In this study, Ahel and colleagues identify a new role for the SNF2 family ATPase ZRANB3 in replication-associated DNA repair. ZRANB3 is found to display a unique structure-specific endonuclease activity that allows ZRANB3 to cleave branched DNA structures with unusual polarity upon its recruitment to stalled replication forks. This study thus elucidates a novel nucleolytic activity by which ZRANB3 processes stalled replication forks in association with the replication machinery.
Keywords: structure-specific endonuclease, DNA repair, DNA replication, PCNA, ubiquitin
Abstract
To efficiently duplicate their genomic content, cells must overcome DNA lesions that interfere with processive DNA replication. These lesions may be removed and repaired, rather than just tolerated, to allow continuity of DNA replication on an undamaged DNA template. However, it is unclear how this is achieved at a molecular level. Here we identify a new replication-associated factor, ZRANB3 (zinc finger, RAN-binding domain containing 3), and propose its role in the repair of replication-blocking lesions. ZRANB3 has a unique structure-specific endonuclease activity, which is coupled to ATP hydrolysis. It cleaves branched DNA structures with unusual polarity, generating an accessible 3′-OH group in the template of the leading strand. Furthermore, ZRANB3 localizes to DNA replication sites and interacts with the components of the replication machinery. It is recruited to damaged replication forks via multiple mechanisms, which involve interactions with PCNA, K63–polyubiquitin chains, and branched DNA structures. Collectively, our data support a role for ZRANB3 in the replication stress response and suggest new insights into how DNA repair is coordinated with DNA replication to maintain genome stability.
SNF2 ATPases are a functionally heterogeneous family of proteins that regulate a number of diverse nuclear functions, including transcription, DNA replication, DNA repair, and recombination (Flaus et al. 2006). They are modular proteins characterized by the presence of a helicase core but ordinarily do not act as classical DNA helicases (Durr et al. 2006). Instead, their helicase core confers a DNA translocase activity. Domain structures of SNF2 ATPases also reveal the presence of accessory domains (Hauk and Bowman 2011), which often mediate interactions with protein partners or post-translational modifications and thus define the proteins' subcellular specificity. Indeed, the large variety of accessory domains identified in members of the SNF2 family seems to reflect their functional diversity.
A phylogenetically distant branch of SNF2 ATPases includes the SMARCAL1 (also known as HARP) and ZRANB3 (zinc finger, RAN-binding domain containing 3, also known as AH2) proteins. SMARCAL1 is a replication stress response protein, which is recruited to the sites of DNA damage through its interaction with replication protein A (RPA) (Bansbach et al. 2009; Ciccia et al. 2009; Postow et al. 2009; Yuan et al. 2009; Yusufzai et al. 2009). Additionally, SMARCAL1 is thought to possess DNA annealing activity that supports genome stability by minimizing the amount of RPA-bound ssDNA at stalled replication forks (Yusufzai and Kadonaga 2008). Stalling of replication forks occurs when the replication machinery encounters DNA lesions caused by endogenous or exogenous agents, which inhibit its progression. Because this jeopardizes complete and accurate duplication of the genome and consequently poses a serious threat to genetic stability, cells employ different strategies that allow them to maintain the continuity of replicated DNA (Friedberg 2005; Chang and Cimprich 2009; Ulrich 2011).
Cellular capacity to deal with replication stress relies on multiple and redundant mechanisms. DNA lesions that block replication can be bypassed without their actual removal in a process known as DNA damage tolerance, which involves two major pathways (Supplemental Fig. 1; Chang and Cimprich 2009; Ulrich 2011). One is translesion synthesis, which uses specialized and often mutagenic DNA polymerases (TLS polymerases) that can accommodate a damaged DNA template and replicate through DNA lesions. The other DNA damage tolerance pathway is error-free template switching, in which the undamaged sister chromatid serves as a temporary replication template. Alternatively, replication-associated DNA damage can be removed (not bypassed) by a process that involves the reversal of the replication forks (Supplemental Fig. 1). This results in the reannealing of the template strands and relocation of DNA damage in the dsDNA region, where it can be processed by the excision-based repair system (Atkinson and McGlynn 2009; Ulrich 2011).
The responses to replication-associated DNA damage involve PCNA, which is conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways (Moldovan et al. 2007; Lee and Myung 2008; Chang and Cimprich 2009; Ulrich 2009). The molecular mechanisms that support translesion synthesis are relatively well explored and are known to involve PCNA monoubiquitination, which recruits TLS polymerases via their ubiquitin-binding domains and motifs (such as UBM and UBZ motifs found in TLS polymerases of the Y family: Polη, Polι, Polκ, and Rev1) (Kannouche et al. 2004; Bienko et al. 2005; Waters et al. 2009). In contrast, the K63–polyubiquitination of PCNA is associated with the error-free pathways, but the underlying molecular mechanism of this association is not well understood (Hoege et al. 2002; Chiu et al. 2006). In yeast, the K63–polyubiquitination of PCNA is catalyzed by the RING domain-containing E3 ubiquitin ligase Rad5 (Hoege et al. 2002). Rad5 also contains a SNF2 ATPase domain that mediates fork regression activity in vitro, and it is believed that this activity might facilitate the resolution of stalled forks and promote template switching (Blastyak et al. 2007; Unk et al. 2010). Mammalian cells contain two Rad5 orthologs, SNF2 histone-linker PHD finger RING finger helicase (SHPRH) and helicase-like transcription factor (HLTF), both of which have been associated with the polyubiquitination of PCNA and the regulation of DNA damage tolerance (Motegi et al. 2008; Unk et al. 2008; Lin et al. 2011).
Despite genetic evidence linking the polyubiquitination of PCNA to the nonmutagenic pathways of replication-associated repair, the molecular insights into how this polyubiquitination supports the cellular resistance to DNA damage is unclear. By analogy with the translesion synthesis, it can be hypothesized that polyubiquitinated PCNA serves as a platform for the recruitment of relevant protein factors that support replication-associated DNA repair processes. However, the key players and events have not yet been identified.
Here we describe the identification of a novel replication-associated SNF2 protein, ZRANB3, and show evidence that supports its role in the replication stress response. ZRANB3 localizes at the sites of DNA replication and interacts with the key replication factor PCNA and subunits of the replicative helicase complex MCM. ZRANB3 is also recruited to DNA breaks and stressed replication forks, and its deficiency leads to an increased susceptibility to the DNA damage induced by MMS. The roles of ZRANB3 in DNA replication and repair are supported by a number of structural domains, which provide specificity for a series of defined substrates, including PCNA, K63–polyubiquitin chains, and branched DNA structures. Most importantly, we demonstrate that ZRANB3 exhibits unusual structure-specific ATP-dependent endonuclease activity that is contained within its C-terminal HNH domain (which derives its name from the three most conserved histidine and asparagine residues in the degenerate motif). Collectively, our results provide the first evidence of a replication-associated nuclease that is linked to PCNA polyubiquitination and are consistent with the role of ZRANB3 in replication-associated DNA repair. This provides important insights into the molecular mechanisms that support genetic stability at replication forks.
Results
ZRANB3 interacts and colocalizes with DNA replication factors
In order to gain insight into the cellular function of ZRANB3, we initially analyzed the expression of YFP-ZRANB3 in U2OS cells by fluorescent microscopy. We observed patterned signals (Supplemental Fig. 2A) reminiscent of replication foci (Poot et al. 2004), which prompted us to test whether YFP-ZRANB3 foci correspond to sites of DNA synthesis. To this end, we immunostained cells that had been pulse-labeled with bromodeoxyuridine (BrdU), and observed an overlap of the YFP-ZRANB3 signal with sites of BrdU incorporation (Fig. 1A). Furthermore, costaining of YFP-ZRANB3-overexpressing cells with endogenous PCNA showed colocalization of their foci (Fig. 1A).
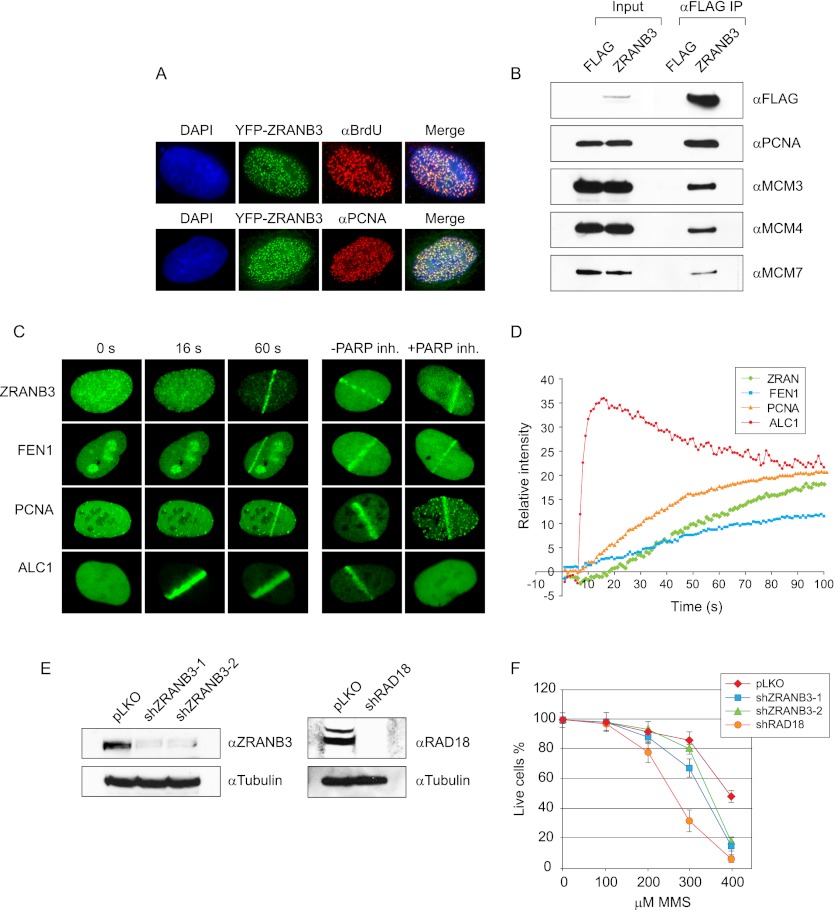
Figure 1.
(A) ZRANB3 targets sites of DNA replication. U2OS cells were transiently transfected with YFP-ZRANB3, pulsed with BrdU for 15 min, and stained with anti-BrdU antibody. Alternatively, cells were transfected with YFP-ZRANB3 and stained against endogenous PCNA. (B) Interaction of ZRANB3 with replication-associated factors. 293T cells were transiently transfected with Flag-tagged ZRANB3 or with empty vector, and extracts were subjected to immunoprecipitation on anti-Flag beads. Immunocomplexes were eluted with 3xFlag peptide and immunoblotted with the indicated antibodies. (C) ZRANB3 is recruited to the sites of laser-induced DNA damage. U2OS cells were transiently transfected with the indicated YFP constructs and analyzed by live-cell imaging. Shown are representative images at the indicated time points post-damage. Recruitment of ZRANB3 is compared with the replication factors PCNA and FEN1 and the SNF2 chromatin remodeling factor ALC1. Cells were also assayed 60 sec after the damage in the presence or absence of a PARP inhibitor. (D) Kinetics of the ZRANB3 recruitment to sites of laser-induced DNA damage, and comparison with the kinetics of PCNA, FEN1 and ALC1. More than 10 cells were analyzed for each construct. (E) Down-regulation of ZRANB3 and RAD18 expression by shRNA. Stable cell lines were created by the expression of empty pLKO-Puro, shZRANB3-1, shZRANB3-2, and shRAD18 vectors. Following selection against puromycin, the expression of ZRANB3 or RAD18 was evaluated by Western blotting. (F) Sensitivity of ZRANB3-deficient cells to MMS.
To examine the potential interactions of ZRANB3 with replication factors, we analyzed ZRANB3 protein complexes by immunoprecipitation. 293T cells were transiently transfected with Flag-tagged ZRANB3, and the extracts were subjected to anti-Flag immunoprecipitation. Western blot analysis of ZRANB3 immunocomplexes revealed the presence of the key replication factors—PCNA and several subunits of the major replicative DNA helicase MCM (MCM3, MCM4, and MCM7) (Fig. 1B). These results demonstrate the association of ZRANB3 with DNA replication.
ZRANB3 is recruited to DNA damage sites
A number of replication factors, such as PCNA, RPA, and flap endonuclease 1 (FEN1), play roles in both DNA replication and the repair of a variety of DNA lesions. To determine whether this is also the case for ZRANB3, we tested its ability to localize to sites of DNA damage. We induced DNA damage by laser microirradiation in cells that expressed YFP-ZRANB3 and tracked changes in the fluorescent signal by live-cell imaging. As shown in Figure 1C, ZRANB3 was recruited to the microirradiated stripes within the first minute following the induction of DNA damage. The kinetics of ZRANB3 recruitment was slower than that observed for the SNF2-type chromatin remodeling factor ALC1, which localized to DNA damage sites instantaneously (Ahel et al. 2009). Interestingly, the recruitment of the replication factors PCNA and FEN1 followed kinetics comparable with ZRANB3 (Fig. 1D), suggesting similarities in the mechanism of their response. Indeed, the recruitment of ALC1 was inhibited by the treatment of cells with a PARP inhibitor, unlike the recruitment of ZRANB3, PCNA, and FEN1 (Fig. 1C), which was unaffected.
We further explored the function of ZRANB3 by assessing the sensitivity of ZRANB3-deficient cells to exogenous DNA damage. ZRANB3 expression was down-regulated in 293T cells by the expression of two different shRNA constructs (Fig. 1E). Analyses of these cells revealed them to be sensitive toward DNA damage induced by MMS but not camptothecin, hydroxyurea, H2O2, or UV (Fig. 1F; Supplemental Fig. 2C).
Functional characterization of structural domains
Our data showed that ZRANB3 is a DNA replication-associated protein that is recruited to sites of DNA damage. To understand the molecular basis of this association, we analyzed the modular structure of the ZRANB3 protein and identified the presence of several functional domains (Fig. 2A). In addition to a helicase core at its N terminus, characteristic for members of the SNF2 ATPase family, ZRANB3 contains a PCNA-interacting protein motif (PIP-box), an unusual zinc finger of the NZF-type (Npl4 zinc finger) (Wang et al. 2003), and a putative HNH-type endonuclease domain at the C terminus.
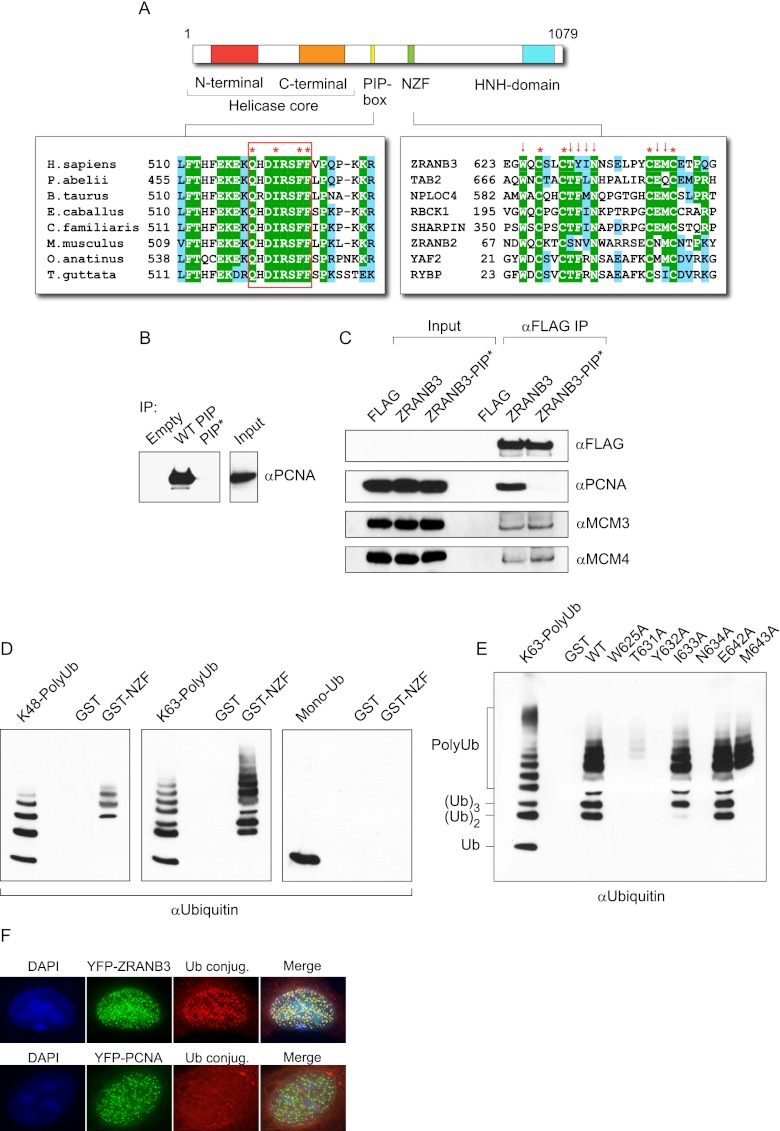
Figure 2.
(A) Modular structure of ZRANB3. (Left) Conservation of the PIP-box residues among ZRANB3 proteins is shown in the alignment. (Right) Conservation of the NZF motif includes several representative human proteins. Conserved cysteine residues are marked by asterisks. Arrows indicate mutated residues Trp 625, Thr 631, Tyr 632, Ile 633, Asn 634, Glu 642, and Met 643. (B) Interaction of the ZRANB3 PIP-box motif with PCNA in vitro. Biotinylated PIP-box peptide was bound to streptavidin beads and incubated with recombinant PCNA. The interaction was assayed by Western blotting with PCNA antibody. Mutations of the conserved PIP-box residues (Q519A, F525A, and F526A in PIP*) abrogated the interaction with PCNA. (C) Interaction of ZRANB3 with PCNA is PIP-box-dependent. 293T cells were transiently transfected with empty vector, Flag-tagged wild-type ZRANB3, and the PIP-box mutant (ZRANB3-PIP*). Following immunoprecipitation on anti-Flag beads, immunocomplexes were eluted by 3xFlag peptide and analyzed by Western blotting. Expression of Flag-ZRANB3 is detectable only after immunoprecipitation. (D) Interaction of the NZF motif with polyubiquitin chains in vitro. The wild-type NZF motif was expressed as a GST fusion protein and bound to the GST beads. The beads were then incubated with the monoubiquitin, polyubiquitin K48(2-7), or polyubiquitin K63(2-7) chains. Interactions were assayed by anti-ubiquitin Western blotting. (E) Interactions of mutant NZF motifs with K63–polyubiquitin chains. The experiment was performed as in D. Mutated positions are indicated by arrows in the alignment in A. (F) Colocalization of ZRANB3 with ubiquitin conjugates. U2OS cells were transiently transfected with YFP-ZRANB3 or YFP-PCNA and immunostained with FK2 antibody, which recognizes ubiquitin conjugates but not free ubiquitin.
The PIP-box motif is defined by a Q-x-x-[VLIM]-[VLIM]-x-x-[YF]-[YF] consensus and mediates interactions with PCNA in a number of PCNA-binding proteins (Warbrick 1998). To assess the PCNA-binding potential of ZRANB3′s PIP motif, we synthesized it as a biotinylated peptide, which was then bound to streptavidin beads and incubated with recombinant PCNA. As shown in Figure 2B, the PIP peptide efficiently interacted with PCNA, which was not retained on the streptavidin beads alone. In addition, this interaction was ablated by mutation of the conserved residues.
Given that our immunoprecipitation experiments demonstrated the interaction of ZRANB3 with PCNA in vivo (Fig. 1B; Supplemental Fig. 2B), we next examined whether this interaction is mediated by the PIP-box. We therefore introduced the PIP-box-inactivating mutations (Q519A, F525A, and F526A) into the Flag-tagged ZRANB3 construct and compared its ability to immunoprecipitate PCNA with the wild-type ZRANB3. Analysis of the ZRANB3 immunocomplexes showed that the PIP-box mutations abrogated the interaction of ZRANB3 with PCNA (Fig. 2C), demonstrating that the interaction of ZRANB3 with PCNA is PIP-box-dependent. Mutation of the PIP-box did not affect the interaction of ZRANB3 with the MCM proteins (Fig. 2C).
We next analyzed the NZF motif, characterized by the conserved cysteine residues and a T-[YF]-x(10)-Φ sequence (Φ represents a hydrophobic residue), as shown in Figure 2A. This type of zinc finger is widespread but appears to be present in a number of proteins that function in ubiquitin-dependent pathways (Alam et al. 2004). Indeed, several isolated NZF domains have been shown to bind monoubiquitin and polyubiquitin in vitro (Meyer et al. 2002; Wang et al. 2003). We therefore wanted to test whether the NZF motif in ZRANB3 possesses the same ability. We expressed NZF as a GST fusion protein, which was bound to GST beads and incubated with monoubiquitin or polyubiquitin chains. Our results revealed a strong interaction of the NZF motif with K63-linked polyubiquitin (Fig. 2D). Binding of K48-linked ubiquitin was less pronounced, whereas an interaction with monoubiquitin could not be detected (Fig. 2D). These observations are in line with the evidence showing that the related NZF motifs in TAB2 and TAB3 specifically bind K63-linked polyubiquitin chains (Kanayama et al. 2004; Komander et al. 2009; Sato et al. 2009). Furthermore, to define the structural requirements for the interaction with polyubiquitin, we introduced mutations in the conserved residues of the NZF motif (Fig. 2A). Our data show that interactions of the NZF motif with K63–polyubiquitin chains were completely abolished by mutations of W625, Y632, or N634 and significantly reduced by mutation of T631 (Fig. 2E; Supplemental Fig. 2D). Additionally, mutation of M643 compromised the affinity of NZF for oligomeric ubiquitin forms (2 –5 ubiquitin units) but not its ability to interact with K63–polyubiquitin chains of higher molecular weight (≥6 ubiquitin units).
Furthermore, we analyzed the colocalization of YFP-ZRANB3 with polyubiquitin conjugates. For this purpose, we used an FK2 antibody that recognizes polyubiquitylated and monoubiquitylated proteins but not free ubiquitin. We observed colocalization of YFP-ZRANB3 with ubiquitin conjugates (Fig. 2F), which could not be detected with transiently expressed YFP-PCNA. This may suggest that overexpression of ZRANB3 potentiates the accumulation of ubiquitinated proteins.
Last, we analyzed the potential functions of the HNH domain (Supplemental Fig. 2E). The HNH domain is a promiscuous and polyphyletic domain associated with a variety of DNA-binding proteins. It is found among homing endonucleases, inteins, group I and group II introns, and free-standing ORFs and has been linked to a variety of cellular functions, including endonuclease restriction, phage packaging, and recombination (Dalgaard et al. 1997; Veluchamy et al. 2009). Available data on the HNH domain suggest that it acts as a nucleic acid-binding and cleavage module (Huang and Yuan 2007), so we sought to investigate whether the HNH domain of ZRANB3 possesses these abilities. We expressed and purified the C-terminal fragment of ZRANB3 containing the HNH domain as a His-tagged protein and evaluated its potential to bind and cleave different DNA substrates. Our results show that the HNH domain formed nucleoprotein complexes with branched DNA substrates, including those that mimic DNA replication intermediates (Supplemental Fig. 2F,G). This concurs with our data supporting the role of ZRANB3 in DNA replication.
ZRANB3 is a structure-specific ATP-dependent endonuclease
Although SNF2 ATPases generally do not act as DNA helicases, we wondered whether ZRANB3 might possess helicase activity. We purified full-length, His-tagged ZRANB3 protein from insect cells (Supplemental Fig. 3A,B) and incubated it with a splayed DNA duplex and ATP in a helicase assay (Fig. 3A). Following the incubation of the enzyme with the fluorescently labeled splayed duplex (as indicated in Fig. 3A), a surplus of unlabeled complementary ssDNA was added to prevent reannealing of the separated DNA strands. Therefore, efficient DNA helicase activity was expected to yield fluorescently labeled ssDNA in this assay. Our data showed that ZRANB3 catalyzed time-dependent conversion of the splayed DNA duplex into a faster-migrating product (Fig. 3A, indicated by an arrow; Supplemental Fig. 3C,D). However, we were surprised to notice that the mobility of this product did not align with the ssDNA form expected to arise as a product of proficient helicase activity (Fig. 3A). Intriguingly, the mobility of the observed product aligned between the ssDNA and dsDNA markers, corresponding to the presumed mobility of a DNA duplex containing a 5′ single-stranded overhang. Given that the formation of such a product would be consistent with the nucleolytic degradation of the splayed duplex used as a substrate in the reaction, we proceeded to investigate the ability of ZRANB3 to act as a structure-specific nuclease.
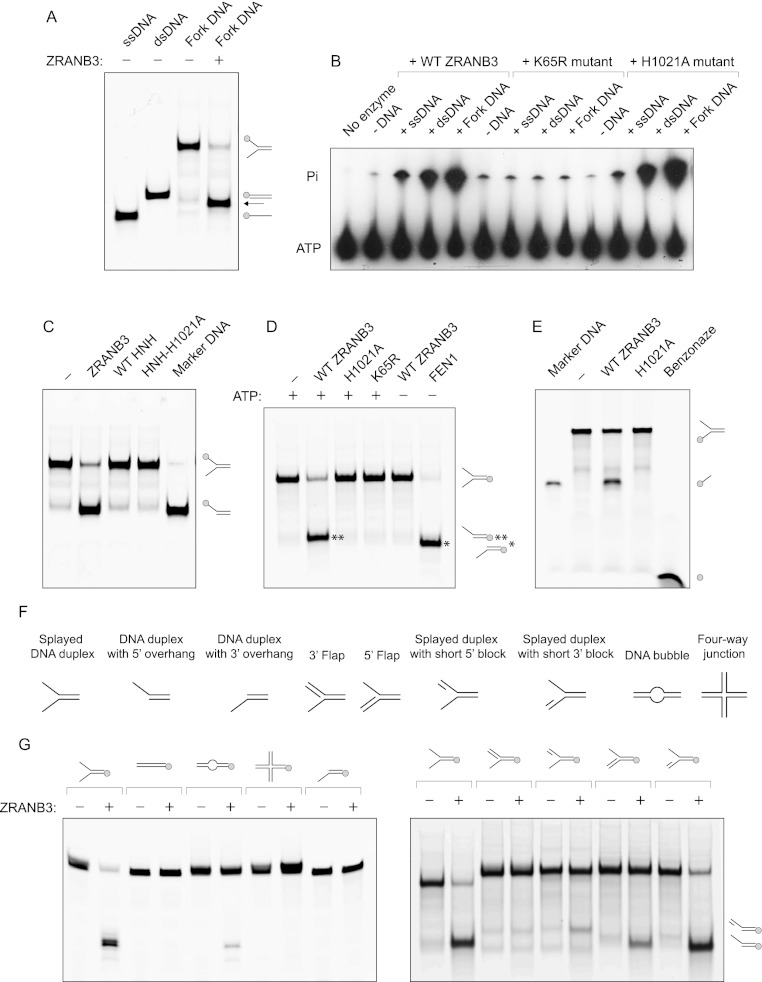
Figure 3.
(A) Helicase assay with ZRANB3. Fork DNA (splayed DNA duplex), fluorescently labeled at the 5′ end (as indicated in the picture), was incubated with ZRANB3 in the presence of ATP. After 30 min, an excess of unlabeled complementary ssDNA was added to prevent reannealing of the displaced oligos. Efficient DNA helicase activity is expected to yield fluorescently labeled ssDNA. Labeled ssDNA and dsDNA oligos were used as markers. Products were analyzed by native polyacrylamide gel electrophoresis. The reaction yielded a product of unexpected mobility (indicated by an arrow). (B) ATPase assay with the wild-type ZRANB3, helicase core mutant K65R, and HNH mutant H1021A. Recombinant proteins were incubated with 32P-labeled ATP in the absence or presence of indicated DNA substrates (40 nM). Reaction products were resolved by thin-layer chromatography. (C) Nuclease assay with full-length ZRANB3, isolated wild- type HNH domain, and HNH domain containing the H1021A mutation. Splayed DNA duplex fluorescently labeled at the 5′ end (shown in the picture) was incubated with the indicated proteins in the presence of ATP. Fluorescently labeled DNA duplex with 5′ overhang was used as a marker. Reactions were analyzed by native polyacrylamide gel electrophoresis. (D) Nuclease assay with the full-length ZRANB3 proteins. Wild-type ZRANB3, ATPase dead K65R mutant, and HNH mutant H1021A were incubated with splayed DNA duplex fluorescently labeled at the 5′ end (shown in the picture) in the presence of ATP. Additionally, wild-type ZRANB3 and FEN1 were incubated with the same substrate in the absence of ATP. Reactions were analyzed by native polyacrylamide gel electrophoresis. (E) Nuclease assay with the 3′-end-labeled splayed DNA duplex. Wild-type ZRANB3 was incubated with 3′-end FITC-labeled DNA substrate (as indicated in the picture). As a control, DNA substrate was incubated with benzonase nuclease to yield fluorescently labeled mononucleotides. Shown is a 26-nt marker DNA. (F) Substrates used in nuclease assays with ZRANB3. (G) The ability of ZRANB3 to cleave different DNA substrates. Wild-type ZRANB3 was incubated with the indicated fluorescently labeled DNA substrates in the presence of ATP. Reactions were analyzed by denaturing (left) or native (right) polyacrylamide gel electrophoresis.
Although the HNH domain acts as a DNA cleavage module in a number of the HNH-containing proteins, we could not detect the degradation of DNA substrates with the isolated HNH domain (Fig. 3C). We therefore considered the possibility that ZRANB3 possesses nucleolytic activity that requires not only the HNH domain, but also the ATPase activity contained within the helicase core. To test this hypothesis, we constructed corresponding ZRANB3 mutants by introducing K65R and H1021A changes into the helicase core and HNH domain, respectively. Lys 65 is a conserved residue in the ATP-binding Walker A motif of ZRANB3, and mutations of the analogous residue in other ATPases typically yield ATPase-deficient enzymes (Smith and Peterson 2005). On the other hand, His 1021 is a conserved catalytic residue in the HNH motif, and its mutation abrogated the nucleolytic activity of several HNH nucleases (Walker et al. 2002). We purified mutant ZRANB3 enzymes (Supplemental Fig. 3B) and first evaluated their ATPase activity.
The ATPase activity of ZRANB3 is barely detectable in the absence of exogenous DNA, but its stimulation can be observed by the addition of DNA—most significantly in the presence of fork DNA (Fig. 3B; Supplemental Fig. 3E,F; Yusufzai and Kadonaga 2010). Importantly, the ATPase activity was abolished by the K65R mutation, whereas the H1021A mutant retained activity comparable with wild-type ZRANB3. We next tested the ability of the K65R and H1021A mutants to cleave fork DNA structures. We used 5′ fluorescently labeled substrate (as indicated in Fig. 3D) and incubated it with the wild-type and mutant ZRANB3 proteins in the presence of ATP. Interestingly, while wild-type ZRANB3 catalyzed the conversion of the splayed duplex into the faster-migrating product, neither K65R, H1021A, nor the ZRANB3 mutant lacking the HNH domain (ΔHNH mutant) displayed such activity (Fig. 3D; Supplemental Fig. 4A,B). Additionally, the cleavage of fork DNA substrate could not be detected with wild-type ZRANB3 in the absence of ATP (Fig. 3D; Supplemental Fig. 4A). Collectively, these results demonstrate that ZRANB3 acts as an ATP-dependent HNH nuclease.
We also compared the activity of ZRANB3 with the activity of FEN1, a structure-specific nuclease that removes 5′ overhanging DNA flaps during DNA replication and repair. Both ZRANB3 and FEN1 cleaved model fork DNA structures; however, while the reaction with FEN1 yielded a DNA duplex containing 3′ overhangs, ZRANB3 cleaved the opposite DNA strand, generating a duplex with 5′ overhangs (Fig. 3D; Supplemental Fig. 4A,C). Such polarity demonstrated that ZRANB3 cleaves the DNA template of the leading strand.
To additionally confirm that ZRANB3 acts as an endonuclease and to exclude a possibility that the mapped 5′-end-labeled products in Figure 3, C and D, arise as a result of 3′-to-5′ exonuclease activity, we incubated ZRANB3 with 3′ fluorescently labeled substrate (as indicated in Fig. 3E). Consistent with the endonucleolytic activity, we observed the formation of a 3′ fluorescently labeled ssDNA (Fig. 3E). Furthermore, we could not detect 3′ fluorescently labeled nucleotides, which would be expected to accumulate as a result of exonuclease activity. We therefore conclude that ZRANB3 acts as an endonuclease.
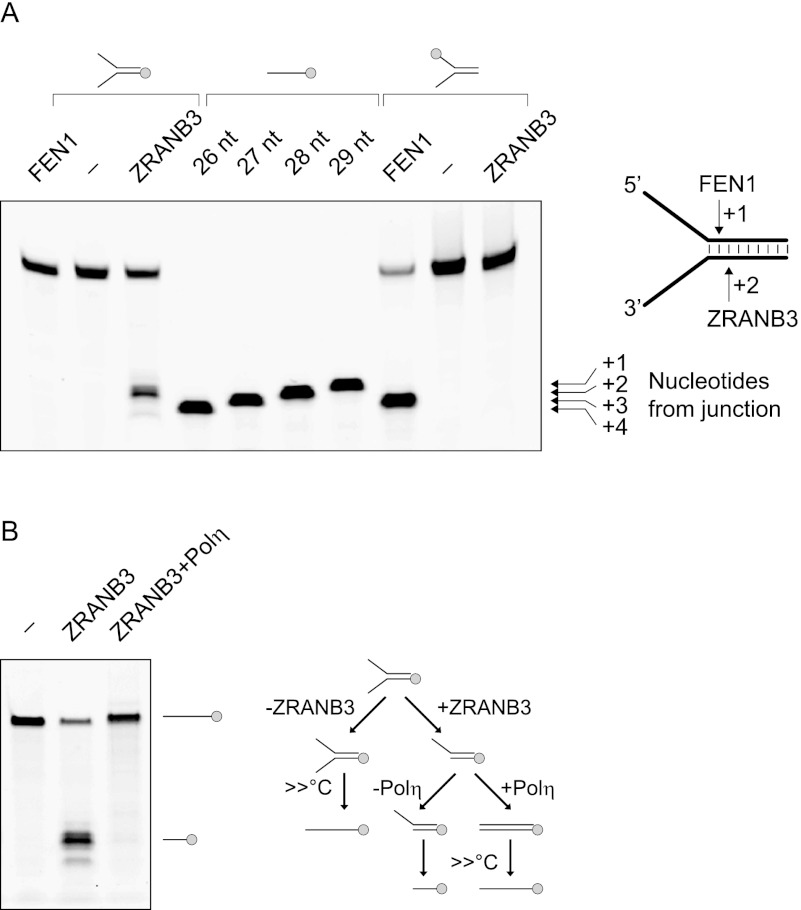
To determine the structural preferences for the endonuclease activity of ZRANB3, we tested a range of DNA substrates (Fig. 3F). As shown in Figure 3G and Supplemental Figure 4, D and E, ZRANB3 efficiently cleaved a splayed DNA duplex and displayed weak activity with the DNA duplex containing a 10-nucleotide (nt) bubble. In contrast, no activity could be observed with ssDNA and dsDNA, a four-way junction, a splayed DNA duplex containing a short 13-nt flap, or a DNA duplex containing a 3′ overhang. Furthermore, ZRANB3 cleaved 5′ flap structures (albeit less efficiently than a splayed duplex) but could not digest 3′ flap structures (Fig. 3G). These results indicate that structural constraints around the branch point of the DNA fork define its suitability for cleavage by ZRANB3. Additional experiments fine-mapped the cleavage site of ZRANB3, which occurred in the double-stranded region of the fork DNA substrate, preferentially 2 nt from the branch point (Fig. 4A). In contrast, FEN1 cleaved the opposite strand of the fork DNA structure, also in the double-stranded region, but only 1 nt from the branch point (Fig. 4A).
Figure 4.
(A) ZRANB3 cleaves DNA forks in the dsDNA region 2 nt from the branch point. ZRANB3 and FEN1 were incubated with the indicated fluorescently labeled DNA substrates. ATP was added to the reactions with ZRANB3. Reactions were resolved by denaturing polyacrylamide gel electrophoresis, and mobility of the obtained products was compared with the fluorescently labeled ssDNA markers. (B) Cleavage of fork DNA by ZRANB3 yields a 3′-OH end that can be extended by DNA polymerase. The splayed DNA duplex fluorescently labeled at the 5′ end (shown in the picture) was incubated with or without ZRANB3 in the presence of ATP. Polη and dNTPs were then added to the reaction to support extension of the cleaved DNA strand. Products were analyzed by denaturing polyacrylamide gel electrophoresis. Experimental outline is shown on the right.
Given that ZRANB3 cleaves the DNA template of the leading strand, we considered whether it created a 3′-OH terminus amenable to extension by DNA polymerase. Following the incubation of a model fork DNA substrate with ZRANB3, TLS polymerase Polη was added to the reaction in conditions that supported DNA polymerization. As shown in Figure 4B, Polη efficiently extended the product created by ZRANB3 digestion.
The role of ZRANB3 in replication-associated DNA repair
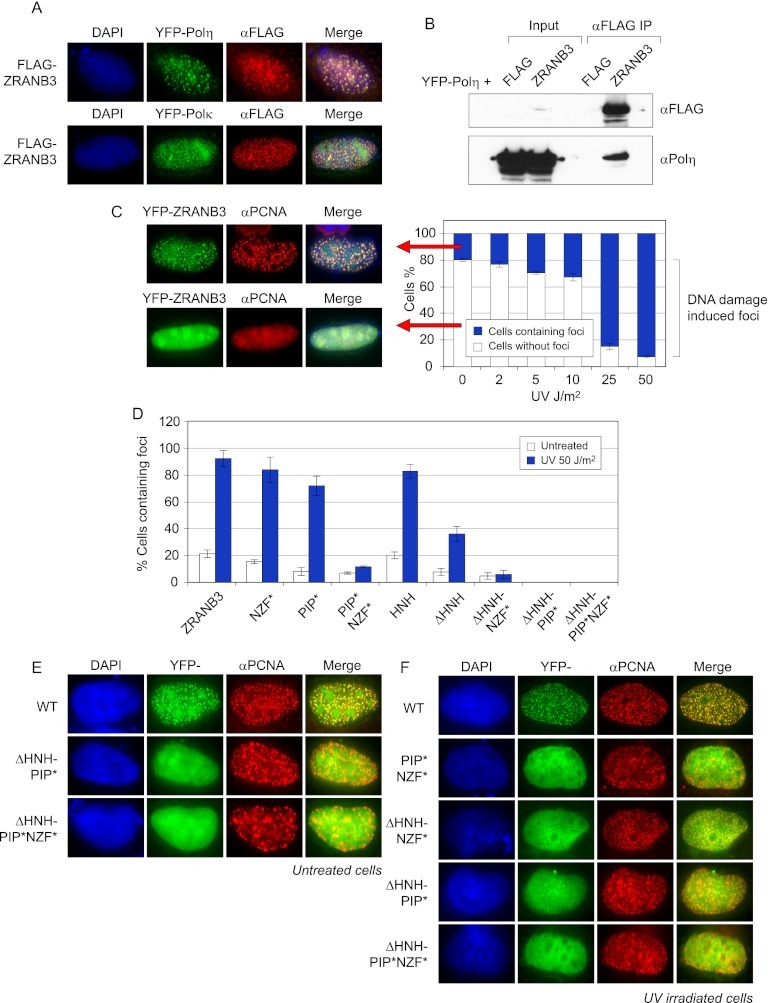
Our functional analyses identified ZRANB3 as a DNA replication-associated protein that is recruited to DNA damage. Furthermore, we showed that it colocalizes and interacts with both PCNA and K63–polyubiquitin chains and that it possesses replication structure-specific endonuclease activity. These observations suggested that ZRANB3 may play a role in replication-associated DNA repair. Additionally, we noted several similarities between ZRANB3 and TLS polymerases of the Y family. Both contain PIP-boxes that mediate interactions with PCNA- and ubiquitin-binding domains and motifs (Bienko et al. 2005; Moldovan et al. 2007). Indeed, the UBZ motif of Polη and NZF motif of ZRANB3 (both of which are zinc finger structures) displayed a similar affinity for K63–polyubiquitin chains (Supplemental Fig. 4F). Moreover, Polη and Polκ colocalized with PCNA and polyubiquitin chains and with ZRANB3 itself (Fig. 5A; Supplemental Fig. 4G). Our experiments also showed that ZRANB3 immunoprecipitated overexpressed YFP-Polη from whole-cell extracts (Fig. 5B). Given these observations, we aimed to investigate whether ZRANB3 and TLS polymerases share any DNA damage-related phenotypes.
Figure 5.
(A) ZRANB3 colocalizes with TLS polymerases Polη and Polκ. U2OS cells were cotransfected with Flag-tagged ZRANB3 and YFP-Polη or YFP-Polκ. Cells were stained with anti-Flag antibody. (B) ZRANB3 immunoprecipitates YFP-Polη from whole-cell extracts. 293T cells were cotransfected with Flag-tagged ZRANB3 and YFP-Polη. Following immunoprecipitation on anti-Flag beads, immunocomplexes were eluted by 3xFlag peptide and analyzed by Western blotting with Polη antibody. (C) ZRANB3 accumulates at replication forks stalled at DNA damage. U2OS cells were transfected with YFP-ZRANB3 and exposed to the indicated doses of UV irradiation. After 6 h, cells were fixed and stained with PCNA antibody. They were analyzed by microscopy, and the percentage of cells containing ZRANB3 foci that colocalized with PCNA was determined. Representative images of cells with and without ZRANB3 foci are shown on the left. (D) Contribution of functional domains within the ZRANB3 structure to its recruitment to sites of DNA replication before and after DNA damage. U2OS cells were transfected with the indicated YFP-ZRANB3 constructs and either untreated or exposed to UV irradiation. After 6 h, cells were fixed and stained with PCNA antibody. The percentages of cells containing ZRANB3 foci that colocalized with PCNA were determined as in C. (E) Representative images of mutant ZRANB3 proteins that do not colocalize with PCNA in the absence of exogenous DNA damage. U2OS cells were transfected with the indicated YFP-ZRANB3 constructs. Cells were stained with PCNA antibody. Merge represents green and red channels. (F) Representative images of mutant ZRANB3 proteins that do not colocalize with PCNA following UV irradiation.
Polη was previously shown to accumulate at stalled replication forks following treatment of cells with UV irradiation (Kannouche et al. 2001). This is observed as a dramatic increase in the number of cells containing Polη foci that colocalize with PCNA. To test whether ZRANB3 displays the same phenotype, we expressed YFP-ZRANB3 in U2OS cells, which were then exposed to UV irradiation and allowed to recover for 6 h. We found that ∼20% of unirradiated cells contained ZRANB3 foci that colocalized with PCNA and that this number increased following UV irradiation in a dose-dependent manner (Fig. 5C). We could also observe the accumulation of ZRANB3 at damaged replication forks following the treatment of cells with MMS and H2O2 (Supplemental Fig. 5B).
To gain insight into the mechanism that targets ZRANB3 to the sites of DNA replication, we analyzed the ability of functional ZRANB3 mutants to colocalize with endogenous PCNA. We introduced inactivating mutations in the conserved residues of the PIP-box (Q519A, F525A, and F526A; PIP* mutant) and the NZF (Y632A; NZF* mutant) motif (Fig. 2C,E) and assessed their potential to accumulate in replication foci. While the NZF* mutant efficiently colocalized with PCNA (Fig. 5D; Supplemental Fig. 5C), the PIP* mutant showed a notable decrease in the efficiency of recruitment to the sites of DNA replication (9% of cells expressing PIP*, compared with ∼20% of cells expressing ZRANB3) (Fig. 5D). This indicates that PIP-box-mediated interaction with PCNA plays an important role in the recruitment of ZRANB3 to replication foci.
We further examined the contribution of the HNH domain to the localization of ZRANB3. Interestingly, in undamaged cells, both wild-type ZRANB3 and the HNH domain alone colocalized with PCNA with comparable efficiencies (in ∼20% of cells) (Fig. 5D; Supplemental Fig. 5C). However, deletion of the HNH domain in ZRANB3 did not abrogate the colocalization with PCNA (Fig. 5D; Supplemental Fig. 5C, ΔHNH mutant). Despite this, the recruitment of ΔHNH to replication foci was less efficient than the recruitment of the wild-type ZRANB3 and could only be observed in 7.4% of cells (Fig. 5D). This highlights the importance of the HNH domain for the efficient recruitment of ZRANB3 to sites of DNA replication.
Our results suggested that the PIP-box and the HNH domain play major roles in the recruitment of ZRANB3 to replication foci. However, both PIP* and the ΔHNH mutant showed reduction, but not a complete loss, of colocalization with PCNA. We therefore inactivated the PIP-box in the ΔHNH mutant and evaluated the efficiency of its recruitment to the sites of DNA replication. Our data demonstrated that the ΔHNH-PIP* mutant could not be recruited to replication foci and did not colocalize with PCNA (Fig. 5D,E). This indicates that the PIP-box and the HNH domain provide alternative and partially redundant mechanisms through which ZRANB3 accumulates at replication foci.
We also wanted to address the contribution of functional domains within the ZRANB3 structure to its recruitment to sites of stalled DNA replication. We therefore examined the colocalization with PCNA of the ZRANB3 mutants following UV damage. Wild-type ZRANB3, NZF*, PIP*, the HNH domain, and the ΔHNH mutant all showed an induction in the number of cells containing focally concentrated ZRANB3 constructs, whose number increased approximately fourfold to sevenfold following UV irradiation (Fig. 5D). In contrast to this, the PIP*NZF* and ΔHNH-NZF* mutants did not show such induction, and their ability to form DNA damage-induced foci was severely compromised (∼12% of cells expressing PIP*NZF* and <6% of cells expressing ΔHNH-NZF* vs. >90% of cells expressing wild-type ZRANB3) (Fig. 5D,F). This indicates that NZF-mediated polyubiquitin binding plays an important role in the recruitment of ZRANB3 to stalled replication forks.
ZRANB3 functional mutants were also assessed for their ability to accumulate at the sites of laser-induced DNA damage. These results support our previous observations and show redundancy in the mechanisms that target ZRANB3 to the DNA damage sites (Supplemental Fig. 5D).
Discussion
To efficiently duplicate genomic content, cells must contend with the challenges imposed by DNA damage that interferes with replication fork progression. Unrepaired lesions that remain in the DNA template after DNA strand separation are particularly problematic and cannot be repaired by the excision repair mechanisms because the complementary DNA strand is not available as a template for DNA synthesis (Ulrich 2011). The number of possible mechanisms by which cells may deal with such lesions is considerable, but is generally thought to involve the bypass of DNA damage without its actual removal (DNA damage tolerance), or processing of DNA damage by the DNA repair machinery (Supplemental Fig. 1). Processing of DNA damage at the replication fork without lesion bypass is likely to involve replication fork regression, which would result in the formation of “chicken foot” structures and reannealing of the DNA template strands (Supplemental Fig. 1; Atkinson and McGlynn 2009). As a consequence, replication-blocking lesions would be placed in the dsDNA region and therefore become amenable to repair by excision-based repair systems. However, this model would necessitate the careful coordination of DNA replication and excision-based repair mechanisms, and it is unclear how this might be achieved at a molecular level. Moreover, the key effector proteins and enzymes that could support such activities have not as yet been identified. Here we describe the identification of a novel replication-associated structure-specific endonuclease, ZRANB3, and propose that it plays an important role in the repair of replication-associated DNA damage.
ZRANB3 as a novel replication-associated DNA repair factor
The association of ZRANB3 with DNA replication is supported by the evidence showing its localization at sites of active DNA synthesis and by its interactions with key replication factors, such as PCNA and subunits of the replicative helicase complex MCM. Moreover, ZRANB3 is recruited to DNA damage sites and accumulates at stalled replication forks. To understand the molecular basis of the role of ZRANB3 in DNA replication and repair, we analyzed the functional relevance of its constituting domains and motifs. We found that ZRANB3 comprises several structural domains that provide specificity for a series of defined substrates, including PCNA, K63–polyubiquitin chains, and fork DNA structures. These domains and motifs support the role of ZRANB3 in DNA replication and repair, and our analyses suggest that recruitment of ZRANB3 to the sites of DNA replication involves multiple and redundant mechanisms. One of the mechanisms seems to involve the HNH domain, and, given that we observed the affinity of this domain for branched DNA structures, we propose that structure-specific recognition of DNA replication intermediates plays an important role in the localization of ZRANB3 to the sites of DNA synthesis. Moreover, the PIP-box-mediated interaction with PCNA additionally supports the recruitment of ZRANB3 to replication foci, whereas NZF mediates the interaction with K63–polyubiquitin induced by DNA damage and contributes to the accumulation of ZRANB3 at sites of stalled DNA replication. These observations link ZRANB3 with both PCNA K63–polyubiquityination and the response to replication stress, indicating a possible role of ZRANB3 in the repair of replication-associated DNA damage.
ZRANB3 is structure-specific ATP-dependent endonuclease
The efficiency of DNA replication and repair relies on various nucleases that serve as tools for processing DNA ends and damaged replication forks into substrates for appropriate pathways. In rare instances, nucleolytic activity can be coupled with the ATPase activity, as observed in bacterial nucleases RecB and AddAB (which are involved in homologous recombination) (Kooistra et al. 1993; Ghatak and Julin 2006) and eukaryotic DNA2 protein (which is involved in Okazaki fragment processing and double-strand break DNA repair) (Fortini et al. 2011). Here we demonstrate that ZRANB3 possesses a highly unusual, ATP-dependent structure-specific HNH endonuclease activity.
The HNH domain is widespread in all walks of life in metal finger endonucleases, which include homing endonucleases, colicins, restriction endonucleases, transposases, and DNA packaging factors (Huang and Yuan 2007). The presence of the HNH domain in vertebrates is restricted to the ZRANB3 protein, and sequence comparisons of ZRANB3 HNH domains indicate that they are most closely related to the self-standing HNH proteins found in several bacterial species (Supplemental Fig. 2E). Given that the nuclease activity of ZRANB3 protein could not be detected in Escherichia coli genomic DNA (Yusufzai and Kadonaga 2010), it has been hypothesized that either the HNH domain has a non-nuclease-related function or its activity is activated in specific conditions. Our data reveal that the latter is the case and, surprisingly, that the activation of the endonuclease function necessitates ATPase activity contained within the helicase core of the ZRANB3 protein. In this respect, it seems interesting that the HNH domain is also found in type III restriction endonucleases, which cleave DNA in an ATP-dependent manner (Szczelkun 2011). Importantly, although the nucleolytic activity of ZRANB3 requires efficient ATP hydrolysis, the reverse is not the case. This is supported by the evidence showing that the ATPase-deficient K65R mutant does not display nuclease function, while the nucleolitically inactive H1021A variant retains efficient ATPase activity (Fig. 3B,D). This may suggest the sequential activation of nucleolytic activity (ATP hydrolysis followed by nuclease cleavage), which we presume is achieved through communication between discrete domains within the ZRANB3 structure.
ZRANB3 is a structure-specific endonuclease and cleaves fork DNA structures such as the splayed DNA duplex and 5′ flap structures but shows no activity toward 3′ flap structures. It requires single-stranded segments around the branch point of DNA (Fig. 3G), which is consistent with the recruitment of ZRANB3 to stalled replication forks (Fig. 5C), where such ssDNA regions would be expected to accumulate. Interestingly, both ZRANB3 and FEN1 cleaved model fork structures in the double-stranded region adjacent to the branch point; however, while FEN1 cleaved the DNA strand on the side of the 5′ flap, ZRANB3 cleaved the opposite DNA strand, generating a duplex with 5′ overhangs (Figs. 3D, 4A). Such polarity of cleavage displayed by FEN1 is consistent with its physiological role; i.e., removal of 5′ flaps in DNA repair and processing of the Okazaki fragments during lagging strand DNA synthesis (Liu et al. 2004). Furthermore, another structure-specific endonuclease, FAN1, cleaves relevant fork DNA structures from the same side as FEN1 (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). Conversely, ZRANB3 cleaves the DNA template of the leading strand, and such polarity seems to be less common among structure-specific endonucleases. This raised the possibility that cleavage of replication forks by ZRANB3 may lead to their collapse and the formation of double-stranded breaks; however, ZRANB3 foci do not colocalize with the double-strand break markers γH2AX and 53BP1 (Supplemental Fig. 5A), and overexpression of ZRANB3 is not associated with an increase in γH2AX levels (Supplemental Fig. 2B). We therefore believe that the replisome does not collapse following the incision by ZRANB3, but that it remains engaged with DNA. This could be explained by the current model of activation of eukaryotic replicative helicase, which suggests that the association of MCM2–7 with its activators, GINS/Cdc45 and ATP, gives rise to two topologically segregated channels involved in tracking the leading and lagging strands at the replication fork (Costa et al. 2011). Therefore, following the cleavage of the template strand by ZRANB3, the Cdc45–MCM–GINS complex could stay attached to the fork via the template of the lagging strand. The 3′-OH group generated by ZRANB3 in the template of the leading strand could subsequently be used for the extension by DNA polymerase (Fig. 4B).
The role of ZRANB3 in replication-associated repair
Replication-blocking DNA lesions that occur in the lagging strand template would not be expected to cause stalling of the replication forks, as they would merely result in unfinished Okazaki fragments (Ulrich 2011). In contrast, lesions in the leading strand template would cause stalling of the replication forks due to the continuous nature of DNA synthesis. Such lesions might compromise the successful replication of genetic material and thus pose threats to the maintenance of genome stability. Our data suggest that ZRANB3 could play an important role in the repair of such lesions by executing its structure-specific endonuclease activity.
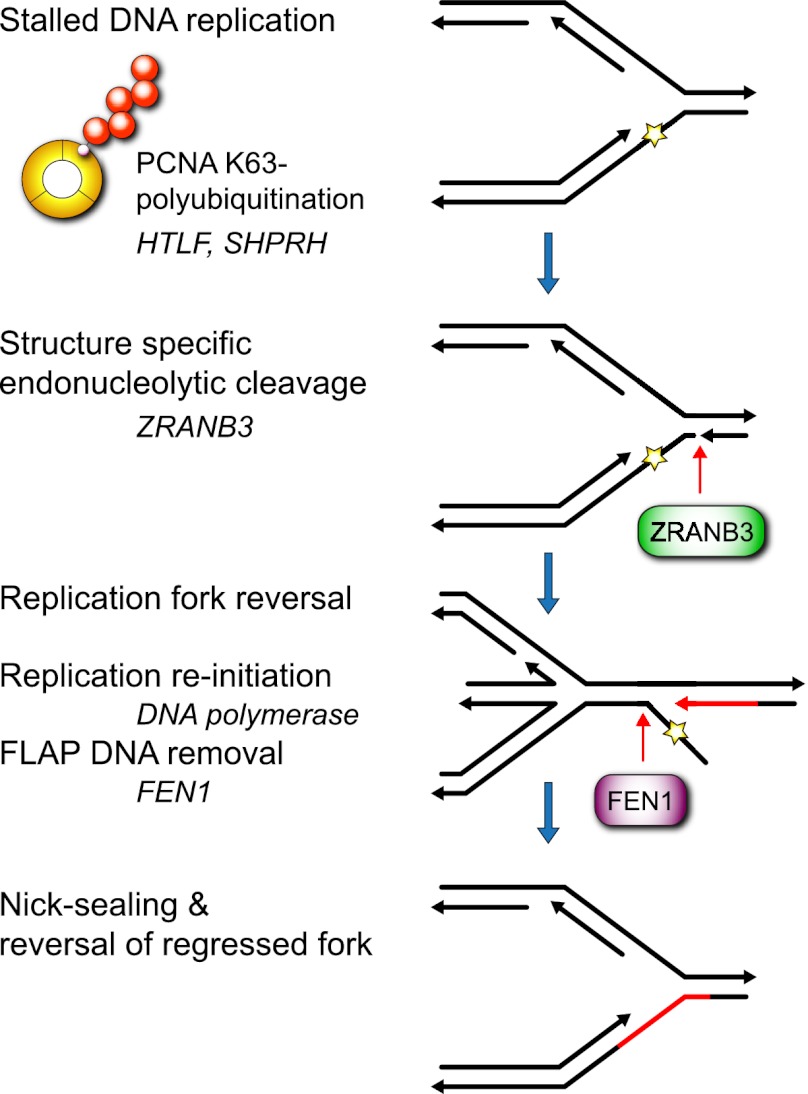
We propose the following model (Fig. 6): Replication-blocking lesions in the leading strand would stall replication forks, which would activate post-translational modifications of PCNA. Monoubiquitination of PCNA recruits TLS polymerases to promote DNA damage tolerance. On the other hand, extension of monoubiquitin to K63–polyubiquitin (possibly stimulated by the inability of TLS polymerases to replicate past DNA damage) would recruit ZRANB3 to the sites of stalled DNA replication. ZRANB3 could then act as a structure-specific endonuclease and cleave the replication fork intermediate, generating an accessible 3′-OH group in the template of the leading strand, which would be amenable to extension by DNA polymerase. Importantly, the nucleolytic cleavage of the DNA template induced by ZRANB3 would have to be coupled to fork regression in order to prevent disintegration of the replication fork and the formation of double-strand breaks. This concurs with previously proposed models (Atkinson and McGlynn 2009; Ulrich 2011) and with evidence showing that reversed replication forks can be visualized by electron microscopy (Lopes et al. 2006; Atkinson and McGlynn 2009). Possibly, fork regression could be mediated by the activities of the mammalian Rad5 orthologs HLTF and SHPRH, which catalyze PCNA polyubiquitination. Indeed, HLTF has been shown to possess fork regression activity in vitro (Blastyak et al. 2010). Alternatively, SMARCAL1 or ZRANB3 itself might actively participate in this process, as a recent report demonstrated that, like SMARCAL1, ZRANB3 possesses DNA annealing activity in vitro (Yusufzai and Kadonaga 2010). Such activity could hypothetically be used to reverse replication forks and support their stabilization. Indeed, a recent report demonstrated that SMARCL1 catalyzes fork regression to maintain genome stability during DNA replication (Betous et al. 2012). Finally, the extension of a 3′-OH terminus would generate a 5′ overhanging DNA flap containing a DNA lesion, which could be processed by the activity of FEN1. Following nick sealing and reversal of regressed forks, DNA replication could resume on an undamaged DNA template. This model suggests how DNA replication and excision-based repair mechanisms could be coordinated to facilitate the repair of replication-blocking lesions.
Figure 6.
Proposed model by which ZRANB3 facilitates replication-associated DNA repair. Separation of DNA strands by replicative helicase exposes the DNA lesion in the leading strand, which blocks processive DNA replication. Unavailability of the complementary DNA strand prevents repair of the exposed lesions by excision repair machinery at this point. E3 ubiquitin ligases HLTF and SHPRH catalyze PCNA K63–polyubiquitination of stalled replication forks, which recruits ZRANB3. At stalled replication forks, ZRANB3 acts as a structure-specific endonuclease and induces a DNA break in the double-stranded region of the replication fork 2 nt from the branching point. This is coordinated with the replication fork regression to prevent its disintegration. Cleavage by ZRANB3 exposes a free 3′-OH group, which can be extended by DNA polymerase to remove the replication-blocking DNA lesion. This leads to the formation of the 5′ overhanging DNA flap, which can be processed by the activity of FEN1. Following nick sealing and reversal of regressed forks, DNA replication resumes on the undamaged DNA template.
Materials and methods
Plasmids
ZRANB3 was amplified from a HeLa cDNA library and cloned via the pDONR211 entry vector (Invitrogen) into Gateway-compatible destination vectors comprising N-terminal YFP or N-terminal 3xFlag sequence. Point mutations were introduced using the QuickChange II site-directed mutagenesis kit (Stratagene). The ΔHNH and HNH variants were subcloned from the ZRANB3 pDONR221 entry vector, and, in the case of the HNH domain, the SV40 large T-antigen nuclear localization signal (NLS; sequence PKKKRKV) was introduced at the N terminus. In addition, for purification of the His-tagged protein, the HNH domain was cloned into the pET28a expression vector, and the NZF-motif was cloned into pGEX-4T1 (GE Healthcare) for the expression of the GST-tagged protein. Biotinylated PIP-box peptide was synthesized by Eurogentec. The pBac2 vector (Novagen) was used for purification of the full-length ZRANB3 with an N-terminal His tag from insect cells. FEN1 and PCNA were amplified from a HeLa cDNA library and cloned into pET28a and pET30a vectors for expression in bacteria. Polη and Polκ clones were purchased from Open Biosystems and subcloned into a destination vector with N-terminal YFP. ALC1 expression plasmids were previously described (Ahel et al. 2009).
Proteins
GST-tagged NZF proteins and His-tagged HNH (residues 871–1079 of the full-length ZRANB3) and PCNA were expressed in Rosetta competent cells (Merck). Expression was induced at OD600 = 0.6 with 0.4 mM IPTG for 3 h. Bacteria were lysed in the appropriate buffers (PBS buffer with 1 mM DTT, protease inhibitor cocktail [Roche] for the GST proteins; 50 mM Tris HCl at pH 8, 500 mM NaCl, 0.2% Triton-X, 10 mM imidazole, protease inhibitor cocktail for the His-tagged proteins) and purified over glutathione-sepharose or Ni-NTA columns, respectively. Wild-type and mutant ZRANB3 proteins were purified using a baculovirus expression system by the Protein Expression Facility at the University of Manchester. Hi5 cells were infected by viral stocks, harvested 72 h post-infection, and resuspended in the lysis buffer (25 mM Tris-HCl at pH 8, 0.3 M NaCl, 10% glycerol, 1%Triton-X, protein inhibitor cocktail, 5 mM imidazole). Protein was bound to nickel resin, washed with an imidazole gradient (10–60 mM), and eluted in 250 mM imidazole. Proteins were then diluted in 50 mM Tris HCl (pH 8), 50 mM NaCl, and 1 mM DTT and applied to a Heparin column. They were eluted with a NaCl gradient (50 mM to 1 M) and additionally purified on a Superose 6 gel filtration column in 50 mM Tris-HCl (pH 8), 300 mM NaCl, and 1 mM DTT. Fractions containing ZRANB3 were pooled for buffer exchange on PD10 columns (GE Healthcare) against 30 mM HEPES (pH 7.8), 50 mM NaCl, 0.25 mM EDTA, 0.01% NP40 1 mM DTT, and 10% glycerol at 4°C and stored at −80°C.
Antibodies
Rabbit ZRANB3 (Bethyl Laboratories, A303-033A), rabbit PCNA (Abcam, ab18197), rabbit MCM3 (Abcam, ab4460), rabbit MCM4 (Abcam, ab4459), rabbit MCM7 (Millipore, MABE188), rabbit Polη (Abcam, ab17725), mouse anti-Flag M2 HRP-conjugated antibody (Sigma-Aldrich, A8592), mouse ubiquitin (Enzo Life Sciences, P4D1), mouse γH2AX (Millipore, JBW301), and rat Tubulin (Abcam, ab6160) antibodies were used for Western blot analyses. Rabbit PCNA (Abcam, ab18197), mouse anti-Flag M2 (Sigma-Aldrich, F1804), and mouse FK2 antibody against monoubiquitinylated and polyubiquitinylated conjugates (Enzo Life Sciences) were used for immunofluorescence. BrdU was detected using the BrdU-Labeling and Detection Kit II (Roche). Unconjugated mouse anti-Flag M2 antibody (Sigma-Aldrich, F1804) was used for immunoprecipitation.
FACS-based sensitivity assay
shRNA oligonucleotides targeting the ORF region of ZRANB3 and RAD18 were designed and cloned into the pLKO(Puro) vector. 293T cells were transfected and selected with puromycin to create stable cell lines, which were then tested for the expression of ZRANB3. ZRANB3 expression was efficiently reduced by shZRANB3-1 (GTGCATAGATTGTGTTTAA) and shZRANB3-2 (TGGAAGACTTTAATACAGA) oligonucleotides. RAD18 expression was reduced by shRAD18 (GAGCATGGATTATCTATTCAA).
A FACS-based LIVE/DEAD Fixable Far-Red Dead Cell Stain kit (Invitrogen) was used to measure cellular sensitivity to DNA damage. Stable control and shZRANB3-expressing cell lines were grown in six-well plates in the presence of indicated concentrations of DNA-damaging agents for 4 d. Medium containing dead cells was collected, and cells were trypsinized and washed with PBS. They were processed according to the specifications in the LIVE/DEAD Fixable Far-Red Dead Cell Stain kit and analyzed by FACS. Sensitivity to UV was assessed as described by Watanabe et al. (2004).
Immunofluorescence
U2OS cells were grown on glass coverslips in a 24-well plate and transfected with the appropriate YFP constructs using Polyfect reagent (Qiagen). After 24 h, they were washed with PBS, treated with a pre-extraction buffer (10 mM PIPES, 300 mM sucrose, 3 mM MgCl2, 20 mM NaCl, 0.5% Triton-X) for 5 min at −20°C, and fixed in 4% paraformaldehyde for 10 min. Following fixation, cells were incubated in a second extraction buffer (10 mM Tris pH 8, 10 mM NaCl, 3 mM MgCl2, 1% Tween 40) for 5 min at −20°C, washed in PBS, and permeabilized with 0.5% Triton-X. They were then blocked in 2% BSA, incubated with the appropriate primary antibodies, washed in PBS, incubated with the secondary antibodies coupled to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen), and washed again in PBS. Finally, they were mounted in ProLong Gold anti-fade reagent with DAPI (Invitrogen) and analyzed by confocal microscopy.
For the accumulation of ZRANB3 at sites of stalled DNA replication, YFP-ZRANB3 transfected U2OS cells were untreated or treated with the indicated does of UV and allowed to recover for 6 h. Alternatively, cells were treated with the indicated concentrations of MMS and H2O2 for 1 h and then allowed to recover for 6 h in fresh medium. Following pre-extraction and staining with PCNA antibody, they were analyzed by confocal microscopy. Over 100 cells were counted for each experiment, and three independent results were pooled for statistical analyses.
For BrdU labeling, U2OS cells were transfected with the YFP-ZRANB3 construct and pulsed with 10 μM BrdU for 15 min. The BrdU Labeling and Detection kit II (Roche) was used to detect sites of BrdU incorporation as specified by the manufacturer, except an Alexa Fluor 594 goat anti-mouse secondary antibody (Invitrogen) was used instead of the anti-mouse alkaline phosphatase-labeled antibody provided in the kit.
Live-cell imaging by laser microirradiation
U2OS cells were grown in glass-bottom dishes (Iwaki) and transfected with the appropriate YFP constructs using Polyfect reagent (Qiagen). They were sensitized with 10 μM BrdU in phenol red-free medium (Invitrogen) for 16 h at 37°C. Laser microirradiation was carried out on a spinning-disk confocal microscope (Roper) equipped with an environmental chamber (Solent Scientific) where CO2/air, temperature of the chamber, and objective lens were maintained at 37°C. To ensure that cells with similar expression levels were assayed, cells showing moderate levels of expression were systematically chosen using identical 488-nm laser settings. To correct for overall bleaching of the signal due to repetitive imaging, fluorescence intensities were normalized against intensities measured in an undamaged nucleus in the same field after background subtraction. Equipment control and image capture were handled by Metamorph (Molecular Devices).
Immunoprecipitation
293T cell lines were grown in 15-cm dishes until subconfluency and transfected with Flag-tagged ZRANB3 constructs using Lipofectamine 2000 (Invitrogen). After 48 h, cells were washed with PBS and solubilized in lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM DTT) supplemented with 50 U/μL benzonase nuclease (Sigma) and protein inhibitor cocktail (Roche). Whole-cell extracts were clarified by centrifugation and incubated with anti-Flag M2 antibody (Sigma-Aldrich) prebound to protein A magnetic beads (Invitrogen). Following repeated washes with lysis buffer, the immunoprecipitates were eluted with 3xFlag peptide and analyzed by Western blotting.
PCNA pull-downs
Wild-type and mutant biotinylated PIP-box peptides were synthesized by Eurogentec and bound to magnetic streptavidin beads (Invitrogen). Unbound peptide was removed by washing with 50 mM Tris HCl (pH 8), 150 mM NaCl, 1 mM DTT, and 0.2% Triton-X. His-tagged recombinant PCNA was then incubated with the beads for 1 h and extensively washed with the same buffer. The beads were then boiled in SDS-PAGE loading buffer and analyzed by anti-PCNA Western blotting.
Polyubiquitin pull-downs
Recombinant GST-tagged proteins were expressed in bacteria and purified on glutathione-sepharose. Approximately 70 μL of the protein-bound beads were incubated with 1.5 μg of monoubiquitin, K48(2-7) polyubiquitin, or K63(2-7) polyubiquitin chains (Enzo Life Sciences) overnight at 4°C in 50 mM Tris HCl (pH 8), 150 mM NaCl, 1 mM DTT, and 0.2% Triton-X. The beads were extensively washed with the same buffer, boiled in SDS-PAGE loading buffer, and analyzed by Western blotting with anti-ubiquitin antibody P4D1 (Enzo Life Sciences).
Nuclease and helicase assays
Oligonucleotides used in the nuclease and helicase assays are shown in Supplemental Figure 6. Cy3 5′-end-labeled, FITC 3′-end-labeled, or [32P] 5′-end-labeled oligonucleotides were incubated with the appropriate unlabeled oligonucleotides in NEB4 buffer for 3 min at 94°C, after which the reactions were allowed to gradually cool down. Purified recombinant proteins (200 nM) were incubated for up to 30 min with the fluorescently labeled annealed DNA substrates (200 nM) at 30°C in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 2 mM ATP, and 0.1 mg/mL BSA. For the radioactivity-based assays, 10 nM DNA and 20 nM enzymes were used. For the helicase assays, a surplus of unlabeled complementary ssDNA (CCACCCGTCCACCCGACGCCACCTCCTG) was added to prevent reannealing of the separated DNA strands. The reaction was stopped by the addition of 0.1% SDS, 20 mM EDTA, and 4 mg/mL proteinase K for 30 min. For the native gel electrophoresis, the samples were analyzed by 10% PAGE. For the denaturing gel electrophoresis, samples were boiled in formamide buffer for 5 min at 95°C and analyzed by denaturing PAGE (10% polyacrylamide, 8 M urea).
Gel mobility shift assays
DNA substrates were annealed by slow cooling of radioactively 5′-32P-labeled oligonucleotide 5′-CCTCGATCCTACCAACCAGATGACGCGCTGCTACGTGCTACCGGAAGTCG-3′ with the relevant unlabeled ones as described in MacKay et al. (2010). Reactions (10 μL) contained 100 nM DNA and 0.5, 1.5, or 3 μg of either isolated ZRANB3 HNH domain or full-length ZRANB3 protein in 50 mM Tris-HCl, (pH 8.0), 30 mM NaCl, 5 mM EDTA, 1 mM DTT, 100 μg/mL bovine serum albumin, and 5% glycerol. Incubation was for 10 min at room temperature. Reactions were then transferred to ice and resolved immediately by 5% nondenaturing PAGE using 0.5× Tris borate-EDTA running buffer at 4°C. Gel mobility shifts were visualized by autoradiography.
ATPase activity assay
The ATPase TLC analysis was performed in a 5-μL reaction containing 10 μM ATP (Sigma), 0.1 μCi of [γ-32P]ATP (3000 Ci/mmol) (Perkin Elmer), 50mM Tris (pH 7.5), 5 mM MgCl2, and 50 ng of ZRANB3 proteins. When indicated, substrate DNA was added to the reaction. The reaction was carried out for 10 min at room temperature and stopped by the addition of 50 mM EDTA. Reaction products were spotted onto polyethyleneimine (PEI)-cellulose plates (Macherey-Nagel, Polygram CEL 300 PEI/UV254) and developed in 0.15 M LiCl and 0.15 M formic acid. Dried plates were exposed on X-ray film.
Acknowledgments
We thank Edward McKenzie and Matthew Ball for purification of the ZRANB3 proteins. We are grateful to Karim Labib and Ivan Ahel for critical reading of the manuscript and helpful discussions. This work is funded by Cancer Research UK.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.193516.112.
References
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI 2004. Ubiquitin interactions of NZF zinc fingers. EMBO J 23: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 37: 3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Hajdu I, Unk I, Haracska L 2010. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol 30: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA 2009. DNA damage tolerance: When it's OK to make mistakes. Nat Chem Biol 5: 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG 2006. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet 2: e116 doi: 10.1371/journal.pgen.0020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM 2011. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18: 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS 1997. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res 25: 4626–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr H, Flaus A, Owen-Hughes T, Hopfner KP 2006. Snf2 family ATPases and DExx box helicases: Differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res 34: 4160–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini BK, Pokharel S, Polaczek P, Balakrishnan L, Bambara RA, Campbell JL 2011. Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J Biol Chem 286: 23763–23770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC 2005. Suffering in silence: The tolerance of DNA damage. Nat Rev Mol Cell Biol 6: 943–953 [DOI] [PubMed] [Google Scholar]
- Ghatak A, Julin DA 2006. Kinetics of ATP-stimulated nuclease activity of the Escherichia coli RecBCD enzyme. J Mol Biol 361: 954–968 [DOI] [PubMed] [Google Scholar]
- Hauk G, Bowman GD 2011. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struct Biol 21: 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Huang H, Yuan HS 2007. The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J Mol Biol 368: 812–821 [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ 2004. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR 2001. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev 15: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D 2009. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra J, Haijema BJ, Venema G 1993. The Bacillus subtilis addAB genes are fully functional in Escherichia coli. Mol Microbiol 7: 915–923 [DOI] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88 [DOI] [PubMed] [Google Scholar]
- Lee KY, Myung K 2008. PCNA modifications for regulation of post-replication repair pathways. Mol Cells 26: 5–11 [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA 2011. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell 42: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kao HI, Bambara RA 2004. Flap endonuclease 1: A central component of DNA metabolism. Annu Rev Biochem 73: 589–615 [DOI] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J 2010. FAN1 acts with FANCI–FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G 2002. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4. EMBO J 21: 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, et al. 2008. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci 105: 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD 2004. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol 6: 1236–1244 [DOI] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H 2009. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 284: 35951–35961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Peterson CL 2005. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol Cell Biol 25: 5880–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD 2011. Translocation, switching and gating: Potential roles for ATP in long-range communication on DNA by Type III restriction endonucleases. Biochem Soc Trans 39: 589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD 2009. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 8: 461–469 [DOI] [PubMed] [Google Scholar]
- Ulrich HD 2011. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett 585: 2861–2867 [DOI] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci 105: 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Blastyak A, Haracska L 2010. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 9: 257–267 [DOI] [PubMed] [Google Scholar]
- Veluchamy A, Mary S, Acharya V, Mehta P, Deva T, Krishnaswamy S 2009. HNHDb: A database on pattern based classification of HNH domains reveals functional relevance of sequence patterns and domain associations. Bioinformation 4: 80–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DC, Georgiou T, Pommer AJ, Walker D, Moore GR, Kleanthous C, James R 2002. Mutagenic scan of the H-N-H motif of colicin E9: Implications for the mechanistic enzymology of colicins, homing enzymes and apoptotic endonucleases. Nucleic Acids Res 30: 3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR, Sundquist WI 2003. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J Biol Chem 278: 20225–20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E 1998. PCNA binding through a conserved motif. Bioessays 20: 195–199 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M 2004. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 23: 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev 23: 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT 2008. HARP is an ATP-driven annealing helicase. Science 322: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT 2010. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc Natl Acad Sci 107: 20970–20973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT 2009. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 23: 2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]