The tumor suppressor p53 is a well-known regulator of gene expression in response to cellular stress. While its mechanism of transcription activation is well studied, the underlying molecular basis for p53-mediated transcriptional repression has, until now, remained elusive. Manfredi and colleagues now find that p53 directly up-regulates a novel target, E2F7, upon DNA damage. E2F7 in turn down-regulates a specific subset of genes that contribute to p53-dependent cell cycle arrest.
Keywords: E2F7, p53, repression, cell cycle, transcription
Abstract
The p53 tumor suppressor protein is a transcription factor that exerts its effects on the cell cycle via regulation of gene expression. Although the mechanism of p53-dependent transcriptional activation has been well-studied, the molecular basis for p53-mediated repression has been elusive. The E2F family of transcription factors has been implicated in regulation of cell cycle-related genes, with E2F6, E2F7, and E2F8 playing key roles in repression. In response to cellular DNA damage, E2F7, but not E2F6 or E2F8, is up-regulated in a p53-dependent manner, with p53 being sufficient to increase expression of E2F7. Indeed, p53 occupies the promoter of the E2F7 gene after genotoxic stress, consistent with E2F7 being a novel p53 target. Ablation of E2F7 expression abrogates p53-dependent repression of a subset of its targets, including E2F1 and DHFR, in response to DNA damage. Furthermore, E2F7 occupancy of the E2F1 and DHFR promoters is detected, and expression of E2F7 is sufficient to inhibit cell proliferation. Taken together, these results show that p53-dependent transcriptional up-regulation of its target, E2F7, leads to repression of relevant gene expression. In turn, this E2F7-dependent mechanism contributes to p53-dependent cell cycle arrest in response to DNA damage.
Molecular sensors respond to DNA damage signals by activating a sequence of events that leads to the execution of cellular stress responses (Levine 1997; Giono and Manfredi 2006; Laptenko and Prives 2006). In mammalian cells, the tumor suppressor p53 is a master regulator of stress response pathways such as cell cycle checkpoints, DNA repair, senescence, and apoptosis (Levine 1997; Vousden and Prives 2009). Its importance in human cancers is emphasized by the fact that most tumors express a mutant p53 or have alterations in the p53 pathway itself (Levine 1997). Most of the activity of p53 as a tumor suppressor has been ascribed to its function as a sequence-specific, DNA-binding transcription factor (Laptenko and Prives 2006; Beckerman and Prives 2010; Brady et al. 2011). While the mechanism of transcriptional activation by p53 is well understood, the molecular basis for transcriptional repression remains unclear. In this study, E2F7 is identified as a new p53 target gene playing a role in DNA damage-induced, transcriptional repression of indirect p53 targets.
The E2F family of transcription factors is known for being important regulators of cellular proliferation (Ren et al. 2002; Saavedra et al. 2002; Zhu et al. 2004; Chen et al. 2009). There are eight known E2Fs, and they can be functionally divided into two groups: activators and repressors (DeGregori et al. 1997; Dyson 1998; DeGregori 2004; Xu et al. 2007; Moon and Dyson 2008; Lammens et al. 2009). E2F1, E2F2, and E2F3a are known activators, whereas E2F3b, E2F4, E2F5, E2F6, E2F7, and E2F8 are considered to be repressors (Lammens et al. 2009). Within the repressor group of E2Fs, there is an atypical subgroup composed of E2F7 and E2F8 (Logan et al. 2004; Lammens et al. 2009). While atypical E2Fs differ structurally from typical E2Fs, all mammalian E2Fs share a conserved DNA-binding domain (Logan et al. 2004; Lammens et al. 2009). However, typical E2Fs contain only one DNA-binding domain and require a DP partner in order to bind DNA (Dyson 1998; DeGregori 2004; Lammens et al. 2009). The Rb family is composed of Rb (p105), p107, and p130 (Dyson 1998; DeGregori 2004; Zhu et al. 2004; Jackson et al. 2005). They have been well-characterized as repressors of E2F-mediated transcription in cells (Dyson 1998). In general, hypophosphorylated Rb (p105) binds to the activator E2Fs (E2F1–3a), and p107 and p130 bind to the repressor E2Fs (E2F3b–5) (Dyson 1998; DeGregori 2004). In contrast, the repressors E2F6, E2F7, and E2F8 have not been shown to interact with any of the Rb family members (Cartwright et al. 1998).
The E2F7 protein is an atypical E2F with two DNA-binding domains. It does not heterodimerize with a DP partner and does not have an Rb family-binding domain (Di Stefano et al. 2003; Logan et al. 2004). Therefore, E2F7 functions as a transcriptional repressor in an Rb family-independent manner. E2F7 recognizes E2F-binding sequences and participates in an autoregulatory loop with E2F1 by repressing E2F1-dependent transcription (Di Stefano et al. 2003; Moon and Dyson 2008). These studies suggest that E2F7 activity is required in order to keep members of the E2F family and their target genes under tight control. For example, E2F7 and E2F8 double-knockout mice die early during development due to widespread E2F1-induced, p53-dependent apoptosis (Li et al. 2008). In addition to playing an important role during embryogenesis, E2F7 has been implicated in the DNA damage response and cell cycle control of E2F target genes (Panagiotis Zalmas et al. 2008). Most recently, E2F7 was shown to regulate proliferation and apoptotic pathways in human keratinocytes (Endo-Munoz et al. 2009; Hazar-Rethinam et al. 2011). These studies further suggest that the deregulation of the E2F1–E2F7 regulatory loop may play a major role in the initiation of cutaneous squamous cell carcinoma, which could have important implications for therapy. Taken together, the importance of E2F7 during development and tumorigenesis is clear. The present study addresses how E2F7 transcription is regulated in response to DNA damage and its function as a downstream effector of p53-dependent repression. These data demonstrate that E2F7 plays a role in transcriptional repression of a subset of indirect p53 target genes involved in DNA replication. DNA damage-induced up-regulation of E2F7 is p53-dependent, with E2F7 being a direct target for transcriptional activation by p53. These findings provide additional insights into E2F7 regulation in response to DNA damage, an important step toward fully understanding the role of E2F7 in cellular processes and in cancer.
Results
DNA damage-induced transcriptional repression of a subset of cell cycle genes is p53-dependent
A large number of genes have been shown to be repressed by p53 in response to DNA damage (Seto et al. 1992; Lee et al. 1999, 2001; Murphy et al. 1999; Badie et al. 2000; Shats et al. 2004; St Clair et al. 2004; Imbriano et al. 2005; Laptenko and Prives 2006). This study presents new insights into the mechanism of DNA damage-induced repression of a select group of genes indirectly repressed by p53. First, DNA damage-induced down-regulation of known p53 repression targets involved in the G2/M phase of the cell cycle (CDC25C, CDK1, CCNB1, and CDC20) was compared with E2F1 and other G1/S genes (RRM2, DHFR, and CDK2). In order to address the p53 dependence, human colon carcinoma isogenic cell lines HCT116 p53+/+ and p53−/− were used. Cells were subjected to treatment with the DNA-damaging agent doxorubicin (Supplemental Fig. 1S). Immunoblot analysis of cell extracts from treated samples showed p53 induction (Supplemental Fig. 1SA). In addition, DNA damage triggered both a limited G1/S arrest and a substantial G2/M cell cycle checkpoint in p53+/+ cells, while p53−/− cells arrested only in G2/M (Supplemental Fig. 1SB). These cells were also analyzed for gene expression by quantitative RT–PCR (qRT–PCR) to compare levels of the G2/M targets (CDC25C, CDK1, and CCNB1) with that of G1/S targets (E2F1, RRM2, and CDK2) in both cell lines. DNA damage-induced transcriptional repression of all targets was observed in p53-replete cells but not in the p53-null cells (Fig. 1A). Likewise, CDKN1A (p21) levels were induced after DNA damage in p53-expressing cells but not in p53-null cells (Fig. 1A). These findings confirm that DNA damage-induced repression of this set of genes is p53-dependent.
Figure 1.
DNA damage induces p53-dependent transcriptional repression of cell cycle genes and up-regulates E2F7 mRNA and protein. (A) qRT–PCR using mRNA extracted from HCT116 p53+/+ and p53−/− cells before and after treatment with the DNA-damaging agent doxorubicin. Expression analysis of a subset of genes involved in cell cycle regulation was evaluated at different time points after treatment. (B) Immunoblot analysis using whole-cell extracts from U2OS (p53+/+) cells before and after treatment with increasing amounts of doxorubicin. Protein expression of E2F7, p53, p21, and Actin was detected with specific antibodies. (C) qRT–PCR expression analysis of E2F6, E2F7, and E2F8 in HCT116 (p53+/+) and U2OS (p53+/+) cells nontreated and treated with doxorubicin. (D) Immunoblot analysis using whole-cell extracts from U2OS (p53+/+) cells before and after treatment with increasing amounts of the DNA-damaging agent etoposide. Protein expression analysis was conducted as in B. (E) qRT–PCR expression analysis of E2F1, E2F7, and E2F8 using mRNA extracted from U2OS cells nontreated and treated with etoposide as in D. Bar graphs represent the average of three independent experiments. (NT) Nontreated; (D) days, Error bars indicate the standard error of the mean (SEM); statistical significance is shown using the Student's t-test analysis. (**) P < 0.01; (*) P < 0.05; (n.s.) not significant.
To validate these results in a different system, p53 was targeted in osteosarcoma-derived U2OS p53+/+ cells using RNAi. Immunoblot analysis verifies the knockdown of p53 in these cells (Supplemental Fig. 1C). Ablation of p53 impaired DNA damage-induced repression of CDC25C, CCNB1, CDC20, RR2, E2F1, and DHFR in U2OS cells (Supplemental Fig. 1SD). Furthermore, CDKN1A induction was also impaired in p53-ablated cells. These results authenticate the p53 dependence of DNA damage-induced repression of select G1/S (replication targets) and G2/M (mitotic targets) genes.
DNA damage induces an up-regulation of E2F7 but not E2F6 or E2F8
E2F7 had been previously reported to be up-regulated in response to DNA damage in cells, but the basis for this was not addressed (Panagiotis Zalmas et al. 2008). Recently, E2F7 was implicated in regulating S-phase progression by repressing a large network of genes (Westendorp et al. 2012). The present study hypothesizes that E2F7 is up-regulated by p53 and that it mediates transcriptional repression of indirect p53 targets in response to DNA damage. Thus, DNA damage-induced up-regulation of E2F7 in response to genotoxic stress was corroborated using multiple tumor cell lines. Cells were subjected to increasing amounts of doxorubicin followed by gene expression analysis of E2F7 (Supplemental Fig. 2S). qRT–PCR studies substantiate that E2F7 up-regulation is dose-dependent in all cell lines tested (Supplemental Fig. 2S). CDKN1A levels were also increased, whereas CDC25C levels were decreased (Supplemental Fig. 2S). In addition, U2OS cells were treated with a single dose of the DNA-damaging agent doxorubicin. qRT–PCR expression analysis at different time points showed a time-dependent increase in E2F7 after treatment (Supplemental Fig. 2SD). E2F7 expression levels peaked at 24 h post-treatment, coinciding with transcriptional repression of E2F1, DHFR, and RRM2 (Supplemental Fig. 2SD). Immunoblotting of cell extracts from U2OS cells treated with increasing amounts of doxorubicin shows a dose-dependent increase in E2F7 levels (Fig. 1B). Taken together, these data support the possibility that in response to DNA damage, E2F7 up-regulation is p53-dependent.
E2F7 and E2F8 have been reported to play compensatory roles for one another in some settings (Logan et al. 2005; Li et al. 2008; Lammens et al. 2009). In one report, E2F7 and E2F8 were reported to be induced after treatment with the genotoxic drug etoposide (Panagiotis Zalmas et al. 2008). Therefore, expression levels of E2F8 were probed together with those of E2F6 and E2F7. In response to doxorubicin treatment, E2F8 levels were down-regulated in U2OS cells (Fig. 1C). Although E2F8 expression in HCT116 cells was reproducibly lower after treatment, this effect was mild and not statistically significant (Fig. 1C). The down-regulation of E2F6 was not statistically significant, but it was not found up-regulated in any of the examined cell lines (Fig. 1C). Of the atypical repressor E2F proteins, only E2F7 was reproducibly up-regulated in response to treatment with doxorubicin. U2OS cells were also treated with increasing amounts of the DNA-damaging agent etoposide. E2F7 protein was again up-regulated in a dose-dependent manner (Fig. 1D). Gene expression analysis of each corresponding sample in Figure 1D confirms a statistically significant dose-dependent decrease in E2F1 and E2F8 (Fig. 1E). E2F7 was the only E2F analyzed in this study found to be consistently up-regulated in response to either doxorubicin or etoposide treatment (Fig. 1B–E).
E2F7 is a p53 target
In order to investigate the p53 dependence of the up-regulation of E2F7, p53 levels were increased in the absence of DNA damage by treating U2OS cells with increasing amounts of the Mdm2 inhibitor Nutlin-3. As expected, p53 and p21 levels increased in response to such treatment (Fig. 2A). This increase in p53 was accompanied by a dose-dependent increase in E2F7 protein levels (Fig. 2A). In addition, Nutlin-3 treatment triggered a cell cycle arrest primarily in the G1 phase of the cell cycle (Fig. 2B). To confirm the p53 dependence of this effect, p53 was targeted with RNAi oligonucleotides. Indeed, ablation of p53 expression abrogates E2F7 up-regulation in response to Nutlin-3 treatment (Fig. 2C), whereas E2F7 and CDKN1A (p21) were up-regulated in response to Nutlin-3 in cells transfected with a nonspecific control RNAi (Fig. 2C). Consistent with the results shown above (Fig. 1), E2F8 was repressed in control RNAi-treated cells after Nutlin-3 treatment, but this was not observed in p53-ablated cells. These results demonstrate that induction of p53 in the absence of genotoxic stress is sufficient to up-regulate E2F7 protein and mRNA. Furthermore, E2F8 is repressed in a p53-dependent manner in response to Nutlin-3 treatment.
Figure 2.
E2F7 is up-regulated in a p53-dependent manner in response to either Nutlin3a treatment or ectopic expression of p53. (A) Immunoblot analysis of E2F7, p53, p21, and Actin expression using cell extracts from U2OS cells nontreated and treated with increasing amounts of the Mdm2 inhibitor Nutlin3a. (B) Cell cycle profile using flow cytometry of corresponding samples in A. (C) qRT–PCR expression analysis of TP53, CDKN1A, E2F6, E2F7, and E2F8 using mRNA extracted from U2OS p53+/+ cells nontreated or treated with increasing amounts of Nutlin3a. (D) Immunoblot analysis of p53 levels using the EJ cell line, which was engineered to ectopically express p53 under the control of a tetracycline-repressible system. (E) Cell cycle profile analysis of each corresponding sample in D before and after removal of tetracycline from cells in culture. (F) qRT–PCR expression analysis of the same target genes used in C from each corresponding EJp53 sample shown in D and E. The average of three independent experiments is shown. (NT) Nontreated; (T) tetracycline. Error bars indicate the standard error of the mean. Statistical significance is shown using the Student's t-test analysis; (**) P < 0.01; (*) P < 0.05; (n.s.) not significant.
To further examine whether up-regulation of p53 is sufficient to affect E2F7 expression, an inducible cell system was used. The bladder carcinoma EJ (p53-null) cell line was engineered to express p53 in a tetracycline-dependent manner and has been previously characterized (Macip et al. 2003). In these cells, ectopic expression of p53 is sufficient to induce a cell cycle arrest in both G1 and G2 phases of the cell cycle (Fig. 2D,E). p53 induces both CDKN1A and E2F7 expression, while E2F6 and E2F8 levels decrease (Fig. 2F). These results indicate that p53 expression alone is sufficient to induce E2F7 expression, but not that of E2F6 or E2F8, and that DNA damage-induced up-regulation of E2F7 is p53-dependent.
To examine the p53 dependence of the increase in E2F7 levels after DNA damage, U2OS cells in which p53 had been silenced using a siRNA approach were used. Consistent with the findings shown in Figure 2, induction of E2F7 and CDKN1A in response to DNA damage was impaired in p53-ablated cells (Fig. 3A). Examination of E2F7 expression in doxorubicin-treated HCT116 p53+/+ and p53−/− cells showed that up-regulation of E2F7 was clearly p53-dependent (Fig. 3B). As shown in Figure 3C, p53 levels increase after DNA damage in p53+/+ cells but not in p53−/− cells. Therefore, in response to DNA damage, p53 is required for the up-regulation of E2F7.
Figure 3.
DNA damage-induced E2F7 up-regulation is p53-dependent. (A) U2OS cells were used to silence p53 expression via a siRNA approach. Cells transfected with either control (Ctl) or p53 siRNA oligonucleotides were treated with doxorubicin 24 h after transfection. mRNA was extracted from each sample and processed for gene expression analysis of E2F7, CDKN1A, and TP53 by qRT–PCR. (B) HCT116 p53+/+ or p53−/− cells were treated with doxorubicin and processed for gene expression analysis of E2F7 and CDKN1A. Immunoblot analysis showing p53 expression levels from each corresponding sample shown in C. (NT) Nontreated; (D) days. Error bars indicate the standard error of the mean. Statistical significance is shown using the Student's t-test analysis; (**) P < 0.01; (*) P < 0.05; (n.s.) not significant. The average of three independent experiments is shown.
p53 is recruited to the E2F7 promoter via a consensus p53 response element
To ascertain whether p53 is recruited to the E2F7 promoter after DNA damage, the sequence of the proximal promoter was inspected for a possible p53 response element, corresponding to the p53 consensus RRRCWWGYYYRRRCWWGYY (el-Deiry et al. 1992; Wei et al. 2006). The cartoon in Figure 4A outlines the region of the E2F7 promoter where a potential element was found. Genomic DNA primers were designed in order to assay for enrichment of p53 protein to this region of the E2F7 promoter by chromatin immunoprecipitation (ChIP) (Fig. 4A). First, ChIP assays were conducted on H1299 (p53-null) cells that were transfected with empty vector or expression constructs for either wild-type or a truncated p53 (Δ24), which lacks the C-terminal 24 amino acids. The truncated p53 (Δ24) was shown recently to be defective in transactivation of CDKN1A (p21) and for binding to the corresponding promoter (Hamard et al. 2012). Ectopic expression of p53 resulted in a significant increase in the signal enriched for p53 at either the E2F7 or CDKN1A promoters but not at the albumin promoter (Fig. 4B). Overexpression of the truncated p53 did not enrich at the E2F7 or CDKN1A promoter even though both wild-type and mutant p53 were expressed at comparable levels (Fig. 4B). This finding was confirmed by ChIP using endogenous p53. HCT116 p53+/+ cells were treated with doxorubicin for 24 h. Analysis of purified ChIP DNA fragments by qPCR showed a significant signal enriched for p53 at the E2F7 promoter in response to DNA damage, but not at the albumin promoter (Fig. 4C, left). As expected, enrichment for p53 was also detected at the CDKN1A promoter (Fig. 4C, right). Taken together, these studies demonstrate that p53 is recruited to the E2F7 promoter during the DNA damage response. This finding strongly supports the observation that E2F7 is a novel p53 target gene induced by DNA damage.
Figure 4.
p53 is recruited to the E2F7 promoter via a putative p53-binding site. (A) Schematic of the E2F7 promoter is shown with a consensus p53 response element. (RE) Response element; (R) purine; (Y) pyrimidine; (W) A or T; (arrows) forward and reversed primers. (B) H1299 (p53-null) cells were transfected with either a control empty vector (CMV), a wild-type p53 (WT), or a mutant p53 (Δ24). Transfected samples were collected 24 h after transfection and processed for ChIP using a p53 monoclonal antibody. Genomic DNA was isolated and analyzed by qPCR using genomic DNA primers surrounding the p53RE as depicted in A. Primers in the albumin and CDKN1A promoters were used as a negative and positive control, respectively. Immunoblot analysis showing the relative expression of p53 after transfection is shown to the right of B. (C) HCT116 (p53+/+) cells in culture were treated with doxorubicin for 24 h and processed for ChIP using a p53 monoclonal antibody or GFP antibody as a control. Genomic DNA was isolated and analyzed as in B. (D) H1299 (p53-null) cells were transfected with an E1b-TATA luciferase reporter (E1b-TATA-p53RE) containing a p53 response element from the E2F7 promoter and increasing amounts of either wild-type p53 (WT) or a DNA-binding mutant p53 (248H). Empty E1b-TATA luciferase and E1b-TATA containing a CDKN1A (p21) minimal promoter with an isolated p53 response element (E1b-TATA-5′ p21P) were used as negative and positive control, respectively. (NT) Nontreated; (Dox) doxorubicin. Error bars indicate the standard error of the mean. Statistical significance is shown using the Student's t-test analysis; (**) P < 0.01; (*) P < 0.05; (n.s.) not significant. The average of three independent experiments is shown.
To determine whether p53 can activate E2F7 via this identified putative p53-responsive element, a luciferase reporter assay was used. First, the 20-base-pair (bp) element from the E2F7 promoter was cloned into luciferase reporter containing a minimal adenoviral E1b promoter. Next, H1299 p53-null cells were transfected with the luciferase reporter (E1b-TATA-p53RE) containing the p53 response element from the E2F7 promoter and increasing amounts of either wild-type p53 (WT) or a DNA-binding mutant p53 (248H). A luciferase reporter lacking this insert and a reporter containing a p53 response element derived from the CDKN1A (p21) promoter (E1b-TATA-5′ p21P) were used as negative and positive control, respectively. Cotransfection of the E1b-TATA-p53RE reporter with wild-type p53 shows a 50-fold increase in luciferase activity compared with an empty E1b-TATA (Fig. 4D). Similarly, cotransfection of the p21 reporter (E1b-TATA-5′ p21P) with wild-type p53 yielded a significant increase (250-fold) in luciferase activity. The 248H DNA-binding mutant failed to activate either reporter (Fig. 4D). These findings support the notion that the identified response element is responsible for p53-dependent up-regulation of E2F7.
E2F7 mediates p53-dependent repression
E2F7 protein and mRNA are induced in a p53-dependent manner during the DNA damage response. In addition, a higher cellular level of E2F7 is accompanied by p53-dependent transcriptional repression of specific targets. Previously, E2F7 had been shown to regulate transcription of E2F1 under certain conditions (Di Stefano et al. 2003; Li et al. 2008; Panagiotis Zalmas et al. 2008). Most recently, E2F7 was shown to regulate S-phase progression by repressing a network of oscillating cell cycle genes (Westendorp et al. 2012). Thus, the E2F7 dependence of the down-regulation of specific targets by p53 during the DNA damage response was then addressed. E2F7 expression was targeted using a siRNA approach in cells treated with a DNA-damaging agent, and gene expression of relevant target genes was analyzed by qRT–PCR. No significant changes in cell cycle distribution were observed in E2F7-ablated cells treated with doxorubicin at 1 d and 3 d post-treatment compared with control cells (Supplemental Fig. 3SA,B). In order to rule out possible off-target effects, E2F7 was targeted using four distinct RNAi oligonucleotides (Supplemental Fig. 3SC). qRT–PCR expression analysis was used to verify E2F7 knockdown efficiency (Supplemental Fig. 3SC, middle). DNA damage-induced transcriptional repression of E2F1 is observed in control RNAi transfected cells but not in samples where E2F7 had been ablated (Supplemental Fig. 3SC, left). Interestingly, p53-dependent repression of G1/S target genes (RRM2 and DHFR) as well as E2F8, but not G2/M targets (CDC25C and CDC20), is abrogated in E2F7-ablated cells (Supplemental Fig. 3SD). Confirming the knockdown, E2F7 protein levels increase after DNA damage in cells transfected with a nonspecific control RNAi but are significantly lower in cells transfected with a pool of RNAi targeting oligonucleotides (Fig. 5A). Ablation of E2F7 impairs the down-regulation of E2F1 but not CDC25C after DNA damage (Fig. 5A). In fact, E2F1 protein expression is higher in E2F7-depleted cells (Fig. 5A). The data in Figure 5B demonstrate the efficiency of E2F7 ablation at the mRNA level with a combination of E2F7 RNAi targeting sequences. As expected, CDKN1A is up-regulated in response to DNA damage, and knockdown of E2F7 does not impair its induction in response to genotoxic stress (Fig. 5B). E2F7 depletion using a pool of RNAi oligonucleotides in cells impairs DNA damage-induced repression of the replication targets E2F1, RRM2, and DHFR (Fig. 5C, bottom). Ablation of E2F7, however, did not affect p53-mediated repression of the mitotic targets CDC25C, CCNB1, and CDK1 (Fig. 5C, top). Cell cycle progression under these conditions was then analyzed by flow cytometry (Supplemental Fig. 3SA). Knockdown of E2F7 by this method did not affect the ability of cells to undergo a cell cycle arrest in response to DNA damage. These studies nevertheless implicate E2F7 in DNA damage-induced transcriptional repression of specific targets involved in DNA replication, but not those associated with mitotic progression. This latter finding suggests that there are at least two distinct mechanisms of p53-dependent repression, one of which involves the up-regulation of E2F7 by p53 in response to DNA damage.
Figure 5.
DNA damage-induced transcriptional repression of replication targets, but not mitotic targets, is E2F7-dependent. (A) Immunoblot analysis of E2F7, E2F1, Cdc25C, and p53 protein levels. E2F7 expression was targeted for degradation in U2OS cells using a siRNA approach. Cells in culture transfected with either control (Ctl) or E2F7 siRNA oligonucleotides were treated with 0.1 μg/mL doxorubicin 24 h after transfection and harvested at the indicated time point; (D) days. qRT–PCR expression analysis of E2F7 and CDKN1A is shown in B, and that of CDC25C, CCNB1, CDK1, E2F1, RRM2, and DHFR is shown in C from each corresponding sample in A. (NT) Nontreated; (D) days. Error bars indicate the standard error of the mean. Statistical significance is shown using the Student's t-test analysis; (*) P < 0.01; (*) P < 0.05; (n.s.) not significant. The average of three independent experiments is shown.
E2F7 binds to G1/S target promoters and inhibits cellular proliferation
In order to ascertain whether E2F7 up-regulation is sufficient to have a biological effect in cells, Flag-tagged E2F7 was transfected into U2OS cells, and the consequence of this was examined in colony formation assays. Transfected Flag-tagged E2F7 expression in U2OS cells was detected by immunoblot analysis using an anti-Flag antibody (Fig. 6A). Figure 6B shows a representative result for the colony assay: Forced expression of E2F7 inhibited colony formation of these cells. These data implicate E2F7 in playing a role in the DNA damage response in part by mediating an anti-proliferative effect. This is consistent with the ability of E2F7 to repress transcription of genes such as E2F1. The p53 dependence of the E2F7 anti-proliferative effects were assayed by cotransfecting a pBabe-puro plasmid with either an empty pcDNA3, Flag-tagged E2F7, or Flag-tagged p53 expression vectors into H1299 p53-null cells. It is shown that overexpression of E2F7 or p53 in a p53-null background inhibited colony formation (Fig. 6C). Together, these studies demonstrate that the anti-proliferative effects of E2F7 are indeed independent of p53 status.
Figure 6.
E2F7 is anti-proliferative and binds specifically to the E2F1 and DHFR promoters. (A) U2OS cells were transfected with increasing amounts of either empty pcDNA3 vector or a Flag-tagged E2F7-expressing pcDNA3 vector. Immunoblot analysis of transfected U2OS cells shows the relative expression levels of Flag-E2F7 in cells. (B) Colony formation assay of U2OS cells cotransfected with pBabe-Puro and either empty pcDNA3 vector or Flag-tagged E2F7-expressing vector. pBabe-Puro was used as a selection marker. The bar graph shows the average of three independent colony formation experiments. (C) Colony formation assay of H1299 (p53-null) cells cotransfected with pBabe-Puro and either empty pcDNA3 vector, Flag-tagged E2F7-expressing vector, or Flag-tagged wild-type p53-expressing vector. pBabe-puro was used as a selection marker. The bar graph shows the average of three independent colony formation experiments. (D) H1299 (p53-null) cells were transfected with 2 μg of pcDNA3-Flag E2F7 or empty pcDNA3. Cells were harvested 24 h after transfection and processed for ChIP using an anti-Flag antibody. Immunopurified genomic DNA fragments were analyzed for Flag-E2F7 enrichment at the indicated promoters by qPCR. (B–D) Error bars indicate the standard error of the mean. Statistical significance is shown using the Student's t-test analysis; (**) P < 0.01; (*) P < 0.05; (n.s.) not significant. The average of three independent experiments is shown. (E) HCT116 (p53+/+) cells were treated with doxorubicin for 24 h and processed for ChIP using a polyclonal E2F7 antibody or no antibody as a control. Immunopurified genomic DNA fragments were analyzed for endogenous E2F7 enrichment at the indicated promoters by qPCR. Primers in the albumin promoter were used as a negative control. (NT) Nontreated. The average of two independent experiments is shown.
Previous studies suggest that E2F7 mediates its effects by binding directly to E2F-responsive promoters (Di Stefano et al. 2003; Moon and Dyson 2008). In order to test this, the ability of E2F7 to bind to the E2F1 and DHFR promoters was analyzed by ChIP. H1299 p53-null cells were transfected with a Flag-tagged E2F7 expression vector and processed for ChIP 24 h after transfection. Analysis of the E2F1 and DHFR promoters by qPCR using purified ChIP DNA fragments shows a significant enrichment for E2F7 (Fig. 6D). Transfected Flag-tagged E2F7 was not detected at the albumin (ALB) promoter or at the promoter of the mitotic target CDC25C (Fig. 6D; Supplemental Fig. 4S). These results were confirmed in wild-type p53-expressing cells after DNA damage using an antibody to the endogenous E2F7. HCT116 cells were treated with doxorubicin for 24 h. Endogenous E2F7 occupancy at the E2F1 and DHFR promoters was demonstrated using purified ChIP DNA fragments (Fig. 6E). This validates that endogenous E2F7 is indeed recruited to the E2F1 and DHFR promoters during the DNA damage response (Fig. 6E).
Discussion
The molecular basis for DNA damage-induced transcriptional repression by p53 has not been well understood. Earlier studies suggested that p53 could inhibit transcription by directly disrupting the basal transcription machinery (Seto et al. 1992; Farmer et al. 1996). Others proposed that p53 could also interfere with specific sequence-specific transcription factors via protein–protein interactions (Imbriano et al. 2005; Di Agostino et al. 2006; Le Gac et al. 2006). For example, p53 has been shown to interact with the NF-Y transcription factor and thereby inhibit transcription (Imbriano et al. 2005; Di Agostino et al. 2006). Thus, two general models of p53-dependent repression were proposed: “direct” and “indirect” (Farmer et al. 1996; Lee et al. 1999, 2001; Badie et al. 2000; St Clair et al. 2004; Estève et al. 2005; Wang et al. 2010). The “direct” model of repression involves the binding of p53 to a putative p53 response element on specific repression target genes (Lee et al. 1999, 2001; Murphy et al. 1999; St Clair et al. 2004; McKenzie et al. 2010; Wang et al. 2010). These studies argued that p53 plays an active role in repression by binding to specific sequences on a target promoter and recruiting corepressors. The “indirect” model of repression involves the activation of specific p53 target genes that in turn mediate transcriptional repression downstream from p53. For example, the up-regulation of the gene encoding the cyclin-dependent kinase inhibitor p21 has been implicated as playing a role in p53-dependent transcriptional repression (Badie et al. 2000; Devgan et al. 2005; Fritah et al. 2005). Additionally, several noncoding microRNAs have also been reported to play such a role in Barsotti and Prives (2010), Huarte et al. (2010), and Liu et al. (2011). It is reasonable to speculate that under certain environmental, genotoxic, or developmental stimuli, distinct p53-dependent mechanisms of repression are activated. Which pathway is activated and how they interact may, in fact, be context-dependent. The present study examines DNA damage-induced transcriptional repression of a subset of indirect p53 target genes. A member of the E2F family of transcription factors, E2F7, is characterized as a p53-induced target gene. E2F7, in turn, is a mediator of DNA damage-induced, p53-dependent transcriptional repression of E2F1, RRM2, DHFR, and E2F8.
The E2F7 protein is an atypical E2F resembling the E2F-like (E2L1–3) in plants (Vlieghe et al. 2005). Atypical E2Fs contain two DNA-binding domains, do not form heterodimers with DP proteins, and repress transcription independently of Rb (Logan et al. 2004; Lammens et al. 2009). Interestingly, E2L proteins may play a role in negative regulation of endoreplication in plants (Vlieghe et al. 2005). In human cells, E2F7 was reported to play a role in the DNA damage response, repress E2F1-mediated transcription, and have anti-proliferative properties (de Bruin 2003; Di Stefano et al. 2003; Panagiotis Zalmas et al. 2008). Most recently, it was reported that E2F7 regulates S-phase progression by regulating an array of oscillating genes during the cell cycle (Westendorp et al. 2012). In mice, E2F7 and E2F8 are essential for embryonic development (Li et al. 2008; Winn et al. 2011). The E2F7/8 homolog EFL-3 in Caenorhabditis elegans was recently implicated in playing a role in repressing cell type-specific cell death (Winn et al. 2011). Interestingly, it was reported that E2F7 plays a role in squamous cell differentiation and that loss of E2F7 may give rise to the initiation of cutaneous squamous cell carcinoma (Hazar-Rethinam et al. 2011). Taken together, these studies bring to light important E2F7 biological functions and stress the need for continued research in order to fully characterize its function and regulation. The present study sheds light on how E2F7 transcription is regulated in response to DNA damage.
A putative p53 response element was identified in the human E2F7 promoter. Therefore, it was predicted that p53 is recruited to the E2F7 promoter in response to DNA damage. ChIP assays confirmed this, demonstrating that p53 binds to the E2F7 promoter when p53 is ectopically expressed or induced by DNA damage in cells. Furthermore, wild-type p53 significantly induced expression of a luciferase reporter containing a putative p53-responsive element from the E2F7 promoter. A DNA-binding p53 mutant, however, failed to drive transcription from this reporter. Induction of p53 with Nutlin-3 increases E2F7 protein and mRNA expression. Likewise, exogenous expression of p53 in cells is sufficient to induce E2F7 mRNA transcription. Additionally, it is shown that DNA damage also induces E2F7 expression in cells treated with doxorubicin in a p53-dependent manner. This occurs in a dose- and time-dependent manner. Together, these results support the notion that the E2F7 gene is a direct p53 target gene that is up-regulated in response DNA damage.
Interestingly, E2F7 is the only E2F family member tested in this study that is transactivated in response to DNA damage and in a p53-dependent manner. In contrast, E2F1, E2F6, and E2F8 are down-regulated in response to DNA damage. Previously, it had been reported that E2F8 levels increase during the DNA damage response (Panagiotis Zalmas et al. 2008). In contrast, it is shown here that E2F8 transcription is repressed in an E2F7-dependent manner. This is corroborated by a recent genome-wide study that suggests E2F8 is a potential E2F7 target (Westendorp et al. 2012). Furthermore, E2F1 down-regulation is E2F7-dependent. DNA damage-induced transcriptional repression of the E2F1 target genes RRM2 and DHFR is also E2F7-dependent. This is consistent with the model that E2F7 is a negative regulator of transcription of E2F-responsive promoters. It is likely that other targets will be identified that are also repressed by E2F7.
It is shown here and by others that ectopic expression of E2F7 in cells is anti-proliferative (Di Stefano et al. 2003). E2F7 overexpression in p53-null cells inhibited growth in colony formation assays. This demonstrates that the anti-proliferative properties of E2F7 are p53-independent, suggesting that its effects are downstream from p53. This finding is in line with other studies cited herein and with results included in this study demonstrating that E2F7 is a transcriptional repressor of cell cycle genes. It is shown here that DNA damage induces transcriptional repression of E2F1, DHFR, RRM2, and E2F8 in an E2F7-dependent manner. In contrast, DNA damage-induced repression of the mitotic targets CDC25C, CDK1, CCNB1, and CDC20 is not E2F7-dependent. Nevertheless, DNA damage-induced transcriptional repression of these mitotic targets is also p53-dependent. This finding suggests that while transcriptional repression of a variety of cell cycle genes is p53-dependent, the molecular basis for repression of specific targets may differ significantly. Hence, not all cell cycle genes repressed by p53 share the same underlying mechanism for their down-regulation. ChIP assays show that E2F7 binds selectively to the E2F1 and DHFR promoter but not to the CDC25C promoter. Thus, E2F7 appears to target genes involved in the G1 and S phases, but not those involved in the G2 and mitotic phases of the cell cycle.
Previous studies have shown that E2F7 plays an important role during development. Its importance is emphasized by the fact that deletion of both E2F7 and E2F8 in mice is embryonic-lethal in a manner likely due to E2F1-induced apoptosis (Li et al. 2008). This observation suggests that during animal development, E2F7 and E2F8 activity is a critical regulator of E2F1 activity, and the Rb family is not capable of compensating for their loss. Furthermore, it suggests that under certain conditions, E2F7 can regulate E2F transcriptional responses independently from the Rb family. It is shown here that E2F7 can regulate DNA damage-induced transcriptional repression of E2F-responsive promoters. It is important to note that while overexpression of E2F7 inhibited cellular proliferation in colony formation assays, this was not sufficient to initiate a DNA damage checkpoint. Previously, it was shown that Rb-deficient cells can undergo Ras-induced senescence in culture but undergo endoreduplication due to derepression of replication target genes (Chicas et al. 2010). Thus, it is not surprising that knockdown of E2F7 in cancer cells does not impair cell cycle arrest in response to DNA damage. Although E2F7 depletion in cancer cells abrogates DNA damage-induced transcriptional repression of replication target genes, a significant increase in endoreduplication was not observed. Perhaps E2F7 is part of a larger DNA damage response network activated to ensure proper maintenance of DNA damage checkpoints. This possibility will require future studies. Ablation of E2F7 expression did not substantially change the cell cycle profile after DNA damage (Supplemental Fig. 3SA,B). As E2F7 does not contribute to transcriptional activation by p53, and its down-regulation does not influence repression of targets that are relevant in the G2-to-M phase transition, it is not necessarily remarkable that this would be the case. While we have not provided data for the role of E2F7 in long-term responses to p53, these experimental results suggest that its ablation will not affect the ability of p53 to inhibit colony formation, for example. Thus, the role of E2F7 is likely to enforce p53-dependent effects on cell cycle progression, rather than be absolutely required for their execution.
The present study identifies E2F7 as a mediator of p53-dependent transcriptional repression and highlights the selective nature of p53-dependent mechanisms of repression. Additionally, the cyclin-dependent kinase inhibitor p21 is also transactivated by p53 during the DNA damage response. Therefore, the p21 dependence of p53-mediated transcriptional repression was examined. As it turns out, p21-ablated cells are defective for transcriptional repression of all of the p53 targets analyzed (Supplemental Fig. 5S). This finding suggests that an important interaction between E2F7 and p21 may exist. For example, it is possible that p21 may regulate E2F7 protein activity via interactions with cyclin-dependent kinases.
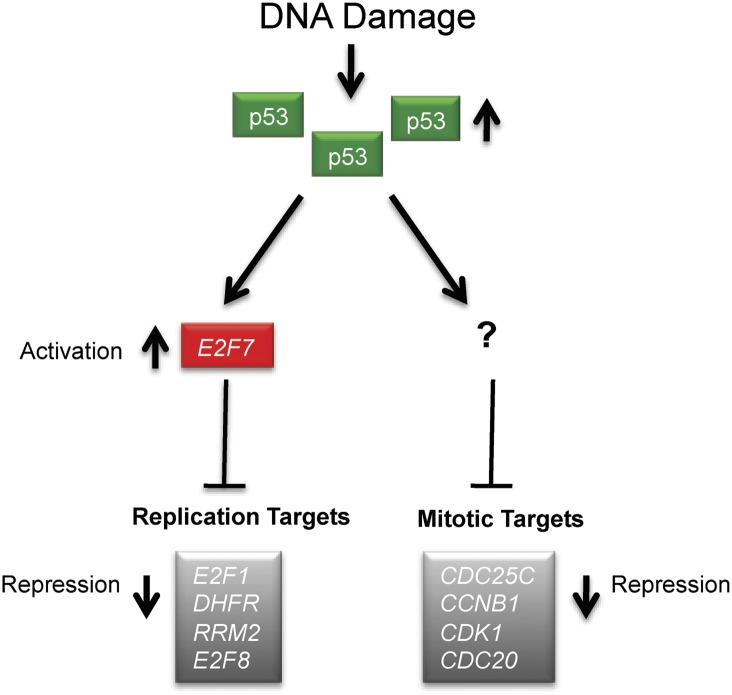
Overall, these results are consistent with the model shown in Figure 7. In response to DNA damage, stabilized p53 binds to the promoter of the E2F7 gene and causes its transactivation. Repression of replication targets by E2F7 is cell cycle-independent, since E2F7-ablated cells are still able to arrest in G1/S. It is thus hypothesized that E2F7 is induced in response to DNA damage by p53 in order to reinforce checkpoint responses.
Figure 7.
Model: E2F7 is a novel p53 target and mediates transcriptional repression of replication target genes in response to DNA damage. DNA damage induces transcriptional repression of genes involved in DNA replication and mitotic progression in a p53-dependent manner. E2F7 is a novel p53 target gene up-regulated in response to DNA damage. E2F7 binds specifically to replication target gene promoters and mediates DNA damage-induced transcriptional repression. Mitotic target genes are repressed via a different mechanism, and E2F7 does not appear to play a role.
Materials and methods
Tissue culture, cell lines, and drug treatments
Cells were grown in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. Medium for EJ cells was supplemented with 50 mg/mL hygromycin. Cell lines were treated with a range of 0.025–0.25 μg/mL doxorubicin (Sigma) in order to determine the appropriate dose for each. U2OS and MCF7 cells were treated with 0.1 μg/mL doxorubicin, while HCT116 cells were treated with 0.25 μg/mL doxorubicin for the indicated times. Ectopic expression of p53 was induced in EJp53 cells by washing cells grown in tetracycline-containing medium five times with 1× PBS. Cells were then grown for at least 1 d in medium minus tetracycline. H1299 and U2OS cells were transfected using Lipofectamine and Plus reagent (Invitrogen) according to the manufacturer's instructions.
ChIP assay
The protocol for this assay was described previously by Espinosa et al. (2003), except that Protein A Dynabeads (Invitrogen) were used for the pull-down. DNA fragments were purified using the QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. Recovered DNA was subjected to qPCR amplification using the Mx3005P QPCR system (Agilent Technologies). Relative values or percent of input was calculated using the comparative cycle threshold method using the following equation: 2−[Ct(x) − Ct(INPUT)]. Primers used for ChIP-qPCR are provided in Supplemental Table 1. The antibodies used in the ChIP assays were anti-p53 (sc-DO1), anti-E2F7 (sc-H300), anti-flag (M2, Sigma), and anti-GFP (JL-8, BD Bioscience).
RNA purification, cDNA synthesis, and qPCR
Cells were treated as described in the figure legends and processed for RNA purification using the RNeasy kit (Qiagen). One microgram of RNA was used for cDNA synthesis using qScript cDNA Super Mix (Quanta Biosciences) according to the manufacturer's instructions. cDNA samples were then diluted to 1:40, and gene expression analysis was carried out by qPCR using the Mx3005P qPCR system (Agilent Technologies). A 40-cycle qPCR reaction was performed using the VeriQuest SYBR Green qPCR Master Mix with ROX according to the manufacturer's instructions. A primer list of sequences used for qRT–PCR is provided in Supplemental Table 1.
Immunoblotting
Whole-cell extracts were prepared by washing cells once with 1× PBS. Cells were scraped into a lysis buffer composed of 50 mM HEPES (pH 7.5), 1% Triton X-100, 150 mM NaCl, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 μg/mL leupeptin, and 50 μg/mL aprotinin. Fifty micrograms of total protein concentration was resolved by SDS-PAGE. Immunoblot analysis was conducted using the following antibodies: anti-p53 (sc-DO1), anti-p21 (SMX30, BD Bioscience), anti-Flag (M2, Sigma), and anti-β-actin (Sigma).
Plasmids
Wild-type p53 constructs were derived from a pCMV-Flag-p53 plasmid, which was a gift from Dr. Wei Gu. The Δ24 deletion was created by inserting a termination codon at the 370-amino-acid position by site-directed mutagenesis. For this purpose, the following primers were used: Forward, 5′-ACTCCAGCCACCTGTAGTCCAAAAAGGGTC-3′, and Reverse, 5′-GACCCTTTTTGGACTACAGGTGGCTGGAGT-3′.
Luciferase reporters
A minimal promoter sequence consisting of a 20-bp p53 consensus sequence from the E2F7 promoter was cloned into a pGL3-E1b-TATA reporter construct. The oligonucleotide sequences 5′-CTCGAGCGGCAAGTTGGACAACTCCCTCGAG-3′ and 5′-CTCGAGGCCGTTCAGACCTGTTGAGGCTCGAG-3′ were annealed and cloned into the pGL3-E1b-TATA, also digested with XhoI.
Colony formation assay
Transfections were carried out as indicated in the figure legends. The pBABE-puromycin vector was cotransfected with the indicated plasmids for selection of puromycin-resistant colonies. Transfections were carried out as described above, and, 24 h after transfection, cells were passaged into medium containing puromycin (1 μg/mL). Colonies were fixed and stained with Giemsa (Sigma) 2–3 wk after transfection.
Propidium iodide staining for flow cytometry analysis
Cell pellets were resuspended and fixed in 1 mL of 70% ethanol and stored for at least 12 h at −20°C. Samples were then spun at 2300 rpm for 5 min, and pellets were resuspended in 1 mL of 1× PBS and spun again as above. Pellets were resuspended in 1 mL of 1× PBS containing 1× propidium iodide and 1 mg/mL RNase A (Sigma) to make ∼106 cells per milliliter. Samples were stored in the dark overnight at 4°C and then analyzed by flow cytometry.
siRNA transfection
ON_TARGET plus siRNA SMART pools were purchased directly from Dharmacon siRNA technologies (Fisher Scientific). Confluent dishes were split the day before transfection at a 1:10 dilution. Transfections were carried out using Optimen (Gibco) and oligofectamine (Invitrogen) according to the manufacturers' instructions. siRNA oligonucleotide sequences will be provided on request.
Acknowledgments
We thank the members of the laboratory—Lois Resnick-Silverman, Melissa Mattia-Sansobrino, Caleb Lee, and Emir Senturk—as well as Matthew O'Connell, James Bieker, and Zhen-Qiang Pan for helpful discussions and advice. We also thank Bert Vogelstein (Johns Hopkins University) for the isogenic HCT116 cell lines, Stuart Aaronson (Mount Sinai School of Medicine) for the EJ-inducible p53 cell line, Wei Gu (Columbia University) for the expression vector for Flag-tagged p53, and Gustavo Leone (Ohio State University) for the expression vector for E2F7. These studies were supported in part by National Cancer Institute grants T32CA078207 and F31CA150539 to L.C. and RO1CA125741 to J.J.M.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.184911.111.
References
- Badie C, Itzhaki JE, Sullivan MJ, Carpenter AJ, Porter AC 2000. Repression of CDK1 and other genes with CDE and CHR promoter elements during DNA damage-induced G(2)/M arrest in human cells. Mol Cell Biol 20: 2358–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsotti AM, Prives C 2010. Noncoding RNAs: The missing ‘linc’ in p53-mediated repression. Cell 142: 358–360 [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C 2010. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2: a000935 doi: 10.1101/cshperspect.a000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Broz DK, Basak S, Park EJ, Mclaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, Müller H, Wagener C, Holm K, Helin K 1998. E2F-6: A novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17: 611–623 [DOI] [PubMed] [Google Scholar]
- Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, Bremner R 2009. Division and apoptosis of E2F-deficient retinal progenitors. Nature 462: 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A 2003. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 278: 42041–42049 [DOI] [PubMed] [Google Scholar]
- DeGregori J 2004. The Rb network. J Cell Sci 117: 3411–3413 [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci 94: 7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP 2005. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev 19: 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G 2006. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10: 191–202 [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 22: 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N 1998. The regulation of E2F by pRB-family proteins. Genes Dev 12: 2245–2262 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49 [DOI] [PubMed] [Google Scholar]
- Endo-Munoz L, Dahler A, Teakle N, Rickwood D, Hazar-Rethinam M, Abdul-Jabbar I, Sommerville S, Dickinson I, Kaur P, Paquet-Fifield S, et al. 2009. E2F7 can regulate proliferation, differentiation, and apoptotic responses in human keratinocytes: Implications for cutaneous squamous cell carcinoma formation. Cancer Res 69: 1800–1808 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Estève P-O, Chin HG, Pradhan S 2005. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci 102: 1000–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer G, Friedlander P, Colgan J, Manley JL, Prives C 1996. Transcriptional repression by p53 involves molecular interactions distinct from those with the TATA box binding protein. Nucleic Acids Res 24: 4281–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritah A, Saucier C, Mester J, Redeuilh G, Sabbah M 2005. p21WAF1/CIP1 selectively controls the transcriptional activity of estrogen receptor α. Mol Cell Biol 25: 2419–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giono LE, Manfredi JJ 2006. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol 209: 13–20 [DOI] [PubMed] [Google Scholar]
- Hamard PJ, Lukin DJ, Manfredi JJ 2012. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J Biol Chem 287: 22397–22407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazar-Rethinam M, Cameron SR, Dahler AL, Endo-Munoz LB, Smith L, Rickwood D, Saunders NA 2011. Loss of E2F7 expression is an early event in squamous differentiation and causes derepression of the key differentiation activator Sp1. J Invest Dermatol 131: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R 2005. Direct p53 transcriptional repression: In vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol 25: 3737–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MW, Agarwal MK, Yang J, Bruss P, Uchiumi T, Agarwal ML, Stark GR, Taylor WR 2005. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J Cell Sci 118: 1821–1832 [DOI] [PubMed] [Google Scholar]
- Lammens T, Li J, Leone G, De Veylder L 2009. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol 19: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C 2006. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ 13: 951–961 [DOI] [PubMed] [Google Scholar]
- Lee KC, Crowe AJ, Barton MC 1999. p53-mediated repression of α-fetoprotein gene expression by specific DNA binding. Mol Cell Biol 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Wernke-Dollries K, Stratton SA, Aronow B, Barton MC 2001. p53 targets chromatin structure alteration to repress α-fetoprotein gene expression. J Biol Chem 276: 42057–42062 [DOI] [PubMed] [Google Scholar]
- Le Gac G, Estève P-O, Ferec C, Pradhan S 2006. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J Biol Chem 281: 24161–24170 [DOI] [PubMed] [Google Scholar]
- Levine AJ 1997. p53, the cellular gatekeeper for growth and division. Cell 88: 323–331 [DOI] [PubMed] [Google Scholar]
- Li J, Ran C, Li E, Gordon F, Comstock F, Siddiqui H, Cleghorn W, Chen H, Kornacker K, Liu C 2008. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell 14: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17: 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan N, Delavaine L, Graham A, Reilly C, Wilson J, Brummelkamp TR, Hijmans EM, Bernards R, La Thangue NB 2004. E2F-7: A distinctive E2F family member with an unusual organization of DNA-binding domains. Oncogene 23: 5138–5150 [DOI] [PubMed] [Google Scholar]
- Logan N, Graham A, Zhao X, Fisher R, Maiti B, Leone G, La Thangue NB 2005. E2F-8: An E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 24: 5000–5004 [DOI] [PubMed] [Google Scholar]
- Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA 2003. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol Cell Biol 23: 8576–8585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie L, King S, Marcar L, Nicol S, Dias SS, Schumm K, Robertson P, Bourdon J-C, Perkins N, Fuller-Pace F, et al. 2010. p53-dependent repression of polo-like kinase-1 (PLK1). Cell Cycle 9: 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon N, Dyson N 2008. E2F7 and E2F8 keep the E2F family in balance. Dev Cell 14: 1–3 [DOI] [PubMed] [Google Scholar]
- Murphy M, Ahn J, Walker KK, Evans RM, Levine AJ, George DL 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev 13: 2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotis Zalmas L, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, La Thangue NB 2008. DNA-damage response control of E2F7 and E2F8. EMBO Rep 9: 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra HI, Wu L, de Bruin A, Timmers C, Rosol TJ, Weinstein M, Robinson ML, Leone G 2002. Specificity of E2F1, E2F2, and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ 13: 215–225 [PubMed] [Google Scholar]
- Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T 1992. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci 89: 12028–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, Kogan I, Braunstein I, Tzukerman M, Ginsberg D, et al. 2004. p53-dependent down-regulation of telomerase is mediated by p21waf1. J Biol Chem 279: 50976–50985 [DOI] [PubMed] [Google Scholar]
- St Clair S, Giono L, Varmeh-Ziaie S, Resnick-Silverman L, Liu W-J, Padi A, Dastidar J, DaCosta A, Mattia M, Manfredi JJ 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: One involves direct binding to the cdc25C promoter. Mol Cell 16: 725–736 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L 2005. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao Z, Ko HL, Ren E-C 2010. The p53 response element and transcriptional repression. Cell Cycle 9: 870–879 [DOI] [PubMed] [Google Scholar]
- Wei C-L, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Westendorp B, Mokry M, Groot Koerkamp MJ, Holstege FC, Cuppen E, de Bruin A 2012. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res 40: 3511–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn J, Carter M, Avery L, Cameron S 2011. Hox and a newly identified E2F co-repress cell death in Caenorhabditis elegans. Genetics 188: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ 2007. A comprehensive ChIP chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 17: 1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J 23: 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]