Rrp6 is an exoribonuclease involved in the quality control of mRNA biogenesis. The authors analyzed the association of Rrp6 with the Balbiani ring pre-mRNPs of Chironomus tentans to obtain insight into the role of Rrp6 in splicing surveillance. Rrp6 was recruited to transcribed genes and its distribution did not correlate with the positions of exons and introns. Rrp6 also bound to both unspliced and spliced transcripts and was released from the mRNPs in the vicinity of the nuclear pore. Hence, Rrp6 was associated with newly synthesized transcripts during all the nuclear steps of gene expression, independent of their splicing status. The quality control of pre-mRNA splicing was not based on the selective recruitment of the surveillance machinery to unprocessed mRNAs.
Keywords: pre-mRNA splicing, surveillance, exosome, Chironomus tentans, nuclear bodies
Abstract
Rrp6 is an exoribonuclease involved in the quality control of mRNA biogenesis. We have analyzed the association of Rrp6 with the Balbiani ring pre-mRNPs of Chironomus tentans to obtain insight into the role of Rrp6 in splicing surveillance. Rrp6 is recruited to transcribed genes and its distribution along the genes does not correlate with the positions of exons and introns. In the nucleoplasm, Rrp6 is bound to both unspliced and spliced transcripts. Rrp6 is released from the mRNPs in the vicinity of the nuclear pore before nucleo-cytoplasmic translocation. We show that Rrp6 is associated with newly synthesized transcripts during all the nuclear steps of gene expression and is associated with the transcripts independently of their splicing status. These observations suggest that the quality control of pre-mRNA splicing is not based on the selective recruitment of the exoribonuclease Rrp6 to unprocessed mRNAs.
INTRODUCTION
Pre-mRNAs are synthesized by RNA polymerase II (Pol-II) and assemble into ribonucleoprotein (RNP) complexes during transcription. The maturation of the pre-mRNPs involves multiple processing reactions, including capping, splicing, and 3′ end formations, that often take place cotranscriptionally (for review, see Neugebauer 2002). The biogenesis of correct, export-competent mRNPs relies on a number of quality control checkpoints. Some of these checkpoints rely on the activity of the RNA exosome, a protein complex with ribonucleolytic activity that is involved in the processing and degradation of many RNA classes (for reviews, see Houseley et al. 2006; Houseley and Tollervey 2008; van Hoof and Wagner 2011). The exosome is evolutionarily conserved from archea to eukaryotes. The eukaryotic exosome is composed of a ring-shaped core made of nine different subunits (for review, see Lorentzen et al. 2008). The core subunits are catalytically inactive and the ribonucleolytic activity of the exosome is provided by additional factors (for review, see Lykke-Andersen et al. 2011). Two active subunits, Rrp6 and Rrp44/Dis3, were identified initially in budding yeast and are conserved in higher eukaryotes (Mitchell et al. 1997; Andrulis et al. 2002). A third active subunit, Dis3L, has recently been identified in human cells (Staals et al. 2010; Tomecki et al. 2010). There is ample evidence for the functional association of the catalytic subunits with the core of the exosome in both yeast and metazoans (Andrulis et al. 2002; Synowsky et al. 2009). Moreover, Rrp6 and Dis3 have other interaction partners than the core exosome subunits and have functions that are independent of the core (Graham et al. 2009; Synowsky et al. 2009; Kiss and Andrulis 2011).
Rrp6 is a central player in the nuclear surveillance of mRNA biogenesis (for review, see Schmid and Jensen 2008). In budding yeast, the exosome and Rrp6 retain and degrade defective transcripts in the nucleus (Hilleren et al. 2001). In metazoans, Rrp6 is needed for correct transcription termination by Pol-II, and unprocessed transcripts are retained at the transcription site in an Rrp6-dependent manner (de Almeida et al. 2010; Eberle et al. 2010). Studies in budding yeast have also suggested that there is a post-transcriptional quality control step that takes place at the nuclear periphery prior to the export of the mRNPs to the cytoplasm. The Mlp1 protein, a protein associated with the nuclear pores, physically retains unprocessed pre-mRNAs in the nucleus through a mechanism that also involves Rrp6 (Galy et al. 2004). Whether Rrp6 plays a role in the surveillance of splicing at the nuclear periphery also in metazoans is an open question.
In spite of recent advances in the study of the structure and catalytic properties of Rrp6 in animal cells, much remains to be discovered about its functions in vivo. We have studied Rrp6 in the dipteran Chironomus tentans to gain further insight into the role of Rrp6 in the nuclear quality control of pre-mRNA splicing in metazoans. The Balbiani ring (BR) genes of C. tentans code for large secretory proteins in the salivary glands of the Chironomus larvae (for review, see Wieslander 1994). The BR pre-mRNAs have all the features of protein-coding transcripts and undergo typical pre-mRNA processing (for review, see Wieslander et al. 1996; Kiesler and Visa 2004). The BR genes can be identified in polytene chromosome preparations, and by using immunoelectron microscopy (immuno-EM) it is possible to study the association of specific proteins with the newly synthesized BR pre-mRNAs at the site of transcription and on the way to the nuclear pores. This makes the BR genes of C. tentans a powerful experimental system for in situ studies of mRNA biogenesis (for review, see Daneholt 2001). We have applied immunocytochemistry methods to analyze the nuclear distribution of Rrp6 and its association with the BR mRNPs at different stages of synthesis, processing, and transport. We have also used immunoprecipitation methods to study the relationships between splicing and Rrp6 recruitment.
RESULTS AND DISCUSSION
The nuclear distribution of Rrp6 in the salivary gland cells of C. tentans
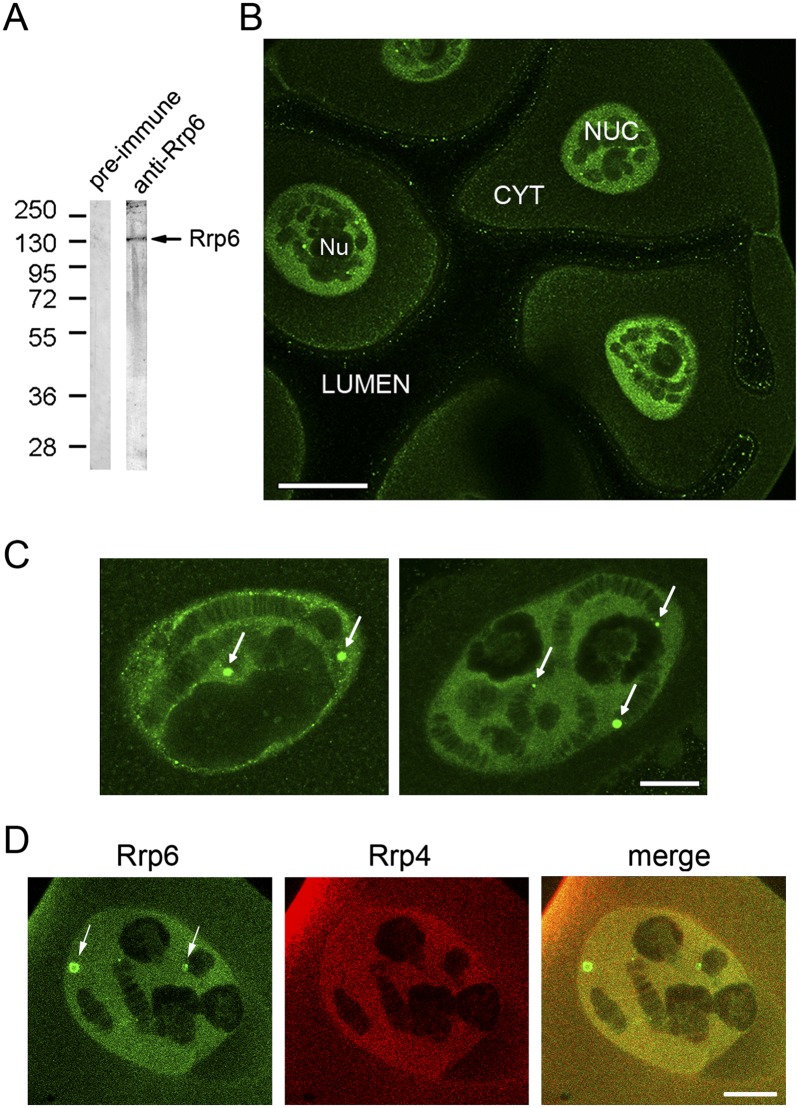
We amplified a partial cDNA for the Rrp6 of C. tentans (Ct-Rrp6, or Rrp6 for simplicity) following a degenerate PCR approach. The predicted protein sequence encoded by this partial cDNA was 84% and 85% similar to the corresponding Rrp6 sequences of Drosophila melanogaster and Homo sapiens, respectively. We expressed the cDNA in Escherichia coli and used the recombinant protein to raise an anti-Ct-Rrp6 antibody (Supplemental Fig. S1). The specificity of the anti-Ct-Rrp6 antibody was analyzed by Western blotting and the antibody was found to be monospecific (Fig. 1A). We used the anti-Ct-Rrp6 antibody for immunocytochemical studies. We first analyzed the general distribution of Rrp6 in the salivary gland cells of C. tentans by immunofluorescence (IF). Rrp6 stained the cytoplasm faintly and gave intense IF staining in the nucleus (Fig. 1B; Supplemental Fig. S2). The nucleoplasm was strongly labeled, and the polytene chromosomes displayed a banded pattern similar to that observed with antibodies against the core exosome subunit Rrp4 (Hessle et al. 2009). Furthermore, a small fraction of Rrp6 was concentrated in discrete nuclear bodies (Fig. 1C). The number of bodies per cell ranged from five to 20 and their diameter ranged between 1 and 5 μm. We carried out double-IF experiments with antibodies against Rrp6 and Rrp4. We could not detect Rrp4 in the Rrp6-positive bodies (Fig. 1D), which suggests that the presence of Rrp6 in these bodies is related to an exosome-independent function of Rrp6. In summary, our results indicate that Rrp6 is associated with many different structures in the nuclei of the salivary gland cells, which is compatible with the many functions of this protein. The nuclear localization of Rrp6 in the salivary glands of C. tentans is consistent with results from previous studies in yeast, flies, and humans (Allmang et al. 1999; Graham et al. 2006).
FIGURE 1.
The distribution of Rrp6 in the salivary gland cells of C. tentans. (A) Western blot showing the specificity of the anti-Ct-Rrp6 antibody. Nuclear protein extracts prepared from C. tentans culture cells were probed with either pre-immune serum or the serum anti-Rrp6. A major band with an apparent molecular mass of ∼110 kDa was obtained (arrow). The mobility of molecular mass standards is shown to the left, in kDa. (B) The general distribution of Rrp6 in the salivary glands analyzed by IF. The image shows a confocal section of a salivary gland stained with the anti-Rrp6 antibody. The lumen of the gland, nucleus (NUC), cytoplasm (CYT), and nucleolus (Nu) are indicated. The staining observed at the cell surface is unspecific as it has been observed with other rabbit sera. The magnification bar represents ∼50 μm. (C) Two salivary gland nuclei stained with the anti-Rrp6 antibody at higher magnification showing the location of Rrp6 in discrete nuclear bodies (arrows). The magnification bar represents ∼10 μm. (D) Salivary glands were double-labeled with anti-Rrp6 (green) and anti-Rrp4 (red) antibodies. The Rrp6-positive bodies are not labeled by the anti-Rrp4 antibody. The bar represents ∼10 μm.
Rrp6 is recruited to transcribed genes and is present along the entire transcription unit
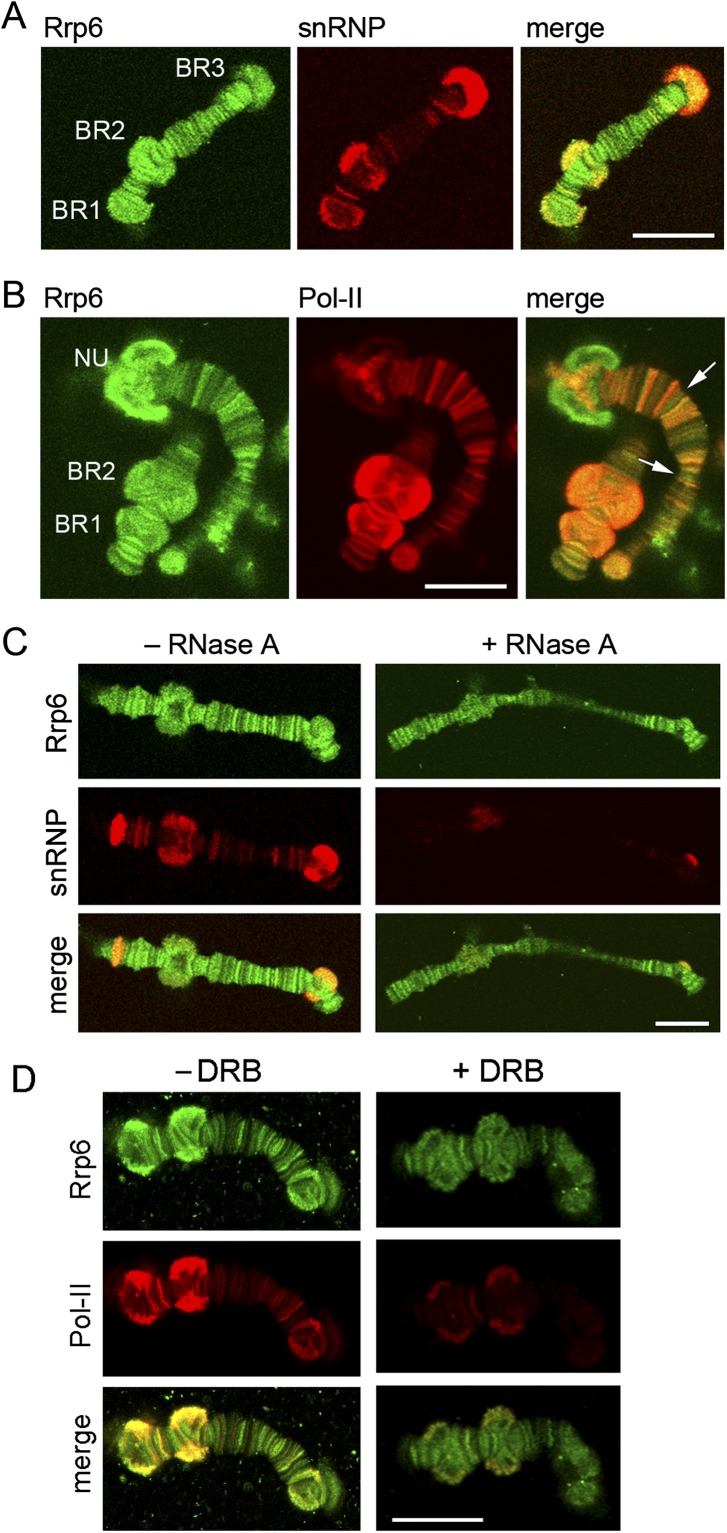
A more detailed analysis of the distribution of Rrp6 in isolated polytene chromosomes revealed that Rrp6 is associated with many loci, including the BR puffs in chromosome IV, and the nucleoli (Fig. 2; Supplemental Fig. S2). The distribution of Rrp6 was compared with those of snRNPs and Pol-II in double-IF experiments. Rrp6 and snRNPs colocalized in numerous loci, including the BR puffs (Fig. 2A). Rrp6 and Pol-II also showed a large extent of colocalization (Fig. 2B). We concluded that Rrp6 is recruited to transcribed genes.
FIGURE 2.
Rrp6 in the polytene chromosomes. (A) Double-IF staining of isolated polytene chromosomes with antibodies against Rrp6 (green) and snRNPs (red). The image shows a confocal section of an isolated chromosome IV. The positions of the three Balbiani ring puffs (BR1, BR2, and BR3) are indicated. (B) Double-IF staining of isolated polytene chromosomes with antibodies against Rrp6 (green) and Pol-II (red). NU, nucleolus. The arrows in the merged image point at loci that are stained by the anti-Rrp6 antibody but that are negative for Pol-II. (C) Isolated chromosomes were digested with (+) or without (−) RNase A before staining with antibodies against Rrp6 (green) and snRNPs (red). The anti-snRNP staining was strongly reduced by the treatment whereas the anti-Rrp6 was partially resistant to the RNase digestion. (D) Isolated chromosomes prepared from larvae treated with DRB for 45 min were double-stained with antibodies against Rrp6 and Pol-II, as indicated. The anti-Pol-II was drastically reduced by the DRB treatment but a significant amount of Rrp6 was still detected on the chromosomes. The magnification bars represent ∼20 μm.
Isolated polytene chromosomes were digested with RNase A before the IF staining to determine whether the Rrp6 that is found at sites of transcription is bound to the chromatin template or to the transcription products (Fig. 2C). The chromosomes were labeled simultaneously with the anti-snRNP antibody to assess the efficiency of the RNase digestion. Anti-Rrp6 IF labeling was somewhat lower in chromosomes treated with RNase, which suggests that a fraction of Rrp6 is bound to the nascent pre-mRNPs. However, the majority of the anti-Rrp6 staining was resistant to RNase digestion. This result shows that a relatively small fraction of Rrp6 is bound to the nascent pre-mRNP and that a large fraction of Rrp6 is associated with either the chromatin or the transcription machinery.
In another series of experiments, the larvae were treated with the transcription inhibitor DRB before the IF staining with antibodies against Rrp6 and Pol-II. The DRB treatment caused a strong reduction of anti-Pol-II staining but had a less pronounced effect on Rrp6 (Fig. 2D). This result suggests that a fraction of Rrp6 is bound to chromatin.
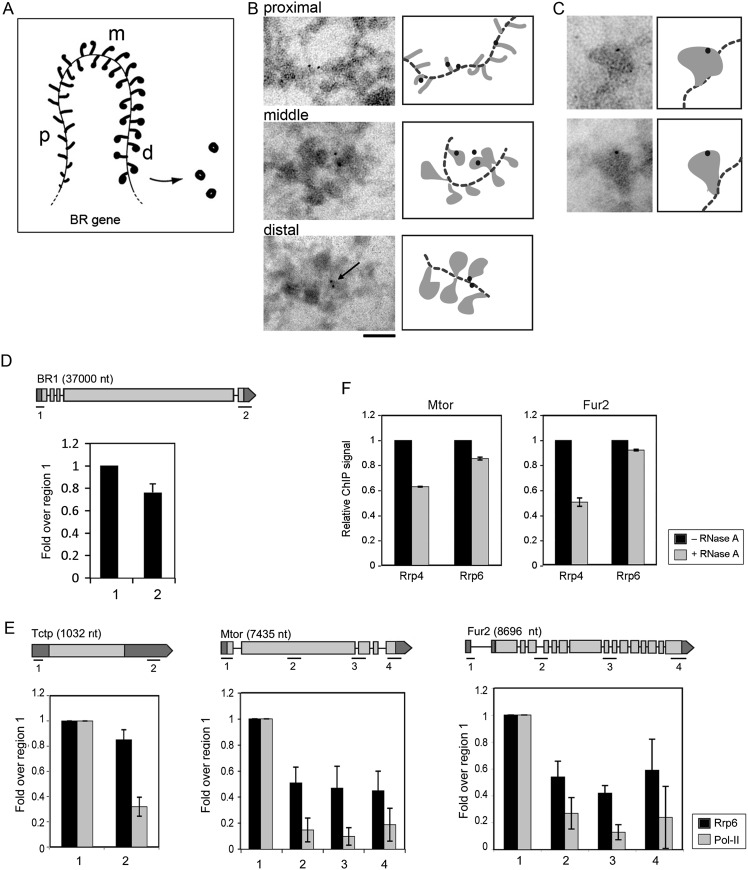
We analyzed also the localization of Rrp6 in the BR genes by immuno-EM. Nascent BR pre-mRNA molecules are rapidly packaged into growing pre-mRNP fibers that can be observed along the BR genes (Fig. 3A). As transcription progresses, the growing RNP fibers gradually increase in length and become packed into globular RNPs (Skoglund et al. 1983). Thin cryosections of salivary glands were immunolabeled with the anti-Rrp6 antibody. Negative control preparations processed in parallel were not significantly labeled (Supplemental Fig. S3). Rrp6 was associated with all the regions of the transcribed BR gene (Fig. 3B). The labeling was often located near the chromatin axis (see arrow in Fig. 3B), which indicates binding of Rrp6 with either the chromatin, the transcription machinery, or the stalk of the nascent pre-mRNPs. A few BR particles were labeled on the RNP fraction, far from the chromatin, which indicates that at least a small fraction of Rrp6 becomes incorporated into the nascent pre-mRNP during transcription (Fig. 3C). The results were confirmed by immuno-EM staining of isolated chromosomes with the anti-Rrp6 antibody (Supplemental Fig. S4).
FIGURE 3.
The association of Rrp6 with transcribed genes studied by immuno-EM and ChIP. (A) Schematic representation of a transcribed BR gene showing the synthesis and assembly of the BR pre-mRNPs. The proximal (p), middle (m), and distal (d) regions of the BR gene are indicated. (B) Immuno-EM analysis of the association of Rrp6 with the nascent BR pre-mRNPs. Salivary gland cryosections were immunostained with the anti-Rrp6 antibody and the labeling was visualized with a gold-conjugated secondary antibody. The figure shows three images that correspond to the proximal, middle, and distal portions of the BR gene, respectively. The arrow shows labeling close to the chromatin axis. Schematic interpretations of the images are provided next to each micrograph. The dotted line represents the chromatin axis. The bar represents 100 nm. (C) Two examples of immunogold labeling decorating the globular portion of BR pre-mRNPs in the distal region of the gene. Schematic interpretations of the images are provided next to each micrograph. (D) The association of Rrp6 with the BR1 gene analyzed by ChIP. The association of Rrp6 with sequences near the promoter (region 1) and in the 3′ UTR (region 2) was quantified. The relative levels of Rrp6 in each region were calculated relative to the input and expressed as the fold change relative to region 1, in order to reveal variations along the gene. The histograms show averages from three independent experiments. The error bars represent standard deviations (n = 3). The exon–intron structure of the gene is represented, and the positions of the regions analyzed are indicated in the cartoon. (E) ChIP analysis of Rrp6 distribution in genes of D. melanogaster (black bars). Parallel experiments were carried out with an anti-Pol-II antibody (gray bars). The histograms summarize the ChIP results obtained for three different genes: Fur2, Mtor, and Tctp. The nucleotide lengths of the pre-mRNAs and their exon–intron structure are depicted. The relative levels of Rrp6 and Pol-II in each region were calculated relative to the input and expressed as the fold changes relative to region 1, in order to reveal variations along the genes. The histograms show averages from three independent experiments. The error bars represent standard deviations (n = 3). (F) ChIP analysis of the association of Rrp6 and Rrp4 with Mtor and Fur2 in the presence of RNase A. ChIP experiments were carried out as above and the bound complexes were treated with RNase A after the immunoprecipitation. The levels of Rrp4 were significantly reduced by the RNase A digestion but the levels of Rrp6 were not. The histograms show averages from two independent experiments, each with two technical replicates. The error bars represent standard deviations (n = 4).
The cotranscriptional association of Rrp6 with the BR genes was confirmed by chromatin immunoprecipitation (ChIP) using chromatin from salivary glands. The immunoprecipitated DNA was analyzed by quantitative real-time PCR (qPCR). Rrp6 was associated with both the proximal and the distal parts of the BR1 gene (Fig. 3D; Supplemental Fig. S2). The association of Rrp6 with the central region of the BR1 gene could not be analyzed by ChIP due to the repetitive nature of the BR sequence (Paulsson et al. 1992).
We extended our analysis and analyzed also the association of Rrp6 with genes of D. melanogaster. We used D. melanogaster S2 cells that expressed a V5-tagged version of Rrp6 (Hessle et al. 2009), and we immunoprecipitated Rrp6 using an anti-V5 antibody. We were interested in determining whether the distribution of Rrp6 correlated with gene features such as exons or introns, and therefore we chose to analyze the occupancy of Rrp6 along three genes that we selected arbitrarily because they differ in length and exon–intron organization: Tctp, Mtor, and Fur2. The expression of these three genes in S2 cells was documented in the FLIGHT database (http://flight.licr.org/) and validated by RT-PCR in a previous study (Hessle et al. 2009). The results are summarized in Figure 3E. Rrp6 was associated with the three genes and was detected in all the regions analyzed. The levels of Rrp6 were significantly higher near the promoter and decreased in the 5′–3′ direction. This trend was less pronounced in the intronless Tctp gene, but also in Rpl40, an intron-containing gene of the same length as Tctp (Supplemental Fig. S5). We concluded that the distribution of Rrp6 along the genes does not correlate with the exon–intron structure of the genes.
The distribution of Pol-II along the Tctp, Mtor, and Fur2 genes was also analyzed in parallel for reference purposes. The levels of Pol-II were higher near the transcription start site than in the rest of the gene. This distribution pattern agrees with previous observations showing higher densities of Pol-II at the 5′ ends of active genes relative to the gene bodies (Min et al. 2011). In summary, both Rrp6 and Pol-II levels were higher at the 5′ ends than at the 3′ ends, but the Pol-II levels dropped in a more pronounced manner. Indeed, the relative levels of Rrp6 were higher than the levels of Pol-II inside the gene, for all the genes analyzed.
Comparison of the present ChIP results with those reported previously for Rrp4 (Hessle et al. 2009) reveals that the levels of Rrp6 do not follow strictly those of Rrp4, which suggests that Rrp6 and the exosome core bind to the transcribed genes independently of each other. The immuno-EM results and the RNase-digestion experiments also revealed differences in the way in which Rrp6 and Rrp4 interact with transcribed genes. A large fraction of Rrp4 is associated with the nascent BR pre-mRNPs, and the association of Rrp4 with the chromosomes is RNase-dependent (Hessle et al. 2009). The RNase digestion had a less pronounced effect on the association of Rrp6 with the chromosomes (Fig. 2C), and the immuno-EM results showed that Rrp6 was mostly located close to the transcribed chromatin (Fig. 3B). Moreover, ChIP experiments combined with RNase A digestions indicated that the association of Rrp4 with the transcribed genes depends on RNA to a large extent, whereas the association of Rrp6 does not (Fig. 3F). Altogether, these observations suggested that, although both exosome subunits associate cotranscriptionally with active genes, they do so through different types of interaction, with Rrp6 being less intimately associated with the pre-mRNPs. This is an interesting observation because it implies that the association of Rrp6 with the transcribed genes is, at least to some extent, independent from its association with Rrp4 or with the core exosome. Moreover, the fact that the association of Rrp6 with the transcribed genes does not depend on the RNA and does not correlate with the distribution of the transcription machinery suggests that a fraction of Rrp6 is bound to the active chromatin.
Rrp6 accompanies the spliced mRNPs to the nuclear pore
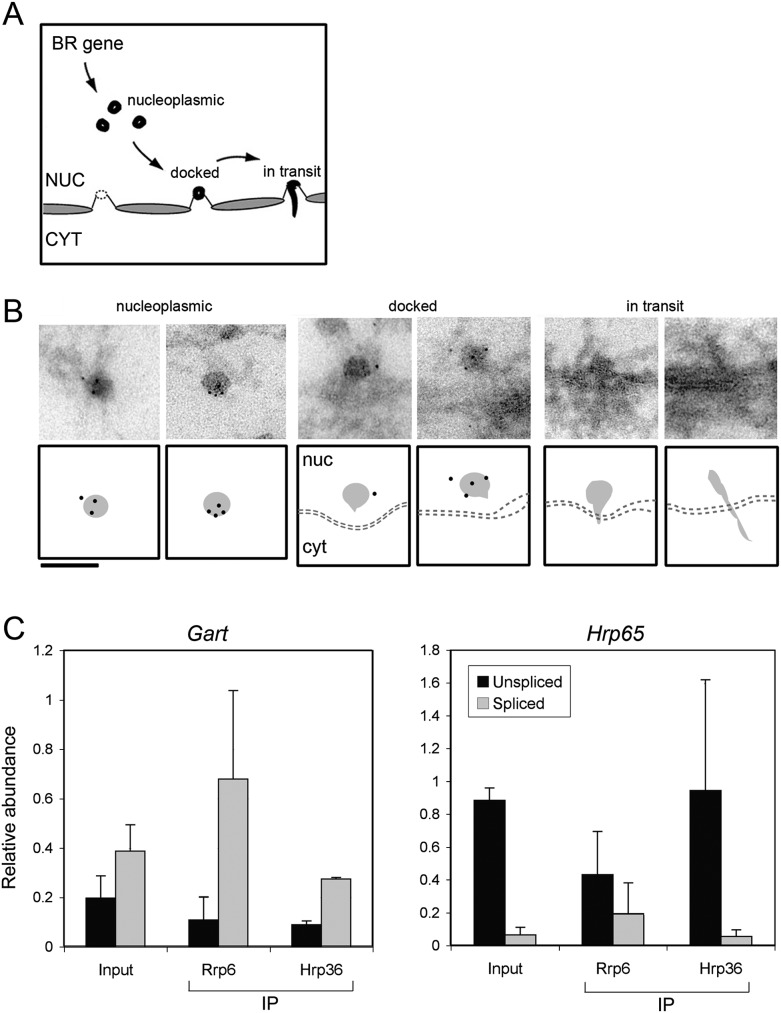
We sought to analyze the association of Rrp6 with BR mRNPs during nucleo-cytoplasmic transport in order to find evidence for the involvement of Rrp6 in a post-transcriptional surveillance step before nuclear export. When the transcription of the BR gene is completed, the BR particles are released from the chromosome and migrate into the nucleoplasm where they are seen as mRNP particles with a diameter of ∼50 nm (Fig. 4A). The BR particles unfold when they reach the nuclear envelope and remain unfolded during translocation through the nuclear pores (Mehlin et al. 1992). We analyzed salivary gland cryosections stained with the anti-Rrp6 antibody to study the association of Rrp6 with BR mRNP particles after transcription. Nucleoplasmic BR particles were labeled with the anti-Rrp6 antibody, as shown in Figure 4B. In contrast, unfolded BR particles in transit through the nuclear pore were devoid of labeling. These observations suggest that Rrp6 is bound to the mRNP during transport from the gene to the nuclear pore and is released from the mRNP before translocation through the nuclear pore. At this stage, the BR mRNPs undergo deep structural rearrangements that result in the unfolding of the mRNP. We propose that this structural remodeling also leads to the release of Rrp6 from the mRNP.
FIGURE 4.
The post-transcriptional association of Rrp6 with mRNPs analyzed by immuno-EM and RIP. (A) Schematic representation of the transport of BR pre-mRNPs through the nucleoplasm, the docking at the NPC, and the translocation to the cytoplasm. (B) Immuno-EM analysis of Rrp6 in BR particles during transport to and translocation through the NPC. The figure shows examples of BR mRNPs at different stages of nucleo-cytoplasmic transport, as indicated. Schematic interpretations are provided under each micrograph. The bar represents 100 nm. (C) The association of Rrp6 with mRNPs in C. tentans cultured cells was analyzed by RIP and RT-qPCR. An antibody against Hrp36 was used in parallel as a reference. The abundances of unspliced and spliced transcripts for two different genes, Gart and Hrp65, were analyzed. For each gene, the abundance of an exonic sequence was also quantified for normalization purposes. The histograms show the relative abundances of spliced and unspliced RNAs relative to the level of the exonic reference sequence for that same gene. The values are averages from three independent experiments, each with two technical replicates (n = 3).
The density of immunogold labeling on the nucleoplasmic BR particles was relatively high with several gold markers decorating each labeled BR particle (Fig. 4B). However, the fraction of labeled BR particles was low, <10% (data not shown). This could reflect a preferential binding of Rrp6 to a subpopulation of BR particles. Based on the role of the yeast Rrp6 in the surveillance of unspliced mRNAs in the nucleus, we asked whether Rrp6 was bound preferentially to unspliced pre-mRNPs. In situ studies of the splicing of the BR1 pre-mRNA have revealed the existence of a small fraction (<5%) of unspliced BR1 transcripts in the nucleoplasm of the salivary gland cells (Baurén and Wieslander 1994). To determine whether Rrp6 was preferentially bound to intron-containing transcripts, we used C. tentans tissue-cultured cells and prepared nuclear extracts containing nucleoplasmic RNPs (Tyagi et al. 2009). The nucleoplasmic extracts were used for RNA immunoprecipitation (RIP) using the anti-Rrp6 antibody. We used for comparison an antibody against Hrp36 and abundant hnRNP protein of C. tentans that binds extensively to both pre-mRNAs and mRNAs (Visa et al. 1996). The immunoprecipitated RNAs were purified, reverse-transcribed, and analyzed by quantitative PCR using primer pairs specific for spliced or unspliced transcripts. We also analyzed the transcripts present in the nucleoplasmic extract (referred to as “Input” in Fig. 4C). We argued that, if Rrp6 binds preferentially to unspliced transcripts, the intron-containing pre-mRNAs should be more enriched in the Rrp6-immunoprecipitated fractions than in the input. We analyzed transcripts from two different genes with known genomic sequences: Hrp65 (AJ404654) and Gart (S43653). Figure 4C shows a compilation of RIP results from three independent experiments. The relative abundances of the unspliced Hrp65 and Gart transcripts in the input differed, with Gart being spliced to a much larger extent. This difference reflects the fact that the alternative splicing of the Hrp65 pre-mRNA (Miralles and Visa 2001) is less efficient than the constitutive splicing of the Gart transcripts. Quantification of the Hrp65 and Gart transcripts in the immunoprecipitated fractions did not reveal any enrichment of the unspliced transcripts bound to Rrp6. Instead, there was a preference of Rrp6 for the spliced mRNAs. These observations argue against the preferential association of Rrp6 with intron-containing transcripts. They suggest instead that Rrp6 is bound to mRNAs that are already spliced and accompanies the mature mRNAs to the nuclear pore.
The analysis of Rrp6 in the salivary glands of C. tentans has revealed several interesting aspects of Rrp6 function. First, Rrp6 is recruited cotranscriptionally to active genes where it associates mostly with the chromatin and with the transcription machinery and, to a lesser extent, with the nascent transcripts as shown by RNase digestion experiments. Second, Rrp6 associates with mRNPs near the 3′ end of the BR genes, which suggests that Rrp6 is recruited to the mRNPs shortly before the release of the transcripts into the nucleoplasm. Third, Rrp6 is bound to nucleoplasmic mRNPs and accompanies the spliced transcripts to the nuclear pore. Interactions of Rrp6 with poly(A)-binding proteins could mediate such recruitment. Interestingly, a physical interaction between Rrp6 and the poly(A)-binding protein PABP2 was detected in a previous study (Hessle et al. 2009). Fourth, Rrp6 does not bind preferentially to unspliced transcripts. This finding indicates that the quality control of splicing by Rrp6 is not based on the selective recruitment of the exosome to the unprocessed transcripts. Our results support a model in which Rrp6 is recruited to newly synthesized mRNAs, independently of their splicing status, and is part of the exported mRNPs in the nucleoplasm. In such a model, the selective degradation of unspliced or defective transcripts is achieved not by the selective recruitment of surveillance factors to unspliced mRNAs, but is instead the result of kinetic or structural differences that render the unprocessed transcripts more susceptible to degradation.
MATERIALS AND METHODS
Culturing conditions
C. tentans was cultured as described by Meyer et al. (1983). Salivary glands were isolated from fourth instar larvae. C. tentans tissue culture cells were cultivated at 24°C as previously described (Wyss 1982). D. melanogaster S2 cells were cultured at 28°C following the Drosophila Expression System manual from Invitrogen. S2 cells stably transfected with plasmids that encode V5-tagged Rrp6 and Rrp4 have been described by Hessle et al. (2009). Stable transfectants were cultured in the presence of 300 μg/mL hygomycin B following the procedure recommended by Invitrogen. Expression of Rrp6–V5 was induced with 400 μM CuSO4 for 24 h.
Cloning and expression of Ct-Rrp6 cDNA
Degenerate primers were designed to match conserved amino acid regions in the Rrp6 sequence. Sequence conservation was based on the Rrp6 sequences of D. melanogaster (NP_001097795), Anopheles gambiae (XP_312669), Bombyx mori (BGIBMGA000626-PA), and Apis mellifera (XP_396975.3). Amino acid sequence alignments were done using the CLUSTALW program at http://workbench.sdsc.edu. The sequence of the forward primer F1 was 5′-GAYBTNGARCAYCAYWSNTAYMG-3′. The reverse primers R2 and R3 were 5′-ARRWARTGNGTRTCYWBNCKNGCRTA-3′ and 5′-GGNCKNATNCKCCARTCNGC-3′, respectively. These primers were used in nested PCR reactions to amplify Rrp6 sequences from cDNA prepared from C. tentans tissue culture cells. A PCR product of length 468 bp was obtained and further amplified with forward and reverse primers containing cloning sites for BamHI and HindIII, respectively. The PCR product was cloned into the pET21b vector (Novagen) to obtain the pET21b–Rrp6 plasmid. For protein expression, the pET21b–Rrp6 was transformed into competent E. coli cells BL21(DE3).
Antibodies
For production of an anti-Ct-Rrp6 antibody, the pET21c–Rrp6 plasmid was transformed into competent E. coli cells, protein expression was induced with IPTG, and the bacterial lysates were resolved by SDS-PAGE. The band corresponding to Rrp6 was cut from the gel and used for immunization of rabbits according to standard protocols at Agrisera AB. The anti-V5 antibody was from Invitrogen. The anti-Pol-II CTD antibody 4H8 (ab5408) was from Abcam and the negative control anti-IgG antibody (Z0456) used for ChIP was from DakoCytomation. The anti-U2B″ antibody was from ICN/CAPPEL (mAb 4G3). A rabbit polyclonal antibody against Ct-Rrp4 and mAb Y12 against snRNP proteins have been characterized in previous studies (Lerner et al. 1981; Hessle et al. 2009). Secondary antibodies for IF and for immuno-EM were from Jackson Immunoresearch Laboratories. Double-IF stainings with Ct-Rrp6 and Ct-Rrp4 antibodies, both produced in rabbit, were carried out using the Zenon Kit (Invitrogen) using Alexa Fluor 488 and 546 for the visualization of Rrp6 and Rrp4, respectively.
SDS-PAGE and Western blotting
Proteins were separated by SDS-PAGE using the Mini-Protean II system (BioRad) and transferred to polyvinylidenefluoride (PVDF) membranes (Millipore) in Tris-glycine buffer with 0.02% SDS and 4 M urea using a semi-dry electrophoretic transfer cell (BioRad). The membranes were probed with antibodies following standard procedures.
Immunofluorescent staining of salivary glands
Immunofluorescent staining of salivary glands was carried out as described by Kiesler et al. (2005). Salivary glands from C. tentans were dissected from fourth instar larvae. Glands were incubated with primary antibodies at 4°C overnight. The working concentrations of the antibodies were 5–10 μg/mL. Sera were diluted 1/500–1/1000. The glands were washed and incubated with fluorescent secondary antibodies, then mounted with Vectashield mounting medium (Vector Laboratories Inc.) and examined in an LSM 510 laser confocal microscope (Carl Zeiss). A more detailed description of the method is provided as Supplemental Data.
Immunofluorescent staining of isolated chromosomes
C. tentans polytene chromosomes were isolated from the salivary glands of fourth instar larvae and fixed essentially as described by Björkroth et al. (1988). The isolated chromosomes were incubated with primary antibody in 0.5% BSA in TKM buffer at 4°C overnight. TKM was 10 mM triethanolamine-HCl, 100 mM KCl, and 1 mM MgCl2. The working concentrations of the antibodies were 5–10 μg/mL. The chromosomes were washed, incubated with fluorescent secondary antibodies, mounted with Vectashield mounting medium (Vector Laboratories Inc.), and examined in an LSM 510 laser confocal microscope (Carl Zeiss). For experiments of RNA digestion, the chromosomes were incubated with 100 μg/mL RNase A for 30 min at 4°C and washed with TKM before being fixed and processed as above. For DRB treatments, the larvae were kept in culture water supplemented with 0.4 mM DRB for 45 min before dissection of the glands and isolation of the chromosomes.
Acquisition and processing of confocal images
Preparations were analyzed and images were taken with a laser scanning microscope (model LSM 510; Carl Zeiss MicroImaging Inc.) equipped with PlanApochromat objectives 40×/1.0 oil and 63×/1.4 oil, using immersion oil Immersol 518F (Carl Zeiss MicroImaging Inc.). The optical sections were 1 μm thick. Photoshop software (Adobe) was used for the preparation of composite images and for adjustment of intensity and contrast.
Immunoelectron microscopy (Immuno-EM)
Immuno-EM was carried out using cryosectioned salivary glands as described by Sjölinder et al. (2005). Salivary glands were fixed for 45 min in 4% paraformaldehyde and cryoprotected with 2.3 M sucrose. Secondary antibodies were conjugated to 6-nm gold particles (Jackson ImmunoResearch Laboratories). After immunolabeling, the sections were stained with 2% aqueous uranyl acetate and embedded in polyvinyl alcohol (Aldrich). The specimens were examined and photographed in a Tecnai G2 Spirit BioTwin electron microscope (FEI Company) at 80 kV. Immuno-EM labeling of isolated chromosomes was essentially carried out as described by Tyagi et al. (2009). More detailed descriptions of the methods are provided as Supplemental Data.
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described by Takahashi et al. (2000) with some modifications (Hessle et al. 2009). When using D. melanogaster S2 cells, ∼1 × 107 cells were used for each immunoprecipitation. The cells were fixed at room temperature by the addition of a fixing solution containing formaldehyde, to give a final concentration of 2%. The chromatin was sheared by sonication to render DNA fragments with a length of 250–900 bp. Immunoprecipitation was performed overnight at 4°C using 10 μg/mL of primary antibody. In the RNase experiments, the washed beads were incubated in the presence of 50 μg/mL RNase A for 30 min at room temperature before elution of the bound complexes and DNA purification. For real-time quantitative PCR (qPCR), the immunoprecipitated DNA was amplified using the KAPA SYBR Fast qPCR Master Mix (KAPA Biosystems) in a RotorGene (Qiagen). ChIP on C. tentans salivary glands was carried out as described by Botelho et al. (2008), except for the analysis of the immunoprecipitated DNA. The immunoprecipitated DNA was quantified by qPCR using a standard curve for each primer pair. A more detailed description of the method is provided as Supplemental Data.
RNA immunoprecipitation (RIP)
C. tentans tissue-cultured cells were used to prepare a soluble nuclear extract containing nucleoplasmic mRNPs as described by Tyagi et al. (2009). The cells were homogenized in PBS buffer containing 0.2% NP-40 and protease inhibitors (Roche Diagnostics) using a glass homogenizer. PBS was 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 2 mM NaH2PO4 at pH 7.2. The homogenate was centrifuged at 1500g for 10 min at 4°C. The pellet containing the nuclei was resuspended in PBS containing protease inhibitors, sonicated three times for 4–5 sec each time, and centrifuged at 16,000g for 10 min at 4°C. The resulting supernatant was the soluble nuclear extract used for immunoprecipitation with either anti-Rrp6, anti-Hrp36, or anti-U2B″ antibodies. A negative control immunoprecipitation was processed in parallel without any primary antibody. Total RNA was purified from the eluates and treated with DNase TURBO (Ambion). Superscript III (Invitrogen) was used for cDNA synthesis using random primers (Invitrogen). The resulting cDNA was quantified by qPCR using the KAPA SYBR Fast qPCR Master Mix (KAPA Biosystems) in a RotorGene (Qiagen). Standard curves were used for quantification.
Primer sequences
The primers used for qPCR quantification of ChIP results for the Fur2, Mtor, and Tctp genes are specified in Hessle et al. (2009). The sequences of the primers used to quantify the BR1 sequences were Proximal-F AAGAGGAGGATTGCCCAAAT, Proximal-R TCAGGTTGTTCGGGTCCTAC, Distal-F GCGATTCACAAAATTTGCCT, and Distal-R AAACTTTCGCTTGCCTTTGA. The sequences of the primers used to quantify the RIP results were Gart-T-F TGGATTACAAATGGTGGTCG, Gart-T-R CGCCACTTTCTTTGTAGGTCA, Gart-U-F was the same as Gart-T-F, Gart-U-R GAGGAAATAAAATGATTGGAATGAG, Gart-S-F was the same as Gart-T-F, Gart-S-R GTCAATGAATGTATTTGGAAAGC, Hrp65-T-F TGTGAATCAACGACCACAAAA, Hrp65-T-R TTTTCTAATATCACATTTGGAGAACG, Hrp65-U-F TTTTTGACTTGTACAAAATGTCCTT, Hrp65-U-R TGCATCCATTCAACTAGCCA, Hrp65-S-F AAGGAAATCAGAGTATTATCGTC, and Hrp65-S-R was the same as Hrp65-U-R.
DATA DEPOSITION
The partial cDNA sequence for Ct-Rrp6 was deposited in the EMBL Nucleotide Sequence Database with accession number HE613798.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank A.B. Eberle for critical reading of the manuscript. Our research is financed by grants from the Swedish Research Council (VR-NT), the Swedish Cancer Society, and the ESF-Eurocores Programme on RNA Quality Control. V.H. was supported by the Lennander's Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032045.111.
REFERENCES
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P 1999. The yeast exosome and human PM–Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev 13: 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841 [DOI] [PubMed] [Google Scholar]
- Baurén G, Wieslander L 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192 [DOI] [PubMed] [Google Scholar]
- Björkroth B, Ericsson C, Lamb MM, Daneholt B 1988. Structure of the chromatin axis during transcription. Chromosoma 96: 333–340 [Google Scholar]
- Botelho SC, Tyagi A, Hessle V, Ostlund Farrants AK, Visa N 2008. The association of Brahma with the Balbiani ring 1 gene of Chironomus tentans studied by immunoelectron microscopy and chromatin immunoprecipitation. Insect Mol Biol 17: 505–513 [DOI] [PubMed] [Google Scholar]
- Daneholt B 2001. Assembly and transport of a pre-messenger RNP particle. Proc Natl Acad Sci 98: 7012–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, García-Sacristán A, Custódio N, Carmo-Fonseca M 2010. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res 38: 8015–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Hessle V, Helbig R, Dantoft W, Gimber N, Visa N 2010. Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes. PLoS ONE 5: e11540 doi: 10.1371/journal.pone.0011540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jaquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Graham AC, Kiss DL, Andrulis ED 2006. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell 17: 1399–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Kiss DL, Andrulis ED 2009. Core exosome-independent roles for Rrp6 in cell cycle progression. Mol Biol Cell 20: 2242–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessle V, Björk P, Sokolowski M, Gonzáles de Valdivia E, Silverstein R, Artemenko K, Tyagi A, Maddalo G, Ilag L, Helbig R, et al. 2009. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol Biol Cell 20: 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2008. The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast. Biochim Biophys Acta 1779: 239–246 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Kiesler E, Visa N 2004. Intranuclear pre-mRNA trafficking in an insect model system. Prog Mol Subcell Biol 35: 99–118 [DOI] [PubMed] [Google Scholar]
- Kiesler E, Hase ME, Brodin D, Visa N 2005. Hrp59, an hnRNP M protein in Chironomus and Drosophila, binds to exonic splicing enhancers and is required for expression of a subset of mRNAs. J Cell Biol 168: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Andrulis ED 2011. The exozyme model: A continuum of functionally distinct complexes. RNA 17: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci 78: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Conti E 2008. Structural organization of the RNA-degrading exosome. Curr Opin Struct Biol 18: 709–713 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A 2011. The eukaryotic RNA exosome: Same scaffold but variable catalytic subunits. RNA Biol 8: 61–66 [DOI] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U 1992. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell 69: 605–613 [DOI] [PubMed] [Google Scholar]
- Meyer B, Mähr R, Eppenberger HM, Lezzi M 1983. The activity of Balbiani rings 1 and 2 in salivary glands of Chironomus tentans larvae under different modes of development and after pilocarpine treatment. Dev Biol 98: 265–277 [DOI] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT 2011. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 25: 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Visa N 2001. Molecular characterization of Ct-hrp65: Identification of two novel isoforms originated by alternative splicing. Exp Cell Res 264: 284–295 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Neugebauer KM 2002. On the importance of being co-transcriptional. J Cell Sci 115: 3865–3871 [DOI] [PubMed] [Google Scholar]
- Paulsson G, Bernholm K, Wieslander L 1992. Conserved and variable repeat structures in the Balbiani ring gene family in Chironomus tentans. J Mol Evol 35: 205–216 [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH 2008. Quality control of mRNP in the nucleus. Chromosoma 117: 419–429 [DOI] [PubMed] [Google Scholar]
- Sjölinder M, Björk P, Söderberg E, Sabri N, Östlund Farrants AK, Visa N 2005. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev 19: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Björkroth B, Lamb MM, Daneholt B 1983. Visualization of the formation and transport of a specific hnRNP particle. Cell 34: 847–855 [DOI] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Shuster G, Heck AJ, Raijmakers R, Pruijn GJ 2010. Dis3-like 1: A novel exoribonuclease associated with the human exosome. EMBO J 29: 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synowsky SA, van Wijk M, Raijmakers R, Heck AJ 2009. Comparative multiplexed mass spectrometric analyses of endogenously expressed yeast nuclear and cytoplasmic exosomes. J Mol Biol 385: 1300–1313 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD 2000. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes Dev 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Drazkowska K, Dziembowski A 2010. Mechanisms of RNA degradation by the eukaryotic exosome. Chembiochem 3: 938–945 [DOI] [PubMed] [Google Scholar]
- Tyagi A, Ryme J, Brodin D, Östlund-Farrants AK, Visa N 2009. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet 5: e1000470 doi: 10.1371/journal.pgen.1000470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Wagner EJ 2011. A brief survey of mRNA surveillance. Trends Biochem Sci 36: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreria J, Daneholt B, Mattaj IW 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol 33: 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L 1994. The Balbiani ring multigene family: Coding repetitive sequences and evolution of a tissue-specific cell function. Prog Nucleic Acid Res Mol Biol 48: 275–313 [DOI] [PubMed] [Google Scholar]
- Wieslander L, Bauren G, Bernholm K, Jiang WQ, Wetterberg I 1996. Processing of pre-mRNA in polytene nuclei of Chironomus tentans salivary gland cells. Exp Cell Res 229: 240–246 [DOI] [PubMed] [Google Scholar]
- Wyss C 1982. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp Cell Res 139: 297–307 [DOI] [PubMed] [Google Scholar]