active form of their cognate receptors [73–77]. Destabilization of the interaction of the arrestin C-terminus with the body of the N-domain, which similarly loosens up the basal arrestin conformation [78] and promotes binding to unphosphorylated receptors [79] also yields mutants that can quench GPCR signalling regardless of receptor phosphorylation [73–75, 80].
The binding of “pre-activated” arrestin mutants to GPCRs that are not phosphorylated, either due to the absence of relevant GRK or because of the mutations eliminating GRK phosphorylation sites, was shown to be tight enough to block G protein activation in vitro [81] and in Xenopus oocytes [73, 74, 77]. These experimental data support the idea that phosphorylation-independent arrestins are viable tools to rein in excessive signaling by overactive GPCRs. The potential of this approach in vivo was recently tested in genetically modified mice lacking rhodopsin kinase (GRK1). The absence of GRK1 precludes normal rhodopsin shutoff, greatly prolonging light-induced signaling [82] and increasing the time of half-recovery from <0.4 sec in WT mouse to ~18 sec in rods expressing normal complement of WT arrestin-1 [83]. Morphologically, the absence of GRK1 leads to the dramatic shortening of the outer segment, the signaling compartment of rod photoreceptors, consequent reduction of light-induced response in rods, and results in progressive photoreceptor death in animals maintained in normal light-dark cycle [82, 83]. It was shown that transgenic expression of enhanced phosphorylation-independent mutant of arrestin-1 instead of WT protein in GRK1-deficient mouse rod photoreceptors alleviates the loss of rod outer segments, increases light-evoked responses, and improves rod survival [83]. Most importantly, enhanced arrestin-1 facilitates rod recovery after light stimuli, reducing the time of half-recovery to ~6 sec. These data suggest that enhanced mutant quenches the signaling by light-activated unphosphorylated rhodopsin much more efficiently than WT arrestin-1 [83]. These proof-of-principle experiments demonstrated the feasibility of compensational approach to gene therapy of gain-of-function mutations in GPCRs in vivo: enhanced phosphorylation-independent arrestin counteracted excessive signalling by unphosphorylated receptor, creating a more normal balance in the cells [83]. Yet the morphology and functional parameters of “compensated” rods did not improve to the level of WT, suggesting that further improvements in mutant design are necessary to achieve a better fit with unphosphorylated Rh* [83]. It should be noted, however, that rods are arguably the most perfect GPCR-driven signaling system, characterized by single photon sensitivity, virtually no noise, and incredibly rapid signal shutoff with sub-second kinetics [67]. All non-visual GPCRs have certain levels of constitutive activity (noise), and their shutoff usually takes minutes, rather than milliseconds. Thus, in other systems the requirements are not as strict, which would make it much easier for enhanced non-visual arrestins to compensate for excessive signaling by other GPCRs.
Considering that activating mutations in different GPCRs underlie a variety of congenital disorders [84], these results have clear therapeutic implications. Interestingly, the formation of the complex of phosphorylation-independent arrestin with unphosphorylated GPCR significantly affects receptor fate: it greatly facilitates recycling, protecting the receptor from down-regulation upon long-term agonist exposure [76]. Since excessive phosphorylation, desensitization and down-regulation of β2-adrenergic receptor significantly contributes to congestive heart failure [44, 45], arrestin mutants that bind unphosphorylated receptor likely have therapeutic potential in this situation.
Unexpectedly, it was shown that the role of receptor-attached phosphates varies widely, depending on a particular arrestin-GPCR combination [85]. Rhodopsin is an extreme case, where the phosphates are crucial for high-affinity arrestin binding in vitro [86] and in vivo [83, 87]. In case of β2-adrenergic receptor (β2AR), which is often used as a model non-visual GPCR [88, 89], phosphates appear to play an important, although less decisive role [85]. In case of M2 muscarinic and D2 dopamine receptors, the role of phosphorylation in arrestin binding [85] and signaling regulation appears to be minimal [90], although phosphorylation plays a role in receptor trafficking [91–93]. Nonetheless, the same activating arrestin mutations that give rise to its phosphorylation-independent binding to β2AR [76] facilitate the interactions with M2 muscarinic receptor [75].
Phosphorylation-independent enhanced versions of all vertebrate arrestins are already available. In case of rhodopsin-specific arrestin-1, the enhanced form was shown to be active in vivo and successfully quench signalling by unphosphorylated receptor [83]. However, the use of enhanced versions of non-visual arrestins in cells is hampered by their broad receptor specificity. Virtually every cell expresses multiple GPCR subtypes. In each congenital disorder only one of these receptors is a mutant that needs to be suppressed to a greater extent than wild type arrestins can achieve. Thus, practical use of phosphorylation-independent non-visual arrestins requires the introduction of additional mutations that will make them specific for particular GPCRs that must be targeted, to avoid unwanted effects on the signaling of perfectly normal other receptors expressed in the same cell.
4.3. Receptor-binding surface and GPCR specificity
Receptor-binding elements of all arrestins studied by several labs using different methods invariably map to the concave sides of the two arrestin domains (Fig 1C) [37, 38, 94–98]. Since both non-visual arrestins are fairly promiscuous, the residues responsible for receptor preference cannot be identified without using arrestin-1, which is naturally selective for rhodopsin and does not bind M2 muscarinic receptor very well [95, 99]. In contrast, arrestin-2 prefers M2 receptor to rhodopsin. The structures of arrestin-1 and -2 are very similar [63], so that all arrestin-1/2 chimeras fold and express normally. Therefore, a simple approach of exchanging various elements between these two arrestins was used. Parts of arrestin-1 that increased arrestin-2 binding to rhodopsin, and parts of arrestin-2 that improved arrestin-1 binding to M2 receptor were identified and further dissected [100]. Two elements encompassing residues 49–90 (β-strands V and VI with adjacent loops) in the N-domain and residues 237–268 (β-strands XV and XVI) in the C-domain of visual arrestin, and homologous elements in arrestin-2 were identified as key players in receptor preference by this approach. This study showed that the exchange of these two elements between arrestin-1 and -2 completely reversed receptor specificity of both [100].
Due to high homology within arrestin family, 35 residues in these two elements are different, only 22 of which are non-conservative substitutions [100]. Interestingly, the replacement of Val90 in arrestin-1, which is highly specific for P-Rh*, with a serine (Ser86 is in homologous position of promiscuous arrestin-2) increases arrestin-1 binding to active phosphorylated M2 muscarinic receptor more than any other point mutation [63, 100, 101]. Remarkably, the side chains of these residues playing crucial role in receptor specificity are not on the surface: both Val90 and Ser86 (as well as homologous Ala87 in arrestin-3 that, like arrestin-2, binds many different GPCRs [102, 103]) are actually buried between the two sheets of the β-strand sandwich of the N-domain [62, 63, 65]. In arrestin-1 Val90 interatcs with several other hydrophobic residues, apparently making the core of the N-domain more rigid [62, 63]. All these potential partners are present in arrestin-2 [63] and arrestin-3 [65], but a much smaller Ser86 and Ala87 cannot reach them. Thus, the simplest explanation of the data is that a bulky hydrophobic residue in this position is necessary to keep the N-domain rigid to ensure high receptor selectivity, whereas Ser or Ala in this position make it more flexible, helping non-visual arrestins to achieve a good fit with a wide variety of GPCRs [26]. The corollary of this conclusion is that any mutants of non-visual arrestins designed for increased receptor specificity must have Val (present in both less promiscuous subtypes, arrestin-1 and arestin-4 [64]) in this position.
Further dissection of the “receptor discriminator” elements by swapping exposed residues showed that the replacement of ten arrestin-2 residues with their arrestin-1 homologues creates an arrestin that binds P-Rh* as well as WT arrestin-1 [101]. Importantly, alanine substitution of these ten residues prevents the binding of arrestin-1 to rhodopsin in vitro, and of arrestin-2 and -3 to β2-adrenergic (β2AR), M2 muscarinic cholinergic (M2 mAChR), and D2 dopamine receptors (D2R) in intact cells [101]. Thus, the elements responsible for the receptor preference of arrestin proteins critically contribute to the energy of the interaction. This finding was subsequently confirmed with D1 dopamine receptor [85]. While these studies narrowed the field to just ten key players (Fig. 1C), if each position can be occupied by any of the 20 amino acids, the number of possible combinations would still be 2010, i.e., too large for experimental testing. However, the evolution comes to the rescue: the comparison of arrestin sequences in different species separated by hundreds of millions of years of independent evolution [59] shows that there were only 2–3 different residues in each of the key positions. If one assumes that only the residues found in each position in actually existing arrestins should be there, this brings the number of combinations to be tested down to manageable [104].
The first attempt to manipulate arrestin receptor-binding surface provided strong support for this approach [105]. First, Ala87Val mutation was introduced into arrestin-3, which appears to be even more promiscuous than the other non-visual subtype, arrestin-2 [102, 103], to create the base mutant with a more rigid N-domain. This mutation per se slightly decreased the binding to M2 mAChR and D2R, did not change the binding to D1 dopamine receptor (D1R), and slightly increased the interaction with β2AR [105]. Four and six exposed residues in the N- and C-domain, respectively, were previously identified as key receptor discriminators [85, 101]. Out of these ten, two positions in the N-domain and all six in the C-domain were mutated on Ala87Val background. The interaction of these mutants with β2AR, M2 mAChR, D1R, and D2R in cells was compared to that of the parental WT arrestin-3 and its A87V mutant, using BRET between luciferase-tagged receptors and Venus-tagged arrestins as a readout [106, 107]. Interestingly, none of the mutations appreciably increased arrestin-3 binding to any of the receptors tested. However, seven out of ten significantly reduced the interaction with some of the receptors, but not with others, changing the selectivity up to 4-fold [105]. This unexpectedly high ~70% success rate clearly shows that the key players in receptor specificity were identified correctly [101]. Importantly, the combination of two mutations that significantly reduced β2AR binding without affecting the interactions with M2 mAChR and D2R (D260K+Q262P; Fig. 1C) yielded an arrestin with ~50-fold preference for these receptors over β2AR [105]. Similarly, combination of two substitutions that reduced the binding to D2R, but not D1R (Y239T+Q256Y; Fig. 1C), generated an arrestin with >5-fold preference for D1R over D2R [105]. Thus, the effects of individual mutations appear to be additive, which paves the way to the construction of non-visual arrestins with high specificity for particular GPCRs. This promising research direction is still in its infancy, and a lot of additional work needs to be done to generate receptor-specific arrestins with high therapeutic potential.
In-cell analysis of the binding of non-visual arrestins with several GPCRs suggests that these interactions have two components: basal, agonist-independent, and agonist-induced, each accounting for about half of maximum observed binding [85]. Interestingly, the manipulation of the receptor-binding surface changed these two components in the same direction to a similar extent, which is reflected in a very good correlation between mutation-induced changes in both components [105]. Thus, a limited set of exposed residues mediates both basal and agonist-induced arrestin binding to GPCRs, and targeted mutagenesis of these elements is a feasible approach to generation of arrestin proteins specifically targeting individual receptor subtypes. Arrestin mutants that combine narrow receptor specificity with increased ability to desensitize GPCRs that cannot be phosphorylated or have excessive activity for other reasons are likely to be effective tools for normalizing GPCR signaling in conditions where its excess underlies the pathology.
4.4. Changing interactions with MAP kinases
Arrestins were reported to scaffold all three major MAP kinase (MAPK) cascades, facilitating the activation of ERK1/2 [108], JNK3 [109], and p38 [110]. The activity of these kinases sends very different messages to the cell that promote or suppress proliferation, differentiation, etc. Thus, directing arrestin-mediated signalling to a particular pathway can affect these important aspects of cell biology.
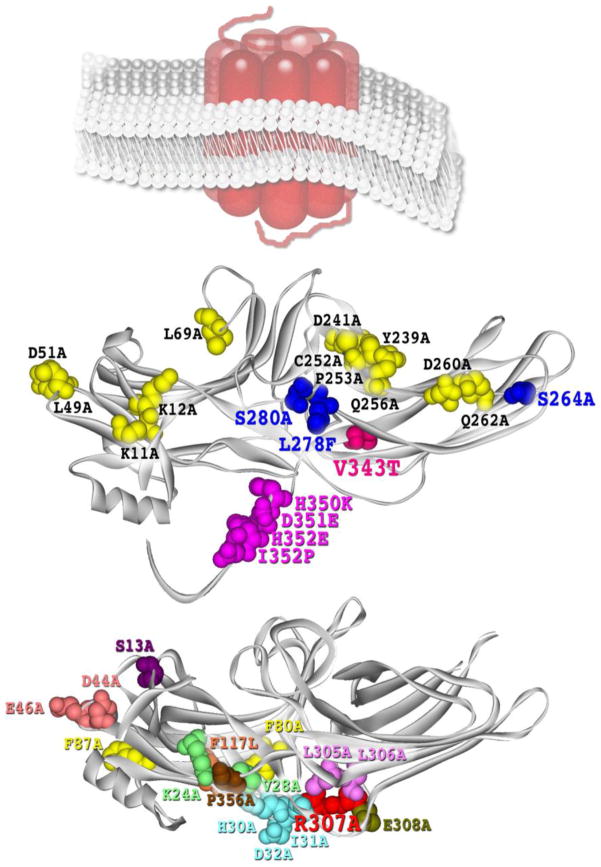
MAP kinase signalling in the cell was first shown to be differentially affected by the expression of different arrestin subtypes several years ago [111]. Over-expressed arrestin-1, -2, and -3 mobilized ERK1/2, but not upstream kinases c-Raf1 or MEK1, to the cytoskeleton, thereby reducing the level of ERK1/2 phosphorylation in the cell, whereas arrestin-4 did not produce this effect [111]. Subsequently, Asp26 and Asp29 were identified as critical MEK1-binding residues in arrestin-2, interacting with Arg47 and Arg49 in the N-terminus of MEK1 [112]. Alanine substitution of Asp26 and Asp29 generated a mutant with impaired MEK1 binding. The expression of this mutant, as well as the peptide mimicking MEK1 interaction site on arrestin-2, reduced the phosphorylation of arrestin-2 by ERK1/2 at the C-terminal Ser-412, thereby facilitating arrestin-2 interaction with clathrin and agonist-induced internalization of β2AR [112]. This is consistent with previous finding that arrestin-2 phosphorylation by ERK1/2 at Ser-412 inhibits clathrin binding and receptor internalization [113]. The next described signaling-biased arrestin-2-R307A mutant demonstrated normal ERK1/2 and MEK1 binding, but impaired ability to interact with c-Raf1, which resulted in its inability to promote ERK1/2 activation [114]. Importantly, out of 15 mutations that did not affect receptor binding (Fig. 2B), only R307A demonstrated an impaired c-Raf1 interaction, and none showed any appreciable changes in the binding to ERK1/2 or MEK1 [114]. These results show that arrestin functions can be modified individually by appropriately targeted mutations.
Fig. 2. Mutations affecting arrestin-mediated MAP kinase signaling.
A. Key players in JNK3 activation. Crystal structure of arrestin-3 [120], the only arrestin subtype that facilitates JNK3 phosphorylation [109, 117]. Substitutions with arrestin-2 residues on the non-receptor-binding side of the molecule, V343T, S264A+L278F+S280A, H350K+D351E+H352E+I353P, that impair the ability of arrestin-3 to promote JNK3 activation in cells are shown as CPK models [119]. The set of 12 alanine substitutions in arrestin-3-KNC mutant (K11A, K12A, L49A, D51A, R52A, L69A, Y239A, D241A, C252A, P253A, D260A, Q262A) that block its binding to GPCRs [101] and eliminate its ability to promote JNK3 activation without blocking its binding to ASK1, MKK4, and JNK3 [121] is also shown as CPK models. Schematic representation of membrane-imbedded GPCR is shown for comparison. B. c-Raf1 binding and ERK1/2 activation. Crystal structure of arrestin-2 [63]. Alanine substitutions on the non-receptor-binding side that do not affect arrestin-2 interactions with GPCRs, c-Raf1, MEK1, or ERK1/2 (F9A, S13A, K24A, V28A, H30A, I31A, D32A, D44A, E46A, F80A, F87A, F117A, L305A, L306A, E308A, and P356A), as well as R307A mutation that significantly reduces c-Raf1 binding and blocks arrestin-2 ability to promote ERK1/2 activation [114] are shown as CPK models. The structures were rendered in ViewerPro; the C-tail that is not resolved in arrestin crystal structures was added in Adobe Photoshop.
Even though all arrestins bind JNK3 [115, 116], arrestin-3 is the only subtype that facilitates its activation [109, 117, 118], which suggests that subtype-specific residues play a critical role. Further dissection of this function by site-directed mutagenesis replacing arrestin-3-specific residues with their arrestin-2 homologues generated multiple arrestin-3 mutants with perfectly normal ability to bind JNK3 and its upstream kinases ASK1 and MKK4, which nonetheless failed to facilitate JNK3 phosphorylation [119]. This study identified several residues on the convex side of the arrestin-3 C-domain, demonstrating that Val343 is the key contributor to JNK3 activation, whereas Leu278, Ser280, His350, Asp351, His352, and Ile353 play supporting roles (Fig. 2A). Interestingly, a single Val343Thr mutation reduced arrestin-3-dependent JNK3 activation ~3-fold, but the reverse Thr350Val mutation in arrestin-2 did not confer any appreciable ability to promote JNK3 phosphorylation [119], demonstrating that it is much easier to destroy individual arrestin functions than to build them. This study clearly demonstrated that the binding of ASK1, MKK4, and JNK3 and productive scaffolding of this cascade are independent functions, even though arrestins act as a simple scaffold by simultaneous binding of the kinases and bringing them together [120]. Collectively, these data suggest that to facilitate signaling in MAPK pathways, arrestins must not only bind appropriate kinases, but also hold them in an optimal orientation relative to each other. These findings suggest that an arrestin that binds the kinases but does not facilitate the signaling should be able to suppress MAPK activity in the cell. Indeed, recent discovery that arrestin-3-KNC (Fig. 2A) mutant binds ASK1 and MKK4 normally, and JNK3 even better than WT arrestin-3, but does not promote JNK3 phosphorylation, acts as a dominant-negative [121] supports this idea. Apparently, arrestin-3-KNC recruits MAPKs, keeping them away from productive scaffolds, including WT arrestin-3. Via this mechanism the mutant prevents JNK3 activation, which makes it a “silent scaffold”, a novel type of molecular tool for suppressing MAPK signaling [121]. This is an extreme case of elimination of one arrestin function while preserving the others, which leads to profound consequences in cellular environment.
All these examples involve disabling of a particular arrestin function. Naturally, to assemble a complete toolbox, we also need mutants with individual functions enhanced, rather than destroyed. This would provide much greater flexibility in using “biased” arrestins as tools for targeted manipulation of cell signalling. So far only receptor binding of arrestins was successfully enhanced by targeted mutations, so a lot of work still needs to be done to identify modifications enhancing other functions.
4.5. Modifying other interactions
Considering how many cellular proteins were reported to interact with non-visual [61] and visual [67] subtypes, it is remarkable how few arrestin elements responsible for individual interactions were identified. Microtubules [111] and calmodulin [122] engage parts of the receptor-binding surface, and therefore compete with GPCRs for arrestins [123]. Clathrin binding involves two elements in the C-tail [39, 124], and appears to be regulated by multiple intra-molecular interactions within arrestin-2 [125] and the phosphorylation of Ser-412 [113]. AP2 binding site is also localized in the C-tail [40, 66]. As the C-tail is released upon receptor binding (Fig. 1B) [37, 38, 126–128], GPCRs, receptor mimics, and “activating” mutations enhance arrestin interactions with these trafficking proteins [66, 129]. Most non-receptor partners of arrestins fall into three broad categories depending on their preference for a particular arrestin conformation. ERK1/2 and c-Raf1 prefer GPCR-associated arrestins [108, 130]. ERK1/2 is particularly selective, showing very transient interaction with free arrestins that can only be detected by immunoprecipitation after cross-linking [117]. E3 ubiquitin ligases Mdm2 [115] and parkin [131] preferentially bind arrestins in basal conformation, whereas MEK1 [130] and JNK3 [115] do not discriminate between the two. Interestingly, ERK1/2 and c-Raf1 also bind well arrestin-2 and -3 mutants with deletions in the inter-domain hinge [130] that are thought to freeze these proteins in basal-like conformation, impairing their ability to bind GPCRs [111, 121, 128]. The introduction of hinge deletions shifts arrestin conformational equilibrium towards basal state, whereas “activating” mutations apparently increase the flexibility [78], promoting arrestin transition into active-like state resembling the conformation of receptor-bound arrestin [72].
At the moment, the interactions with most partners could only be channelled by mutations that globally modify the conformational equilibrium, which change the binding of several signalling proteins simultaneously. This approach has an additional drawback: these mutations also significantly affect receptor binding. Thus, to enable more targeted manipulation of arrestin-mediated signalling via the majority of pathways, it is very important to identify key residues responsible for arrestin interactions with individual partners.
4.6. The potential of combinatorial approach
Spatial separation of receptor-binding elements, located on the concave side of the two arrestin domains, with the interaction sites for the majority of other signalling proteins localized on the surface that remains accessible in the arrestin-receptor complex, and the binding sites of trafficking proteins clathrin and AP2 on the arrestin C-tail (Fig. 1B) creates an opportunity to construct arrestins with different functions simultaneously modified by precisely targeted mutations. For example, a mutant that selectively binds certain receptors and does not promote the activation of ERK1/2 would block ERK1/2 signaling initiated by that particular receptor, but not others, which can still act via endogenous WT arrestins without competition from receptor-specific mutant. Similarly, receptor-specific arrestin that does not facilitate JNK3 activation would selectively block this branch of signalling by the receptors it targets, but not other GPCRs expressed in the same cell. In both cases independent modification of clathrin and/or AP2 sites on the C-tail would generate additional versions of these mutants that can or cannot internalize via coated pits. The combination of any of these traits with the ability to bind non-phosphorylated receptors would further increase the number of “flavours” in which a signalling-biased mutant can be produced. Ultimately, this approach would enable channelling the signalling from any desired GPCR subtype to the signalling pathway of choice.
5. Conclusions
Faulty signaling underlies a variety of congenital and acquired human disorders. The most striking examples are neurodegenerative diseases, such as Alzheimer’s and Parkinson’s, where neurons necessary for proper function simply die out, or the opposite situation in cancer, the hallmark of which is excessive proliferation of cells that escape from normal control by the signals from the rest of the body. Many other disorders, such as diabetes, heart failure, high or low blood pressure, etc., are caused by various signaling imbalances. Current therapies too often aim either at eliminating the most damaging consequences (e.g., surgery, radio- and chemotherapy in cancer) or at managing the symptoms (e.g., the use of antidepressants or antipsychotics), without even attempting to actually cure the disease by correcting the underlying problem. More creative approaches, which will likely include the use of gene therapy to deliver WT or reengineered proteins, have a better chance of getting the signaling back into balance, which is a pre-requisite for curing multiple disorders. This is particularly important for chronic conditions, where the patients remain on medication for years or even decades. Gene delivery techniques are improving at a rapid pace, and recent successful gene therapy trials [6, 8] confirm the feasibility of this approach.
Highlights.
Targeted manipulation of cell signaling pathways has high therapeutic potential
Changed expression of wild type proteins can change cell function
Reengineered signaling proteins change the flow of information in the cell
Novel proteins constructed from existing domains modify cell signaling
Acknowledgments
This work was supported by NIH grants NS065868, DA030103 (EVG), EY011500, GM077561, GM081756 (VVG), and Vanderbilt University Medical Center.
Footnotes
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons its gene is called “arrestin 3” in the HUGO database).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elowitz M, Lim WA. Nature. 2010;468:889–890. doi: 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Annu Rev Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo J, Bourre L, Soden DM, O’Sullivan GC, O’Driscoll C. Biotechnol Adv. 2011;29:402–417. doi: 10.1016/j.biotechadv.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.DeVincenzo JP. Antivir Ther. 2012;17(1 Pt B):213–225. doi: 10.3851/IMP2064. [DOI] [PubMed] [Google Scholar]
- 5.Kawase Y, Ladage D, Hajjar RJ. J Am Coll Cardiol. 2011;57:1169–1180. doi: 10.1016/j.jacc.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AJ, Bainbridge JW, Ali RR. Trends Genet. 2009;25:156–165. doi: 10.1016/j.tig.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Berger W, Kloeckener-Gruissem B, Neidhardt J. Prog Retin Eye Res. 2010;29:335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Roy K, Stein L, Kaushal S. Hum Gene Ther. 2010;21:915–927. doi: 10.1089/hum.2010.041. [DOI] [PubMed] [Google Scholar]
- 9.Kiser PD, Golczak M, Maeda A, Palczewski K. Biochim Biophys Acta. 2012;1821:137–151. doi: 10.1016/j.bbalip.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Travis GH, Golczak M, Moise AR, Palczewski K. Annu Rev Pharmacol Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagali PS, Balya D, Awatramani GB, Münch TA, Kim DS, Busskamp V, Cepko CL, Roska B. Nat Neurosci. 2008;11:667–675. doi: 10.1038/nn.2117. [DOI] [PubMed] [Google Scholar]
- 12.Jones BW, Marc RE. Exp Eye Res. 2005;81:123–137. doi: 10.1016/j.exer.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Mancuso K, Hauswirth WW, Li Q, Connor TB, Kuchenbecker JA, Mauck MC, Neitz J, Neitz M. Nature. 2009;461:784–787. doi: 10.1038/nature08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albin RL, Young AB, Penney JB. Trends Neurosci. 1989;12:366–375. doi: 10.1016/0166-2236(89)90074-x. [DOI] [PubMed] [Google Scholar]
- 15.DeLong MR. Trends Neurosci. 1990;13:281–285. doi: 10.1016/0166-2236(90)90110-v. [DOI] [PubMed] [Google Scholar]
- 16.Hamani C, Richter E, Schwalb JM, Lozano AM. Neurosurgery. 2005;56:1321–1324. doi: 10.1227/01.neu.0000159714.28232.c4. [DOI] [PubMed] [Google Scholar]
- 17.LeWitt PA, Rezai AR, Leehey MA, Ojemann SG, Flaherty AW, Eskandar EN, Kostyk SK, Thomas K, Sarkar A, Siddiqui MS, Tatter SB, Schwalb JM, Poston KL, Henderson JM, Kurlan RM, Richard IH, Van Meter L, Sapan CV, During MJ, Kaplitt MG, Feigin A. Lancet Neurol. 2011;10:309–319. doi: 10.1016/S1474-4422(11)70039-4. [DOI] [PubMed] [Google Scholar]
- 18.Luo J, Kaplitt MG, Fitzsimons HL, Zuzga DS, Liu Y, Oshinsky ML, During MJ. Science. 2002;298:425–429. doi: 10.1126/science.1074549. [DOI] [PubMed] [Google Scholar]
- 19.Azzouz M, Martin-Rendon E, Barber RD, Mitrophanous KA, Carter EE, Rohll JB, Kingsman SM, Kingsman AJ, Mazarakis ND. J Neurosci. 2002;22:10302–10312. doi: 10.1523/JNEUROSCI.22-23-10302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarraya B, Boulet S, Ralph GS, Jan C, Bonvento G, Azzouz M, Miskin JE, Shin M, Delzescaux T, Drouot X, Hérard AS, Day DM, Brouillet E, Kingsman SM, Hantraye P, Mitrophanous KA, Mazarakis ND, Palfi S. Sci Transl Med. 2009;14:2ra4. doi: 10.1126/scitranslmed.3000130. [DOI] [PubMed] [Google Scholar]
- 21.Rompler H, Staubert C, Thor D, Schulz A, Hofreiter M, Schoneberg T. Mol Interv. 2007;7:17–25. doi: 10.1124/mi.7.1.5. [DOI] [PubMed] [Google Scholar]
- 22.Jenner P. Nat Rev Neurosci. 2008;9:665–677. doi: 10.1038/nrn2471. [DOI] [PubMed] [Google Scholar]
- 23.Fisone G, Bezard E. Int Rev Neurobiol. 2011;98:95–122. doi: 10.1016/B978-0-12-381328-2.00004-3. [DOI] [PubMed] [Google Scholar]
- 24.Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mushegian A, Gurevich VV, Gurevich EV. PLoS One. 2012;7:e33806. doi: 10.1371/journal.pone.0033806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurevich VV, Gurevich EV. Pharm Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carman CV, Benovic JL. Curr Opin Neurobiol. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 28.Gurevich VV, Gurevich EV. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 29.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 30.Gurevich EV, Benovic JL, Gurevich VV. J Neurochem. 2004;91:1404–1416. doi: 10.1111/j.1471-4159.2004.02830.x. [DOI] [PubMed] [Google Scholar]
- 31.Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed MR, Berthet A, Bychkov E, Porras G, Li Q, Bioulac BH, Carl YT, Bloch B, Kook S, Aubert I, Dovero S, Doudnikoff E, Gurevich VV, Gurevich EV, Bezard E. Sci Transl Med. 2010;2:28ra28. doi: 10.1126/scitranslmed.3000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed MR, Bychkov E, Gurevich VV, Benovic JL, Gurevich EV. J Neurochem. 2008;104:1622–1636. doi: 10.1111/j.1471-4159.2007.05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezard E, Gross CE, Qin L, Gurevich VV, Benovic JL, Gurevich EV. Neurobiol Dis. 2005;18:323–335. doi: 10.1016/j.nbd.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Gold SJ, Hoang CV, Potts BW, Porras G, Pioli E, Kim KW, Nadjar A, Qin C, LaHoste GJ, Li Q, Bioulac BH, Waugh JL, Gurevich E, Neve RL, Bezard E. J Neurosci. 2007;27:14338–14348. doi: 10.1523/JNEUROSCI.4223-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell ID, Baron M. Philos Trans R Soc Lond B Biol Sci. 1991;332:165–170. doi: 10.1098/rstb.1991.0045. [DOI] [PubMed] [Google Scholar]
- 37.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Proc Natl Acad Sci U S A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vishnivetskiy SA, Francis DJ, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, Gurevich VV. J Mol Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Nature. 1996;383(6599):447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 40.Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson sSG, Caron MG, Barak LS. Proc Nat Acad Sci USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orsini MJ, Benovic JL. J Biol Chem. 1998;273:34616–34622. doi: 10.1074/jbc.273.51.34616. [DOI] [PubMed] [Google Scholar]
- 42.Koch WJ, Inglese J, Stone WC, Lefkowitz RJ. J Biol Chem. 1993;268:8256–8260. [PubMed] [Google Scholar]
- 43.Koch WJ, Hawes BE, Inglese J, Luttrell LM, Lefkowitz RJ. J Biol Chem. 1994;269:6193–6197. [PubMed] [Google Scholar]
- 44.Iaccarino G, Tomhave ED, Lefkowitz RJ, Koch WJ. Circulation. 1998;98:1783–1789. doi: 10.1161/01.cir.98.17.1783. [DOI] [PubMed] [Google Scholar]
- 45.Perrino C, Rockman HA. Curr Opin Cardiol. 2007;22:443–449. doi: 10.1097/HCO.0b013e3282294d72. [DOI] [PubMed] [Google Scholar]
- 46.Rengo G, Lymperopoulos A, Leosco D, Koch WJ. J Mol Cell Cardiol. 2011;50:785–927. doi: 10.1016/j.yjmcc.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akhter SA, Eckhart AD, Rockman HA, Shotwell K, Lefkowitz RJ, Koch WJ. Circulation. 1999;100:648–653. doi: 10.1161/01.cir.100.6.648. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan T, Kodali K, Rex TS. Neurochem Res. 2011;36:613–618. doi: 10.1007/s11064-010-0272-6. [DOI] [PubMed] [Google Scholar]
- 49.Sonnhammer EL, Kahn D. Protein Sci. 1994;3:482–492. doi: 10.1002/pro.5560030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 51.Yadav SS, Yeh BJ, Craddock BP, Lim WA, Miller WT. Biochemistry. 2009;48:10956–10962. doi: 10.1021/bi900978f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim WA. Nat Rev Mol Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peisajovich SG, Garbarino JE, Wei P, Lim WA. Science. 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson GL, Dohlman HG, Graves LM. Curr Opin Chem Biol. 2005;9:325–331. doi: 10.1016/j.cbpa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Dhanasekaran DN, Kashef K, Lee CM, Xu H, Reddy EP. Oncogene. 2007;26:3185–3202. doi: 10.1038/sj.onc.1210411. [DOI] [PubMed] [Google Scholar]
- 56.Good MC, Zalatan JG, Lim WA. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 58.Chatel G, Fahrenkrog B. Cell Signal. 2011;23:1555–1562. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 59.Gurevich EV, Gurevich VV. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng WT, Dinculescu A, Li Q, Boye SL, Li J, Gorbatyuk MS, Pang J, Chiodo VA, Matthes MT, Yasumura D, Liu L, Alkuraya FS, Zhang K, Vollrath D, Lavail MM, Hauswirth WW. Invest Ophthalmol Vis Sci. 2012;53:1895–1904. doi: 10.1167/iovs.11-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, Lefkowitz RJ. Proc Natl Acad Sci U S A. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. Cell. 1999;97(2):257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 63.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Structure. 2001;9(9):869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 64.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 65.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim YM, Benovic JL. J Biol Chem. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- 67.Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. Prog Retin Eye Res. 2011;30:405–430. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurevich VV, Gurevich EV. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 70.Gurevich VV, Benovic JL. J Biol Chem. 1995;270(11):6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 71.Gurevich VV, Benovic JL. Mol Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 72.Gurevich VV, Gurevich EV. TIPS. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 73.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. J Biol Chem. 2002;277(11):9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 74.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. J Biol Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 75.Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ. J Biol Chem. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- 76.Pan L, Gurevich EV, Gurevich VV. J Biol Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 77.Celver J, Lowe J, Kovoor A, Gurevich VV, Chavkin C. J Biol Chem. 2001;276:4894–4900. doi: 10.1074/jbc.M007437200. [DOI] [PubMed] [Google Scholar]
- 78.Carter JM, Gurevich VV, Prossnitz ER, Engen JR. J Mol Biol. 2005;351:865–878. doi: 10.1016/j.jmb.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 79.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. J Biol Chem. 2000;275(52):41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 80.Gurevich VV. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 81.Gray-Keller MP, Detwiler PB, Benovic JL, Gurevich VV. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 82.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Proc Nat Acad Sci USA. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schöneberg T, Schulz A, Biebermann H, Hermsdorf T, Römpler H, Sangkuhl K. Pharmacol Ther. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 85.Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. J Biol Chem. 2012;287 doi: 10.1074/jbc.M111.311803. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. J Biol Chem. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, Chen J. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 88.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GFWIW, Kobilka BK. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 89.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Namkung Y, Dipace C, Urizar E, Javitch JA, Sibley DR. J Biol Chem. 2009;284:34103–34115. doi: 10.1074/jbc.M109.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Namkung Y, Dipace C, Javitch JA, Sibley DR. J Biol Chem. 2009;284:15038–15051. doi: 10.1074/jbc.M900388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pals-Rylaarsdam R, Hosey MM. J Biol Chem. 1997;272(22):14152–14158. doi: 10.1074/jbc.272.22.14152. [DOI] [PubMed] [Google Scholar]
- 93.Lee KB, Ptasienski JA, Pals-Rylaarsdam R, Gurevich VV, Hosey MM. J Biol Chem. 2000;275:9284–9289. doi: 10.1074/jbc.275.13.9284. [DOI] [PubMed] [Google Scholar]
- 94.Ohguro H, Palczewski K, Walsh KA, Johnson RS. Protein Sci. 1994;3:2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 96.Hanson SM, Gurevich VV. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sommer ME, Farrens DL, McDowell JH, Weber LA, Smith WC. J Biol Chem. 2007;282:25560–25568. doi: 10.1074/jbc.M702155200. [DOI] [PubMed] [Google Scholar]
- 98.Kirchberg K, Kim TY, Möller M, Skegro D, Dasara Raju G, Granzin J, Büldt G, Schlesinger R, Alexiev U. Proc Natl Acad Sci U S A. 2011;108:18690–18695. doi: 10.1073/pnas.1015461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gurevich VV, Richardson RM, Kim CM, Hosey MM, Benovic JL. J Biol Chem. 1993;268(23):16879–16882. [PubMed] [Google Scholar]
- 100.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. J Biol Chem. 2004;279(2):1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 101.Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barak LS, Ferguson SS, Zhang J, Caron MG. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 103.Kohout TA, Lin FS, Perry SJ, Conner DA, Lefkowitz RJ. Proc Nat Acad Sci USA. 2001;98:1601–1606. doi: 10.1073/pnas.041608198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gurevich VV, Gurevich EV. Expert Rev Mol Med. 2010;12:e13. doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gimenez LE, Gurevich VV. J Biol Chem. 2012:287. doi: 10.1074/jbc.M112.366674. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walther C, Nagel S, Gimenez LE, Mörl K, Gurevich VV, Beck-Sickinger AG. J Biol Chem. 2010;285:41578–41590. doi: 10.1074/jbc.M110.162156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klewe IV, Nielsen SM, Tarpø L, Urizar E, Dipace C, Javitch JA, Gether U, Egebjerg J, Christensen KV. Neuropharmacology. 2008;54:1215–1222. doi: 10.1016/j.neuropharm.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Proc Nat Acad Sci USA. 2001;98(5):2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Science. 2000;290(5496):1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 110.Bruchas MR, Macey TA, Lowe JD, Chavkin C. J Biol Chem. 2006;281:18081–18089. doi: 10.1074/jbc.M513640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song S, Nair KS, Slepak VZ, Klug CS, Gurevich VV. J Mol Biol. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meng D, Lynch MJ, Huston E, Beyermann M, Eichhorst J, Adams DR, Klusmann E, Houslay MD, Baillie GS. J Biol Chem. 2009;284:11425–11435. doi: 10.1074/jbc.M806395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin FT, Krueger KM, Kendall HE, Daake Y, Fredericks ZL, Pitcher JA, Lefkowitz RJ. J Biol Chem. 1997;272:31051–31057. doi: 10.1074/jbc.272.49.31051. [DOI] [PubMed] [Google Scholar]
- 114.Coffa S, Breitman M, Spiller BW, Gurevich VV. Biochemistry. 2011;50:6951–6958. doi: 10.1021/bi200745k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Song X, Gurevich EV, Gurevich VV. J Neurochem. 2007;103:1053–1062. doi: 10.1111/j.1471-4159.2007.04842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song X, Coffa S, Fu H, Gurevich VV. J Biol Chem. 2009;284(1):685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, Lefkowitz RJ. Journal of Biological Chemistry. 2001;276(30):27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- 119.Seo J, Tsakem EL, Breitman M, Gurevich VV. J Biol Chem. 2011;286:27894–27901. doi: 10.1074/jbc.M111.260448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhan X, Kaoud TS, Dalby KN, Gurevich VV. Biochemistry. 2011;50:10520–10529. doi: 10.1021/bi201506g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, Gurevich VV. J Biol Chem. 2012;287 doi: 10.1074/jbc.M112.358192. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, Gurevich VV. J Mol Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, Benovic JL. J Biol Chem. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kern RC, Kang DS, Benovic JL. Biochemistry. 2009;48:7190–7200. doi: 10.1021/bi900369c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhuang T, Vishnivetskiy SA, Gurevich VV, Sanders CR. Biochemistry. 2010;(49):10473–10485. doi: 10.1021/bi101596g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. J Biol Chem. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 128.Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. J Biol Chem. 2002;277(46):43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 129.Nobles KN, Guan Z, Xiao K, Oas TG, Lefkowitz RJ. J Biol Chem. 2007;282:21370–21381. doi: 10.1074/jbc.M611483200. [DOI] [PubMed] [Google Scholar]
- 130.Coffa S, Breitman M, Hanson SM, Callaway K, Kook S, Dalby KN, Gurevich VV. PLoS One. 2011;6:e28723. doi: 10.1371/journal.pone.0028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Biochemistry. 2011;50:3749–3763. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]