Malat1 is an abundant long noncoding RNA that localizes to nuclear bodies known as nuclear speckles, which contain a distinct set of pre-mRNA processing factors. Previous studies in cell culture have demonstrated that Malat1 interacts with pre-mRNA splicing factors, including the serine- and arginine-rich (SR) family of proteins, and regulates a variety of biological processes, including cancer cell migration, synapse formation, cell cycle progression, and responses to serum stimulation. To address the physiological function of Malat1 in a living organism, the authors generated Malat1-knockout (KO) mice using homologous recombination. Unexpectedly, the Malat1-KO mice were viable and fertile, showing no apparent phenotypes.
Keywords: nuclear speckles, Malat1, paraspeckles
Abstract
Malat1 is an abundant long, noncoding RNA that localizes to nuclear bodies known as nuclear speckles, which contain a distinct set of pre-mRNA processing factors. Previous studies in cell culture have demonstrated that Malat1 interacts with pre-mRNA splicing factors, including the serine- and arginine-rich (SR) family of proteins, and regulates a variety of biological processes, including cancer cell migration, synapse formation, cell cycle progression, and responses to serum stimulation. To address the physiological function of Malat1 in a living organism, we generated Malat1-knockout (KO) mice using homologous recombination. Unexpectedly, the Malat1-KO mice were viable and fertile, showing no apparent phenotypes. Nuclear speckle markers were also correctly localized in cells that lacked Malat1. However, the cellular levels of another long, noncoding RNA—Neat1—which is an architectural component of nuclear bodies known as paraspeckles, were down-regulated in a particular set of tissues and cells lacking Malat1. We propose that Malat1 is not essential in living mice maintained under normal laboratory conditions and that its function becomes apparent only in specific cell types and under particular conditions.
INTRODUCTION
Evidence from a growing number of studies has revealed the functional importance of long, non-protein-coding RNAs (lncRNAs), which constitute a large proportion of the transcriptional output of the mammalian genome (for review, see Mercer et al. 2009; Hung and Chang 2010). Although chromatin modification is a characteristic function of lncRNAs (for review, see Hung and Chang 2010), a group of lncRNAs has been shown to be retained within the nucleus and to localize to specific nuclear bodies (for review, see Prasanth and Spector 2007; Ip and Nakagawa 2012). Malat1 (metastasis-associated lung adenocarcinoma transcript 1) was originally identified as a gene that was specifically up-regulated in metastatic non-small-cell lung cancer cells (Ji et al. 2003), but has subsequently been recharacterized as one of the two long, noncoding RNAs that accumulate in the nucleus and is referred to as NEAT2 (nuclear-enriched noncoding transcript 2) (Hutchinson et al. 2007). The nucleus of higher eukaryotes is functionally divided into multiple nuclear bodies that contain a specific group of proteins and nucleic acids that are involved in particular nuclear processes (for review, see Cremer et al. 2004; Platani and Lamond 2008; Hubner and Spector 2010). Malat1 localizes to one type of these nuclear bodies, known as nuclear speckles, which contain various pre-mRNA splicing regulators, including uridine-rich small nuclear RNA–protein complexes (UsnRNPs) and the serine- and arginine-rich (SR) family of splicing factors, which are involved in exon recognition and alternative splicing (for review, see Hall et al. 2006; Spector and Lamond 2011). MALAT1 interacts with several SR splicing factors, including SRSF1, 2, and 3, and is required for their localization to nuclear speckles (Tripathi et al. 2010). In cultured mouse hippocampal neurons, Malat1 modulates synaptogenesis by regulating the expression of genes involved in synaptogenesis (Bernard et al. 2010). Cultured human cancerous cell lines depleted with MALAT1 contain increased levels of SR proteins, including the dephosphorylated forms, which display more homogeneous nuclear distribution (Tripathi et al. 2010). Interestingly, several alternative splicing events are dysregulated in cells lacking MALAT1 (Tripathi et al. 2010; Lin et al. 2011). MALAT1 also influences the migratory behavior of several human cell lines by regulating the expression of motility-related genes (Tseng et al. 2009; Tano et al. 2010). Furthermore, a recent study has demonstrated that MALAT1 is essential for serum-stimulated gene expression through its interaction with the nonmethylated form of the Polycomb 2 protein (Pc2) in coactivator complexes (Yang et al. 2011). Although all of these studies clearly indicate that MALAT1 has important functions in a variety of biological processes, the exact nature of those functions in living organisms remains unknown.
To examine the physiological roles of Malat1 in living animals, we used homologous recombination to create a knockout (KO) mouse of Malat1. Surprisingly, the KO mice were fertile and viable, and no apparent abnormality was observed. We suggest that Malat1 is not essential in mouse cells under normal physiological conditions but that it becomes essential in particular cell types, such as metastatic cancer cells.
RESULTS
Malat1 knockout mice are viable and fertile
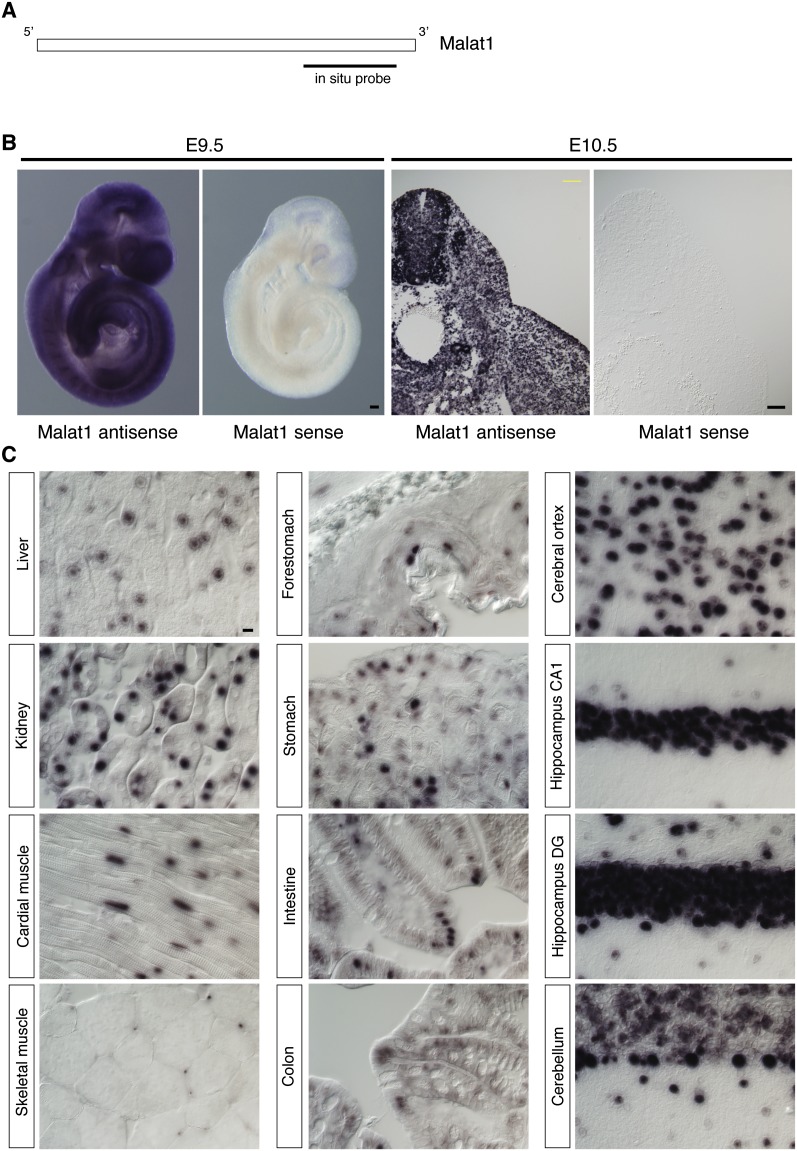
We initially examined the expression pattern of Malat1 during early embryonic development and in adult tissues using in situ hybridization. Consistent with previously reported Northern blot and RT-PCR results (Ji et al. 2003; Hutchinson et al. 2007; Bernard et al. 2010), Malat1 displayed ubiquitous expression in all of the cell types during the early embryonic stages, including embryonic days 9.5 (E9.5) and E10.5 (Fig. 1B). In the adult tissues, Malat1 expression varied markedly between tissue types, with the highest levels of expression in the brain (Fig. 1C). However, essentially all of the cells expressed some level of Malat1.
FIGURE 1.
The expression pattern of Malat1 during early embryonic development and in adult tissues. (A) A schematic of the positions of the probes. (B) Whole-mount and section in situ hybridization of E9.5 and E10.5 mouse embryos with Malat1 antisense and sense probes. Note that strong and uniform expression was detected using the antisense probes, whereas no expression was detected using the sense probes. (C) In situ hybridization of adult organs using Malat1 antisense probes. (CA1) Hippocampus CA1; (DG) dentate gyrus. Scale bars, (B) 100 μm; (C) 10 μm.
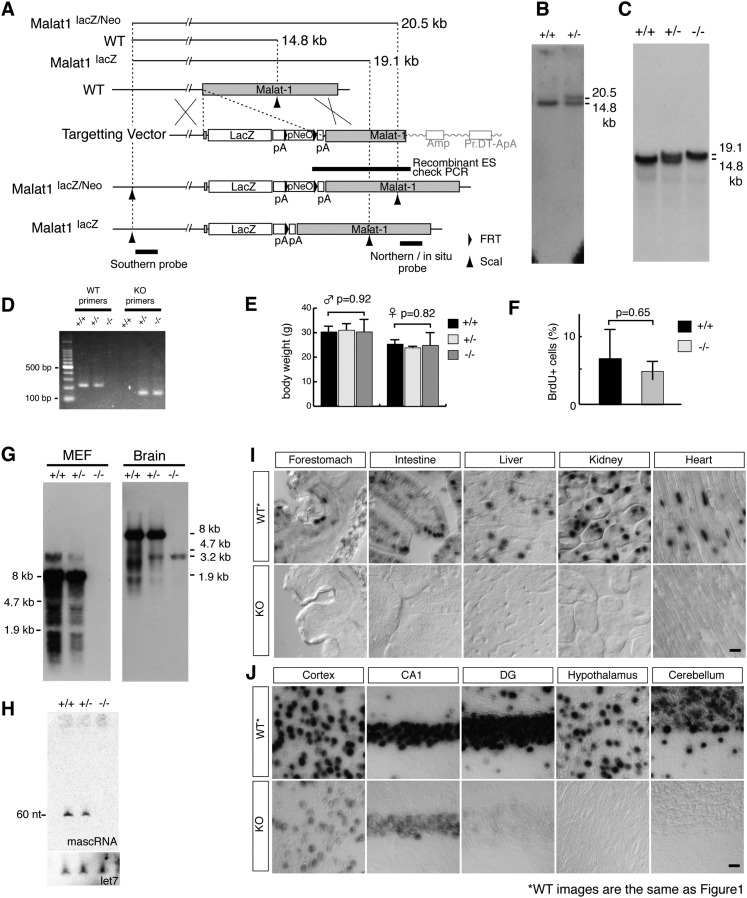
To address the physiological function of Malat1 at the organismal level, we used a gene-targeting strategy and inserted the β-galactosidase (lacZ) gene followed by polyadenylation signals immediately downstream from the transcriptional start site of Malat1 (Fig. 2A–D). Unexpectedly, the homozygous mutant mice were born at an expected Mendelian ratio (χ-square value = 0.48) (Table 1); we found no apparent abnormality in their external morphology or body weight; and the knockout mice were fertile (Fig. 2E; Table 1). In addition, mouse embryonic fibroblast cells (MEFs) obtained from the KO mice responded to serum stimulation at similar levels relative to wild-type MEFs (Fig. 2F). A Northern blot analysis using RNA samples from MEFs and brain tissues confirmed the complete loss of full-length Malat1 transcripts (Fig. 2G). The Malat1 locus also codes for a small ∼60-nt mascRNA (MALAT1-associated small cytoplasmic RNA), which is synthesized through the 3′ processing of the long Malat1 RNA (Wilusz et al. 2008). The Northern hybridization results confirmed the absence of this mascRNA in the Malat1-KO MEFs (Fig. 2H). Notably, Northern hybridization using a Malat1 RNA probe from the brains of heterozygous and KO mice showed a weak but specific band at 3.2 kb (Fig. 2G, +/− and −/−). RNA in situ hybridization analyses of various tissues from the KO mice also confirmed the complete loss of Malat1 expression in non-neural tissues (Fig. 2I). In contrast, residual levels of the Malat1 signal were observed in specific neural tissues of Malat1-KO mice, with relatively high expression in the cerebral cortex and the hippocampal CA1 region, whereas no expression was observed in the hypothalamus or the cerebellum (Fig. 2J). Based on the probes that were used for the Northern blot analysis and the in situ hybridizations (Figs. 1A, 2A), the neural-specific 3.2-kb RNA was thought to be produced from the 3′ region of Malat1, which suggests that a specific internal promoter is artificially activated in certain neural tissue of the KO mice. At the subnuclear level, the distribution of the 3.2-kb Malat1 fragment overlapped with Srsf1 in the cortical neurons of KO mice (Fig. 4D, below), suggesting that the transcripts generated from the 3′ portion of Malat1 are localized to nuclear speckles. A similar observation was reported previously, in which the 3′ fragment of Malat1 RNA localized to nuclear speckles when expressed exogenously in human cancerous cells (Tripathi et al. 2010). Standard histological analyses using adult tissues showed no differences between the wild-type and KO mice (Fig. 3A). In the brain, a synaptic marker, synapsin, was normally distributed in the brain of Malat1-KO mice (Fig. 3B).
FIGURE 2.
The generation of Malat1-KO mice. (A) A schematic of the targeting strategy for the disruption of the Malat1 gene. The lacZ and polyadenylation signals were inserted 69 bp downstream from the transcription start site of Malat1 (NR_002847). The founder heterozygous mice (Malat1lacZ/Neo allele) were crossed to Flp-expressing mice, and the Malat1lacZ allele without the promoter neomycin cassette (pNeO) was generated. (B) Southern blot analysis of ScaI-digested DNA from targeted ES cells. (C) Southern blot analysis of ScaI-digested DNA from tails of wild type (+/+), heterozygous (+/−), and homozygous (−/−) Malat1lacZ littermates. (D) A gel image of PCR products amplified by the genotyping primers. (E) The average body weight of wild-type (+/+), heterozygous (+/−), and homozygous (−/−) Malat1lacZ mice (n = 5 for each genotype). (F) The percentage of BrdU+ cells after serum stimulation. Two sets of wild-type (WT) and knockout (KO) MEFs were obtained from two independent siblings, and the percentage of BrdU+ cells was calculated using three replicate cultures for each MEFs. The error bars in E and F indicate the standard deviations, and the P-values were calculated from a two-tailed, nonequal variance t-test. (G) Northern blot analysis of Malat1 expression in the MEFs and whole-brain tissues of Malat1-KO mice. Note the complete loss of full-length Malat1, whereas weak 3.2-kb signals of the targeted allele were detected in the brain. (H) Northern blot analysis of mascRNA expression in MEFs. (I,J) The in situ hybridization of Malat1 using antisense probes in non-neural (I) and neural (J) adult tissues of wild-type and Malat1-KO mice. Note the complete loss of Malat1 expression in the KO mice in non-neural tissues, whereas a significant level of expression was detected in particular areas of the brain, including the cerebral cortex, hippocampus CA1 (CA1), and dentate gyrus (DG). Essentially no signals were detected in certain brain regions, such as the hypothalamus and cerebellum. The wild-type images are the same as those in Figure 1. Scale bar, 10 μm.
TABLE 1.
The genotypes of the offspring from the MALAT1-KO mice
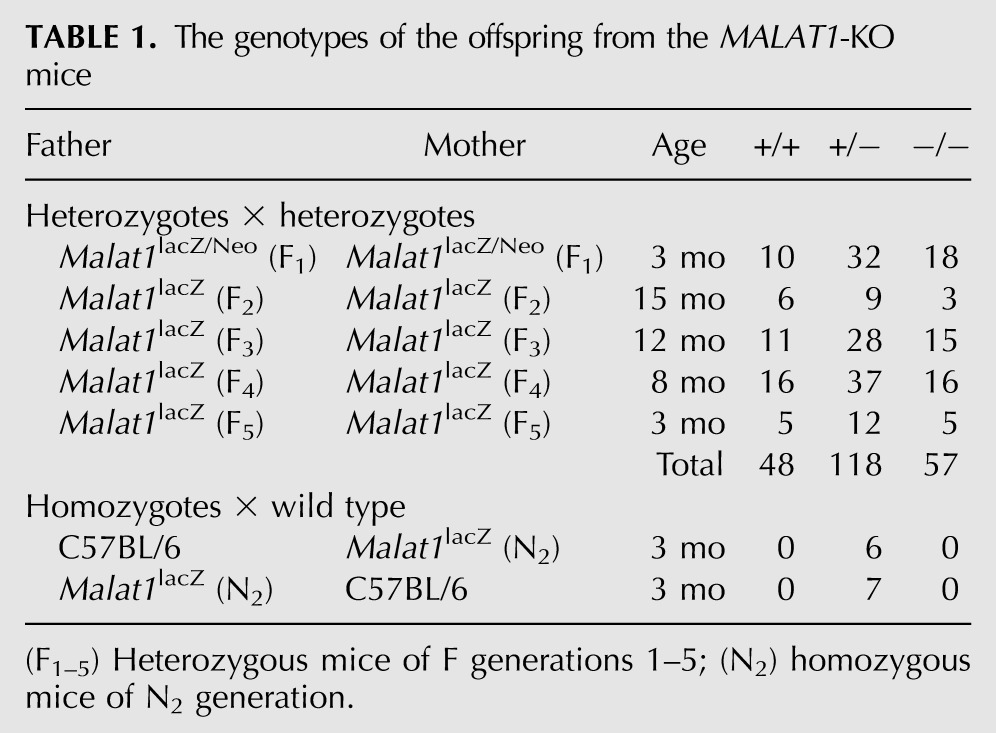
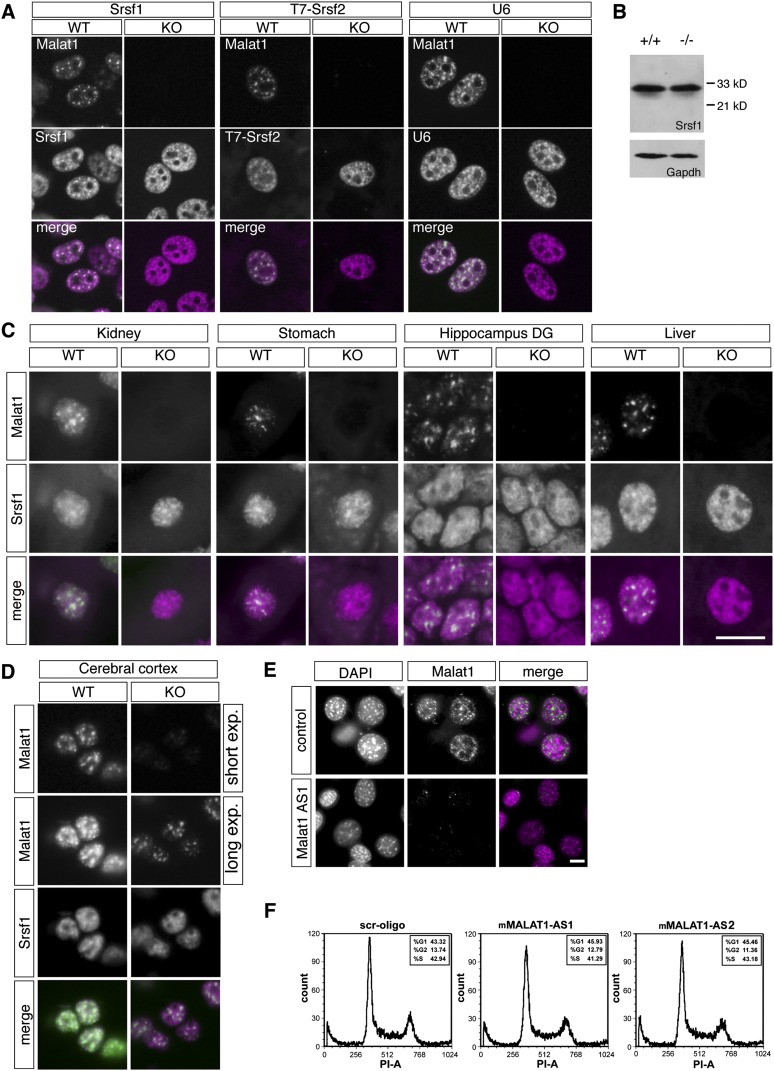
FIGURE 4.
Nuclear speckle marker expression in MEFs and various tissues and the normal proliferation of Malat1-depleted cells. (A) The simultaneous detection of Malat1 and nuclear speckle markers in MEFs from the WT and homozygous KO mice. To examine the distribution of Srsf2, MEFs were transfected with T7-Srsf2, and the tagged proteins were visualized using immunostaining against T7. An uneven and speckled distribution of the nuclear speckle components was observed in the MEFs from KO mice. (B) Western blot analysis of Srsf1 expression in MEFs from the mutant mice. (C) The expression pattern of Malat1 and Srsf1 in the kidney, stomach, hippocampus dentate gyrus (DG), liver, and cerebral cortex. The speckled distribution of Srsf1 was observed in the Malat1-KO mice. (D) The residual expression of Malat1 in the cerebral cortex. In the cerebral cortex of the KO mice, the short form of Malat1 (Fig. 1B) was correctly localized to the nuclear speckles and overlapped with the intense signals of Srsf1. Note that the expression level of the short transcript in the Malat1-KO mice was much weaker than the full-length transcript (Fig. 2G) in the WT mouse and could only be detected only in overexposed images. (E) The depletion of Malat1 in NIH3T3 cells using antisense oligonucleotides. The knockdown of Malat1 did not cause fragmentation of the cell nuclei. Note that this AS oligonucleotide is complementary to a region in MALAT1 that is completely conserved between mouse and human. HeLa cells depleted of MALAT1, using the same oligo displayed nuclear breakdown phenotype (Tripathi et al. 2010). (F) The cell cycle distribution of NIH3T3 cells depleted of Malat1. Scale bars, 10 μm.
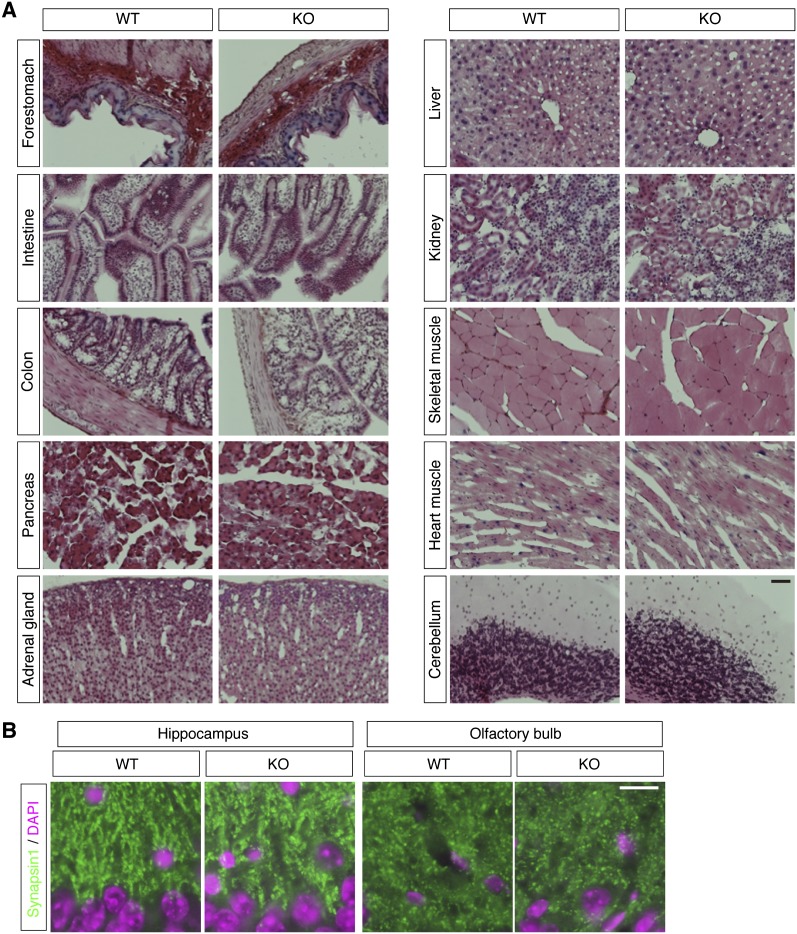
FIGURE 3.
Histological analyses of Malat1 knockout mice. (A) Hematoxylin-eosin staining of frozen sections from various adult tissues from wild-type (WT) and Malat1 knockout (KO). No apparent abnormalities were observed in the tissues from Malat1-KO mice. (B) Expression pattern of a synaptic marker protein synapsin (green) in the hippocampus and the olfactory bulb of WT and Malat1-KO brain. Nuclei were counterstained with DAPI (magenta). Scale bars, (A) 100 μm; (B) 10 μm.
Malat1 does not influence the localization of nuclear speckle components in mice
Previous studies using human cancerous cells have demonstrated that the depletion of MALAT1 using antisense oligonucleotides dramatically increases the cellular pool of SR splicing factors (SRSF1 and 2), including the hypophosphorylated forms, which are evenly dispersed throughout the nucleoplasm (Tripathi et al. 2010; Lin et al. 2011). Therefore, we examined the localization of Srsf1 and Srsf2 in MEFs prepared from KO mice that completely lacked Malat1 (Fig. 4A). Surprisingly, all of these nuclear speckle markers exhibited similar localization patterns in wild-type and Malat1-KO MEFs (Fig. 4A). Distribution of U6 snRNAs, which are known to localize to nuclear speckles (Carmo-Fonseca et al. 1992), was also not affected by the loss of Malat1 expression (Fig. 4A). In addition, we found no increase in the hypophosphorylated form of Srsf1 in the MEFs from the KO mice (Fig. 4B). We next examined the distribution of Srsf1 in various tissues from the KO mice (Fig. 4C,D). Compared with the MEFs, the enrichment of Srsf1 in nuclear speckles was not prominent in mouse tissues, even in the wild-type animals; however, the speckle distribution was clearly recognizable in both the wild-type and KO mice (Fig. 4C,D). These results were largely different from previous knockdown experiments that showed the essential roles of Malat1 in a variety of biological processes in human cell lines (Tano et al. 2010; Tripathi et al. 2010; Lin et al. 2011; Yang et al. 2011). To further investigate whether the lack of an obvious phenotype in the KO mice was due to the differential experimental approach (i.e., KO vs. knockdown using siRNA or antisense [AS] oligonucleotides), we depleted Malat1 in NIH3T3 cells using Malat1-specific AS oligonucleotides (Fig. 4E). The AS oligonucleotide-mediated depletion of Malat1 did not lead to any apparent defects, and the growth of the Malat1-depleted cells was comparable to the control oligonucleotide-treated cells (Fig. 4F). One of the Malat1-AS oligonucleotides targets a region of Malat1 that is completely conserved between mouse and human transcripts (Tripathi et al. 2010). Importantly, the depletion of MALAT1 in human-derived HeLa cells using the same Malat1-AS oligonucleotide showed changes in the cellular distribution of SR proteins, resulting in the abnormal alternative splicing of pre-mRNAs and mitotic catastrophe (Tripathi et al. 2010). These observations suggest that, in contrast to MALAT1 in HeLa cells, Malat1 in mouse fibroblasts does not influence the distribution and activity of pre-mRNA splicing factor and may not be essential for cell cycle progression.
Malat1 controls Neat1 expression and paraspeckle formation in particular cell types
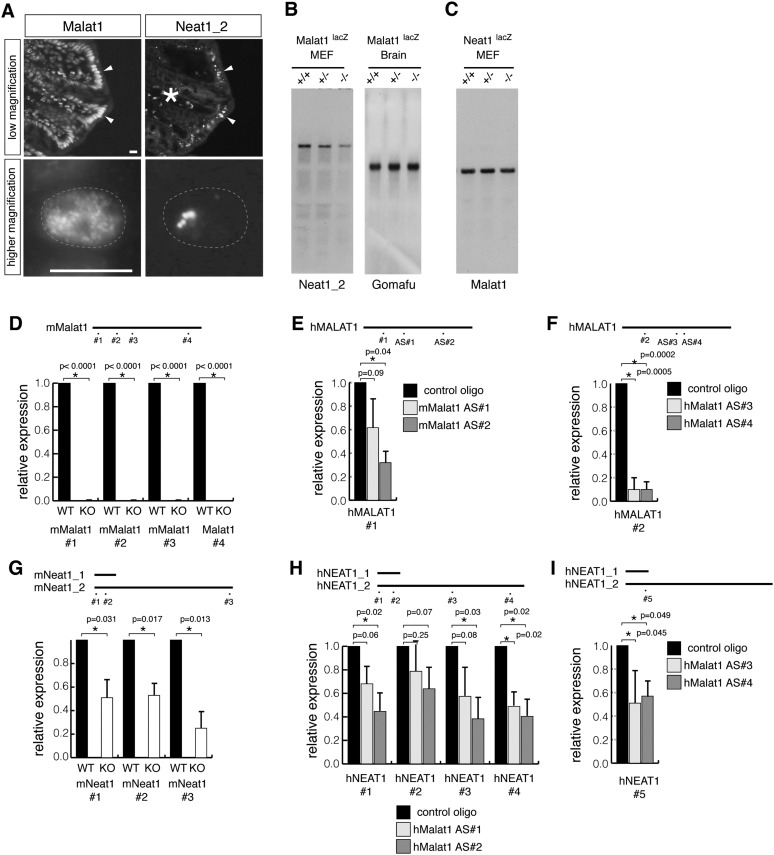
During the course of these studies, we noticed that, in certain tissues such as intestine and colon, cell types that displayed increased levels of Malat1 also expressed higher levels of the lncRNA Neat1, an architectural component of the nuclear bodies known as paraspeckles (Fig. 5A; Fox et al. 2002; Bond and Fox 2009). Accordingly, we speculated that Malat1 might control the expression of Neat1, at least in certain cell types in the KO mice. The Neat1 locus generates two isoforms, Neat1_1 and Neat1_2, and the longer isoform, Neat1_2, primarily regulates the formation of paraspeckles (Sasaki et al. 2009; Sunwoo et al. 2009). Interestingly, Neat1_2 expression was significantly decreased in the Malat1-KO MEFs (Fig. 5B, left), whereas the expression of another nuclear noncoding RNA, Gomafu (Sone et al. 2007), was unchanged (Fig. 5B, right). In addition, Malat1 levels were not different in MEFs prepared from Neat1-KO mice (Fig. 5C; Nakagawa et al. 2011), suggesting that the regulation of these two abundant nuclear noncoding RNAs is not reciprocal. Reverse transcription–quantitative PCR (RT-qPCR) analysis confirmed an ∼50% reduction in Neat1_1 and Neat1_2 levels in MEFs obtained from Malat1-KO mice (Fig. 5D,G). Because the Malat1 and Neat1 genes are located next to each other in the same chromosomal region, it is possible that the insertion of the β-galactosidase cassette into the Malat1 locus disrupted enhancer sequences required for Neat1 expression, leading to the down-regulation of the neighboring gene. To test this possibility, we examined NEAT1 levels in several human cell lines that were depleted of MALAT1 using MALAT1-specific antisense oligonucleotides, and we found that NEAT1 levels were decreased in A549 (adenocarcinoma) and HCT116 (colorectal carcinoma) upon MALAT1 depletion (Fig. 5E,F,H,I).
FIGURE 5.
Reduction of Neat1 expression in cells lacking Malat1. (A) The simultaneous detection of Malat1 and Neat1_2 in the colon. NEAT1_2 is specifically expressed in colonic surface epithelial cells facing the lumen (the arrowheads in low magnification), which also express higher levels of Malat1. (*) The deep region of the colon epithelium with auto-fluorescent signals. (B) Northern blot analysis of the expression of Neat1_2 and Gomafu in the MEFs and brain, respectively. RNAs were obtained from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) Malat1lacZ littermates. Note the marked decrease in Neat1_2 in the MEFs prepared from the KO mice. (C) Northern blot analysis of the expression of Malat1 in MEFs obtained from NEAT1lacZ littermates. Malat1 expression was not affected by the loss of NEAT1 expression. (D–F) RT-qPCR analysis of the expression of Malat1/MALAT1 in MEFs from Malat1-KO mice (D), A549 cells (E), and HCT116 cells (F). (G–I) RT-qPCR analysis of the expression of NEAT1_1/2 in MEFs from Malat1-KO mice (G), A549 cells (H), and HCT116 cells (I). In A–F, the positions of the primers used for the RT-qPCR analysis are indicated in the schematic. The error bars show the standard deviations, and the P-values were calculated from a two-tailed, nonequal variance t-test using two biological replicates (n = 2) for D, E, G, and H and triplicates (n = 3) for F and I. (*) p < 0.05.
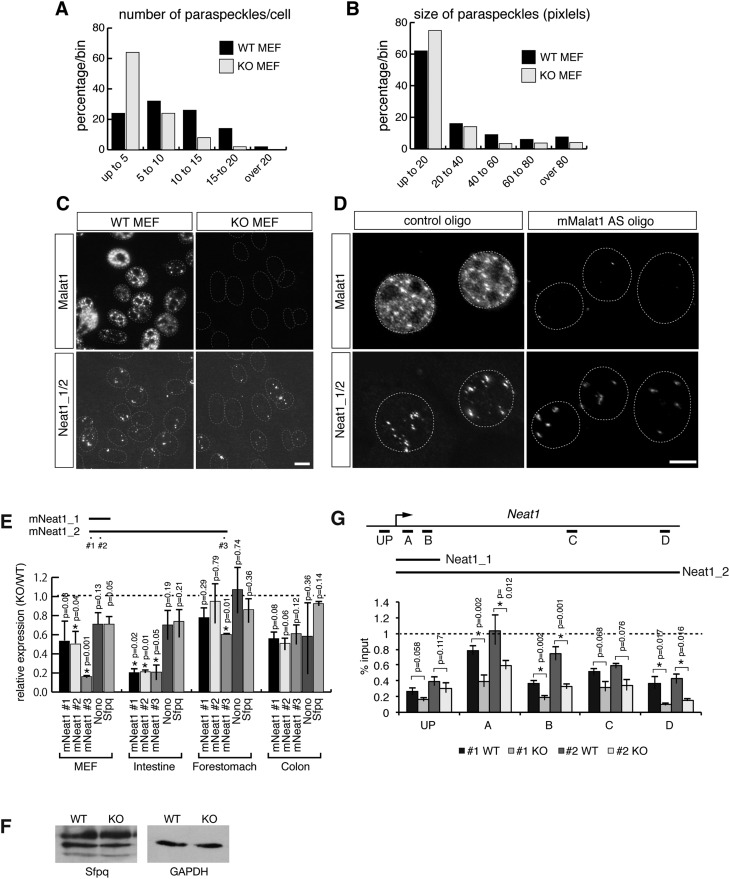
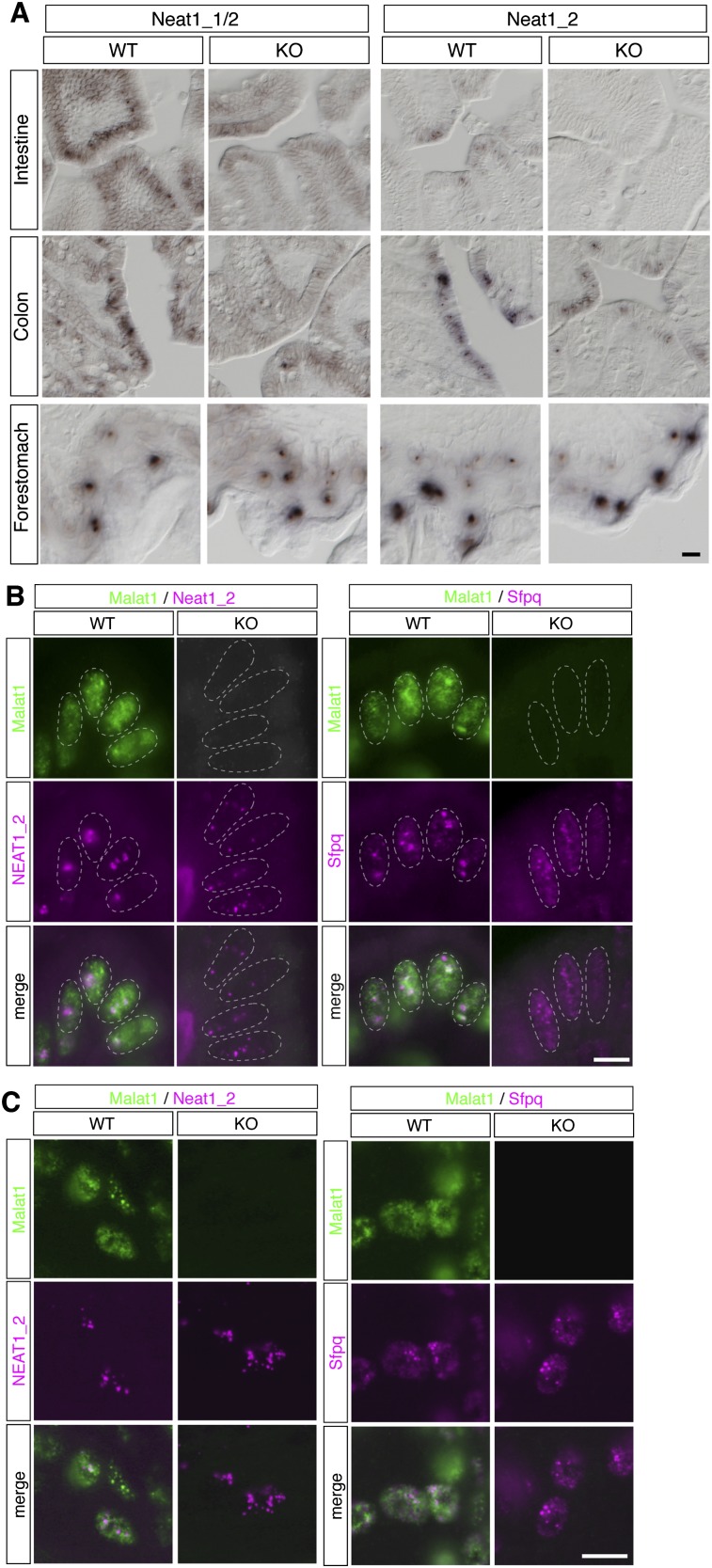
Because Neat1 is a structural component of paraspeckles (Chen and Carmichael 2009; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009), we then examined whether the number or size of the paraspeckles was affected in cells lacking Malat1. Two pairs of MEFs from wild-type and KO mouse embryos were obtained from two independent siblings, and we quantified the size and number of the paraspeckles by determining the overlap of Neat1 signals with the paraspeckle marker Sfpq (Fig. 6A–C; Fox et al. 2002; Prasanth et al. 2005). As expected, the MEFs from the Malat1-KO mice contained fewer paraspeckles (Fig. 6A; average number WT = 9.9 and KO = 5.4, p = 0.000003, two-tailed, nonequal variance t-test), and the size of the paraspeckles was also decreased (Fig. 6B; average size WT = 35.3 pixels and KO = 25.5 pixels, p = 0.0004, two-tailed, nonequal variance t-test). Similar decreases in the number and size of the paraspeckles were also observed in NIH3T3 cells depleted of Malat1 (Fig. 6D). We also examined the expression pattern of Neat1_1 and Neat1_2 in intestinal and colonic epithelial cells, cell types that contain prominent paraspeckles (Nakagawa et al. 2011). Consistent with the results obtained in MEFs and NIH3T3 cells, both Neat1_1 and Neat1_2 were down-regulated in the intestine and colon (Figs. 6E, 7A), and the reduced formation of paraspeckles was confirmed by the expression of Sfpq (Fig. 7B). Importantly, the effect of Malat1 knockout on Neat1 expression was tissue-specific. The intestines of Malat1-KO mice showed particularly low levels of Neat1, whereas cells from other tissues including forestomach showed normal levels of Neat1 (Figs. 6E, 7A) and formed distinct paraspeckles (Fig. 7C).
FIGURE 6.
Reduction of the formation of paraspeckles in cells lacking Malat1. (A,B) Histograms of the size and number of paraspeckles in the wild-type and Malat1-KO MEFs. (C) The expression of Malat1 and Neat1_1/2 in MEFs from WT and homozygous (KO) Malat1 lacZ mice. (D) The expression of Malat1 and NEAT1_1/2 in NIH 3T3 cells treated with control and antisense oligonucleotides against Malat1. Scale bar, 10 μm. (E) The relative expression of Neat1_1/2, Neat1_2, Sfpq, and Nono in MEFs and various tissues from Malat1-KO mice compared with the wild-type animals. Two sets of WT and KO samples were obtained from independent siblings. (F) Western blot analysis of Sfpq expression in MEFs. (G) The reduced occupancy of Pol II on the Neat1 locus. Chromatin immunoprecipitation (ChIP) of Pol II was performed using the anti-Pol II antibody 4H8 and cross-linked chromatin samples from MEFs from two different siblings (#1 and #2) of WT and Malat1-KO mice. Primers specific for the indicated regions of the NEAT1 locus were used to determine the binding of Pol II to the DNA. The results are represented as the percentage of input chromatin in each ChIP. The error bars show the standard deviations, and the P-values were calculated from a two-tailed, nonequal variance t-test using two biological replicates (n = 2) for E and triplicate qPCR reactions for G. (*) p < 0.05.
FIGURE 7.
The decreased formation of paraspeckles in specific cell types in Malat1-KO mice. (A) The expression of Neat1_1 and Neat1_2 in the intestine, colon, and forestomach of WT and homozygous (KO) Malat1lacZ mice. Note the decreased intensity of the Neat1_1 and Neat1_2 signals in the intestine and colon of Malat1-KO mice, which was not apparent in the forestomach of Malat1-KO mice. (B,C) The simultaneous detection of Malat1/Neat1_2 and Malat1/Sfpq in the intestine (B) and forestomach (C). Note the decreased formation of paraspeckles in the intestine cells of Malat1-KO mice. Scale bars, 10 μm.
To further explore the mechanism leading to the decrease in paraspeckles in particular cells, we examined the mRNA expression of two paraspeckle core protein components (Sfpq and Nono) in Malat1-KO cells (Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009). The RT-qPCR analysis revealed slight down-regulation of Sfpq and Nono in both cultured MEFs and the intestine from Malat1-KO mice (Fig. 6E); however, the differences were not statistically significant, and the total cellular levels of the Sfpq protein remained unaltered in wild-type and KO MEFs (Fig. 6F). This finding indicates that the decrease in Neat1 transcripts primarily contributes to the reduced formation of paraspeckles in Malat1-KO MEFs and specific tissues. To confirm whether Malat1 regulates Neat1 expression at the transcriptional level, we examined the occupancy of RNA polymerase II (Pol II) on the Neat1 using chromatin immunoprecipitation (ChIP) in control and Malat1-KO MEFs. As shown in Figure 6G, Pol II occupancy at the NEAT1 locus was lower in the MEFs from the Malat1-KO mice, suggesting that the decrease in Neat1 expression in Malat1-KO mice was due to a decrease in transcription.
DISCUSSION
In this study, we have demonstrated that Malat1-KO mice are viable and fertile, without apparent phenotypes. We also show that nuclear speckle components are correctly localized in the nuclei of the KO mice. These results are different from those of other studies that used antisense oligonucleotides to show that the depletion of Malat1 leads to a dramatic increase in the hypophosphorylated forms of SR family proteins and the abnormal localization of nuclear speckle components in human cancerous cell lines (Tripathi et al. 2010; Lin et al. 2011). In addition, we found that MEFs from Malat1-KO mice responded to serum stimulation, which is also different from the results obtained in a recent study showing the role of Malat1 in cell cycle progression in human cells (Yang et al. 2011). These inconsistent results may not be derived from different experimental approaches for the depletion of Malat1 expression because the knockdown of Malat1 in primary MEFs and NIH3T3 cells using antisense oligonucleotides also did not show any apparent cellular phenotype. It is possible that Malat1 has acquired a function in some human cells that is not easily detected in cell lines from other organisms. A similar scenario also exists in the case of Hotair, another lncRNA that plays vital roles in human cells but has no obvious function in mice (Rinn et al. 2007; Gupta et al. 2010; Kogo et al. 2011; Schorderet and Duboule 2011). Considering that Malat1 is up-regulated in various tumor tissue samples and transformed cell lines (Ji et al. 2003; Lin et al. 2007; Perez et al. 2008; Silva et al. 2010), it is also possible that a particular group of transformed cells may show increased susceptibility to the loss of Malat1 through a function or molecular pathway that does not play significant roles in living animals under normal physiological conditions. It would be intriguing to investigate whether transformed tumor cells derived from Malat1-KO mice show decreased/increased malignancy or proliferative capacity compared with cells from wild-type animals. The lack of apparent phenotypes in the Malat1-KO mice can also be explained by the gradual induction of as-yet-unidentified lncRNA(s) that compensate for the function of Malat1, although microarray analysis has failed to detect the up-regulation of any such lncRNAs.
Although we detected no histological abnormalities in the Malat1-KO mice, we observed a reduction in Neat1 expression and subsequent paraspeckle formation in specific cells in Malat1-KO mice. Consistent with our previous observation that the loss of paraspeckles did not lead to obvious phenotypes in mice under normal laboratory conditions (Nakagawa et al. 2011), we observed no visible effects caused by the reduction in paraspeckles in Malat1-KO tissues. Considering that Malat1 homologs are specifically found in all vertebrates and that Neat1 homologs are further restricted to mammalian species (Stadler 2010), these abundant nuclear lncRNAs may control higher-order biological processes, including the functional maintenance of large and complex nervous systems with higher degrees of plasticity, which are not easily addressed using in vitro studies. Interestingly, Malat1 is highly expressed in the nervous system compared with other organs in adult mice, and the acute depletion of Malat1 leads to a decrease in synaptic density in cultured hippocampal cells (Bernard et al. 2010). Further studies, such as a behavioral analysis, will be necessary to further clarify the function of Malat1 in living animals.
Although Malat1 controls the level of Neat1 in specific cell types such as intestinal epithelial cells, the precise molecular mechanism(s) of this regulation remains to be investigated. Considering the close spatial association of paraspeckles with neighboring nuclear speckles (Fox et al. 2002), the association of the Neat1 locus with Malat1-containing nuclear speckles might be necessary for efficient activation of the Neat1 expression. Because a reduction in NEAT1 was also observed in particular cell lines treated with antisense oligonucleotides against MALAT1, it is less likely that the changes in the Neat1 RNA levels observed in the Malat1-KO cells are due to artifactual alterations of the local chromosomal environment caused by the insertion of the lacZ cassette. It should be stressed that the reduction in Neat1 levels in our Malat1-KO mice is restricted to certain cell types such as MEFs and intestine cells and is not necessarily found in all cell types. Together with the fact that Neat1 expression was highly dynamic in different cell populations (Nakagawa et al. 2011), Malat1 may regulate the expression of Neat1 in a cellular context-dependent manner. Whatever the case, further studies may reveal unexpected communication between the two nuclear bodies, paraspeckles and nuclear speckles.
MATERIALS AND METHODS
The generation of Malat1-KO mice
The Malat1-KO mouse was generated using homologous recombination in embryonic stem (ES) cells using previously described protocols (http://www.cdb.riken.jp/arg/Methods.html). Briefly, the left and right arms were amplified with PCR, using the BAC clone RP23-209P9 as a template, and digested with SalI/NotI and EcoRV/SalI. These fragments were then subsequently subcloned into the SalI/NotI and NheI (blunted)/XhoI (right arm) sites of DT-ApA/LacZ/NeO to generate the targeting vector. The vector was subsequently linearized using SalI digestion and electroporated into TT2 ES cells (Yagi et al. 1993), and G418-resistant clones were screened using PCR followed by Southern blot analysis for homologous recombination. Chimeric mice were generated with the recombinant ES clone and mated with C57BL/6 females to generate heterozygous animals (Malat1lacZ/Neo/+). These heterozygotes were then mated with Gt (ROSA)26Sortm1(FLP1)Dym (Jackson Laboratory) to remove the PGK-Neo cassette, and the resulting heterozygous mice (Malat1lacZ/+) were maintained on the C57BL/6 genetic background. PCR-mediated genotyping was performed using DNA obtained from adult or embryonic tails using the following conditions: predenaturation for 1 min at 96°C and 30 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 62°C, and extension for 30 sec at 72°C. All of the animal protocols were approved by the safety division of RIKEN (#H22-EP056).
Northern blotting and RT-qPCR analysis
RNAs were extracted using TRIzol (Invitrogen), and 5 μg of total RNAs was separated on 1.0% agarose gels containing 10% formaldehyde in MOPS buffer, and blotted on positively charged nylon membranes (Roche) according to standard protocols. The DIG-labeled RNA probes against Malat1 and Neat1 were synthesized using the RIKEN Fantom cDNA clones AK141413 and AV089414 as templates, respectively. The labeled probes were then hybridized in Easy Hyb buffer (Roche), and the hybridized signals were detected using CSPD-star (Roche). For the qPCR analysis, 1 μg of RNA was first reverse-transcribed using the ReverTra Ace qPCR RT kit (Toyobo) according to the manufacturer's instructions. The cDNA was then diluted 10-fold, and 2.5 μL of the diluted cDNA was used as the input in a 30-μL qPCR reaction prepared using the Thunderbird SYBR qPCR Mix (Toyobo). All of the reactions were performed using the 7900HT Sequence Detection System (Applied Biosystems) under the following conditions: 5 min at 95°C followed by 40 cycles of 10 sec at 95°C and 45 sec at 60°C. The data were generated using the software SDS2.3 (Applied Biosystems) and analyzed using the comparative CT method. The expression of 18S rRNA was used for normalization except for Figure 6C, where GAPDH was used instead.
In situ hybridization, immunohistochemistry, and Western blotting
In situ hybridization and immunostaining were performed as described previously (Sone et al. 2007). The antibodies used were mouse monoclonal anti-PSF (P2860, Sigma-Aldrich), mouse monoclonal anti-ASF/SF2 (32-4600, Invitrogen), alkaline phosphatase-conjugated sheep anti-DIG (11 093 274 910, Roche), rabbit polyclonal anti-FITC (ab73831, Abcam), Cy3-conjugated goat anti-mouse (AP124C, Millipore), and Alexa Fluor 488-conjugated anti-rabbit (A11029, Invitrogen). The fluorescent and differential interference contrast (DIC) images were obtained using an epifluorescence microscope (BX51, Olympus) equipped with a CCD camera (DP70). Western blotting was performed according to standard protocol with anti-Gapdh antibody (MAB374, Millipore) and the antibodies against PSF and ASF/SF2 mentioned above.
Image processing and statistical analysis
All of the images were processed using the Image J application. The images were initially converted into binary data using the same thresholds, and the area and number of paraspeckles for each cell were quantitated using the “analyze particle” function. In total, 100 cells from two batches of MEFs from different embryos were analyzed. For all of the statistical analyses, the P-values were calculated from two-tailed, nonequal variance t-test.
Cell cultures
All of the cells were cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham F12 (#048-29785, Wako, Japan) supplemented with 10% fetal calf serum (DH10). The mouse embryonic fibroblasts (MEFs) were prepared from E13.5 embryos, and all of the KO and wild-type pairs were prepared from the same littermates. Typically, each embryo was plated into one 6-cm culture dish and further expanded into 10-cm dishes on the following day. All of the MEFs were maintained for up to 2 wk. For the serum stimulation, MEFs were cultured in a medium containing 0.1% serum for 48 h and subsequently cultured in DH10 for 16 h. The cells were labeled with 10 μM BrdU for the last 4 h.
Chromatin immunoprecipitation (ChIP) using anti-RNA polymerase II
ChIP was performed using the EZ-ChIP Chromatin Immunoprecipitation Kit (17-371 Millipore) according to the manufacturer's instructions. The anti-Pol II antibody 4H8 (05-623B Millipore) was used at 1 μg per IP reaction. The DNA was analyzed using qPCR as described above, except that the ChIP input was 2 μL of purified DNA. The comparative CT method was used to calculate the percent input.
Microarray analysis
Standard microarray analyses were performed using Affymetrix Mouse Genome 430 2.0 array for RNA samples from MEFs and Affymetrix Mouse Exon 1.0 ST array for RNA samples from adult hippocampus.
Primers used in this study
Target vector construction and KO mouse genotyping
Left arm FW: gaattctgtcgacGGCCTCAGGGTAGTGCAGCTGAGTCTC
Left arm RV: gaattcgcggccgcGCTGAGTCTCCTGCCTCACGAGCTCAG
Right arm FW: gaattcgatatcCCGAGGAAATCGCAGATAAGTTTTTAA
Right arm RV: gaattcgtcgacGGCATTAATCATGTAGGCTTTTGTGTTG
Southern probe FW: GAGCCAGGGTCTGTGCTAAG
Southern probe RV: AGACCCAGCAGGACTGAGAA
Recombinant ES check primer FW: ACCCGTGATATTGCTGAAGAGCTTGG
Recombinant ES check primer RV: AATGATCCCTTTCATGGGGTCTTCAAG
Genotyping PCR WT primer FW: AGACTCAGCCCGAGGAAATC
Genotyping PCR WT primer RV: GCTCTGGTCAGCCTCCATTA
Genotyping PCR KO FW: TTGAAGTGGCGAGCGATAC
Genotyping PCR KO FW: AGATCCCAGCGGTCAAAAC
qPCR analysis
mMalat1 #1 FW: TCGGCCTTGTAGATTAAAACGAA
mMalat1 #1 RV: AACGGCCGTCAACTTAACCT
mMalat1 #2 FW: AGCAGGCATTGTGGAGAGGA
mMalat1 #2 RV: ATGTTGCCGACCTCAAGGAA
mMalat1 #3 FW: CGGCTTTGGTTCACAGTCAC
mMalat1 #3 RV: ACACAAGGCCACAGCCAACT
mMalat1 #4 FW: CCCCAGCTTTTCCAGAATCC
mMalat1 #4 RV: GCATCAAGGTGAGGGGTGAA
mNeat1 #1 FW: TTGGGACAGTGGACGTGTGG
mNeat1 #1 RV: TCAAGTGCCAGCAGACAGCA
mNeat1 #2 FW: GATCGGGACCCCAGTGACCT
mNeat1 #2 RV: AGCTTTCCCCAACACCCACA
mNeat1 #3 FW: GCTCTGGGACCTTCGTGACTCT
mNeat1 #3 RV: CTGCCTTGGCTTGGAAATGTAA
hMalat1 #1 FW: GATAATCAGACCACCACAGGTTTACAG
hMalat1 #1 RV: AAAGAGTAACTACCAGCCATTTCTCCA
hMalat1 #2 FW: GACGGAGGTTGAGATGAAGC
hMalat1 #2 RV: ATTCGGGGCTCTGTAGTCCT
hNeat1 #1 FW: AGCTGCGTCTATTGAATTGGTAAAGTAA
hNeat1 #1 RV: GACAGAAAGATCCCAACGATAAAAATAA
hNeat1 #2 FW: GATCTTTTCCACCCCAAGAGTACATAA
hNeat1 #2 RV: CTCACACAAACACAGATTCCACAAC
hNeat1 #3 FW: TGTGTGTGTAAAAGAGAGAAGTTGTGG
hNeat1 #3 RV: AGAGGCTCAGAGAGGACTGTAACCTG
hNeat1 #4 FW: TGTGTGTGTAAAAGAGAGAAGTTGTGG
hNeat1 #4 RV: AGAGGCTCAGAGAGGACTGTAACCTG
hNeat1 #5 FW: TCGGGTATGCTGTTGTGAAA
hNeat1 #5 RV: TGACGTAACAGAATTAGTTCTTACCA
mSfpq FW: ATTGTGGATGATCGCGGAAG
mSfpq RV: GGCGAGGAGTCGTTGTCAGT
mNono FW: TTGTGGATGACCGAGGAAGG
mNono RV: GCCGAGGAAATGTAGTCAGCA
mSrsf1 FW: TCTGGACCTCCGAGTGGAAG
mSrsf1 RV: CACGACACCAGTGCCATCTC
18S rRNA FW: CCCAGTAAGTGCGGGTCATAA
18S rRNA RV: GATCCGAGGGCCTCACTAAC
ChIP analysis
Neat1UP FW: AACCGCCACTCCCTCAGATT
Neat1UP RV: CTTATCCCCGGTGGTGTTCA
Neat1A FW: AGCGCCTTTAACAATCCACAAA
Neat1A RV: GTTAGGACACTGCCCCCATGTA
Neat1B FW: TGCATTGGTCAGAGGGATTCTT
Neat1B RV: ACCCATTTTACAACCAGCTCCA
Neat1D FW: CTGGTTTATCCCAGCGTCATTC
Neat1D RV: ATTCCTTACCAGACCGCTGACA
Neat1G FW: CCTGCAGACAAGATGCTGAGTT
Neat1G RV: GTGGCCCTCTAAGGAAACATCC
GapdhA FW: GCAGTGGCAAAGTGGAGATTGT
GapdhA RV: CTTGACTGTGCCGTTGAATTTG
GapdhC FW: GTGTTCCTACCCCCAATGTGTC
GapdhC RV: GTAGCCCAAGATGCCCTTCAGT
Antisense oligonucleotides
Control: 5′-mUmCmAmCmCTTCACCCTCTmCmCmAmCmU-3′
hMalat1-AS1: 5′-mCmCmUmCmCTCCCATCCCTmCmCmAmAmA-3′
hMalat1-AS2: 5′-mCmCmCmUmCTCTCTTCCCTmGmUmUmAmA-3′
Backbone: All phosphorothioate-converted
hMalat1-AS3/mMalat1-AS1: GGGAGTTACTTGCCAACTTG
hMALAT1-AS4: ATGGAGGTATGACATATAAT
mMalat1-AS2: CGGTGCAAGGCTTAGGAATT
DATA DEPOSITION
Microarray data have been deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE37707. Malat1-KO mouse data have been deposited at http://www.cdb.riken.jp/arg/mutant%20mice%20list.htmlunder accession number CDB0837K.
ACKNOWLEDGMENTS
We thank Dr. Shinichi Aizawa, Dr. Kenryo Furushima, and Dr. Hiroshi Kiyonari for helpful comments on the production of the KO mouse and Dr. Kentarou Ishida, Ms. Chieko Nashiki, Hiromi Ito, and Kaori Yanaka for technical assistance. This work was supported by Grants-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT) to S.N. and an American Cancer Society RSG-11-174-01-RMC and NIH GM088252 grants to K.V.P.
NOTE ADDED IN PROOF
While our manuscript was in press, two novel publications came to our attention describing similar phenotypes for the Malat1 knockout mice from the laboratories of Sven Diederichs (Eiβmann et al. 2012) and David Spector (Zhang et al. 2012).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033217.112.
REFERENCES
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH 2009. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI 1992. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 117: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-L, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Kupper K, Dietzel S, Fakan S 2004. Higher order chromatin architecture in the cell nucleus: On the way from structure to function. Biol Cell 96: 555–567 [DOI] [PubMed] [Google Scholar]
- Eiβmann M, Gutschner T, Ha¨mmerle M, Gu¨nther S, Caudron-Herger M, Groβ M, Schirmacher P, Rippe K, Braun T, Zo¨rnig M, et al. 2012. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AKL, Lyon CE, Andersen J, Mann M, Lamond AI 2002. Paraspeckles: A novel nuclear domain. Curr Biol 12: 13–25 [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB 2006. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner MR, Spector DL 2010. Chromatin dynamics. Annu Rev Biophys 39: 471–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Chang HY 2010. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol 7: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39 doi: 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Nakagawa S 2012. Long non-coding RNAs in nuclear bodies. Dev Growth Differ 54: 44–54 [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. 2011. Long noncoding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71: 6320–6326 [DOI] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS 2007. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26: 851–858 [DOI] [PubMed] [Google Scholar]
- Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS 2011. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett 585: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T 2011. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS, Smith DI 2008. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet 17: 642–655 [DOI] [PubMed] [Google Scholar]
- Platani M, Lamond AI 2008. Nuclear organisation and subnuclear bodies. In RNA trafficking and nuclear structure dynamics (ed. P Jeanteur), pp. 1–22. Springer, New York [Google Scholar]
- Prasanth KV, Spector DL 2007. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity' conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YTF, Ideue T, Sano M, Mituyama T, Hirose T 2009. MENɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D 2011. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet 7: e1002071 doi: 10.1371/journal.pgen.1002071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Perez DS, Pritchett JR, Halling ML, Tang H, Smith DI 2010. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics 95: 355–362 [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120: 2498–2506 [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646 doi: 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler PF 2010. Evolution of the long non-coding RNAs MALAT1 and MENβ/ε. Adv Bioinform Comput Biol 6268: 1–12 [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N 2010. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584: 4575–4580 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng JJ, Hsieh YT, Hsu SL, Chou MM 2009. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod 15: 725–731 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S 1993. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem 214: 70–76 [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG 2011. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Reports (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]