Oncogene-induced senescence is an anti-proliferative stress response program that acts as a fail-safe mechanism to limit oncogenic transformation and is regulated by the RB and p53 tumor suppressor pathways. In this study from Lowe and colleagues, E2F7, a poorly understood member of the E2F gene family, is identified as a key regulator of cellular senescence. The authors show that E2F7 is the only member of the E2F family of proteins that is consistently up-regulated during oncogene-induced senescence and that disruption of E2F7 and RB promotes escape from senescence, immortalization, and oncogenic transformation. ChIP-seq and gene expression profiling experiments identified genes regulated by E2F7 under different conditions, and the results indicate that E2F7 inactivation leads to complete derepression of mitotic E2F targets, thereby allowing cells to proliferate normally and bypass senescence. Thus, this study provides novel insight into the role of E2F7 in senescence, identifying it as a novel p53 target gene and showing that it cooperates with RB during the senescence program.
Keywords: p53 target, mitosis, checkpoint, compensation, RB, E2F7
Abstract
Oncogene-induced senescence is an anti-proliferative stress response program that acts as a fail-safe mechanism to limit oncogenic transformation and is regulated by the retinoblastoma protein (RB) and p53 tumor suppressor pathways. We identify the atypical E2F family member E2F7 as the only E2F transcription factor potently up-regulated during oncogene-induced senescence, a setting where it acts in response to p53 as a direct transcriptional target. Once induced, E2F7 binds and represses a series of E2F target genes and cooperates with RB to efficiently promote cell cycle arrest and limit oncogenic transformation. Disruption of RB triggers a further increase in E2F7, which induces a second cell cycle checkpoint that prevents unconstrained cell division despite aberrant DNA replication. Mechanistically, E2F7 compensates for the loss of RB in repressing mitotic E2F target genes. Together, our results identify a causal role for E2F7 in cellular senescence and uncover a novel link between the RB and p53 pathways.
Oncogene-induced senescence is a stable cell cycle arrest program that limits the proliferation of cells that have sustained oncogenic mutations, and as such provides an important barrier to tumorigenesis (Mooi and Peeper 2006; Campisi and d'Adda di Fagagna 2007). Initially described in fibroblasts expressing oncogenic Ras (Serrano et al. 1997), oncogene-induced senescence can be triggered by a variety of oncogenic signals and appears to limit the proliferation of developing tumor cells. Indeed, senescent cells have been identified in a variety of premalignant human tumors (Braig et al. 2005; Chen et al. 2005; Collado et al. 2005; Michaloglou et al. 2005), and mutations in genes required for the successful execution of senescence are frequently associated with tumor progression. Like other senescence programs, oncogene-induced senescence is accompanied by unique changes in cellular morphology and physiology that are associated with increased expression of senescence-associated β-galactosidase (SA-β-gal), as well as dramatic changes in gene expression associated with an overall repressive chromatin environment (Narita et al. 2003). In addition to the cell cycle arrest program, senescent cells secrete factors that modulate their microenvironment and trigger immune surveillance, and these processes may contribute to the tumor-suppressive properties of the program in a non-cell-autonomous manner (Campisi and d'Adda di Fagagna 2007; Coppe et al. 2008; Kuilman et al. 2008).
The p53 and retinoblastoma protein (RB) tumor suppressor pathways control the cell-autonomous components of the oncogene-induced senescence program, and disruption of these pathways alone or in combination is sufficient to bypass senescence and promote oncogenic transformation (Shay et al. 1991; Voorhoeve and Agami 2003; Chicas et al. 2010). p53 is a sequence-specific DNA-binding transcription factor that promotes senescence in part through its ability to induce the cyclin-dependent kinase (CDK) inhibitor p21CDKN1A and the anti-proliferative microRNA miR-34 (Pantoja and Serrano 1999; He et al. 2007; Chicas et al. 2010). In contrast, RB promotes senescence by stimulating a repressive chromatin environment that leads to the stable suppression of proliferative genes, perhaps through the production of senescence-associated heterochromatic foci (SAHF) (Narita et al. 2003; Zhang et al. 2005) and/or by facilitating the accumulation of repressive histone marks on proliferation-responsive genes (Narita et al. 2003; Chicas et al. 2012).
Interestingly, RB is a member of a multigene family consisting of RB, p107, and p130 (Burkhart and Sage 2008), and these three proteins act to inhibit gene expression partly through binding and modulation of the activity of certain members of the E2F family of transcription factors. Although RB family members can compensate for each other under some circumstances, RB plays a nonredundant role in its ability to promote E2F target gene repression and cell cycle arrest during senescence, particularly on E2F-responsive genes required for DNA replication (Chicas et al. 2010). As RB is the only member of this family that is frequently mutated or lost in cancer (Burkhart and Sage 2008), it has been argued that RB exerts its tumor suppressor function in part by controlling cellular senescence (Narita et al. 2003; Chicas et al. 2010).
The E2F family of transcription factors consists of eight proteins that bind to the consensus E2F motif (TTTCGCGC) (Zheng et al. 1999) that exists in many genes involved in DNA synthesis, cell cycle progression, and mitosis (Cam and Dynlacht 2003). Although in vivo studies indicate that the roles and regulation of these factors are complex (Trimarchi and Lees 2002), E2F1–3 are most commonly associated with transcriptional activation of genes involved in normal cell cycle transitions, where their activities are restrained by their association with RB family members in a manner that is relieved by CDK-mediated hyperphosphorylation of RB (Dyson 1998). E2F4 and E2F5 are most strongly linked to transcriptional repression during quiescence (Chen et al. 2009), whereas E2F6 has been linked to polycomb-mediated gene regulation (Trimarchi et al. 2001; Attwooll et al. 2005). E2F7/8 are transcriptional repressors with an atypical structure, having two DNA-binding domains and lacking a dimerization domain, which is required for association with dimerization partner (DP) proteins that appear to be important for the sequence-specific binding capacity of other E2Fs (Di Stefano et al. 2003; Logan et al. 2004, 2005). Although little is known about their activity, mice null for both E2f7 and E2f8 die during embryonic development with phenotypes similar to Rb-deficient animals, suggesting a relationship between these proteins (Li et al. 2008). Whether the effect is due to a direct association with RB (Chen et al. 2009) or some parallel activity is not known.
p53 and RB act in a cooperative way to promote senescence, and there are many complex interactions between these pathways. The p53 target gene CDKN1A encodes the protein p21, which, by inhibiting CDKs, prevents phosphorylation of the retinoblastoma family proteins, leading to the activation of the RB family and repression of E2F-driven transcription. On the other hand, inactivation of the retinoblastoma proteins leads to up-regulation of ARF, an E2F target gene, and ARF subsequently stabilizes the p53 protein through MDM2 inhibition (Iaquinta et al. 2005). Interestingly, E2F3b, an isoform of E2F3 that represses ARF transcription, may be crucial in this regulation (Aslanian et al. 2004). The importance of this interplay for the execution of the senescence program can vary depending on context, and in some instances may be a compensatory response to pathway perturbation. For example, loss of RB can trigger p53 up-regulation via ARF or other pathways, thereby providing a safeguard that prevents evasion of senescence and malignant transformation (White 1994; Chicas et al. 2010). Thus, in the context of senescence, the nature of the cross-talk appears to promote and reinforce the cell cycle arrest. In this study, we identify E2F7 as a key senescence regulator with tumor-suppressive activity that provides a novel link between the RB and p53 pathways during cellular senescence.
Results
E2F7 levels increase during cellular senescence
We previously performed a large series of transcriptional profiling experiments in order to identify genes that might be selectively influenced by various RB family members in oncogene-induced senescence relative to other growth states (Chicas et al. 2010). IMR90 human normal diploid fibroblasts, a paradigm model for the study of senescence (Shay et al. 1991; Narita et al. 2003), were triggered to senesce by oncogenic Ras in the presence of potent shRNAs capable of individually repressing each RB family member, and the resulting cells were subjected to transcriptional profiling by Affymetrix U133 Plus 2.0 microarrays. In that study, we noted that RB was unique among RB family members based on its strict requirement for the repression of a subset of E2F target genes, including many involved in DNA synthesis. In contrast, p107 and p130 could compensate for RB in repressing these genes during normal proliferation or upon cell cycle exit into quiescence.
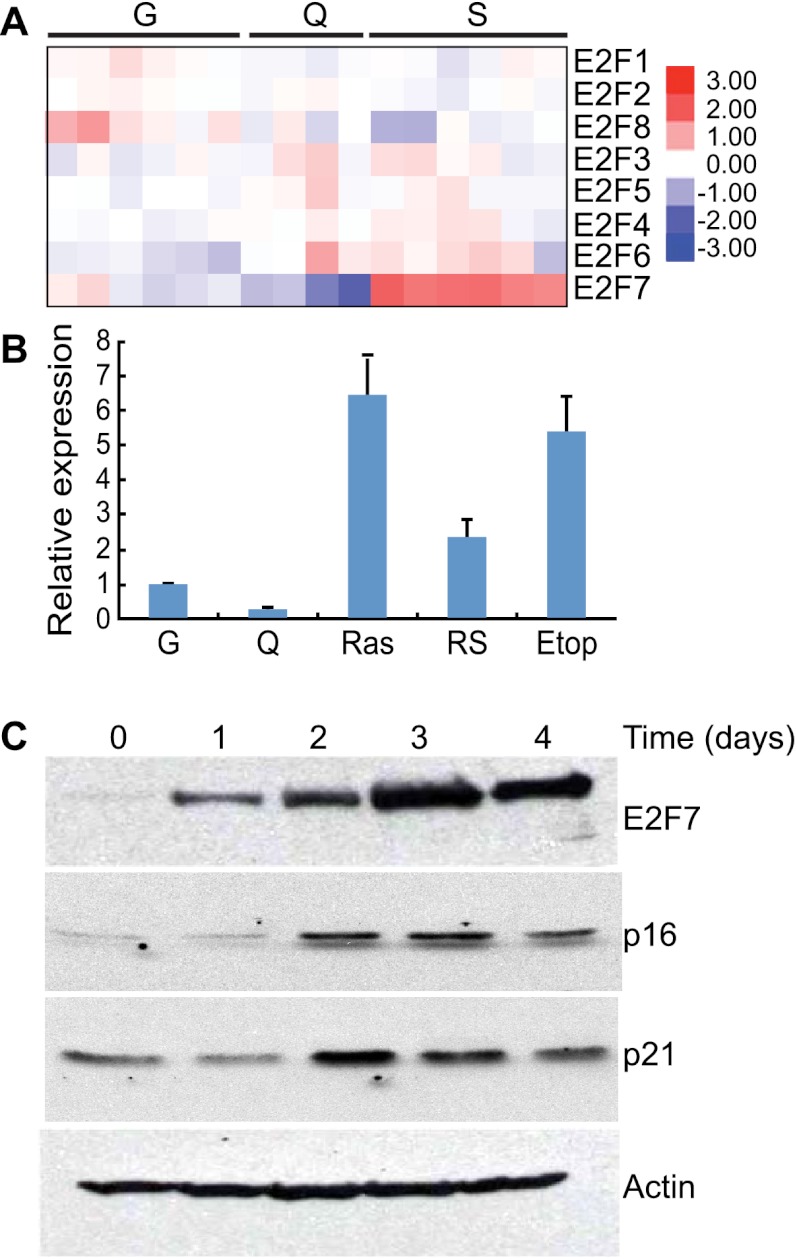
In examining this transcriptional profiling data for the expression of E2F family members, we noticed a marked up-regulation of the atypical E2F family member E2F7 that was distinct for the senescent state (Fig. 1A). These observations were confirmed by quantitative RT–PCR (qRT–PCR) analysis using primers specific to each E2F family member (Fig. 1B; Supplemental Fig. 1) and by immunoblotting using antibodies specific for E2F7 (Fig. 1C). E2F7 transcription levels were also increased in cells triggered to undergo senescence by replicative exhaustion or treatment with the DNA-damaging agent etoposide (Fig. 1B). In contrast, E2F7 was decreased as cells entered quiescence through growth factor depletion, implying that E2F7 up-regulation is not a general consequence of cell cycle arrest. These results raise the possibility that E2F7 plays an active role in the senescence program.
Figure 1.
E2F7 is up-regulated during cellular senescence. (A) Heat map of the expression patterns of E2F family members in growing (G), quiescent (Q), and senescent (S) IMR90 cells. Each row represents a biological replicate. (B) Histogram showing the levels of E2F7 RNA in either growing (G) or quiescent (Q) cells or cells triggered to undergo senescence by either RasV12 (Ras), replicative exhaustion (RS, passage 34), or etoposide (Etop, 100 uM). The quantity of RNA was measured by real-time qPCR analysis (qPCR), normalized to the expression of β-actin, and expressed as fold change relative to growing IMR90 cells. The data represent the mean (±the standard deviation) of three separate experiments. (C) Immunoblots documenting the increase in E2F7 protein in cells undergoing senescence. Protein lysates were from ER-Ras-infected IMR90 cells at the indicated time after 4-hydroxy-tamoxifen (4-OHT) treatment. Accumulation of p16INK4A and p21CDKN1A are shown as an indication of Ras activation upon tamoxifen treatment. Actin probing of a parallel blot was used as a loading control.
E2F7 is a direct p53 target gene
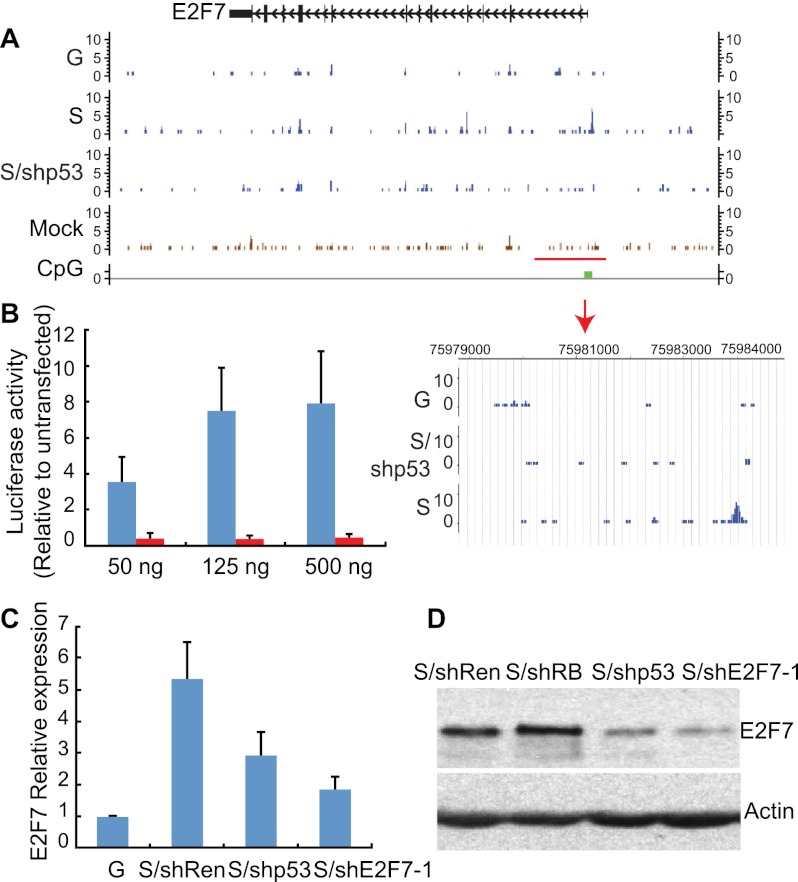
The most well-established transcriptional activator that participates in senescence is p53, which is a sequence-specific DNA-binding factor that binds to two copies of the 10-base-pair (bp) motif 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ separated by 0–13 bp (el-Deiry et al. 1992; Wei et al. 2006). Interestingly, E2F7 has a p53-binding site in its promoter (Supplemental Fig. 2), and indeed, analysis of chromatin immunoprecipitation (ChIP)/next-generation sequencing (ChIP-seq) data from IMR90 cells under different growth conditions revealed a marked and specific binding of p53 to this site in senescence but not in growing conditions (Fig. 2A). No specific p53 binding was detected in any of the other E2F target genes, although E2F4 and E2F8 also have potential p53-binding sites (data not shown). Thus, p53 binds to the E2F7 promoter specifically during cellular senescence.
Figure 2.
E2F7 is a bona fide p53 target gene. (A) Binding patterns of p53 to the E2F7 gene promoter shown as custom tracks on the University of California at Santa Cruz (UCSC) Genome Browser. p53-specific peaks are detected in the promoter of the E2F7 gene (see magnified image of the highlighted area below) specifically in senescent cells (S) but not growing cells (G) or senescent cells expressing a p53 shRNA (S/shp53). Peaks detected outside the promoter are insensitive to suppression of p53, suggesting that they are not specific. (B) Histogram showing the luciferase activity induced by the indicated amount of p53 on the reporter gene driven by the p53 response element from the E2F7 gene. Luciferase activity was plotted as the induction relative to basal luciferase activity. The data represent the mean (±the standard deviation) of three separate experiments. (C) Histogram showing the levels of E2F7 RNA in growing or senescent cells expressing the indicated shRNA. Expression is normalized to the expression of GAPDH and is presented as the fold change relative to growing IMR90 cells. Values are means ± standard deviation of three independent samples. (D) Immunoblot showing the levels of the E2F7 protein in senescent cells expressing the indicated shRNA. Actin was used as loading control. The figure is a representative blot of at least three independent experiments.
To determine whether p53 binding to the promoter of the E2F7 gene influences transcription, we examined whether the p53-binding site in the promoter of the E2F7 gene could confer p53 responsiveness to a heterologous promoter and whether suppression of p53 would compromise E2F7 induction during senescence. The p53-binding site in the E2F7 promoter or a variant in which the p53-binding site was mutated was cloned upstream of a luciferase reporter and transfected together with a p53 expression construct in p53-null HCT116 cells, and luciferase expression was assessed using bioluminescence. As expected, cells transfected with the wild-type reporter showed a marked increase in luciferase expression, whereas those with the mutant did not (Fig. 2B). Similarly, p53 was required for E2F7 induction in cells undergoing senescence. While senescent IMR90 cells harboring a neutral shRNA were capable of inducing E2F7 mRNA and protein in response to oncogenic Ras, those expressing a p53 shRNA were impaired in this effect (Fig. 2C,D). Together, these data validate E2F7 as a bona fide p53 target gene.
E2F7 cooperates with RB to promote senescence
Control of oncogene-induced senescence in IMR90 cells involves a complex interplay between the RB and p53 pathways such that cosuppression of the RB and p53 network components are required to fully bypass the program (Shay et al. 1991; Voorhoeve and Agami 2003; Chicas et al. 2010). Given our observation that E2F7 is a direct p53 target gene that is up-regulated during senescence, we set out to test whether E2F7 contributes to the output of p53 signaling in this setting. IMR90 cell populations were engineered to express two independent shRNAs capable of suppressing E2F7 protein alone or in tandem with a potent RB shRNA (Supplemental Fig. 3A,B) and were examined for their ability to undergo oncogene-induced senescence. Acute induction of senescence was assessed by the ability of cells to express senescence markers or prevent DNA replication as assessed by incorporation of 5-bromo-2′-deoxyuridine (BrdU) into DNA. Long-term proliferative capacity was also assessed by the ability of these cell populations to form colonies when plated at low density.
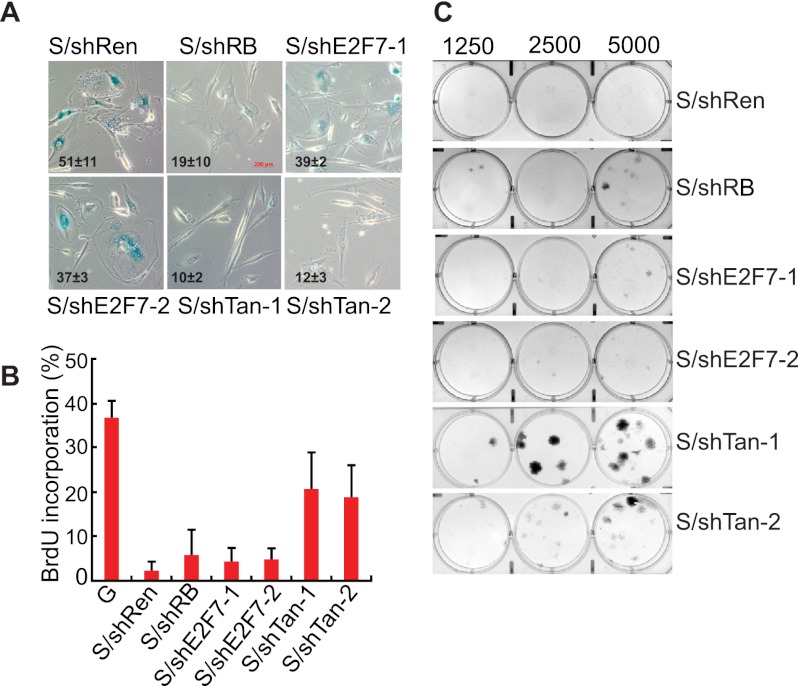
As previously reported, suppression of RB impaired senescence, as measured by the disappearance of senescence markers such as SA-β-gal activity and SAHF formation (Fig. 3A; Chicas et al. 2010), yet these cells still arrest, as assessed by a reduction in BrdU incorporation and an inability to form colonies (Fig. 3B,C). Suppression of E2F7 alone had little impact on certain senescence phenotypes, although, in principle, the failure of E2F7 shRNAs to produce a phenotype in this assay may reflect their inability to completely suppress E2F7 protein expression. Still, in marked contrast, cosuppression of E2F7 together with RB was sufficient to bypass senescence, as measured by various proliferation assays such as BrdU incorporation, colony formation, and cell counting (Fig. 3B,C; Supplemental Fig. 3C). Similar results were observed in another human fibroblast line (WI-38) (Supplemental Fig. 3D). On the other hand, in human BJ fibroblasts, a cell type where senescence is mainly dependent on the p53 pathways due to weak p16INK4a induction (Beausejour et al. 2003), suppression of E2F7 alone was sufficient to override Ras-induced senescence (Supplemental Fig. 3E). Thus, E2F7 cooperates with the RB pathway to promote cellular senescence.
Figure 3.
E2F7 cooperates with RB to promote senescence. (A) Micrographs showing SA-β-gal staining in senescent cells expressing the indicated shRNAs. The numbers shown at the bottom left of each micrograph represent the quantitation of SA-β-gal-positive cells. The data represent means ± standard deviation of three independent experiments. shE2F7-1 and shE2F7-2 are two independent shRNAs targeting E2F7. The same two shRNAs were used in combination with shRB (shTan1 and shTan2). An shRNA targeting Renilla is a neutral control shRNA. Bar, 200 μm. (B) Quantification of BrdU incorporation in senescent IMR90 cells expressing the indicated shRNAs. The data represent means ± standard deviation of three independent experiments. Growing (G) cells were used as a control. (C) Colony formation assay documenting the effect of suppressing RB and E2F7 on the ability of H-RasV12 expressing IMR90 cells to grow at low density. Shown are representative crystal violet stainings of 6-cm plates 2 wk after plating.
Cosuppression of RB and E2F7 transforms primary mouse embryonic fibroblasts (MEFs)
p53 can also control the senescence of MEFs in a manner that directly restricts the ability of Ras to promote malignant transformation (Serrano et al. 1997; Azzoli et al. 1998). To examine the participation of E2F7 in this process, we developed multiple shRNAs targeting mouse E2f7 (Supplemental Fig. 4A) and introduced these into early-passage MEFs in combination with oncogenic Ras. The resulting cell populations were subsequently assessed for their ability to undergo senescence in vitro or form tumors in vivo following subcutaneous injection into immunocompromised mice. Unlike MEFs expressing p53 shRNAs, MEFs expressing E2f7 shRNAs still underwent senescence in response to oncogenic Ras, as assessed by the accumulation of SA-β-gal activity, reduced BrdU incorporation, and an inability to form colonies (Fig. 4B; Supplemental Fig. 4B). Accordingly, these cell populations were incapable of forming tumors in vivo. Of note, these negative results are not simply due to insufficient E2F7 knockdown, as MEFs derived from E2f7-null mice were also arrested in response to oncogenic Ras (Supplemental Fig. 4C). Thus, in this setting, disruption of E2F7 is unable to fully recapitulate the effects of p53 loss on promoting bypass of senescence in MEFs.
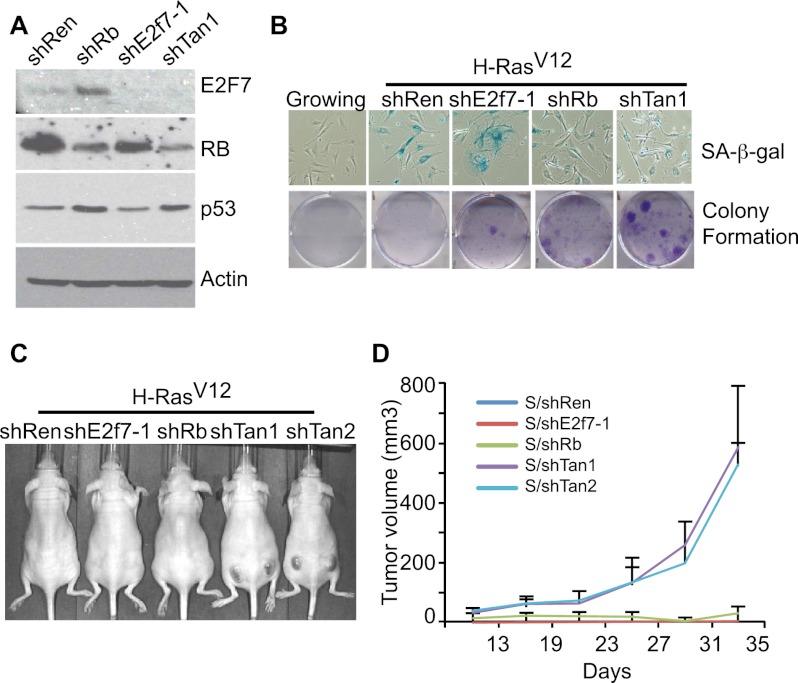
Figure 4.
Cosuppression of RB and E2F7 promotes transformation. (A) Immunoblots from lysates of mouse liver progenitor cells documenting the efficiency of the indicated shRNAs in suppressing RB and E2F7. shTan is the bicistronic shRNA targeting RB and E2F7, and shRen is a neutral control shRNA. Actin was used as a loading control. (B, top) Micrographs showing SA-β-gal staining in MEFs undergoing senescence in the presence of the indicated shRNAs. Growing cells were used as a control. (Bottom) Colony formation assays documenting the effect of suppressing RB and E2F7 on the ability of H-RasV12-expressing MEFs to grow at low density. Shown are representative crystal violet stainings of 6-cm plates 2 wk after plating. (C) Representative images of immunocompromised mice 20 d after subcutaneous injections of 1 × 106 MEFs expressing H-Ras and the indicated shRNA. Tan1 and Tan2 are two different hairpins targeting both RB and E2F7. (D) Quantification of tumor volumes. Each point represents mean tumor volume ± SEM (n = 6). Tumor size was measured once a week.
Acute RB inactivation enables bypass of an immediate Ras-induced cell cycle arrest but, in contrast to p53 loss, does not enable cellular immortalization, and Rb-deficient MEFs are not transformed by Ras alone (Sage et al. 2003). To assess whether, as occurs in human fibroblasts, E2F7 cooperates with RB to promote oncogene-induced senescence in MEFs, we constructed a series of bicistronic shRNAs capable of simultaneously targeting mouse E2f7 and Rb (Fig. 4A; Supplemental Fig. 4D) and examined their ability to modulate the cellular response to oncogenic Ras in the assays described above. As previously reported (Sage et al. 2003), suppression of RB alone was sufficient to impair senescence in MEFs, as measured by the lack of senescent markers (e.g., a flat morphology and SA-β-gal activity) and an increase in BrdU incorporation shortly after introduction of oncogenic Ras (Fig. 4B; Supplemental Fig. 4E). Nonetheless, these cells did not show enhanced clonogenic capacity or form tumors in mice (Fig. 4B–D), indicating that they eventually arrest.
Similarly, MEFs coexpressing E2f7 and Rb shRNAs did not display any signs of senescence and continued to incorporate BrdU. However, in contrast to RB-deficient cells, cells expressing shRNAs targeting both Rb and E2f7 showed a marked increase in long-term proliferative capacity and were potent at producing tumors in vivo (Fig. 4B–D). Similar results were observed in MEFs derived from E2f7-null mice after transduction with retroviruses expressing shRb and Ras (Supplemental Fig. 4F). Together, these data indicate that E2F7 cooperates with RB to control cellular senescence in MEFs, where it contributes to a tumor-suppressive program.
E2F7 binds and represses a subset of E2F target genes
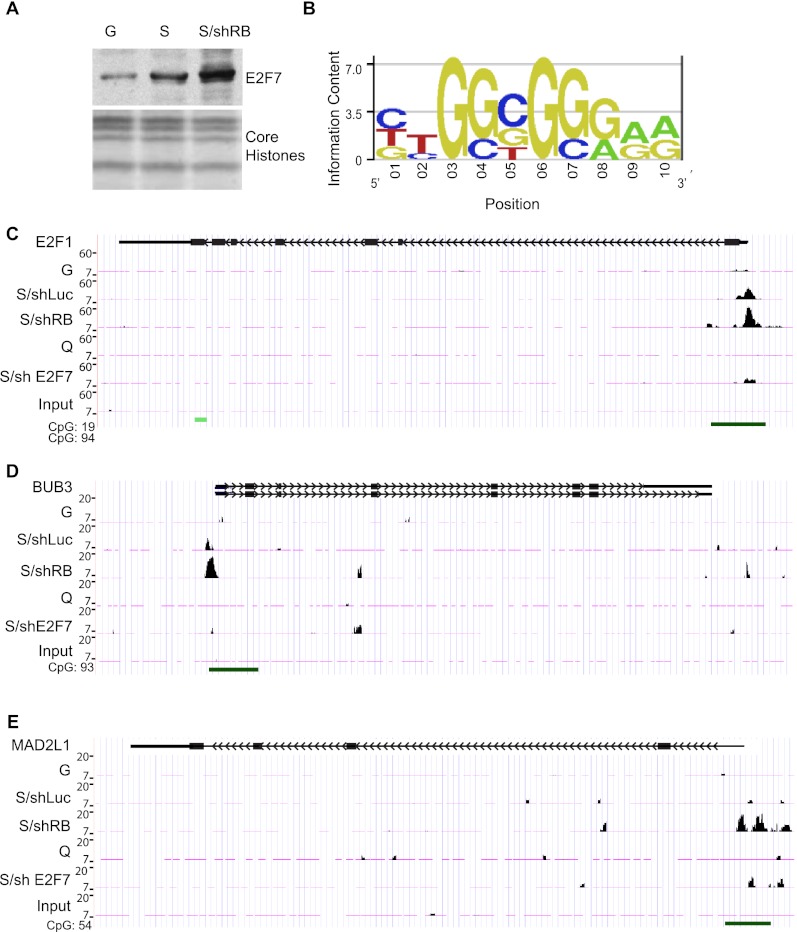
RB inactivation is unable to bypass oncogene-induced senescence in part because RB loss triggers a second senescence barrier involving p53 and its transcriptional target, p21CDKN1A (Chicas et al. 2010). Our results identify E2F7 as a key target of p53 in senescence and suggest that it, too, may participate in this second immortalization barrier. Consistent with this view, E2F7 is hyperinduced in IMR90 cells expressing RB shRNAs that were triggered to senesce, leading to a corresponding increase in the association of E2F7 with chromatin (Fig. 5A). To identify specific genes targeted by E2F7 during senescence, we performed ChIP-seq using an anti-E2F7 antibody in growing, quiescent, or senescent IMR90 cells expressing a control shRNA or an shRNA targeting RB. Sequence reads were mapped back to the human genome, and the number of reads corresponding to different genomic regions was determined. We used MACS with a default P-value cutoff of 1 × 10−5 to identify genomic regions (ChIP-peaks) that showed an enrichment of E2F7-specific binding. Although the total number of called peaks was relatively similar among growing, quiescence, and senescence, we noticed a marked increase in E2F7-binding sites in senescent IMR90 cells expressing RB shRNAs (Supplemental Fig. 5A).
Figure 5.
E2F7 binds to a subset of E2F targets during senescence. (A) Immunoblot showing E2F7 protein levels in chromatin-bound fractions of growing (G), senescent (S), or shRB-expressing senescent (S/shRB) IMR90 cells. Core histones were used as loading control. (B) Position weight matrix of the motif significantly enriched in the DNA associated with E2F7. This sequence is highly similar to the E2F motif (TTTCGCGC). (C) Binding patterns of E2F7 to the E2F1 gene promoter shown as custom tracks on the UCSC Genome Browser. The green bar at the bottom of the figure corresponds to CpG islands in the promoter of the E2F1 gene. Notice the increase in E2F7 binding in senescent cells with or without RB expression compared with growing and quiescent conditions. (D,E) Binding patterns of E2F7 to the BUB3 and MADL2 gene promoters shown as custom tracks on the UCSC Genome Browser. Notice the increase in E2F7 binding in senescent cells suppressed for RB (shRB). (G) Growing; (S) senescent; (shRB) shRNA against RB.
To determine whether E2F7 was associated with known transcription factor-binding sites, we performed a de novo motif analysis on the 300 most significant E2F7-enriched regions. This analysis revealed a consensus motif highly similar to the E2F consensus motif (P < 0.001) (Fig. 5B). Accordingly, as has been reported by others (Di Stefano et al. 2003), we detected E2F7 binding to known E2F target genes (Supplemental Table 1), such as the E2F1 gene promoter (Fig. 5C), a previously reported target of E2F7 (Panagiotis Zalmas et al. 2008; Westendorp et al. 2012). Also consistent with our global analyses, E2F7 binding to the E2F1 promoter was substantially higher in senescent cells (Fig. 5C). These signals were specific to E2F7, were not detected in parallel assays using a nonspecific antibody, and were substantially reduced in samples derived from cells expressing E2F7 shRNAs. Nonetheless, we noted other sites harboring apparently specific binding, raising the possibility that E2F7 binds other genomic regions as well (data not shown).
Based on the known binding of E2F7 to E2F target sites and the nonbiased analysis described above, we focused our subsequent analyses on 283 known E2F target genes, as determined from having an E2F-binding site in the promoter and/or experimental verification of promoter sequences from most of these target genes (Zhao et al. 2005). Compared with growing and quiescent conditions, E2F7 binding to E2F target genes was generally increased during senescence and further enhanced in RB-deficient cells (Supplemental Fig. 5B). In addition, more genes were bound under these conditions; hence, while ∼50 E2F target genes showed significant association under growing and quiescent conditions, this number was doubled in senescent cells and tripled in cells triggered to senesce in the absence of RB (Supplemental Fig. 5C). Approximately 30 E2F target genes were bound under all four conditions, with very few scoring as unique to growing (four), quiescent (five), or senescent (three) conditions. However, a substantial number of known E2F target genes (47) showed significant binding of E2F7 only in RB-deficient senescent cells (Supplemental Fig. 5C). While this increase may reflect qualitative differences in E2F7 binding between conditions, it is possible that the increase in the signal to noise ratio produced by more E2F7 binding enhanced the statistical power for calling a particular region as a peak. Regardless, since E2F7 exerts its most prominent effects when RB is suppressed, we focused more closely on the subset of E2F targets that showed the binding of E2F7 to their promoters in these conditions.
Gene ontology (GO) analysis revealed that E2F target genes bound by E2F7 in cells harboring compromised RB were enriched for those participating in mitosis (Fig. 5D,E; Supplemental Fig. 5D). For instance, we identified binding of E2F7 to the MAD2L1 and BUB3 genes (Fig. 5D,E), each of which encodes proteins involved in the spindle checkpoint (Musacchio and Salmon 2007). Unlike binding to the E2F1 gene, which is detected in senescent cells with or without RB, binding of E2F7 to MAD2L1 and BUB3 promoters only achieved significance in RB-repressed cells triggered to senesce. Similar patterns were observed at the promoters of other “mitotic” genes (Supplemental Table 2) and were different from those observed on DNA replication genes where E2F7 binds equally to these promoters in both conditions (Supplemental Fig. 5E,F). This result raises the possibility that E2F7 might repress genes that are involved in mitosis, particularly in cells compromised for RB function.
E2F7 compensates for RB loss in repressing mitotic cell cycle targets
Our previous work indicated that, during senescence, RB is uniquely required for the repression of E2F targets involved in DNA replication, as this class of genes remains derepressed in RB-deficient cells triggered to senesce; in contrast, other E2F targets (e.g., many involved in cell cycle progression and mitosis) were silenced (Chicas et al. 2010). To identify genes whose expression was influenced by E2F7, we performed transcriptional profiling analysis on the cell populations described above, paying particular attention to genes synergistically impacted by the combined inactivation of E2F7 with RB. RNA was extracted from growing or senescent cells expressing shRNAs targeting either luciferase as control, RB, E2F7, or both RB and E2F7 and hybridized to the Affymetrix U133 Plus 2.0 microarrays. Genes identified as differentially expressed between conditions (Supplemental Table 3) were then subjected to GO analysis to identify putative functional categories of genes controlled by E2F7.
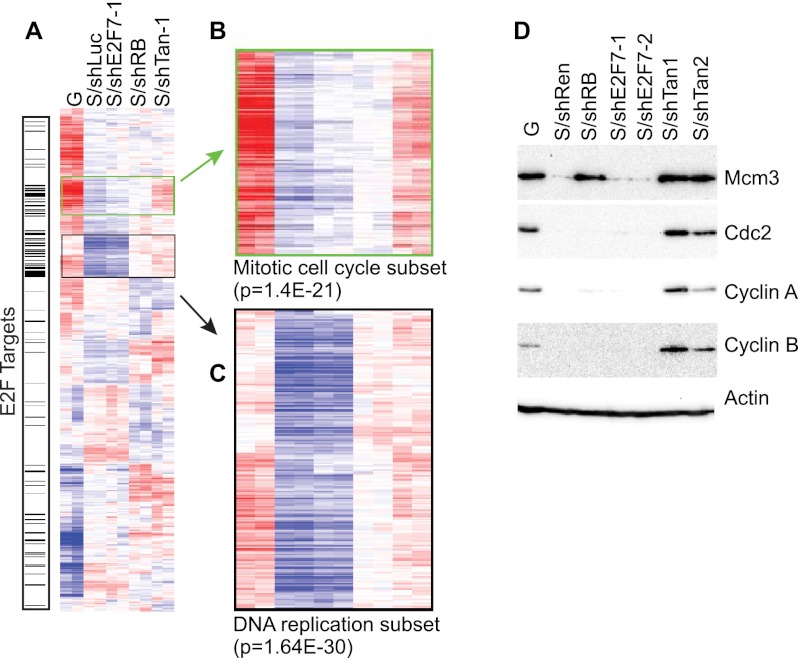
Consistent with our previous work, suppression of RB led to the up-regulation of genes categorized by their role in the “cell cycle” (P = 2.1 × 10−27) and “DNA replication” (P = 7.1 × 10−27) (Supplemental Table 4), but also genes with functions in mitosis (P = 1.1 × 10−15 for the GO term “mitosis,” and P = 8.3 × 10−16 for the GO term “M phase of mitotic cell cycle”). In contrast, many genes categorized by a role in DNA replication were not affected by suppression of E2F7 (hence, “replication” was not detected in the top 25 GO categories), suggesting that the requirement for E2F7 in repression of this subset of genes is distinct from that imposed by RB (Supplemental Table 4). However, genes up-regulated by suppression of E2F7 showed enrichment in GO categories such as “mitotic cell cycle” (P = 7.7 × 10−9), “M phase of mitotic cell cycle” (P = 2.7 × 10−8), and “mitosis” (P = 2.0 × 10−8) (Supplemental Table 4). The expression of the mitotic genes in RB-deficient or E2F7-deficient cells was minimal, but became robust when both RB and E2F7 were inactivated (Fig. 6B). Thus, while E2F7 is dispensable for the repression of DNA replication genes, it appears to cooperate with RB in the repression of mitosis-related genes.
Figure 6.
E2F compensates for RB in controlling gene expression during senescence. (A) Heat map of the expression patterns derived from average-linked clustering of differentially expressed genes among various groups (P < 0.001; 2486 probes) highlighting the E2F target genes. (B,C) Magnification of gene clusters. (B) Mitotic cell cycle gene clusters. (C) DNA replication factors clusters. (D) Immunoblots for representative DNA replication (MCM3) or mitotic cell cycle (CDC2, Cyclin A, and Cyclin B) proteins. Actin was used as a loading control. (G) Growing; (S) H-RasV12-expressing cells.
Similar conclusions were obtained by hierarchical clustering of our gene expression data (Fig. 6A–C) and by direct immunoblotting for E2F target proteins (Fig. 6D). Hence, cells with suppressed RB were clearly distinguishable from normal senescent cells by virtue of their up-regulation of a cluster of genes linked to DNA replication. In marked contrast, cosuppression of E2F7 with RB produced a gene expression state characterized by the up-regulation of both the mitotic gene cluster and the DNA replication gene cluster. These effects were underscored by the changes in sample proteins corresponding to the members of each gene category. Thus, while the DNA replication factor MCM3 was highly expressed in RB-deficient cells, its expression is not affected by suppression of E2F7 (Fig. 6D). Components of the mitotic cell cycle machinery such as Cyclin A, Cyclin B, and CDC2 were highly expressed only in cells deficient of both RB and E2F7 (Fig. 6D). Of note, this association between E2F7 and mitotic cell cycle gene repression is not an indirect consequence of cell cycle proliferation, as our ChIP-seq analysis shows that these genes are direct E2F7 targets (see Fig. 5). Together, these results indicate that E2F7 compensates for loss of RB in the repression of mitotic cell cycle genes.
E2F7 prevents normal cell cycle progression in RB-compromised cells
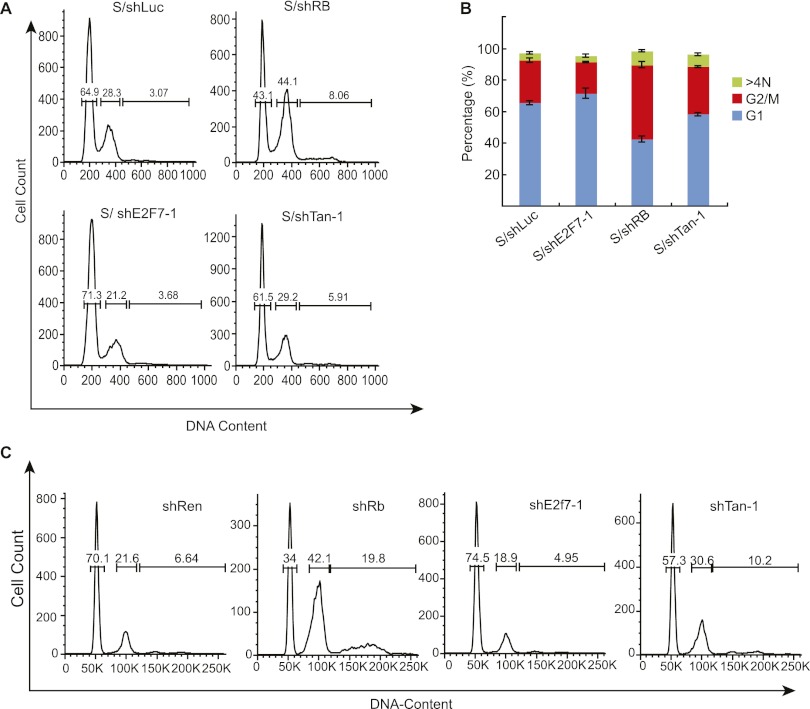
Our combined ChIP-seq and gene expression profiling suggests that E2F7 represses the expression of a subset of E2F target genes that would otherwise enable cell cycle progression and mitosis in cells with compromised RB function. To determine the impact of E2F7 suppression on cell cycle progression, we examined the effect of E2F7 and/or RB shRNAs on the DNA content of IMR90 cells following induction of senescence by oncogenic Ras (Fig. 7A,B). In addition, the impact of E2F7 and RB suppression on cell cycle distribution in response to oncogenes was examined in WI38 diploid fibroblasts (Supplemental Fig. 6A) and a Ras-transformed hepatoblast line expressing a regulatable p53 shRNA that can be triggered to senesce by re-establishing wild-type p53 expression (Fig. 7C; Xue et al. 2007).
Figure 7.
E2F7 enforces a second cell cycle checkpoint that backs up RB. (A) Cell cycle profiles of IMR90 cells undergoing senescence in the presence of the indicated shRNA. The X axis shows the DNA content, whereas the Y axis shows the number of cells. (B) Bar graph showing the percentages for G1 and G2/M cells. The data shown are the mean value ± standard deviation from three independent experiments. (C) Cell cycle profiles of hepatoblasts triggered to undergo senescence by reactivation of p53 in the presence of the indicated shRNAs. The DNA content was measured by flow cytometry 8 d after doxycycline treatment.
Consistent with our previous results (Chicas et al. 2010), suppression of RB caused a greater than a twofold accumulation of cells with a 4N and 8N DNA in senescing IMR90 cells (Fig. 7A,B). Interestingly, the cells that eventually arrest retained high levels of cyclin E with low levels of cyclin B and phospho-histone H3, a phenotype consistent with a mitotic failure and endoreplication rather than a G2 or M arrest (Supplemental Fig. 6B–D; see also Chicas et al. 2010). In marked contrast, simultaneous suppression of RB and E2F7 alleviated this effect, preventing the appearance of polyploid cells and re-establishing a normal cell cycle profile (Fig. 7A,B). Suppression of E2F7 alone also reduced the percentage of cells with 4N DNA content that accumulate in the presence of RB (Fig. 7A,B), suggesting that it normally restricts mitotic progression in a manner that is exacerbated by RB loss. Similar results were also observed in WI38 fibroblasts (Supplemental Fig. 6A) and Ras-transformed hepatoblasts following reactivation of p53 (Fig. 7C). Therefore, in both human and mouse cells, E2F7 controls a cell cycle checkpoint that compensates for RB loss by preventing aberrant proliferation.
Discussion
E2F family members are involved in various cellular processes, including cell cycle progression, DNA replication, DNA repair mechanisms, apoptosis, differentiation, and development (Rowland and Bernards 2006). In our study, we identified a role for E2F7 in cellular senescence, a function that was not previously appreciated. We show that E2F7 is a direct p53 target gene that cooperates with RB in the repression of E2F target genes during oncogene-induced senescence. In cells with compromised RB, E2F7 is required to repress genes involved in mitosis and as such acts as a final safety net to prevent uncontrolled proliferation, immortalization, and, in at least murine cells, oncogenic transformation.
In fibroblasts and most other cell types, cellular senescence is triggered by the combined action of the RB and p53 tumor suppressor pathways, although the precise contribution of each network to the execution and maintenance of the program can be context-dependent (Courtois-Cox et al. 2008). Here we see that in human fibroblasts, E2F7 acts as a key effector of p53 in the program, since like disruption of p53, suppression of E2F7 cooperates with RB to bypass oncogene-induced senescence. E2F7 can also be induced acutely by cytotoxic agents in osteosarcoma cells with intact p53 (Panagiotis Zalmas et al. 2008) and in cells triggered to senesce by DNA-damaging agents or replicative exhaustion (see Fig. 1B), suggesting that its roles and regulation may extend beyond the oncogene-induced senescence program described here. Of note, suppression of p21, a canonical p53 target, also cooperates with RB loss to bypass senescence, thus displaying a phenotype similar to cells with suppressed E2F7. During senescence and more acute DNA damage checkpoints, p21 acts to inhibit the activity of various CDKs, thereby preventing RB hyperphosphorylation and the release of more canonical E2Fs to drive induction of S-phase genes. In contrast, E2F7 apparently acts further downstream directly on E2F target promoters, perhaps to reinforce the effects of p53. Both p53 effectors are crucial for the program, highlighting how p53 can coordinate cellular processes through its action on multiple effectors.
Although our studies clearly show that E2F7 is a direct p53 target gene in human cells, it seems likely that its regulation is more complex. Hence, we do not detect E2F7 activation in response to Ras in MEFs, although it is induced in MEFs following RB suppression (data not shown). We also noted consensus NF-κB- and E2F-binding sites in the E2F7 promoter, and accordingly, studies demonstrate that E2F7 can be transcriptionally activated by E2F1 (Di Stefano et al. 2003). Whether NF-κB sites contribute in a meaningful way to E2F7 remains to be determined, but it is noteworthy that our group and others have recently described a role for NF-κB in mediating various aspects of cellular senescence, including the senescence-associated secretory phenotypes that influence the impact of senescent cells on the tissue microenvironment (Chien et al. 2011; Jing et al. 2011). Moreover, E2F7 may also reinforce p53 activation during senescence, as we noted a modest reduction in p53 phosphorylation levels following E2F7 depletion in IMR90 cells triggered to senescence (data not shown). Similar results have been observed for the p53 target genes miR-34a and PML during senescence (de Stanchina et al. 2004; He et al. 2007), further underscoring the compensatory mechanisms at play in this tumor suppressor network.
Our studies suggest that E2F7 can regulate gene expression during senescence and is most important for the control of E2F target genes involved in cell cycle progression and mitosis. Specifically, we see that E2F7 associates with various E2F target gene promoters in a manner that is substantially enhanced by loss of RB and that, under these circumstances, the direct targets of E2F7 correspond to genes that are significantly enriched for those controlling cell cycle progression and mitosis. Accordingly, disruption of E2F7 under circumstances where RB is compromised prevents repression of various cell cycle progression and mitotic genes and is sufficient to abrogate a checkpoint that otherwise prevents proper mitosis in cells undergoing oncogene-induced senescence. Further studies will be required to define the precise nature of this checkpoint.
Although E2F7 and RB bind overlapping gene sets, the consequence of this binding differs. Hence, while RB is strictly required for the suppression of replication-associated E2F targets during senescence, the requirement for E2F7 in gene repression only becomes evident after suppression of RB. Apparently, E2F7 and RB compensate for each other in repressing genes involved in mitosis, but not those involved in DNA replication, explaining why only the mitotic gene cluster remains silenced after suppression of RB or E2F7 alone. In this manner, E2F7 functionally behaves like a fourth RB family member involved in E2F target gene repression.
RB represses gene expression by sequestering activated E2Fs and recruiting histone-modifying activities that produce a repressive chromatin context (Morris and Dyson 2001; Nijwening et al. 2011; Chicas et al. 2012). How E2F7 acts to repress gene expression remains to be determined. In contrast to the regulation of canonical E2Fs, it seems unlikely that E2F7 acts by corecruitment of RB family members to E2F target promoters because E2F7 lacks the “pocket protein”-binding domain required for RB family member association. Also, E2F7 binds and represses its targets in the absence of functional RB. Furthermore, we noted that cosuppression of p107 and p130 are not able to cooperate with RB loss to bypass oncogene-induced senescence (A Chicas and SW Lowe, unpubl.) Perhaps, instead, E2F7 competes with the activating E2Fs (E2F1–3) for promoter occupancy of E2F target genes to prevent the transcriptional activation. This model is consistent with observations in Drosophila, where the activator E2F (dEF2F1) and the repressor E2F (dE2F2) act antagonistically (Frolov et al. 2001). Further studies will be required to test this possibility.
The crucial role of E2F7 during senescence, and the now established role of senescence as a barrier to tumorigenesis, raises the possibility that E2F7 is a tumor suppressor gene. Consistent with such an activity, E2F7 inactivation can, together with RB loss, promote malignant transformation of murine fibroblasts. E2F7 is located in chromosome 12q21, a region that is frequently deleted in pancreatic cancer, and patients harboring this deletion are associated with a poor prognosis (Kimura et al. 1998). Interestingly, the genetics of pancreatic cancer suggest that this is a setting where oncogene-induced senescence restricts tumorigenesis, as activating KRAS mutations frequently co-occur with mutations targeting the INK4A/ARF locus or p53 in later stages of the disease. Underexpression of E2F7 is also associated with platinum resistance and reduced survival in ovarian cancer patients (Reimer et al. 2007). Still, somatic mutations in E2F7 have yet to be described, raising the possibility that its activity is not rate-limiting for tumorigenesis. Studies in mouse models will further test its tumor-suppressive role. Regardless of their outcome, our studies establish a clear functional role for E2F7 in cellular senescence and highlight the importance of compensatory mechanisms in modulating the RB tumor suppressor network.
Materials and methods
Vectors, cell culture, and gene transfer
The following retroviral vectors were used in this study: pWZL-Hygro (H-rasV12) (Serrano et al. 1997) and pLNCX2-neo (ER:RasV12) (Young et al. 2009). shRNAs targeting RB were previously described (Chicas et al. 2010). shRNAs targeting mouse and human E2F7 were generated using the method previously described (Zuber et al. 2011). Briefly, the 10 top-scoring siRNA predications were obtained using BIOPREDsi, and the siRNA was incorporated into the miR-30 backbone (Silva et al. 2005). The polycistronic shRNA vectors were cloned in two steps as described (Chicas et al. 2010). Luciferase and RB shRNAs were described before (Chicas et al. 2010). Human E2F7 hairpins were as follows: shE2F7-1 corresponds to shE2F7.789, and shE2F7-2 corresponds to shE2F7.1190. Mouse E2f7 hairpins were as follows: shE2f7-1 corresponds to shE2f7.771, and shE2f7-2 corresponds to shE2f7.5220. The human RB shRNA that was used throughout the study corresponds to shRB.698. The sequences of all shRNAs will be provided on request.
Human diploid IMR90 fibroblasts and WI38 (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. MEFs were isolated from embryonic day 13.5 (E13.5) embryos from E2f7−/− mice (O Aksoy, A Chicas, and SW Lowe, unpubl.) derived from embryonic stem cells generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (http://www.komp.org). At least two littermate controls were used for comparison. Retroviruses were packed using Phoenix cells (G. Nolan, Stanford University, CA) and infections were performed as described elsewhere (Narita et al. 2003). The infected population was selected using either 2 μg/mL puromycin (Sigma) for 2 d, 500 μg/mL neomycin for 3 d, or 75 μg/mL hygromycin B (Roche) for 3 d. For coinfection, cells were sequentially selected with puromycin, and then hygromycin or neomycin. Post-selection day 7 refers to 7 d after the conclusion of puromycin selection.
Senescence assays
Senescence was induced by etoposide (100 μM; Sigma) treatment or infection of IMR90 cells with oncogenic H-RasV12 as described (Narita et al. 2003). Replicative senescent IMR90 cells were established by culturing the early-passage cells until passage 34. At this point, the cells were not proliferating and showed all of the senescent markers, including being SA-β-gal-positive.
Senescent IMR90 cells (post-selection day 7) were plated on coverslips coated with 0.1% gelatin and fixed in 4% formaldehyde. For cell cycle arrest analyses, cells were labeled with BrdU (100 μg/mL; Sigma) and 5-fluor-2′-deoxyuridine (10 mg/mL; Sigma) for 4 h. Nuclei incorporating BrdU were visualized by immunostaining with anti-BrdU antibodies (1:400; BD Pharmingen) as described previously (Narita et al. 2003). For colony formation assays, 1250, 2500, or 5000 cells were plated in each well of a six-well plate, cultured for 14 d, fixed with 4% formaldehyde, and stained with crystal violet. Detection of SA-β-gal activity was performed as described previously at pH 6.0 for IMR90 cells and pH 5.5 for mouse cells. Briefly, adherent cells were fixed with 0.5% gluteraldehyde in phosphate-buffered saline (PBS) for 15 min, washed with PBS supplemented with 1 mM MgCl2, and stained for 5–6 h in PBS containing 1 mM MgCl2, 1 mg/mL X-Gal (Roche), and 5 mM each potassium ferricyanide (Sigma) and potassium ferrocyanide (Sigma).
Immunoassays and flow cytometry
Immunoblotting was carried out as described previously (Narita et al. 2003). Isolation of chromatin-bound proteins was performed as described (Chicas et al. 2010). The following antibodies were used: anti-E2F7 (1:500; gift from Kristian Helin), anti-p16INK4a (1:1000; H-156, Santa Cruz Biotechnology), anti-p53 (1:1000; Do-1, Oncogene), anti-Cyclin A (1:1000; Sigma), anti-Cyclin B (1:1000; Cell Signaling), anti-actin (1:5000; ac-15, Sigma), anti-Rb antibody (1:500; G3-245, Pharmingen) together with XZ-55 hybridoma supernatant (1:50), anti-MCM3 (1:1000; gift from Bruce Stillman), anti-p21 (1:200; C-19, Santa Cruz Biotechnology), and anti-cdc2 (1:500; B-6, Santa Cruz Biotechnology).
For flow cytometry, cells were collected, washed with PBS, resuspended in 100 μL of PBS plus 900 uL of cold methanol, and stored overnight at +4°C. Cells were washed in PBS with bovine serum albumin (0.5%) and Tween-20 (0.5%) and resuspended in 500 μL of PI/RNase staining buffer (BD Pharmingen). Data were collected on an LSRII flow cytometer (BD Bioscience) and analyzed with Flowjo software (TreeStar).
Gene expression analysis and RT–qPCR
Total RNA was isolated using the RNeasy minikit (Qiagen), and cDNA was obtained using the TaqMan reverse transcription reagents (Applied Biosystems), used for qPCRs or for cRNA preparation (Message AmpII [Ambion]), and hybridized to U133 Plus 2.0 microarray (Affymetrix) according to the manufacturer's instructions. Data analyses were performed as previously described (Chicas et al. 2010). Gene-specific primer sets were designed using Primer Express 1.5 (sequences available on request). Real-time PCR was carried out in triplicate using SYBR Green PCR Master Mix (Applied Biosystems) on the Roche IQ5 iCycler. GAPDH or β-actin served as endogenous normalization controls.
Microarray data processing
We used the GCRMA function implemented in the R Bioconductor Affymetrix package (Robust Multi-Array Averaging with GC content correction) to subtract background, normalize intensities, and summarize gene expression levels (Wu and Irizarry 2004), and the R program combat was used to perform batch effect normalization. The absolute probe set intensity differences between replicates were calculated and ranked according to descending order, and the difference values of the top 0.1% were used as the threshold to select differentially expressed probe sets that were used for hierarchical clustering analysis to study the global gene expression patterns. Gene expression values were clustered and visualized using the Cluster and TreeView packages (Eisen et al. 1998).
Known E2F targets and GO analysis
Known E2F target genes were extracted from TRED (Zhao et al. 2005). For this study, we used 283 genes with confident evidence (“known” or “likely,” corresponding to evidence of binding or additional functional assays). The significance of overlap between E2F target genes and other gene lists was evaluated by Fisher's exact test. The enrichment of GO terms in a particular gene list was performed using the online tool DAVID (Dennis et al. 2003). P-values reported were corrected using the Benjamini-Hochberg approach.
ChIP-seq analysis
The ChIP experiments were done as previously described (Chicas et al. 2010). The immunoprecipitated DNA was prepared for Illumina sequencing as described (Chicas et al. 2010). The Illumina GA sequencing tags were mapped to the unmasked human reference genome (NCBI version 36, hg18) using the program Bowtie with the parameter setting “-t -q -a –best –strata -m 1.” ChIP-seq peak regions were determined using MACS (Zhang et al. 2008) to find ChIP-enriched regions compared with the input DNA control with default parameter settings and a significance threshold (P ≤ 1 × 10−5). We associated ChIP-enriched regions to a target gene if it localized within the region from 3 kb upstream of to 1 kb downstream from the transcription start site of the gene. The gene coordinates were extracted according to RefSeq gene annotation (hg18) downloaded from the University of California at Santa Cruz Genome Browser.
Acknowledgments
We are indebted to Kristian Helin for kindly providing us with the E2F7 antibody used throughout this study. We thank Charles Sherr for helpful scientific discussions and editorial suggestions, as well as Jessica Bolden for editorial help, Thomas Kitzing for help with the preparation of the figures, and Meredith J. Taylor for technical support. We also thank current and former members of the Lowe laboratory for helpful comments and technical support. This work was supported by funds from NBRPC (2012CB316503) and the NSF of China (60905013, 91019016, 31061160497) to X.W., a post-doctoral fellowship from NIH to A.C. (5F32AG027631), and an RO1 grant (AG16379) from the NIH to S.W.L. O.A. was supported by George A. and Marjorie H. Anderson Fellowship. S.W.L. is an investigator in the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.196238.112.
References
- Aslanian A, Iaquinta PJ, Verona R, Lees JA 2004. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev 18: 1413–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, Wagener C, Sardet C, Moroni MC, Helin K 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem 280: 1199–1208 [DOI] [PubMed] [Google Scholar]
- Azzoli CG, Sagar M, Wu A, Lowry D, Hennings H, Morgan DL, Weinberg WC 1998. Cooperation of p53 loss of function and v-Ha-ras in transformation of mouse keratinocyte cell lines. Mol Carcinog 21: 50–61 [DOI] [PubMed] [Google Scholar]
- Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J 2003. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J 22: 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dörken B, Jenuwein T, Schmitt CA 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Sage J 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Dynlacht BD 2003. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 3: 311–316 [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F 2007. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G 2009. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer 9: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Kapoor A, Wang X, Aksoy O, Everrtts AG, Zang MQ, Garcia BA, Bernstein E, Lowe SW 2012. H3K4 demethylation by Jarid1a and Jarid1b contributes to RB-mediated gene silencing during cellular senescence. Proc Natl Acad Sci 109: 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, et al. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 25: 2125–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, et al. 2005. Tumour biology: Senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cichowski K 2008. Many roads lead to oncogene-induced senescence. Oncogene 27: 2801–2809 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3 doi: 10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW 2004. PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 22: 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N 1998. The regulation of E2F by pRB-family proteins. Genes Dev 12: 2245–2262 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49 [DOI] [PubMed] [Google Scholar]
- Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ 2001. Functional antagonism between E2F family members. Genes Dev 15: 2146–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, Hannon GJ 2007. MicroRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer 7: 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaquinta PJ, Aslanian A, Lees JA 2005. Regulation of the Arf/p53 tumor surveillance network by E2F. Cold Spring Harb Symp Quant Biol 70: 309–316 [DOI] [PubMed] [Google Scholar]
- Jing H, Kase J, Dorr JR, Milanovic M, Lenze D, Grau M, Beuster G, Ji S, Reimann M, Lenz P, et al. 2011. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev 25: 2137–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Furukawa T, Abe T, Yatsuoka T, Youssef EM, Yokoyama T, Ouyang H, Ohnishi Y, Sunamura M, Kobari M, et al. 1998. Identification of two common regions of allelic loss in chromosome arm 12q in human pancreatic cancer. Cancer Res 58: 2456–2460 [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, Cleghorn W, Chen HZ, Kornacker K, Liu CG, et al. 2008. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell 14: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan N, Delavaine L, Graham A, Reilly C, Wilson J, Brummelkamp TR, Hijmans EM, Bernards R, La Thangue NB 2004. E2F-7: A distinctive E2F family member with an unusual organization of DNA-binding domains. Oncogene 23: 5138–5150 [DOI] [PubMed] [Google Scholar]
- Logan N, Graham A, Zhao X, Fisher R, Maiti B, Leone G, La Thangue NB 2005. E2F-8: An E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 24: 5000–5004 [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724 [DOI] [PubMed] [Google Scholar]
- Mooi WJ, Peeper DS 2006. Oncogene-induced cell senescence–halting on the road to cancer. N Engl J Med 355: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ 2001. Retinoblastoma protein partners. Adv Cancer Res 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Nijwening JH, Geutjes EJ, Bernards R, Beijersbergen RL 2011. The histone demethylase Jarid1b (Kdm5b) is a novel component of the Rb pathway and associates with E2f-target genes in MEFs during senescence. PLoS ONE 6: e25235 doi: 10.1371/journal.pone.0025235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotis Zalmas L, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, La Thangue NB 2008. DNA-damage response control of E2F7 and E2F8. EMBO Rep 9: 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja C, Serrano M 1999. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene 18: 4974–4982 [DOI] [PubMed] [Google Scholar]
- Reimer D, Sadr S, Wiedemair A, Stadlmann S, Concin N, Hofstetter G, Müller-Holzner E, Marth C, Zeimet, AG 2007. Clinical relevance of E2F family members in ovarian cancer–an evaluation in a training set of 77 patients. Clin Cancer Res 13: 144–151 [DOI] [PubMed] [Google Scholar]
- Rowland BD, Bernards R 2006. Re-evaluating cell-cycle regulation by E2Fs. Cell 127: 871–874 [DOI] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424: 223–228 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Shay JW, Pereira-Smith OM, Wright WE 1991. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res 196: 33–39 [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. 2005. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 37: 1281–1288 [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA 2002. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Wen J, Lees JA 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci 98: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, Agami R 2003. The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4: 311–319 [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Westendorp B, Mokry M, Groot Koerkamp MJ, Holstege FC, Cuppen E, de Bruin A 2012. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res 40: 3511–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E 1994. Tumour biology. p53, guardian of Rb. Nature 371: 21–22 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA 2004. Preprocessing of oligonucleotide array data. Nat Biotechnol 22: 656–658 [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM 2009. Autophagy mediates the mitotic senescence transition. Genes Dev 23: 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Xuan Z, Liu L, Zhang MQ 2005. TRED: A Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res 33: D103–D107 doi: 10.1093/nar/gki004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Fraenkel E, Pabo CO, Pavletich NP 1999. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW 2011. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]