The nuclear exosome and the nonsense-mediated mRNA decay (NMD) pathways have been implicated in the degradation of distinct unspliced transcripts in Saccharomyces cerevisiae. In this study, the authors show that these two systems can cooperate to limit the accumulation of unspliced mRNAs. Using steady-state and decay analyses, they show that while specific unspliced transcripts rely solely on NMD or on the nuclear exosome for their degradation, some unspliced RNAs are degraded efficiently only when both the nuclear exosome and NMD are inactivated. The specificity in the pre-mRNAs degradation pathways can be manipulated by changing the rate of export or retention of these mRNAs.
Keywords: exosome, nonsense-mediated decay, RNA export, RNA quality control, unspliced RNAs
Abstract
The nuclear exosome and the nonsense-mediated mRNA decay (NMD) pathways have been implicated in the degradation of distinct unspliced transcripts in Saccharomyces cerevisiae. In this study we show that these two systems can act sequentially on specific unspliced pre-mRNAs to limit their accumulation. Using steady-state and decay analyses, we show that while specific unspliced transcripts rely mostly on NMD or on the nuclear exosome for their degradation, some unspliced RNAs are stabilized only when both the nuclear exosome and NMD are inactivated. We found that the mechanism of degradation of these unspliced pre-mRNAs is not influenced by promoter identity. However, the specificity in the pre-mRNAs degradation pathways can be manipulated by changing the rate of export or retention of these mRNAs. For instance, reducing the nuclear export of pre-mRNAs mostly degraded by NMD results in a higher fraction of unspliced transcripts degraded by the nuclear exosome. Reciprocally, inactivating the Mlp retention factors results in a higher fraction of unspliced transcripts degraded by NMD for precursors normally targeted by the nuclear exosome. Overall, these results demonstrate that a functional redundancy exists between nuclear and cytoplasmic degradation pathways for unspliced pre-mRNAs, and suggest that the degradation routes of these species are mainly determined by the efficiency of their nuclear export rates. The presence of these two sequential degradation pathways for unspliced pre-mRNAs underscores the importance of limiting their accumulation and might serve as a fail-safe mechanism to prevent the expression of these nonfunctional RNAs.
INTRODUCTION
Eukaryotic organisms have developed compartmentalized degradation systems to ensure the timely turnover of numerous classes of RNA molecules (for review, see Houseley et al. 2006; Chang et al. 2007). The major pathways that function during RNA degradation are the nuclear exosome (Mitchell et al. 1997), which is stimulated by the activity of the TRAMP complex (LaCava et al. 2005; Wyers et al. 2005), and the deadenylation and decapping of mRNAs and degradation by the cytoplasmic exonuclease, Xrn1p (for review, see Tucker and Parker 2000). After initial removal of the poly-A tail, turnover can also be promoted by the cytoplasmic 3′→5′exosome (Tucker and Parker 2000). Recent studies have shown that the promoter identity can influence the decay route of RNA molecules (Bregman et al. 2011; Trcek et al. 2011).
Degradation of unspliced (intron-retained) mRNAs is a major quality control mechanism that prevents the translation of unprocessed or partially processed transcripts that would result in the production of truncated proteins due to the likelihood of encountering a premature termination codon (PTC) in the intronic sequences. Because of the prevalence of PTC in intronic sequences, many unspliced RNAs are targeted by nonsense-mediated decay in a variety of eukaryotes (He et al. 1993; Mitrovich and Anderson 2000; Jaillon et al. 2008; Sayani et al. 2008). However, the degradation of unspliced RNAs is not limited to the NMD system. Previous studies have shown the involvement of nuclear degradation systems involving multiple components such as the nuclear exosome (Bousquet-Antonelli et al. 2000; Moore et al. 2006; Egecioglu et al. 2012), the RNase III endonuclease Rnt1p (Danin-Kreiselman et al. 2003; Egecioglu et al. 2012), and the nuclear poly(A)-binding protein (Lemieux et al. 2011) in the degradation of unspliced pre-mRNAs. A role for retention factors that could putatively function in the nuclear sequestration of unspliced transcripts was proposed (Galy et al. 2004), which would increase the likelihood of nuclear degradation. In addition to these nuclear degradation pathways, unspliced pre-mRNAs can also be degraded in the cytoplasm in pathways that are dependent on the 5′→3′ exonuclease Xrn1p but independent from NMD (Hilleren and Parker 2003; Dong et al. 2007). In the case of transcripts degraded by the cytoplasmic RNA turnover machinery, the importance of proper nuclear export for efficient pre-mRNA degradation was demonstrated by showing the involvement of Mex67p for the export and subsequent Edc3p-dependent degradation of the YRA1 pre-mRNA (Dong et al. 2007). All of these studies suggest that the degradation routes for unspliced pre-mRNAs are relatively specific, as most unspliced pre-mRNAs accumulate mostly in the absence of either nuclear or cytoplasmic degradation pathways.
In this study, we show that certain unspliced precursors are degraded efficiently only when both the nuclear exosome and nonsense-mediated decay pathways are active. The nuclear exosome can fulfill a larger role in the degradation of unspliced transcripts normally degraded by NMD when these transcripts are sequestered in the nucleus upon inhibition of RNA export. We show that the perinuclear factors, Mlp1p and Mlp2p can have a role in the retention of an endogenous unspliced transcript that appears to be predominantly degraded by the nuclear exosome. Replacement of the native promoter sequences of intron-containing genes with a galactose inducible promoter did not lead to changes in the accumulation profile of unspliced transcripts, nor did it redirect the degradation routes for the turnover of these unspliced precursors. Overall, these results show that the degradation pathways for unspliced pre-mRNAs are largely nonspecific and are mostly determined by their nuclear export efficiency. The fact that two sequential pathways act on unspliced pre-mRNAs suggests the existence of a fail-safe mechanism that limits the accumulation and expression of nonfunctional RNAs.
RESULTS
Higher accumulation of specific unspliced transcripts in strains lacking the nuclear exosome component Rrp6p and NMD factors
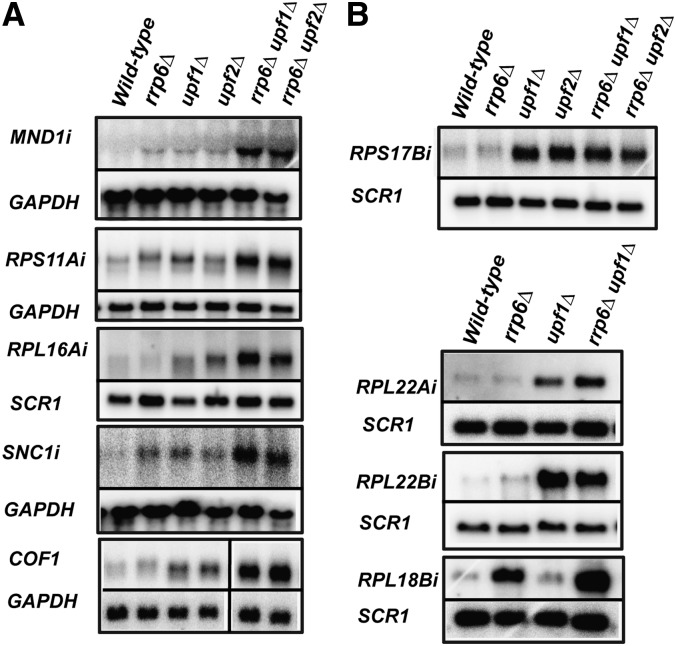
Our previous work revealed that the Nonsense-Mediated Decay (NMD) pathway targets and degrades a large number of unspliced transcripts in Saccharomyces cerevisiae (Sayani et al. 2008). Accumulation of unspliced pre-mRNAs was mostly observed for transcripts exhibiting suboptimal splicing signals. However, we noticed that numerous transcripts that showed suboptimal splicing sequences (splice sites or branchpoints) did not reveal any major accumulation of unspliced transcripts in strains lacking NMD factors (Sayani et al. 2008). Two hypotheses could explain this phenomenon. First, it is possible that these intron-containing genes are efficiently spliced despite their suboptimal splicing signals, and therefore do not accumulate unspliced precursors to detectable levels. On the other hand, we reasoned that some of these intron-containing genes might produce unspliced transcripts, but that the turnover of these precursors could be dependent on more than one degradation pathway. The nuclear exosome has been implicated in degrading some unspliced transcripts (Bousquet-Antonelli et al. 2000; Moore et al. 2006), suggesting that it might cooperate with NMD to degrade some of these transcripts. To test this hypothesis, we constructed yeast strains that lacked the nuclear exosome component Rrp6p, as well as the NMD factors Upf1p or Upf2p. Using intronic riboprobes, we investigated the accumulation of unspliced transcripts in wild-type, rrp6Δ, upf1Δ, upf2Δ, rrp6Δ upf1Δ, or rrp6Δ upf2Δ for the intron-containing genes COF1, MND1, RPS11A, RPL16A, RPL22A, RPL18B, RPS17B, RPL22B, and SNC1 (Fig. 1). The use of these intronic riboprobes allowed the specific detection of unspliced pre-mRNAs without interference of signal from the mature mRNAs, which typically accumulate at much higher levels. In addition, the use of these intronic riboprobes allowed us to detect specific isoforms of ribosomal protein mRNAs, as opposed to both isoforms in the case of highly similar duplicated genes. We did not observe any strong accumulation of unspliced precursors in the wild-type, rrp6Δ, upf1Δ, or upf2Δ single-mutant strains for MND1, SNC1, RPS11A, RPL16A, and COF1. However, we detected a higher accumulation of unspliced transcripts in the rrp6Δ upf1Δ and rrp6Δ upf2Δ double-mutant strains for these genes (Fig. 1A). In the case of RPL22A, unspliced transcripts were not detected in the wild-type or rrp6Δ strains. However, an increase in the levels of unspliced precursors was observed in the upf1Δ strain, and this accumulation was exacerbated in the rrp6Δ upf1Δ strain (Fig. 1B). These observations suggested that the NMD and the nuclear exosome can play functionally redundant roles to promote the turnover of unspliced species of these transcripts. In contrast, we did not detect high levels of unspliced precursors in wild-type or in the upf1Δ strains for the RPL18B transcript (Fig. 1B). However, high levels of unspliced transcript were present in the rrp6Δ strain, and these levels further increased in the rrp6Δ upf1Δ strain (Fig. 1B). This result suggests that while most of the degradation of unspliced RPL18B species is mediated by the nuclear exosome, NMD can also promote the degradation of these unspliced species when the nuclear exosome is inactivated. Unspliced transcripts that were found to accumulate to higher levels in yeast strains lacking both Rrp6p and NMD components are referred to hereby as RNMD (Rrp6p and Nonsense Mediated Decay)-sensitive transcripts.
FIGURE 1.
Steady-state analysis of unspliced transcripts in single and double mutants of RNA degradation pathways. (A) Analysis of RNMD-dependent transcripts. (B) Analysis of transcripts whose turnover depends mostly on one degradation pathway. Northern blots were probed with an intronic (i) riboprobe and either SCR1 or GAPDH was utilized as a loading control. Both the SCR1 and the GAPDH probes were generated via random priming.
The greater accumulation of unspliced species in strains lacking both degradation pathways was observed only for specific genes. For instance, unspliced precursors of RPS17B could be detected in upf1Δ or upf2Δ strains, but the extent of accumulation in the rrp6Δ upf1Δ or rrp6Δ upf2Δ double mutants was similar to single mutants (Fig. 1B). Similarly, very high levels of unspliced precursors were detected in the upf1Δ strain for RPL22B, but these levels were not increased in the double-mutant rrp6Δ upf1Δ (Fig. 1B). While we cannot rule out the existence of other degradation pathways, this steady-state analysis of unspliced transcripts suggested that some unspliced precursor transcripts are degraded through a complex turnover route, potentially involving two sequential pathways, while others seem to be targeted mostly through a single mode of degradation.
Analysis of the rate of turnover of unspliced transcripts in NMD and rrp6-deficient strains
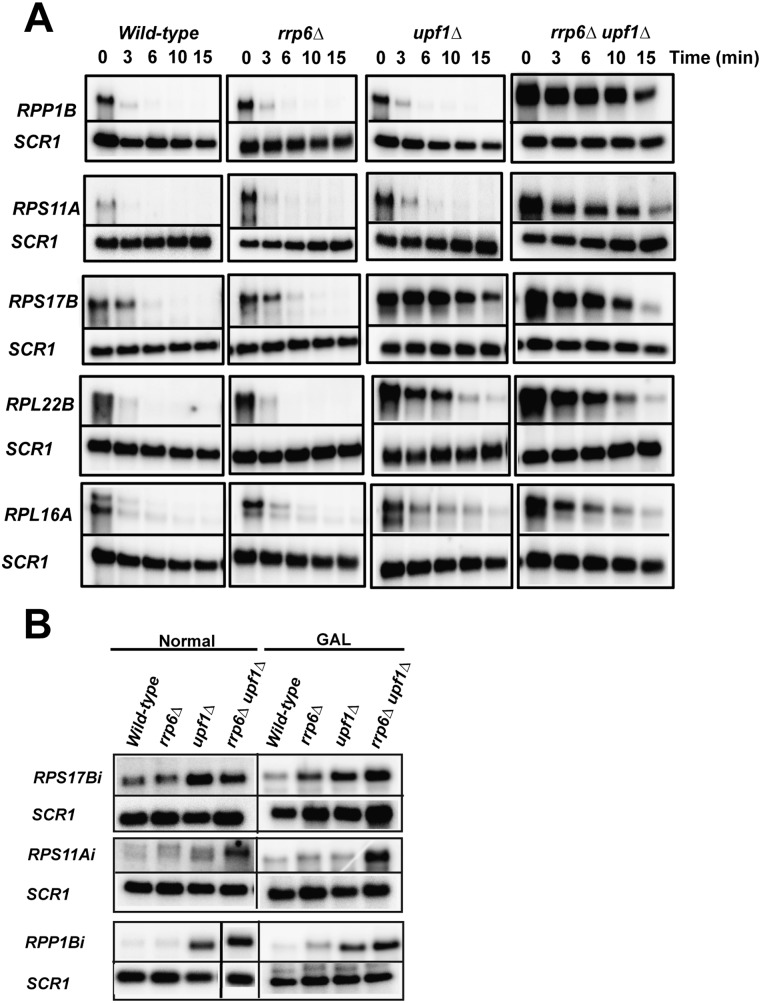
The previous observations based on steady-state analysis suggested that the nuclear exosome and NMD pathways might act sequentially to limit the accumulation of unspliced transcripts. However, we could not rule out that indirect splicing defects might have resulted in the higher accumulation of unspliced precursors observed in these double mutants. In order to demonstrate that the effects observed above are directly due to RNA turnover, we analyzed the rates of decay of some of these unspliced precursors in wild-type, rrp6Δ, upf1Δ, and rrp6Δupf1Δ strains. To this end, we used a conditional galactose promoter that allowed us to control the expression of some of these intron-containing genes as a function of the carbon source. We replaced the endogenous promoters of five intron-containing genes (RPP1B, RPS11A, RPL16A, RPS17B, and RPL22B) by the galactose promoter, and then derived yeast strains that lacked either Rrp6p, Upf1p, or both components. Each strain was grown in galactose-containing medium until logarithmic growth phase (OD600 nm = ∼0.4–0.8), and then switched to glucose-containing medium. The kinetics of turnover of unspliced RNAs were then assessed after the switch to glucose-containing medium. As shown in Figure 2A, all five intron-containing genes (RPP1B, RPS11A, RPL16A, RPS17B, and RPL22B) exhibited a rapid turnover of the unspliced transcripts in wild-type as well as in cells lacking the nuclear exosome component Rrp6p, such that unspliced transcripts were undetectable after ∼3 min. A similar pattern was observed for RPP1B, RPS11A, and RPL16A in upf1Δ strains. Strikingly, the turnover of the unspliced precursors of RPP1B, RPS11A, and RPL16A exhibited a considerable delay in the rrp6Δ upf1Δ strain compared with the single-mutant strains, which, in general, was in good agreement with the steady-state accumulation profiles observed in Figure 1 (Fig. 2B for RPP1B) (we note that for RRP1B the steady-state accumulation seems to be mostly NMD dependent). Because of the time points used and the very short half-lives of these unspliced pre-mRNAs, the individual contributions of NMD and the nuclear exosome on turnover are difficult to estimate. However, this result directly demonstrates that the nuclear exosome and NMD can be functionally redundant to promote the turnover of unspliced precursors for these transcripts. In contrast, the decay of RPS17B and RPL22B unspliced precursors exhibited a considerable lag in turnover in the upf1Δ single mutant, and the decay rate of these transcripts was very similar in the upf1Δ and the rrp6Δ upf1Δ strains (Fig. 2A), suggesting that NMD is the major turnover route for unspliced RPS17B and RPL22B transcripts. We also analyzed the steady-state levels of some of these unspliced transcripts when expressed from the GAL promoter in galactose medium, and showed that the differences observed between strains when these genes are expressed from their endogenous promoters can be reproduced when these intron-containing genes are expressed from the GAL promoter (Fig. 2B). This observation suggests that replacement of the promoter of these genes by the GAL promoter does not perturb the degradation pathway for these unspliced species. Taken together, the results obtained from the steady-state and turnover studies show that the unspliced pre-mRNAs species of RPS11A, RPL16A, and RPP1B can be degraded by two turnover systems, which are likely to act sequentially to limit their accumulation. However, some unspliced transcripts such as RPS17B and RPL22B seem to be degraded predominantly by NMD.
FIGURE 2.
Decay analysis of unspliced transcripts and effect of promoter replacement on steady-state levels of unspliced transcripts. (A) Decay analysis. Each of the intron-containing ribosomal protein genes was placed under the control of the conditional galactose promoter ∼50 bp upstream of the translation start site, leading to the creation of five conditionally expressed ribosomal protein genes: GAL::RPP1B, GAL::RPS11A, GAL::RPL16A, GAL::RPS17B, GAL::RPS22B. Each of the knockouts (rrp6Δ, upf1Δ, rrp6Δ upf1Δ) was derived from a single isogenic wild-type strain. Each of the timepoints is indicated in minutes. SCR1 was utilized as a loading control. (B) Comparison of the levels of unspliced pre-mRNAs for intron-containing genes expressed under the control of their endogenous promoters or from the GAL promoter.
Transposition of the RPL16A RNMD-sensitive intron is not sufficient to provide sensitivity of chimeric unspliced GOT1::RPL16A transcripts to both degradation systems
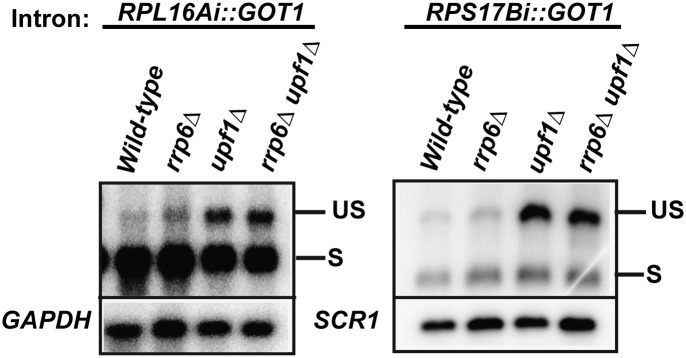
In order to investigate the determinants of targeting of unspliced transcripts to a specific degradation pathway, we undertook an intron-replacement strategy similar to the one used in a previous study (Sayani et al. 2008). We chose the GOT1 locus, which contains a small intron that does not lead to the production of unspliced transcripts (Sayani et al. 2008). We replaced the native GOT1 intron with an NMD-sensitive intron (RPS17B) or an RNMD-sensitive intron (RPL16A) and then derived rrp6Δ, upf1Δ, or rrp6Δ upf1Δ strains. Transposition of the NMD-sensitive RPS17B intron did not lead to the detection of appreciable chimeric unspliced transcripts in the wild-type or rrp6Δ strains (Fig. 3). However, a large accumulation of unspliced precursor was detected in the upf1Δ, and similar levels were observed in the rrp6Δ upf1Δ strain. This result was consistent with our previous studies (Sayani et al. 2008) showing that inclusion of an NMD-sensitive intron into a NMD-insensitive locus leads to the effective production of chimeric unspliced transcripts that can be targeted and degraded by the NMD pathway. Next, we analyzed unspliced transcript accumulation for the transposed RPL16A intron in wild-type, rrp6Δ, upf1Δ, or rrp6Δ upf1Δ strains. Consistent with the pattern of RPL16A unspliced transcript accumulation at its normal locus (Fig. 1A), we did not observe high levels of chimeric unspliced transcripts in trans in the wild-type or rrp6Δ strains (Fig. 3). Surprisingly, however, chimeric unspliced RPL16Ai::GOT1 transcripts were now observed in the upf1Δ single-mutant strain, but the accumulation was not increased in the rrp6Δ upf1Δ double-mutant strain. This result shows that the transposition of the RPL16A RNMD-sensitive intron into an RNMD-insensitive locus is not sufficient to lead to an RNMD-dependent mode of degradation of chimeric unspliced RPL16Ai::GOT1 transcripts. While it is clear that intron identity is important to confer suboptimal splicing and targeting to NMD (Fig. 3; Sayani et al. 2008), this result suggests that, at least in the case of RPL16A, features of the pre-mRNA other than the intronic sequence are limiting to determine the degradation pathway of unspliced pre-mRNAs.
FIGURE 3.
Transposition of the RPL16A RNMD-sensitive intron is not sufficient to provide sensitivity of GOT1::RPL16A chimeric unspliced transcripts to both degradation systems. The native GOT1 intron was replaced by the RPL16A or the RPS17B introns and the wild-type, rrp6Δ, upf1Δ, or rrp6Δ upf1Δ strains were derived. A GOT1 exon 2 antisense RNA probe was used to assay for the spliced and chimeric unspliced transcripts. SCR1 or GAPDH was used as a loading control.
Inactivating the mRNA export factor Mex67p changes the turnover routes of unspliced transcripts
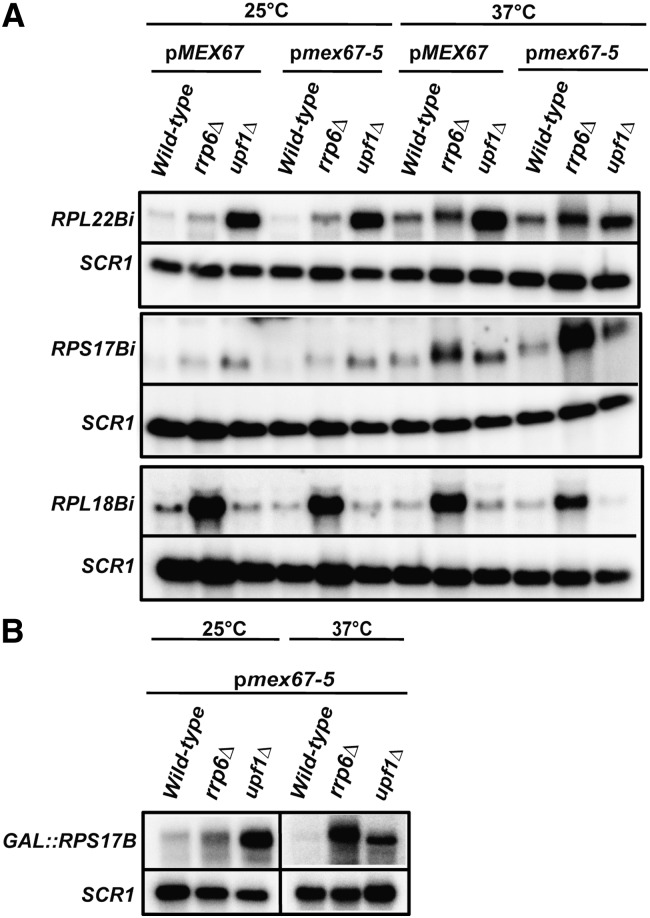
The previous results showed that at least two independent systems exist to degrade unspliced transcripts, one in the cytoplasm and one in the nucleus. These two degradation systems may act as a fail-safe mechanism when the other pathway is inactive or fails to degrade these unspliced species (Figs. 1, 2). These observations also suggested that the mode of degradation of unspliced transcripts might be directly influenced by their rate of export. Specifically, we predicted that the turnover of an unspliced transcript, which is predominantly dependent on NMD, might rely on the nuclear exosome if the rate of nuclear export of this unspliced pre-mRNA is reduced. In order to test this hypothesis, we utilized a yeast strain that carries a deletion of the essential RNA export factor Mex67p at the genomic locus and contains a plasmid expressing either a wild-type version or an allelic version expressing a thermosensitive point mutant (mex67-5) (Segref et al. 1997). We then derived strains that lacked the exosome component Rrp6p, or the NMD factor Upf1p, and analyzed the accumulation of unspliced transcripts in these strains upon inactivation of mRNA export in the mex67-5 thermosensitive strain. We analyzed two NMD-sensitive intron-containing genes (RPS17B or GAL::RPS17B and RPL22B) and one intron-containing gene whose unspliced transcript is degraded predominantly by the nuclear exosome (RPL18B) to investigate the potential cytoplasmic-to-nuclear shift in unspliced transcript turnover upon the induction of an export defect after the shift to the nonpermissive temperature of 37°C. As expected, we did not observe a significant accumulation of unspliced transcripts for the strains grown at the permissive temperature of 25°C expressing MEX67 in a wild-type or an RRP6-null background for both RPS17B and RPL22B, but unspliced precursors were observed for the RPL18B transcript in the rrp6Δ strain (Fig. 4A). On the other hand, unspliced transcripts were detected for RPS17B and RPL22B in the MEX67 upf1Δ strain. A similar pattern of unspliced transcript accumulation was observed for strains expressing mex67-5 in the wild-type, rrp6Δ, or upf1Δ background at the permissive temperature (Fig. 4A). When the cells were shifted to nonpermissive temperature (37°C), a general increase in unspliced transcripts was detected in the MEX67, MEX67 rrp6Δ, and the MEX67 upf1Δ strains for both RPS17B and RPL22B, but not for RPL18B (Fig. 4A). This might be the result of a mild heat-shock resulting in a partial inhibition of splicing. In the case of RPL22B, most of these unspliced precursors were degraded by NMD, as these species were found to accumulate much more dramatically in the MEX67 upf1Δ background. However, in the case of RPS17B, a larger fraction of these unspliced pre-mRNAs was degraded in an Rrp6p-dependent manner, even in the presence of fully functional Mex67p during this temperature shift. This effect might have resulted from a reduction of the export of RPS17B pre-mRNAs, since inhibition of mRNA export has been documented to occur in heat-shock conditions (Liu et al. 1996). Strikingly, in the mex67-5 strain shifted to 37°C, a large fraction of unspliced transcripts were now found to be degraded by the exosome as revealed by the reduction of unspliced transcripts observed in the upf1Δ background and the increase of unspliced transcripts in the rrp6Δ strain. This increase is apparent for RPL22B, but is even more pronounced for RPS17B, as shown by the dramatic accumulation of unspliced RNAs in the mex67-5 rrp6Δ strain (Fig. 4A). In the case of RPL18B, we did not detect any further increase of unspliced RNAs in the mex67-5 rrp6Δ strain in these conditions (Fig. 4A), showing that the increase of unspliced precursors observed upon inhibition of export in the rrp6Δ background for RPS17B and RPL22B is specific to transcripts normally degraded by NMD. This shift in the degradation pathway of unspliced precursors from NMD to the nuclear exosome upon genetic inhibition of nuclear export could also be detected when the RPS17B mRNA was expressed from a galactose promoter (Fig. 4B). Thus, this shift in the degradation route upon inhibition of mRNA export does not depend on the promoter from which these transcripts are being generated. Taken together, these results indicate that when mRNA export is inhibited in the mex67-5 strain at the nonpermissive temperature of 37°C, a large fraction of unspliced transcripts normally degraded by NMD are now retained in the nucleus and targeted for turnover by the nuclear exosome. This observation might be linked to the activation of nuclear RNA surveillance by Rrp6p that was previously shown to occur in RNA export mutants (Jensen et al. 2001; Libri et al. 2002; Strasser et al. 2002; Zenklusen et al. 2002; Rougemaille et al. 2007; Assenholt et al. 2008). These results also demonstrate that the export of several unspliced pre-mRNAs out of the nucleus is dependent on the mRNA export factor, Mex67p, as shown previously for the YRA1 pre-mRNA (Dong et al. 2007).
FIGURE 4.
RNA export inhibition can result in a higher fraction of unspliced transcripts targeted by the nuclear exosome. RPL22B and RPS17B (A) (or GAL::RPS17B; B) are NMD targets, and RPL18B is predominantly degraded by the nuclear exosome. Cells were grown at a permissive temperature (25°C) until they reached logarithmic phase and shifted to nonpermissive temperature (37°C) for ∼3 h. Northern blot analysis was performed with an intron-spanning antisense RNA probe to detect the accumulation of each of the unspliced transcripts. SCR1 was used as a loading control.
Mlp proteins function to retain the nuclear-degraded RPL18B unspliced transcript
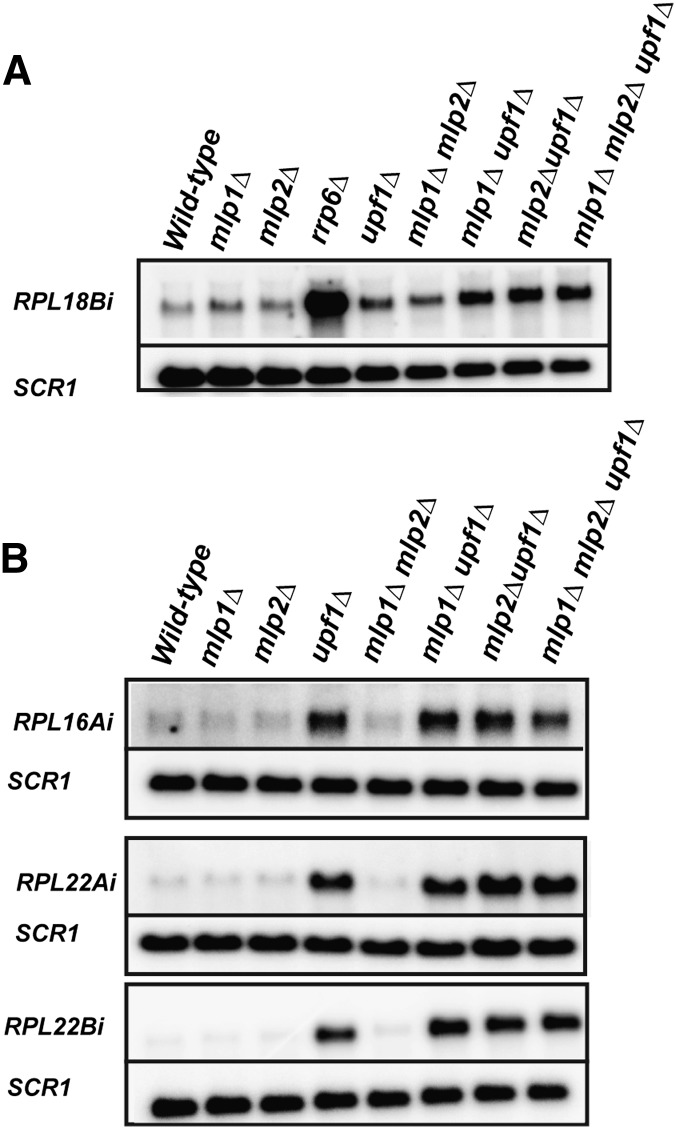
It was previously reported that the nuclear factor Mlp1p functions in the retention of unspliced RNAs (Galy et al. 2004). However, these studies were based on the utilization of reporter constructs that contained a small, synthetic, inefficiently spliced intron. We reasoned that Mlp1p might function in regulating the retention of unspliced RNAs that are predominantly degraded by the nuclear exosome. We hypothesized that in the absence of the potential retention-exerting activity of Mlp1p, these unspliced transcripts would be present at a higher concentration in the cytoplasm, which would then be targeted and degraded by the activity of the nonsense-mediated decay pathway. In order to investigate this hypothesis, we chose RPL18B as a candidate, since this transcript accumulates high levels of unspliced precursors upon inactivation of the nuclear exosome (Figs. 1, 4). We created and utilized yeast strains deleted for the genes encoding the similar (but not identical) retention factors Mlp1p (mlp1Δ) and Mlp2p (mlp2Δ), and also generated combinatorial double-delete mutants of MLP1 and UPF1 (mlpΔ1 upf1Δ), MLP2 and UPF1 (mlp2Δ upf1Δ), and a triple-delete mutant comprising of deletions in MLP1, MLP2, and UPF1 (mlp1Δ mlp2Δ upf1Δ). As expected and shown in Figure 5A, the single mutants mlp1Δ, mlp2Δ, or upf1Δ did not display high levels of RPL18B unspliced transcript compared with the wild-type strain. However, large amounts of unspliced transcript were detected in the rrp6Δ strain. Strikingly, we observed higher amounts of unspliced precursors in the mlp1Δupf1Δ double mutant compared with the upf1Δ or mlp1Δ single-deletion strains. This observation was further supported by an increase in unspliced transcript accumulation in the mlp2Δupf1Δ double mutant and in the mlp1Δmlp2Δupf1Δ triple mutant. We further expanded our investigation and tested other intron-containing genes that exhibited an RNMD behavior (RPL16A, RPL22A) or an NMD-specific unspliced accumulation profile (RPL22B). As shown in Figure 5B, we did not observe any appreciable accumulation of unspliced precursor in wild-type, mlp1Δ, or mlp2Δ strains for RPL16A, RPL22A, and RPL22B. As expected, a strong accumulation of unspliced transcripts was detected in the upf1Δ strain. However, we did not observe an exacerbation in the accumulation of RPL16A, RPL22A, and RPL22B unspliced precursors for the mlp1Δupf1Δ, mlp2Δupf1Δ, or mlp1Δmlp2Δupf1Δ double or triple mutants. We conclude that the Mlp proteins can function in the retention of endogenous unspliced RNAs, as shown for the RPL18B transcript, but that the role of these proteins might be limited to a few transcripts exhibiting a primarily nuclear degradation pathway.
FIGURE 5.
Mlp1p and/or Mlp2p function in retaining the nuclear-degraded unspliced RPL18B transcripts, but not unspliced precursors that are cooperatively degraded by exosome/NMD or NMD. Northern analysis was performed on the RPL18B unspliced transcript (predominantly degraded by the nuclear exosome; A); or RPL22A and RPL16A (cooperatively degraded by exosome and NMD), or RPL22B (predominantly degraded by NMD) in B. SCR1 was used as a loading control.
DISCUSSION
In this study, we have investigated the role of sequential, functionally redundant degradation pathways in unspliced RNAs turnover and have implicated a novel, dual-targeting strategy that is dependent on the activity of the nuclear exosome component, Rrp6p, and the NMD pathway. We found seven intron-containing genes that exhibit such a sequential mode of degradation. This bipartite strategy reveals that the turnover of certain unspliced transcripts is more complex than previously thought and is controlled by the exosome in the nucleus and the NMD in the cytoplasm. In our study, we used a strain carrying a deletion of RRP6 as a tool to monitor nuclear exosome inactivation. However, this particular deletion strain might not be sufficient to reveal the extent of function of the nuclear exosome on unspliced precursors degradation, as the degradation of some unspliced transcripts might rely on the integrity of core subunits of the nuclear exosome other than Rrp6p. Therefore, it is possible that the degradation of unspliced pre-mRNAs by the nuclear exosome is more widespread than revealed by the rrp6Δ deletion. In addition, we cannot rule out that degradation pathways other than the nuclear exosome and NMD are also active in the degradation of other unspliced pre-mRNAs. This is the case, for instance, for the ACT1 and YRA1 pre-mRNAs, which have been shown to rely on NMD-independent cytoplasmic pathways for turnover (Hilleren and Parker 2003; Dong et al. 2007). Regardless of the nature of the degradation pathways involved, the existence of multiple sequential degradation pathways for unspliced pre-mRNA suggests the existence of a fail-safe mechanism that prevents the expression of these nonfunctional RNAs, underscoring the importance of the degradation of these species.
Intronic sequences have been shown to be essential in autoregulation of intron-containing RNA transcripts as well as their degradation (Barta and Iggo 1995; Preker and Guthrie 2006; Dong et al. 2007; Sayani et al. 2008). Therefore, based on these findings, we reasoned that the dependence on NMD and on the nuclear exosome for the degradation of certain unspliced transcripts could lay within their intronic sequences. We found that intron identity is important for NMD-sensitive introns in conferring inefficient splicing in cis and trans (Sayani et al. 2008; this study). However, the results of another intron transposition experiment showed that transposing the intron of an RNMD-sensitive gene, RPL16Ai, does not render the chimeric unspliced RNA sensitive to both degradation pathways (Fig. 3). Thus, although intron identity can determine the turnover route of unspliced transcripts, it is not always the determining factor. It remains to be determined whether this is a general feature or whether this is specific to the example studied here. Contrary to recent studies (Bregman et al. 2011; Trcek et al. 2011), our data show that replacement of upstream promoter sequences as well as regulatory elements does not lead to a change in the post-transcriptional degradation fate of an unspliced transcript. For example, replacement of the native promoter with a galactose-inducible promoter for the intron-containing RPS11A did not lead to a change in the levels of the unspliced transcript. The RPS11A unspliced precursor transcribed from the native or the galactose-inducible promoters was degraded in a sequential manner, requiring the involvement of the nuclear exosome and NMD. Similarly, replacement of the RPS17B promoter sequence with the GAL promoter did not lead to changes in unspliced transcript accumulation nor in the switch in the decay pathway of the NMD-sensitive RPS17B gene. Thus, it appears that the promoter identity does not play a major function in determining the degradation route of specific unspliced pre-mRNAs.
Our results support a model suggesting that the balance between nuclear and cytoplasmic degradation of unspliced transcripts is dependent on the rate of export of these precursors from the nucleus into the cytoplasm. A kinetic delay in the rate of RNA export would result in a higher fraction of unspliced transcripts in the nucleus, which would now be targeted for turnover by the nuclear exosome. This model is supported by earlier studies showing that unspliced species generated in a prp2-ts mutant are mostly degraded by the nuclear exosome (Bousquet-Antonelli et al. 2000). The importance of the function of the nuclear exosome for unspliced precursors degradation in this particular mutant background might be explained by the fact that defects in Prp2p function have been shown to result in nuclear retention of unspliced species (Gencheva et al. 2010). In our study, we found that upon genetic inhibition of mRNA export, a large proportion of unspliced precursors are now retained within the nucleus and degraded by the action of the nuclear exosome. This observation might reflect the higher nuclear retention of unspliced transcripts in the nucleus, but could also be due to the activation of nuclear surveillance that was previously shown to occur upon inhibition of mRNA export in a variety of mutant backgrounds (Libri et al. 2002; Rougemaille et al. 2007; Assenholt et al. 2008). Such shifts in the turnover pathways of unspliced transcripts reveal the dynamic nature of RNA turnover that is modulated and/or controlled by the activity of the conserved RNA export factor, Mex67p. It would be of interest to determine whether, in addition to unspliced transcripts, Mex67p also plays a role in the export of other short-lived RNAs.
To investigate the involvement of putative factors that function in a manner antagonistic to RNA export, we found a physiologically relevant target for the peripheral nuclear retention factors, Mlp1p and Mlp2p. We show that these factors are active in retaining a nuclear-degraded unspliced transcript (RPL18B), but that they do not seem to act on unspliced transcripts that are degraded by the combinatorial action of the nuclear exosome and NMD (RPL16A and RPL22A) or solely through NMD (RPL22B). These results show that unspliced transcripts that are degraded either through NMD alone or by the nuclear exosome and NMD are not actively retained by the Mlp factors Mlp1p and/or Mlp2p. It is likely that RPL18B is not the only transcript that is directly retained by the Mlp factors. However, microarray studies of gene expression in the mlp2 mutant did not reveal any major change in unspliced RNA accumulation (Vinciguerra et al. 2005). Additional genome-wide studies should reveal the global extent of Mlp1p and/or Mlp2p retention functions for unspliced RNAs and other endogenous transcripts.
Taken together, these results show that the degradation of unspliced transcripts is largely generic and likely rule out the possibility of particular sequences in making a transcript directly susceptible to the enzymatic activity of a specific nuclear or cytoplasmic degradation system. Instead, sensitivity of different transcripts to different degradation systems might be the result of the equilibrium between the rates of export or nuclear retention, making subcellular localization the major determinant in deciding on the degradation route for these species. While export or retention efficiency might depend on specific signals within the unspliced transcripts, it is unlikely that these sequences make the transcripts intrinsically prone to the action of the exosome or NMD. This sequential mechanism of RNA quality control has far-reaching implications beyond the surveillance and degradation of unspliced transcripts in yeast. A number of noncoding RNAs are known to be under an active RNA surveillance circuit that involves the degradation of unspliced species. Studies on the Air noncoding (nc) RNA showed that it is an atypical RNA pol II transcript that undergoes splicing and export. However, the unspliced transcript is mostly sequestered in the nucleus (Seidl et al. 2006). Although this study did not directly reveal a role for the nuclear exosome in the degradation of nuclear-retained Air ncRNA, the observation that these RNA species are highly unstable strongly suggests that the nuclear exosome might be involved. A complex involvement of NMD and the nuclear exosome has also been implicated in the regulation of the Xist ncRNA in mammalian cells (Ciaudo et al. 2006). Thus, it appears that modulation of export rate and sequential involvement of nuclear and cytoplasmic degradation activities is not limited to unspliced transcripts degradation in yeast, but can also affect other classes of transcripts, making it a general strategy to regulate RNA expression and turnover.
MATERIALS AND METHODS
Yeast strains utilized in this study
Strain genotype source
ySSC001 BY4741 or BY4742 Open Biosystems
ySSC002 rrp6Δ::KAN Open Biosystems
ySSC003 upf1Δ::KAN Open Biosystems
ySSC004 upf2Δ::KAN Open Biosystems
ySSC004 rrp6Δ::KAN upf1Δ::HIS (Sayani et al. 2008)
ySSC005 rrp6Δ::HIS upf2Δ::TRP (This study)
ySSC006 KAN-GAL::RPP1B (This study)
ySSC007 same as ySSC006 but with rrp6::HIS (This study)
ySSC008 same as ySSC006 but with upf1::HIS (This study)
ySSC009 same as ySSC006 but with rrp6::HIS upf1::HYGR (This study)
ySSC010 KAN-GAL::RPS11A (This study)
ySSC011 same as ySSC010 but with rrp6::HIS (This study)
ySSC012 same as ySSC010 but with upf1::HIS (This study)
ySSC013 same as ySSC010 but with rrp6::HIS upf1::HYGR (This study)
ySSC014 KAN-GAL::RPL16A (This study)
ySSC015 same as ySSC014 but with rrp6Δ (This study)
ySSC016 same as ySSC014 but with upf1Δ (This study)
ySSC017 same as ySSC014 but with rrp6Δ upf1Δ (This study)
ySSC018 KAN-GAL::RPS17B (This study)
ySSC019 same as ySSC018 but with rrp6Δ (This study)
ySSC020 same as ySSC018 but with upf1Δ (This study)
ySSC021 same as ySSC018 but with rrp6Δ upf1Δ (This study)
ySSC022 KAN-GAL::RPL22B (This study)
ySSC023 same as ySSC022 but with rrp6Δ (This study)
ySSC024 same as ySSC022 but with upf1Δ (This study)
ySSC025 same as ySSC022 but with rrp6Δ upf1Δ (This study)
ySSC026 pMEX67, mex67Δ (Segref et al. 1997)
ySSC027 pmex67-5, mex67Δ (Segref et al. 1997)
ySSC028 same as ySSC026 but with rrp6Δ (This study)
ySSC029 same as ySSC026 but with upf1Δ (This study)
ySSC030 same as ySSC027 but with rrp6Δ (This study)
ySSC031 same as ySSC027 but with upf1Δ (This study)
ySSC032 same as ySSC027 but with KAN-GAL::RPS17B (This study)
ySSC033 same as ySSC032 but with rrp6Δ (This study)
ySSC034 same as ySSC032 but with upf1ΔI (This study)
ySSC035 mlp1Δ Open Biosystems
ySSC036 mlp2Δ Open Biosystems
ySSC037 mlp1Δ upf1Δ (Sayani et al. 2008)
ySSC038 mlp1Δ mlp2Δ (This study)
ySSC039 mlp2Δ upf1Δ (This study)
ySSC040 mlp1Δ mlp2Δ upf1Δ (This study)
ySSC041 GOT1 gene with a transposed RPL16A intron (This study)
ySSC042 same as ySSC041 but with rrp6Δ (This study)
ySSC043 same as ySSC041 but with upf1Δ (This study)
ySSC044 same as ySSC041 but with rrp6Δ upf1Δ (This study)
ySSC045 GOT1 gene with a transposed RPS17B intron (This study)
ySSC046 same as ySSC045 but with rrp6Δ (This study)
ySSC047 same as ySSC045 but with upf1Δ (This study)
ySSC048 same as ySSC045 but with rrp6Δ upf1Δ (This study)
Yeast strains and culture
Yeast strains were constructed by a one-step PCR strategy using standard yeast molecular genetics protocols. All strains were genotyped through the purification of clonal genomic DNA using primers flanking the cassette region upstream of and downstream from the ORF of interest. All yeast cultures were grown in either YPD or YPGal medium to an OD600 of ∼0.3–0.8. Typically, 50-mL culture volumes were harvested (with the exception of the decay time-point aliquots, which were ∼25 mL), and harvested cells were utilized for total RNA extraction.
RNA extraction and quantitation
Total RNA was extracted by resuspending cells (from either an ∼50-mL or ∼25-mL culture volume) in ∼400 μL of RNA buffer (50 mM Tris-HCl at pH 7.4; 100 mM NaCl; 10 mM EDTA) and vortexed with glass beads for 2 min at room temperature, followed by an incubation on ice for an equal amount of time. This was followed by the addition of ∼100 μL of 10% SDS. Next, ∼500 μL of RNA-based phenol-chloroform was added and vortexed for ∼1 min and incubated at 65°C for 6 min, followed by a revortexing step for 1 min. The tubes were spun at 13,200 rpm for 5 min and the supernatant layer was transferred to a new tube followed by two additional phenol-chloroform extraction steps as above (1-min vortexing followed by centrifugation, but without the 65°C step). After the final phenol–chloroform extraction, ∼1 mL of 95% ethanol was added with 200 μL of 3.2 M sodium acetate (pH5.2), and total RNA was precipitated at −20°C overnight. Tubes were spun at 13,200 rpm for 20 min at room temperature, resultant supernatant was discarded, and 200 μL of 70% ethanol was added to wash the pelleted total RNA sample. After a brief centrifugation step at 13,200 rpm, the 70% ethanol was discarded and the total RNA pellets were resuspended in sterile water and vortexed until the pellets were completely resuspended. RNA quantitation was carried out in duplicate at λ = 260 nm (normally, RNAs were diluted 1:1000 or 1:500). Approximately 10 μg of RNA was utilized and combined with denaturing buffer (formamide and formaldehyde) and incubated at 65°C for ∼15 min, followed by addition of 2 μL of loading dye (EDTA, glycerol, bromophenol blue, xylene cyanol).
Synthesis of probes
With the exception of the SCR1 probe (which was generated through random priming), all probes were antisense intronic riboprobes that spanned the intronic region of each of the target intron-containing genes that were studied. Riboprobe generation encompassed utilization of an amplified intron containing the T3 RNA Polymerase promoter at the 3′ end. Briefly, 3 μL of product was utilized as a template with 1 μL each of rATP, rCTP, and rGTP, and 2–3 μL of [α-32-P]UTP; 4 μL of 5× Promega transcription buffer; 2 μL of 100 mM DTT; and 1 μL of T3 RNA polymerase (Promega). The in vitro transcription reaction was incubated at 37°C for 1 h, followed by the addition of 1 μL of DNase (to degrade template DNA), and further incubated at 37°C for 15 min. The probe was then added to Church hybridization buffer.
Northern blotting
Approximately 10 μg of total RNA (quantitated in duplicate at λ = 260 nm) was loaded onto 1.2% denaturing agarose gels (7.5% formaldehyde, 1× MOPS buffer), and gels were transferred through capillary action onto positively charged nylon membrane (purchased from GE Healthcare/Life Sciences, catalog no. RPN 303B) overnight. The blots were cross-linked with a Stratagene UV-cross-linker. Northern blots were hybridized overnight at ∼65°C–70°C with desired probes in Church buffer (0.5 M sodium phosphate at pH7.2; 7% SDS; and 1% BSA). After hybridization, blots were washed twice between 65°C and 70°C with 2× SSPE, 0.1% SDS buffer; and once with 0.1× SSPE, 0.1% SDS buffer. Blots were exposed at least overnight (or longer if needed) and scanned with a BioRad scanner.
Decay experiments
The galactose promoter was amplified from a plasmid obtained from Mark Longtine (Longtine et al. 1998). It was inserted ∼50 bp upstream of the translation start site. Strains expressing a desired intron-containing ribosomal protein gene under the control of the galactose promoter were grown overnight in a large culture (∼200 mL) in YPGalactose medium between 25°C and 30°C. When cells were in logarithmic phase (typically OD600 nm of ∼0.3–0.8), an aliquot (∼25 mL) of the cells were removed and added to a 50-mL falcon tube containing 95% ethanol that was cooled at −80°C and spun down at 3000 rpm for 1.5 min. The liquid was decanted and the tubes containing the spun cells were stored at −20°C until they were ready for further use. The first timepoint (t = 0 min) was treated as the steady state. The remainder of the culture grown in YPGal was spun down, decanted, and immediately resuspended in YPD medium to induce transcriptional repression, and subsequently, four additional timepoints were collected (exactly as described above) at 3, 6, 10, and 15 min post-transcriptional shutoff.
ACKNOWLEDGMENTS
This work was supported by a National Institutes of Health Grant to G.F.C. (5R01GM061518-11). We thank Kevin Roy for advice and help with the construction of yeast strains, sharing reagents, and unpublished data; and Dr. Cesar Fernandez for advice and sharing reagents.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033779.112.
REFERENCES
- Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH 2008. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA 14: 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta I, Iggo R 1995. Autoregulation of expression of the yeast Dbp2p ‘DEAD-box' protein is mediated by sequences in the conserved DBP2 intron. EMBO J 14: 3800–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Ciaudo C, Bourdet A, Cohen-Tannoudji M, Dietz HC, Rougeulle C, Avner P 2006. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet 2: e94 doi: 10.1371/journal.pgen.00200904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G 2003. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell 11: 1279–1289 [DOI] [PubMed] [Google Scholar]
- Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell 25: 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Kawashima TR, Chanfreau GF 2012. Quality control of MATa1 splicing and exon skipping by nuclear RNA degradation. Nucleic Acids Res 40: 1787–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Gencheva M, Lin TY, Wu X, Yang L, Richard C, Jones M, Lin SB, Lin RJ 2010. Nuclear retention of unspliced pre-mRNAs by mutant DHX16/hPRP2, a spliceosomal DEAH-box protein. J Biol Chem 285: 35624–35632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci 90: 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, et al. 2008. Translational control of intron splicing in eukaryotes. Nature 451: 359–362 [DOI] [PubMed] [Google Scholar]
- Jensen TH, Boulay J, Rosbash M, Libri D 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol 11: 1711–1715 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell 44: 108–119 [DOI] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liang S, Tartakoff AM 1996. Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)+ RNA export. EMBO J 15: 6750–6757 [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast14: 953–961. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev 14: 2173–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Schwartzfarb EM, Silver PA, Yu MC 2006. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell 24: 903–915 [DOI] [PubMed] [Google Scholar]
- Preker PJ, Guthrie C 2006. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA 12: 994–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J 26: 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 31: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl CI, Stricker SH, Barlow DP 2006. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J 25: 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldón A, Query CC, Singer RH. 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell147: 1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Parker R 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem 69: 571–595 [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J 24: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]