Abstract
Mosquitoes are the major arthropod vectors of human diseases such as malaria and viral encephalitis. However, each mosquito species does not transmit every pathogen, due to reasons that include specific evolutionary histories, mosquito immune system structure, and ecology. Even a competent vector species for a pathogen displays a wide range of variation between individuals for pathogen susceptibility, and therefore efficiency of disease transmission. Understanding the molecular and genetic mechanisms that determine heterogeneities in transmission efficiency within a vector species could help elaborate new vector control strategies. This review discusses mechanisms of host-defense in Anopheles gambiae, and sources of genetic and ecological variation in the operation of these protective factors. Comparison is made between functional studies using Plasmodium or fungus, and we call attention to the limitations of generalizing gene phenotypes from experiments done in a single genetically simple colony.
INTRODUCTION
Serial blood-feeding behavior allows hematophagous arthropods to access a rich protein resource for egg production, but also exposes them to distinct microbes, thereby making them potential disease vectors. A combination of pre-existing genetic variation, pathogen selective pressure and fitness trade-offs have shaped the ways that vectors respond to their co-evolved pathogens by innate immunity and other mechanisms of host-defense, leading to vector-pathogen specificity. Even a competent vector species displays a wide range of infection phenotypes, including complete blockade of pathogen development in genetically resistant individuals [1]. It is difficult to measure fitness trade-offs in nature, and fitness measures made in the laboratory probably have little relevance for nature, but the transmitted pathogens are also under constraints to mitigate the effects of pathogenesis in order to maximize their own transmission. A pathogen that makes the vector too sick to fly and transmit has a reproductive fitness of zero, and pathogens that impose high fitness cost may also promote the evolution and spread of specific pathogen resistance in the host population. Identifying the molecular components of natural resistance, and understanding how they are genetically structured in wild A. gambiae populations, could help to improve applied strategies for surveillance and control of malaria transmission.
Genetics of Plasmodium resistance
A field-based genetic mapping survey of the wild population revealed that at least 40% of the A. gambiae s.s. tested carried a significant locus controlling susceptibility to natural P. falciparum, either for parasite numbers (P. falciparum intensity loci, Pfin) or melanization (P. falciparum melanization, Pfmel). About half of those loci (~20% of mosquitoes tested) are strong-effect resistance factors that can completely protect mosquitoes from infection [2,3]. This latter genetically resistant fraction of the A. gambiae population therefore probably does not transmit malaria, highlighting an important heterogeneity within the malaria transmission system that could potentially be manipulated for purposes of malaria control. A significant cluster of independently mapped resistance loci on chromosome 2L was termed the Plasmodium Resistance Island (PRI) in the A. gambiae genome [3]. This same genomic region controlling P. falciparum development was also mapped in East African A. gambiae [4,5], suggesting that the genetic mechanisms of malaria resistance in A. gambiae are widespread and shared throughout sub-Saharan Africa, rather than a local geographic adaptation.
The genes and the specific mutations underlying the QTLs mapped in nature have not yet been identified. Efforts are underway to identify nucleotide variation association with resistance to P. falciparum infections [6,7], however confirmation of causative mutations associated with the outcome of natural infection will involve large, carefully designed studies that control for population substructure and, as far as possible, identify effects that are clearly independent of genetic background. Nevertheless, candidate gene studies within a region genetically identified by QTL or association can be profitable, even if experimentally demonstrating the phenotype of a gene in the interval is, of course, not evidence that it is the causative agent. For example, in the PRI locus, candidate gene filtering by bioinformatics and function highlighted a paralogous family of genes, APL1A, APL1B and APL1C, encoding leucine-rich repeat (LRR) proteins that share ≥50% peptide identity with each other [8] and that demonstrate function in the laboratory, discussed further below.
Genetic mapping in wild pedigrees indicated that variants with significant effect on parasite development are collectively frequent in the field population [3], but the allelic spectrum of the variants is not known. The observed resistance loci could be due to either a few individually frequent variants of strong effect, or conversely due to many distinct, individually rare variants with functionally equivalent effects. As a model, the APL1A and TEP1 genes display high genetic diversity in the natural population [9,10]. If polymorphism of these genes has phenotypic consequences for malaria resistance in nature (which is not yet known), there could be many individually rare causative mutations in the genes. This is reminiscent of the case of human hemoglobinopathies such as sickle cell anemia and the thalassemias, which result from a large number of independent and individually rare mutations in different genes, and overall yield a similar phenotype of protection against severe malaria [11,12].
LRR regulation and protective function
In A. gambiae, orthologs and homologs of intracellular Drosophila factors belonging to the Toll and Imd innate immune pathways have been identified [13]. At the terminus of the A. gambiae Toll pathway, the NF-κB transcription factor Rel1 (also known as Gambif1) serves as the functional analogue of Drosophila Dif transcription factor, while for the Imd pathway, Rel2 is orthologous to the Drosophila transcription factor Relish [14]. The A. gambiae Nf-KB Rel2 transcript has further been shown to produce, through alternative mRNA splicing, a full-length (Rel2-F) and a shorter (Rel2-S) protein isoform [15]. Both isoforms are involved in defense against Gram-positive and Gram-negative bacteria.
An important part of anti-P. falciparum protection in A. gambiae was shown to be mediated by the Imd/Rel2-S pathway, while reduction of P. berghei parasites is largely based on Toll/Rel1 signaling [8,15–17]. These are probably not the only protective mechanisms, because after inactivating the relevant pathway for the parasite, the infection distribution shifts towards higher oocyst prevalence and/or intensity, but nevertheless there are still uninfected and/or low-infected mosquitoes, suggesting the operation of other lines of host defense. Functional analysis showed that, of the APL1 paralogs, only paralog APL1A protected A. gambiae against infection with P. falciparum, whether from wild parasites or in vitro gametocyte culture, while only paralog APL1C protected against the rodent malaria parasites, P. berghei and P. yoelii [8,16]. Furthermore, protection against P. falciparum correlates with the transcriptional regulation of APL1A by the isoform Rel2-S but not Rel-2F [16], whereas protection against P. berghei correlates with the transcriptional regulation of APL1C by the Toll/Rel1 pathway [8]. The APL1C protein was shown to form a heterodimer with another LRR protein, LRIM1, as part of a complex with the complement-like protein TEP1 [18–20]. LRIM1 displayed protective activity against the rodent parasites P. berghei and P. yoelii [21,22] and is also physically located in the PRI locus. Indeed, the PRI locus holds the largest genomic cluster of LRR genes in A. gambiae [3].
Because A. gambiae is not a natural vector of rodent malaria and there is no shared evolutionary history that could have selected specific resistance [23], the host-defense activity encoded in the PRI locus by at least APL1C and LRIM1 probably also protects mosquitoes against other yet unknown natural pathogens, which would have been the actual selective agents. The criteria of specificity are not yet known for APL1C- and LRIM1-mediated protection against the presumptive natural pathogen targets. Taken together, these data indicate quite fine pathogen specificity, where the host can discriminate between different members of the genus Plasmodium via APL1 paralogs A and C, with each protein required for protection against a distinct class of malaria parasite, and without detectable reciprocal effect upon the other class. These observations together suggest the operation of a gene-for-pathogen-class protection system in A. gambiae, where the functions of LRR proteins are required for fine orientation of host-defense towards distinct classes of pathogens that include but are probably not limited to Plasmodium [16]. The pathogen classes were termed A-class pathogens, protected against by APL1A (currently P. falciparum), and C-class pathogens, protected against by APL1C (currently the rodent malaria parasite species P. berghei and P. yoelii).
Immune specificity
The basis of specificity of such pathogen classes is unknown, nor the identities of other pathogens that might be similarly recognized within a class. The APL1 paralog system distinguishes two kinds of eukaryotic pathogen that otherwise share broadly similar cellular morphology and pathogenic behavior, which would seem to require finer power of discrimination than, for example, the known ability of insects to differentiate between broad groups such as Gram-negative bacteria, Gram-positive bacteria, and fungus [24]. In the rodent malaria model, parasite killing is a collaboration between an LRR dimer that appears to confer target specificity, and a putative effector subunit, TEP1, which in the absence of the LRR dimer binds non-specifically to tissues of self [18]. There are numerous other LRR and TEP genes in the A. gambiae genome, some of which interact in protein complexes to yield protective function [25,26]. There are also important protective molecules that are not yet integrated with the LRR-TEP protein complex model, for example AgDscam and fibrinogen-domain proteins [27,28]. Overall, the observations to date suggest the capacity for considerable combinatorial complexity of immune specificity, and emphasize how little is currently known.
The meaning of the term specificity also needs to be interrogated in the context of Plasmodium protection. The bacterial elicitors that are recognized as pathogen-associated molecular patterns (PAMPs, [29]), and corresponding host pattern-recognition receptors, are reasonably well known, less so for fungi [30], but are largely unknown for protozoa [31]. Indeed, there is no current direct evidence that host-defense against Plasmodium is PAMP-based. It was recently reported that rodent malaria ookinetes, during their passage through the midgut epithelium in a storm of reactive oxygen and cell damage, are surface-labeled by protein nitration, and that the nitration is an important determinant of subsequent binding of at least TEP1 and then ookinete death [32]. It was proposed that a proportion of ookinetes stochastically escape surface nitration, and that this unlabeled fraction therefore evades TEP1 binding and can develop unhindered. The work was done with rodent malaria, and validity for P. falciparum was not reported. However, the findings raise the interesting possibility that nitration could serve as a danger- or damage-associated molecular pattern (DAMP, [33]), thus potentially simplifying the immune challenge of recognizing various parasite PAMPs to, instead, a simpler requirement to merely sense surface nitration. However, this hypothesis might predict that the host requires only minimal complexity of host immune receptors to sense the density of nitrates on the ookinete surface, which may not be consistent with the observed complexity of host molecules that protect against malaria midgut infection [16,25–28]. Similarly, host defense against extracellular hemocoel sporozoites prior to salivary gland invasion [34] probably requires an explanation other than intracellular nitration.
Probing vector immunity with entomopathogenic fungi
In addition to adult female mosquitoes, APL1 paralogs are also expressed by adult males and during larval stages [25], which are never in contact with malaria parasites, indicating a more general protective role for these factors. Both male and female A. gambiae feed on plant nectar [35], which is a source of exposure to a shared pathogen repertoire such as bacteria and fungi. In addition, the female mosquito contacts aquatic and soil pathogens during oviposition. Entomopathogenic fungi such as Metarhizium anisopliae and Beauveria bassiana are environmentally ubiquitous, found in soil, air, and plants that mosquitoes feed on for sugars [36–38].
Fungi are being examined in the lab and field as a potential new vector control tool [39–43]. Reducing vector longevity is an effective approach to reduce transmission of malaria parasites, and is the basis of transmission control by indoor residual insecticide spraying. The oocyst stage in the vector has a long period of latency, producing infectious sporozoites in the salivary glands only just before the demographic end of the mosquito lifespan [44]. Thus, killing the vector at any point in that extended period is equally effective in reducing the proportion of infected mosquitoes that become infectious. One argument in favor of fungal control of vectors is that, as a delayed killing agent, the cost in reproductive fitness to the mosquito is lowered and pressure for the development of resistance mechanisms is lessened [39].
Understanding the genetics and functional biology of resistance to fungal pathogens may reveal new features of A. gambiae immunity. While these fungi are evolutionarily distant from Plasmodium, they are also eukaryotes, and aspects of mosquito immunity to both pathogens may not be completely independent. As a practical question, it should be determined whether widespread use of fungal entomopathogens to control malaria, including the eventual selection of genetic resistance to fungus, could have collateral consequences, either beneficial or detrimental, for mosquito susceptibility to and transmission of Plasmodium. Interestingly, fungus infection can preferentially kill mosquitoes infected with rodent malaria, although the observation needs to be confirmed for P. falciparum [43]. If so, it might be possible to use a conditionally lethal fungus, which preferentially kills malaria-infected mosquitoes, to drive natural malaria-resistance alleles to higher levels in the population, by imposing selection against infected and therefore genetically susceptible mosquitoes in the population [3,45].
Immunity of mosquito larvae
Larval stages of anophelines are aquatic, and the water in breeding sites harbors a repertoire of microbes, including bacteria, fungi, and protozoans [46,47]. A subset of these microbes may be incorporated into a stable gut microbiome in larvae and subsequent adult mosquitoes [48]. Larval-derived exposure to bacteria [49] or protozoa [50] can influence adult mosquito susceptibility to malaria parasites. Of the few genes tested to date, mosquito immune genes regulated transcriptionally in adults were also regulated in larvae [51,52], although larval immunity has not been systematically studied. In adult mosquitoes, functional genomic studies are performed by injection of double stranded RNA (dsRNA). In mosquito larvae this method is more difficult, at least because of the need to perform injections out of the water, with consequent higher mortality, small size, and the tough and elastic larval cuticle. A non-invasive RNAi method was reported for A. gambiae using self-assembled dsRNA nanoparticles to mediate gene silencing through larval feeding [53]. Entomopathogenic fungi have been proposed for larval mosquito control because larval exposure with the right formulation of spores causes high larval mortality [54], and thus fungi can also be used for functional studies with larval stages of A. gambiae.
Ecological differences between larval pools may also be an aspect of niche differentiation by A. gambiae population subgroups, such as the reproductively isolated natural strains referred to as M and S molecular forms [46,55]. Preferential larval site selection by ovipositing females probably exposes their progeny to particular microbial communities and perhaps differential immune pressure, although little is known. It was recently reported that wild M-form mosquitoes display significantly reduced genetic diversity at two unlinked immune gene loci, TEP1 and APL1, as compared to S-form mosquitoes [9,56]. The most likely explanation may be that specific and costly pathogen exposure of M-form larvae, based on larval site ecology, has selected for only a few alleles of the protective immune genes, but the hypothesis requires empirical evidence.
Immune phenotypes and study systems
Wild A. gambiae populations display high genetic diversity [57], which is partitioned between reproductively isolated population subgroups, and between diverged long allelic sequence blocks locked within polymorphic chromosome inversions [58–61]. Laboratory-reared A. gambiae colonies have been necessarily used for almost all reported functional studies of mosquito-pathogen interactions, because such experiments require controlled conditions (mosquito numbers, age, etc.) that are difficult to produce in nature. However, all colonies display low genetic diversity, and therefore segregate only a subset of alleles [62,63]. Thus, the cost of the common practice of working with just one colony in a given laboratory is that resulting genetic claims, including determination of the phenotype of the gene, are based on an experiment that queried only a small number of alleles in a particular genetic background. It is possible that some reported mosquito gene phenotypes would give a different result in a different colony.
The best way to collect high quality phenotypic data that is comparable between laboratories might be to make phenotypic determinations across a panel of independent colonies initiated from nature, modeled on the Genetic Reference Panel of Drosophila [64]. Functional tests of a mosquito gene would be replicated in a panel of reference strains, compensating for genetic simplicity in any one strain by the summation of the same observation repeated across strains. The result would define the gene phenotype not as a single point, but instead as many points that differ according to target gene allelic variation and genetic background of each test strain, and overall would better approximate the true range of the phenotype in the natural genetic system. In fact, otherwise protective genes may even enhance pathogen development in some genetic contexts [22]. One challenge with this approach is that Anopheles culture is more difficult than Drosophila, and there is no method of strain storage other than continuous culture, which requires insectary space and resources. A panel of reference strains initiated from nature could also serve as a useful genetic resource for mapping, because of their individually simplified allelic spectra and extended linkage.
The other important aspect of malaria vector study systems is the parasite model. The rodent malaria laboratory model (P. berghei, P. yoelii, etc.) has been extraordinarily productive, and its robustness and easy availability is largely responsible for the emergence of A. gambiae as a model organism for host-pathogen interactions. However, the accuracy of rodent malaria interactions in A. gambiae as a model for vector response to the naturally transmitted parasite, P. falciparum, has not been systematically examined or validated. When comparisons have been made, the theme is often more about differences than similarities. For example, functional tests show different signaling pathways, and different outcomes for many protective genes [16,17,21,65]. The mapped A. gambiae QTLs that control resistance, even though the overall number is relatively small, are also different between rodent parasites in the laboratory and human malaria in the field [3]. Mosquito transcriptional profiling shows a combination of both overlap and distinct responses between the two parasite classes [21,65]. It was suggested that rodent malaria infection in the mosquito A. stephensi, rather than A. gambiae, may be more representative of P. falciparum interaction with A. gambiae [21]. Overall, these differences encourage caution in generalizing about mechanisms of “protection against Plasmodium,” in particular against P. falciparum, from results obtained only in the rodent malaria model, and mitigate in favor of studies that include direct comparison using challenge with P. falciparum gametocyte culture whenever possible.
CONCLUSION
Mosquitoes are the most important arthropod vectors of diseases. Understanding the molecular and genetic components of vector competence for transmission of a given pathogen could help generate new vector control tools. In addition, the sheer quantity of mosquito-malaria research coupled with genomic technologies has now made A. gambiae a model system for the study of host-pathogen interactions. The A. gambiae – P. falciparum system results from an old and specific evolutionary history due to the obligatory nature of human malaria transmission by Anopheles. This is unlike other model organisms such as Drosophila or Arabidopsis, which are infected by facultative combinations of more generalist pathogens [66]. Thus, the mosquito-malaria study system may be especially advantageous for detecting a clear genetic signal resulting from an evolutionary interplay between pathogen virulence and host defense.
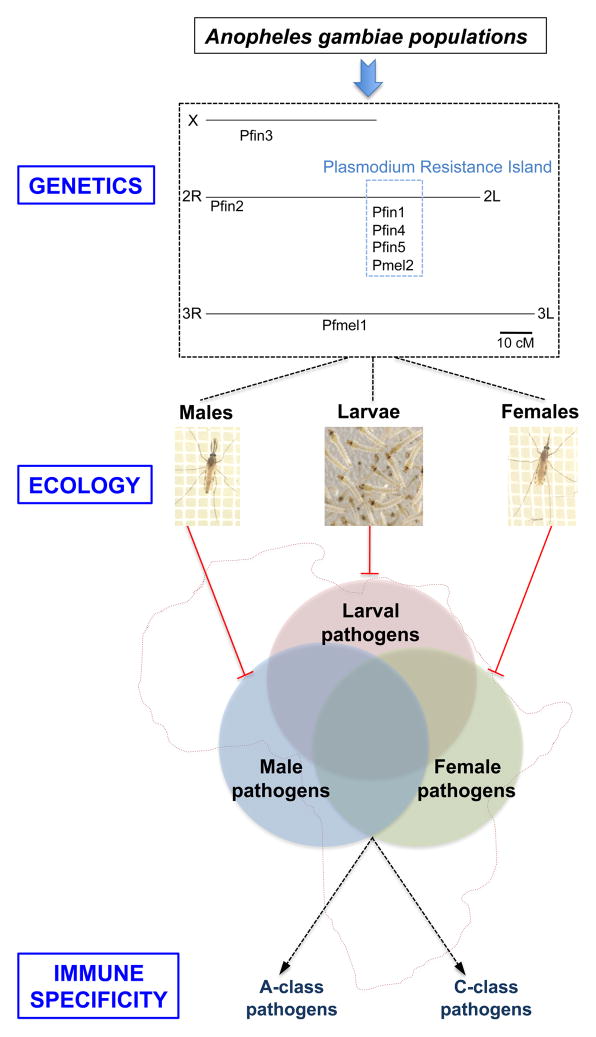
Figure.
Synthetic model depicting the intersection of genetics, ecology and immunity on Anopheles gambiae pathogen susceptibility. Upper panel (Genetics): The genetic location of loci that control P. falciparum infection in the natural A. gambiae population are indicated [3][4,5]. Pfin loci control numbers of normal oocysts following an infected blood meal, Pfmel loci control ookinete melanization, and the Plasmodium Resistance Island comprises a cluster of protective loci on chromosome 2L. Middle panel (Ecology): Genetic and functional mechanisms of Plasmodium resistance can only be tested in female mosquitoes, but some of the same host-defense mechanisms or close variants may be deployed against other pathogens in adult male mosquitoes and larvae of both sexes. Males and larvae are subject to distinct microbial repertoires and immune selective pressure, but microbial repertoires common to all forms may shape shared components of immunity. Lower panel (Immune Specificity): Classes of pathogens that are distinguished by mosquito immunity can be hypothesized, based on differential modes of fine specificity mediated by either APL1A (A-class pathogens, including P. falciparum but not rodent malaria), or APL1C (C-class pathogens, including rodent malaria parasite species but not P. falciparum). Functional tests of other pathogens and immune genes will be necessary to fully define the protective spectra of these and other immune genes, and the network of their interactions [Mosquito pictures: CEPIA, Institut Pasteur].
Highlights.
Mosquito vectors of disease vary between individuals for pathogen infection.
Pathogen susceptibility is controlled by immunogenetics and ecological factors.
Studies of pathogen susceptibility in a single mosquito colony are oversimplified.
Acknowledgments
We thank Michelle M. Riehle, Karin Eiglmeier, and Emmanuel Bischoff for valuable comments and suggestions. We acknowledge the support of NIH/NIAID grants AI073685, AI062995 and ANR Anochamp to KDV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beerntsen BT, James AA, Christensen BM. Genetics of mosquito vector competence. Microbiol Mol Biol Rev. 2000;64:115–137. doi: 10.1128/mmbr.64.1.115-137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niare O, Markianos K, Volz J, Oduol F, Toure A, Bagayoko M, Sangare D, Traore SF, Wang R, Blass C, et al. Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science. 2002;298:213–216. doi: 10.1126/science.1073420. [DOI] [PubMed] [Google Scholar]
- 3••.Riehle MM, Markianos K, Niare O, Xu J, Li J, Toure AM, Podiougou B, Oduol F, Diawara S, Diallo M, et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. The only genetic survey to date of wild A. gambiae for QTLs controlling P. falciparum infection. [DOI] [PubMed] [Google Scholar]
- 4.Menge DM, Zhong D, Guda T, Gouagna L, Githure J, Beier J, Yan G. Quantitative trait loci controlling refractoriness to Plasmodium falciparum in natural Anopheles gambiae mosquitoes from a malaria-endemic region in western Kenya. Genetics. 2006;173:235–241. doi: 10.1534/genetics.105.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riehle MM, Markianos K, Lambrechts L, Xia A, Sharakhov I, Koella JC, Vernick KD. A major genetic locus controlling natural Plasmodium falciparum infection is shared by East and West African Anopheles gambiae. Malar J. 2007;6:87. doi: 10.1186/1475-2875-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton AA, Lee Y, Coulibaly CA, Rashbrook VK, Cornel AJ, Lanzaro GC, Luckhart S. Identification of three single nucleotide polymorphisms in Anopheles gambiae immune signaling genes that are associated with natural Plasmodium falciparum infection. Malaria journal. 2010;9:160. doi: 10.1186/1475-2875-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris C, Lambrechts L, Rousset F, Abate L, Nsango SE, Fontenille D, Morlais I, Cohuet A. Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS pathogens. 2010;6:e1001112. doi: 10.1371/journal.ppat.1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riehle MM, Xu J, Lazzaro BP, Rottschaefer SM, Coulibaly B, Sacko M, Niare O, Morlais I, Traore SF, Vernick KD. Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS One. 2008;3:e3672. doi: 10.1371/journal.pone.0003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Rottschaefer SM, Riehle MM, Coulibaly B, Sacko M, Niare O, Morlais I, Traore SF, Vernick KD, Lazzaro BP. Exceptional diversity, maintenance of polymorphism, and recent directional selection on the APL1 malaria resistance genes of Anopheles gambiae. PLoS biology. 2011;9:e1000600. doi: 10.1371/journal.pbio.1000600. The APL1 locus is highly polymorphic and has undergone a coordinate selective sweep with the TEP1 locus (also White et al, 56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obbard DJ, Callister DM, Jiggins FM, Soares DC, Yan G, Little TJ. The evolution of TEP1, an exceptionally polymorphic immunity gene in Anopheles gambiae. BMC evolutionary biology. 2008;8:274. doi: 10.1186/1471-2148-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M, et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 14.Barillas-Mury C, Charlesworth A, Gross I, Richman A, Hoffmann JA, Kafatos FC. Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae. The EMBO journal. 1996;15:4691–4701. [PMC free article] [PubMed] [Google Scholar]
- 15.Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, Hoa NT, Clayton JR, White KP, Kafatos FC, Christophides GK, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2005;102:11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Mitri C, Jacques JC, Thiery I, Riehle MM, Xu J, Bischoff E, Morlais I, Nsango SE, Vernick KD, Bourgouin C. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5:e1000576. doi: 10.1371/journal.ppat.1000576. APL1 paralogs are required for control of mutually exclusive pathogen specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25:677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 18••.Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, Hoffmann JA, Blandin SA, Levashina EA. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe. 2009;5:273–284. doi: 10.1016/j.chom.2009.01.005. A pair of articles (Fraiture 18, and Povelones19) showed that APL1C, LRIM1 and TEP1 form a functional complex that controls rodent malaria infection. [DOI] [PubMed] [Google Scholar]
- 19••.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;324:258–261. doi: 10.1126/science.1171400. A pair of articles (Fraiture 18, and Povelones19) showed that APL1C, LRIM1 and TEP1 form a functional complex that controls rodent malaria infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baxter RH, Chang CI, Chelliah Y, Blandin S, Levashina EA, Deisenhofer J. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0704967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaramillo-Gutierrez G, Rodrigues J, Ndikuyeze G, Povelones M, Molina-Cruz A, Barillas-Mury C. Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC microbiology. 2009;9:154. doi: 10.1186/1471-2180-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 23.Killick-Kendrick R. Parasitic protozoa of the blood of rodents: a revision of Plasmodium berghei. Parasitology. 1974;69:225–237. doi: 10.1017/s0031182000048071. [DOI] [PubMed] [Google Scholar]
- 24.Lemaitre B, Hoffmann J. The Host Defense of Drosophila melanogaster. Annu Rev Immunol. 2007 doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 25.Mitri C, Vernick KD. Unpublished observations. [Google Scholar]
- 26•.Povelones M, Upton LM, Sala KA, Christophides GK. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS pathogens. 2011;7:e1002023. doi: 10.1371/journal.ppat.1002023. High degree of combinatorial complexity describes the interactions between LRR and TEP proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. The Journal of biological chemistry. 2009;284:9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS biology. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor symposia on quantitative biology. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Current topics in microbiology and immunology. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arrighi RB, Debierre-Grockiego F, Schwarz RT, Faye I. The immunogenic properties of protozoan glycosylphosphatidylinositols in the mosquito Anopheles gambiae. Dev Comp Immunol. 2009;33:216–223. doi: 10.1016/j.dci.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 32••.Oliveira GD, Lieberman J, Barillas-Mury C. Epithelial Nitration by a Peroxidase/NOX5 System Mediates Mosquito Antiplasmodial Immunity. Science. 2012 doi: 10.1126/science.1209678. Rodent malaria ookinetes are nitrated as they traverse the epithelial cell, which marks them for subsequent TEP1-mediated killing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallucci S, Matzinger P. Danger signals: SOS to the immune system. Current opinion in immunology. 2001;13:114–119. doi: 10.1016/s0952-7915(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 34.Hillyer JF, Barreau C, Vernick KD. Efficiency of salivary gland invasion by malaria sporozoites is controlled by rapid sporozoite destruction in the mosquito haemocoel. International journal for parasitology. 2007;37:673–681. doi: 10.1016/j.ijpara.2006.12.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gouagna LC, Poueme RS, Dabire KR, Ouedraogo JB, Fontenille D, Simard F. Patterns of sugar feeding and host plant preferences in adult males of An. gambiae (Diptera: Culicidae) Journal of vector ecology: journal of the Society for Vector Ecology. 2010;35:267–276. doi: 10.1111/j.1948-7134.2010.00082.x. [DOI] [PubMed] [Google Scholar]
- 36.Gurulingappa P, Sword GA, Murdoch G, Mcgee PA. Colonization of crop plants by fungal entomopathogens and their effects on two insect pests when in planta. Biological Control. 2010;55:34–41. [Google Scholar]
- 37.Vega FE, Goettel MS, Blackwell M, Chandler D, Jackson MA, Keller S, Koike M, Maniania NK, Monzon A, Ownley BH, et al. Fungal entomopathogens: new insights on their ecology. Fungal Ecology. 2009;2:149–159. [Google Scholar]
- 38.Darbro JM, Thomas MB. Spore persistence and likelihood of aeroallergenicity of entomopathogenic fungi used for mosquito control. The American journal of tropical medicine and hygiene. 2009;80:992–997. [PubMed] [Google Scholar]
- 39.Thomas MB, Read AF. Can fungal biopesticides control malaria? Nat Rev Microbiol. 2007 doi: 10.1038/nrmicro1638. [DOI] [PubMed] [Google Scholar]
- 40•.Blanford S, Shi W, Christian R, Marden JH, Koekemoer LL, Brooke BD, Coetzee M, Read AF, Thomas MB. Lethal and pre-lethal effects of a fungal biopesticide contribute to substantial and rapid control of malaria vectors. PLoS One. 2011;6:e23591. doi: 10.1371/journal.pone.0023591. Entomopathogenic fungi can efficiently kill mosquitoes before they transmit malaria, including mosquitoes resistant to chemical insecticides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farenhorst M, Knols BG, Thomas MB, Howard AF, Takken W, Rowland M, N’Guessan R. Synergy in efficacy of fungal entomopathogens and permethrin against West African insecticide-resistant Anopheles gambiae mosquitoes. PLoS One. 2010;5:e12081. doi: 10.1371/journal.pone.0012081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholte E, Ng’habi K, Kihonda J, Takken W, Paaijmans K, Abdulla S, Killeen G, Knols B. An entomopathogenic fungus for control of adult African malaria mosquitoes. Science. 2005 Jun 10;308:1641– 1642. doi: 10.1126/science.1108639. [DOI] [PubMed] [Google Scholar]
- 43.Blanford S, Chan B, Jenkins N, Sim D, Turner R, Read A, Thomas M. Fungal pathogen reduces potential for malaria transmission. Science. 2005 Jun 10;308:1638– 1641. doi: 10.1126/science.1108423. [DOI] [PubMed] [Google Scholar]
- 44.Charlwood JD, Smith T, Billingsley PF, Takken W, Lyimo EOK, Meuwissen JHET. Survival and infection probabilities of anthropophagic anophelines from an area of high prevalence of Plasmodium falciparum in humans. Bulletin of Entomological Research. 1997;87:445–453. [Google Scholar]
- 45.Hahn MW, Nuzhdin SV. The fixation of malaria refractoriness in mosquitoes. Curr Biol. 2004;14:R264–265. doi: 10.1016/j.cub.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Fillinger U, Sombroek H, Majambere S, van Loon E, Takken W, Lindsay SW. Identifying the most productive breeding sites for malaria mosquitoes in The Gambia. Malaria journal. 2009;8:62. doi: 10.1186/1475-2875-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Service MW. Mortalities of the immature stages of species B of the Anopheles gambiae complex in Kenya: comparison between rice fields and temporary pools, identification of predators, and effects of insecticidal spraying. J Med Entomol. 1977;13:535–545. doi: 10.1093/jmedent/13.4-5.535. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Gilbreath TM, 3rd, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One. 2011;6:e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. A bacteria cultured from larval pools can render adult mosquitoes resistant to P. falciparum infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bargielowski I, Koella JC. A possible mechanism for the suppression of Plasmodium berghei development in the mosquito Anopheles gambiae by the microsporidian Vavraia culicis. PLoS One. 2009;4:e4676. doi: 10.1371/journal.pone.0004676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimopoulos G, Richman A, della Torre A, Kafatos FC, Louis C. Identification and characterization of differentially expressed cDNAs of the vector mosquito, Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13066–13071. doi: 10.1073/pnas.93.23.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae) Insect molecular biology. 2010;19:683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- 54.Bukhari T, Middelman A, Koenraadt CJ, Takken W, Knols BG. Factors affecting fungus-induced larval mortality in Anopheles gambiae and Anopheles stephensi. Malaria journal. 2010;9:22. doi: 10.1186/1475-2875-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diabate A, Dabire RK, Heidenberger K, Crawford J, Lamp WO, Culler LE, Lehmann T. Evidence for divergent selection between the molecular forms of Anopheles gambiae: role of predation. BMC evolutionary biology. 2008;8:5. doi: 10.1186/1471-2148-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.White BJ, Lawniczak MK, Cheng C, Coulibaly MB, Wilson MD, Sagnon N, Costantini C, Simard F, Christophides GK, Besansky NJ. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:244–249. doi: 10.1073/pnas.1013648108. The APL1 locus is highly polymorphic and has undergone a coordinate selective sweep with the TEP1 locus (also Rottschaefer et al, 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilding CS, Weetman D, Steen K, Donnelly MJ. High, clustered, nucleotide diversity in the genome of Anopheles gambiae revealed through pooled-template sequencing: implications for high-throughput genotyping protocols. BMC Genomics. 2009;10:320–330. doi: 10.1186/1471-2164-10-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 59.Riehle MM, Guelbeogo WM, Gneme A, Eiglmeier K, Holm I, Bischoff E, Garnier T, Snyder GM, Li X, Markianos K, et al. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science. 2011;331:596–598. doi: 10.1126/science.1196759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toure YT, Petrarca V, Traore SF, Coulibaly A, Maiga HM, Sankare O, Sow M, Di Deco MA, Coluzzi M. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia. 1998;40:477–511. [PubMed] [Google Scholar]
- 61.Lawniczak MK, Emrich SJ, Holloway AK, Regier AP, Olson M, White B, Redmond S, Fulton L, Appelbaum E, Godfrey J, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riehle MM, Vernick KD. Unpublished observations. Edited by. [Google Scholar]
- 63.Norris DE, Shurtleff AC, Toure YT, Lanzaro GC. Microsatellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae ss (Diptera: Culicidae) Journal of medical entomology. 2001;38:336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- 64.Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS pathogens. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]