The class IA phosphatidylinositide 3 kinases (PI3Ks) signal downstream from activated receptor tyrosine kinases (RTKs), including EGFR and HER2. However, the specific functional roles of p110α and p110β in mediating growth factor signaling, development, and tumorigenesis in vivo are not clear. In this study, Zhao and colleagues investigate the functions of p110α and p110β in mammary gland development and tumor initiation. Knockouts of both PI3K isoforms result in dramatically different effects: p110α is essential for both development and tumorigenesis in the breast, while p110β is not. Instead, p110β ablation results in hypermorphic ductal development and enhanced tumorigenesis. In addition, the authors found that p110β competes with the more active p110α for binding sites on RTKs. Thus, this study delineates a novel mechanism controlling PI3K signaling whereby p110β can act as a negative regulator.
Keywords: PI3K, HER2, genetic mouse model, mammary tumorigenesis
Abstract
Class Ia phosphatidylinositol 3 kinase (PI3K) is required for oncogenic receptor-mediated transformation; however, the individual roles of the two commonly expressed class Ia PI3K isoforms in oncogenic receptor signaling have not been elucidated in vivo. Here, we show that genetic ablation of p110α blocks tumor formation in both polyoma middle T antigen (MT) and HER2/Neu transgenic models of breast cancer. Surprisingly, p110β ablation results in both increased ductal branching and tumorigenesis. Biochemical analyses suggest a competition model in which the less active p110β competes with the more active p110α for receptor binding sites, thereby modulating the level of PI3K activity associated with activated receptors. Our findings demonstrate a novel p110β-based regulatory role in receptor-mediated PI3K activity and identify p110α as an important target for treatment of HER2-positive disease.
The class Ia phosphatidylinositol 3 kinases (PI3Ks) are key components of growth factor signaling pathways regulating cell proliferation, survival, and death. Class Ia PI3Ks are heterodimeric lipid kinases composed of a p85 regulatory subunit and a p110 catalytic subunit. In response to growth factor stimulation and the subsequent activation of receptor tyrosine kinases (RTKs), PI3K is recruited to the membrane via interaction of its p85 subunit with tyrosine phosphate motifs on activated receptors or to adaptor proteins associated with the receptors; e.g., insulin receptor substrate 1 (IRS1). Other receptors such as G protein-coupled receptors (GPCRs), as well as oncogenes including Ras and polyoma middle T antigen (MT), can also activate the p110 subunit (Liu et al. 2009; Schaffhausen and Roberts 2009; Vanhaesebroeck et al. 2010). The activated p110 catalytic subunit of PI3K then catalyzes the phosphorylation of phosphatidylinositol 4,5-bisphosphate (PIP2) to form PIP3, which elicits multiple downstream signaling pathways. There are three highly homologous p110 catalytic isoforms (p110α, p110β, and p110δ) encoded by three distinct genes: Pik3ca, Pik3cb, and Pik3cd, respectively. In mammals, expression of p110δ is largely restricted to the immune system, while p110α and p110β are ubiquitously expressed (Engelman et al. 2006; Liu et al. 2009; Vanhaesebroeck et al. 2010). The tumor suppressor PTEN, a lipid phosphatase, reduces the cellular pool of PIP3 by converting it back to PIP2 and therefore functionally antagonizes PI3K activity (Parsons 2004).
The two ubiquitously expressed PI3K isoforms p110α and p110β are very similar in their catalytic and regulatory domains. They use the same substrates and generate the same lipid products. The structural and enzymatic overlap of these two PI3K isoforms might suggest that they are functionally redundant. However, recent studies have demonstrated that the p110α and p110β isoforms play distinct roles in cellular signaling, growth, and oncogenic transformation. It has been shown in mice, for example, that both p110α and p110β contribute to insulin action in the liver (Foukas et al. 2006; Ciraolo et al. 2008; Jia et al. 2008; Sopasakis et al. 2010), whereas angiogenesis and vascular endothelial growth factor (VEGF) signaling require p110α but not p110β (Graupera et al. 2008). Interestingly, while p110α is essential for lung adenocarcinoma induced by Kras (Gupta et al. 2007), p110β is required for prostate tumorigenesis driven by Pten loss in mice (Jia et al. 2008).
Aberrant activation of RTKs often leads to malignant transformation, and PI3K—a key effector of most RTKs—has emerged as a major therapeutic target for this class of cancers (Engelman 2009; Liu et al. 2009). While we and others have previously shown that p110α is the dominant PI3K isoform involved in the response to acute growth factor stimulation and RTK activation in cultured cells (Knight et al. 2006; Zhao et al. 2006; Jia et al. 2008), p110β's role in mediating RTK signaling remains unclear. Moreover, the specific functional roles of p110α and p110β in mediating oncogenic RTK-driven signaling and tumorigenesis have not been examined in vivo. This question is becoming increasingly critical given the growing recognition of isoform-specific functions, as mentioned above, as well as the advent of isoform-specific PI3K inhibitors for cancer therapy. In the present study, we investigate the roles of these two isoforms in mammary gland development as well as in MT- and HER2/Neu-induced mammary tumorigenesis and explore the underlying mechanism defining their distinct roles in RTK-mediated signaling and oncogenic transformation.
Results
Ablation of p110α impairs normal mammary gland development, whereas ablation of p110β leads to mammary gland hyperplasia
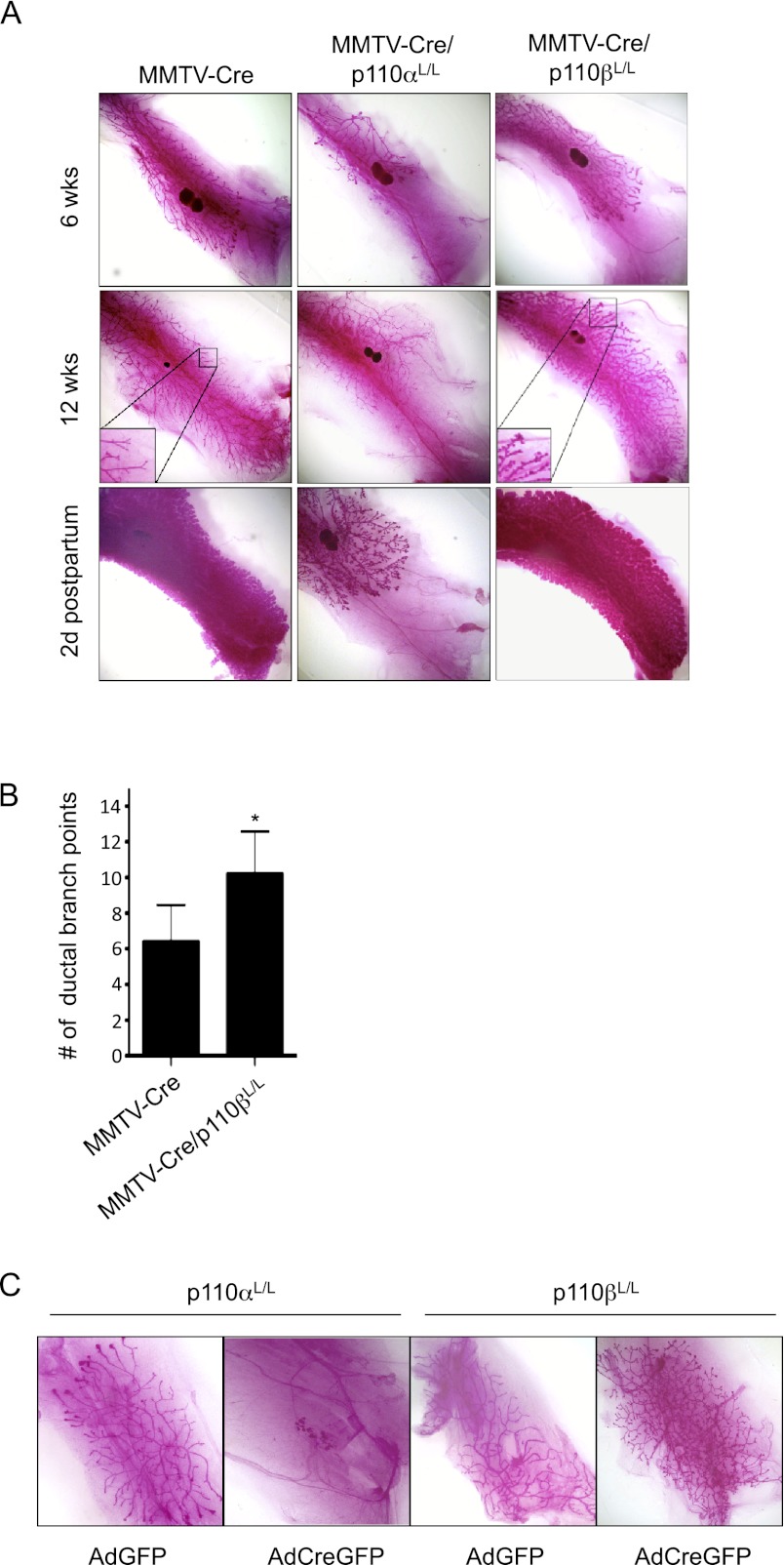
Before investigating the effects of knocking out p110 isoforms in tumorigenesis, we first examined their effects on normal mammary gland development. To determine whether the targeted ablation of p110α or p110β interferes with normal mammary gland development, we crossed mice bearing floxed alleles of p110α (Zhao et al. 2006) or p110β (Jia et al. 2008) with transgenic mice in which Cre expression is driven by the mouse mammary tumor virus (MMTV) promoter (MMTV-Cre) (Wagner et al. 1997). The resulting mice were heterozygous or homozygous for loss of p110α or p110β in the mammary glands. We performed whole-mount analyses of inguinal mammary glands isolated from females with the relevant genotypes at 6 and 12 wk of age and at 2 d post-partum. Mammary glands heterozygous for the loss of p110α had apparently normal duct outgrowth as well as post-partum lactation indistinguishable from that of wild-type littermates (Supplemental Fig. 1). Consistent with this finding, mice carrying one allele of p110α were able to nurse their pups normally. However, homozygous ablation of p110α dramatically impaired mammary duct outgrowth and branching during puberty and significantly decreased post-partum lactation (Fig. 1A).
Figure 1.
Effects of p110α or p110β ablation on mammary gland development. (A) Mammary gland whole mounts prepared from MMTV-Cre, MMTV-Cre/p110αL/L, and MMTV-Cre/p110βL/L mice at 6 and 12 wk of age and 2 d post-partum. (B) Bar graph representing the mean number of ductal branch points in mammary glands of MMTV-Cre or MMTV-Cre/p110βL/L animals at 12 wk of age. Bars are representative of the mean of five fields of five mammary glands of each genotype shown. (*) P < 0.05 (Student's t-test). (C) Mammary gland outgrowth produced by injection of dissociated control (AdGFP) or p110α- or p110β-ablated (AdCreGFP) MMECs into a cleared fat pad.
The impairment of mammary duct outgrowth in MMTV-Cre/p110αL/L mice varied significantly from one gland to another. As shown in Supplemental Figure 1B, a contralateral pair of fourth mammary fat pads isolated from a single MMTV-Cre/p110αL/L female at 6 wk of age displayed dramatic differences in their degree of duct outgrowth. Notably, while one of them had only moderately impaired outgrowth (Supplemental Fig. 1B, left panel), the other had only a few rudimentary ducts emerging in the nipple area and almost no postnatal outgrowth (Supplemental Fig. 1B, right panel). This variation from gland to gland within a single animal suggests a cell-autonomous defect. One possible explanation relates to the mosaic nature of MMTV-Cre-mediated deletion (Wagner et al. 1997). Such mosaicism would cause mammary glands in MMTV-Cre/p110αL/L females to contain varying numbers of mammary epithelial cells that have escaped Cre-mediated p110α ablation and hence are capable of supporting the limited ductal outgrowth observed. To test this hypothesis, we isolated epithelial cells from mounted MMTV-Cre/p110αL/L mammary tissue using laser capture microdissection. To enrich the ductal epithelial cells for this experiment, donor mammary glands were harvested from MMTV-Cre/p110αL/L females at 2 d post-partum (Supplemental Fig. 1C,D). Genomic PCR analysis of these isolated mammary epithelial cells showed the presence of both recombined and unrecombined alleles of the Pik3ca gene encoding p110α (Supplemental Fig. 1E,F), indicating that ablation of p110α was not complete. Together, these data suggest that p110α is essential for mammary gland development.
Our analysis of p110β in mammary gland development showed that p110β, in contrast to p110α, is dispensable for the development of a functional mammary gland. In fact, mammary glands from virgin mice lacking both copies of p110β displayed a modestly precocious lobulo–alveolar development with increased ductal branching, phenotypes normally seen in mammary glands of wild-type mice in early pregnancy (Fig. 1A,B). This hypermorphic phenotype was not readily observed in mammary glands from virgin female MMTV-Cre/p110β+/L mice (Supplemental Fig. 1, right panels).
The effects of p110α or p110β loss on mammary gland development are cell-autonomous
To determine whether the effects of p110α/β deletion on mammary gland development are cell-autonomous, we isolated primary mouse mammary epithelial cells (MMECs) from p110αL/L or p110βL/L mice and infected them with adenovirus expressing Cre-IRES-GFP (AdCreGFP) or GFP alone (AdGFP) as a control. GFP-expressing cells were collected by FACS to ensure a pure population of primary MMECs expressing either CreGFP or GFP alone. We then introduced paired groups of knockout and control MMECs into the contralateral cleared mammary fat pads of 3-wk-old nude mice and allowed them to develop for a period of 8 wk. After infection with AdGFP, floxed MMECs of both types generated mammary epithelial outgrowths upon transplantation (Fig. 1C, first and third panels). In contrast, after infection with AdCreGFP, only a small cluster of cells with little ductal outgrowth was observed in fat pads transplanted with p110α knockout MMECs (Fig. 1C, second panel), while regenerated mammary glands from p110β knockout MMECs displayed increased side branching similar to the mammary development observed in MMTV-Cre/p110β mice (Fig. 1C, fourth panel). These data suggest that the effects of mammary tissue-specific ablation of p110α or p110β are intrinsic to the mammary epithelium.
The effects of p110α or p110β loss on MT-induced mammary tumorigenesis
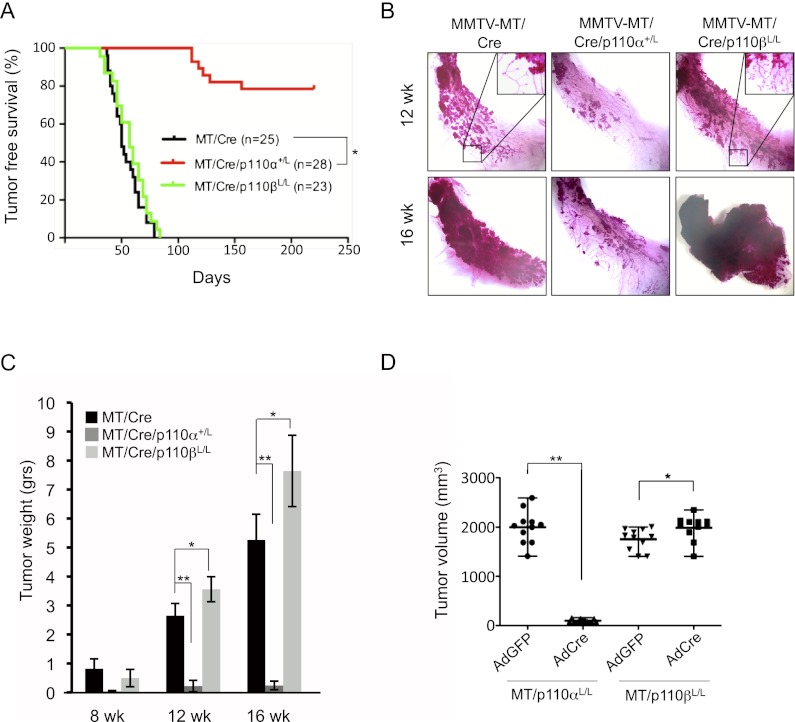
We next examined the roles of p110α and p110β in mammary tumorigenesis. One widely used transgenic mouse model of mammary tumor formation is driven by expression of MT transcribed from the MMTV promoter (Guy et al. 1992). MT is a surrogate for an activated RTK and is dependent on both the PI3K and Ras/Raf pathways for transformation (Whitman et al. 1985; Ichaso and Dilworth 2001; Schaffhausen and Roberts 2009). By examining the effect of heterozygous ablation of p110α on MT-induced tumorigenesis, we found that the onset of palpable tumor formation was markedly delayed upon loss of one copy of p110α (Fig. 2A). In agreement with these data, examination of MMTV-MT/Cre or MMTV-MT/Cre/p110α+/L mammary glands isolated from virgin females using either carmine red staining or cumulative tumor weight analyses revealed a greatly reduced tumor burden at 12 and 16 wk of age in MMTV-MT/Cre animals heterozygous for loss of p110α compared with control littermates (Fig. 2B,C). Tumor burdens were further evaluated using CellVigene automated image analysis (Supplemental Fig. 2A). Histological analysis of mammary glands from 16-wk-old MMTV-MT/Cre animals showed tumor masses composed of solid sheets of epithelial cells with some remaining acinar structures (Supplemental Fig. 2B, left panel), similar to other reports for typical MT-induced mammary tumors in mice of this age (Guy et al. 1992; Lin et al. 2003). In contrast, the primary lesions found in 16-wk-old MMTV-MT/Cre/p110α+/L mice consisted of clusters of hyperplastic acinar structures, resembling early premalignant lesions typically found in MMTV-MT mice at 4–6 wk of age (Supplemental Fig. 2B, middle panel; Lin et al. 2003). We also generated cohorts of MMTV-MT/Cre/p110αL/L mice that are homozygous for p110α. Analysis of mammary glands from these females showed tremendous variation from gland to gland; e.g., no gland development and hence no tumor formation, or partial gland formation with tumor lesions (Supplemental Fig. 2C). Western blot assays of tumor lesions revealed the presence of p110α (Supplemental Fig. 2D), suggesting that MT tumors arose from epithelial cells that escaped Cre-mediated deletion, indicating that p110α is critical for MT-mediated oncogenic transformation.
Figure 2.
Effects of p110α or p110β ablation on mammary tumorigenesis in MMTV-MT mice. (A) Kaplan-Meier curve of tumor-free survival of mouse cohorts of the indicated genotypes. The median latency (T50) is ∼50 d for both MT/Cre and MT/Cre/p110βL/L mice. (*) P < 10 × 10−10 (log-rank test). (B) Representative mammary gland whole mounts of MT/Cre, MT/Cre/p110α+/L, and MT/Cre/p110βL/L mice at 12 and 16 wk of age. (C) Average cumulative tumor weight (±SEM) for each cohort of mice at 8, 12, and 16 wk of age. MT/Cre (8 wk [n = 4], 12 wk [n = 6], 16 wk [n = 6]); MT/Cre/p110α+/L (8 wk [n = 6], 12 wk [n = 8], 16 wk [n = 11]); MT/Cre/p110βL/L (8 wk [n = 6], 12 wk [n = 8], 16 wk [n = 7]). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (D) Volumes of transplanted tumors at day 21 post-injection. Tumors were derived from injection of MT/p110αL/L or MT/p110βL/L MMECs after treatment in vitro with AdCreGFP or AdGFP (n = 11 for both). (*) P < 0.05; (**) P < 0.0001 (Student's t-test).
In contrast to the effects of p110α ablation, homozygous ablation of p110β did not delay MT-induced tumor onset (Fig. 2A). Instead, enhanced tumor growth and increased ductal branching were observed (Fig. 2B,C). Histological analysis of mammary tumors from 16-wk-old MMTV-MT/Cre/p110βL/L females revealed that these tumors were less differentiated (Supplemental Fig. 2B, right panel). These results clearly indicate that, while p110α is important for MT-induced mammary tumor formation, genetic ablation of p110β enhanced tumorigenesis driven by MT.
Targeted ablation of the p110α isoform impairs the growth of established tumors driven by MT
To characterize the effects of p110α or p110β ablation on cells from established MT-driven tumors, we harvested tumors from MMTV-MT/p110αL/L and MMTV-MT/p110βL/L mice at 12–14 wk of age, dissociated the tumor cells, cultured them briefly to allow in vitro infection with AdCreGFP or AdGFP as a control, and then selected productively infected cells by FACS. This approach overcomes the limitation of not being able to assess homozygous MMTV-Cre-mediated deletion of p110α on MT-induced tumorigenesis as described above. These primary MT mammary tumor cells were transplanted into the mammary fat pads of athymic mice. Three weeks post-transplantation, the control MT tumor cells had formed large tumors, but no tumor masses were found at sites injected with cells ablated for both alleles of p110α (Fig. 2D). In contrast, loss of p110β in MT-driven tumor cells resulted in a small but significant increase in tumor volume compared with control tumors (Fig. 2D). These data are consistent with the results described above that, while p110α is critical in mediating MT-induced mammary tumor formation and maintaining the tumorigenic state, p110β can actually down-modulate MT-induced oncogenic transformation.
The effects of p110α or p110β loss on HER2/Neu-induced mammary tumorigenesis
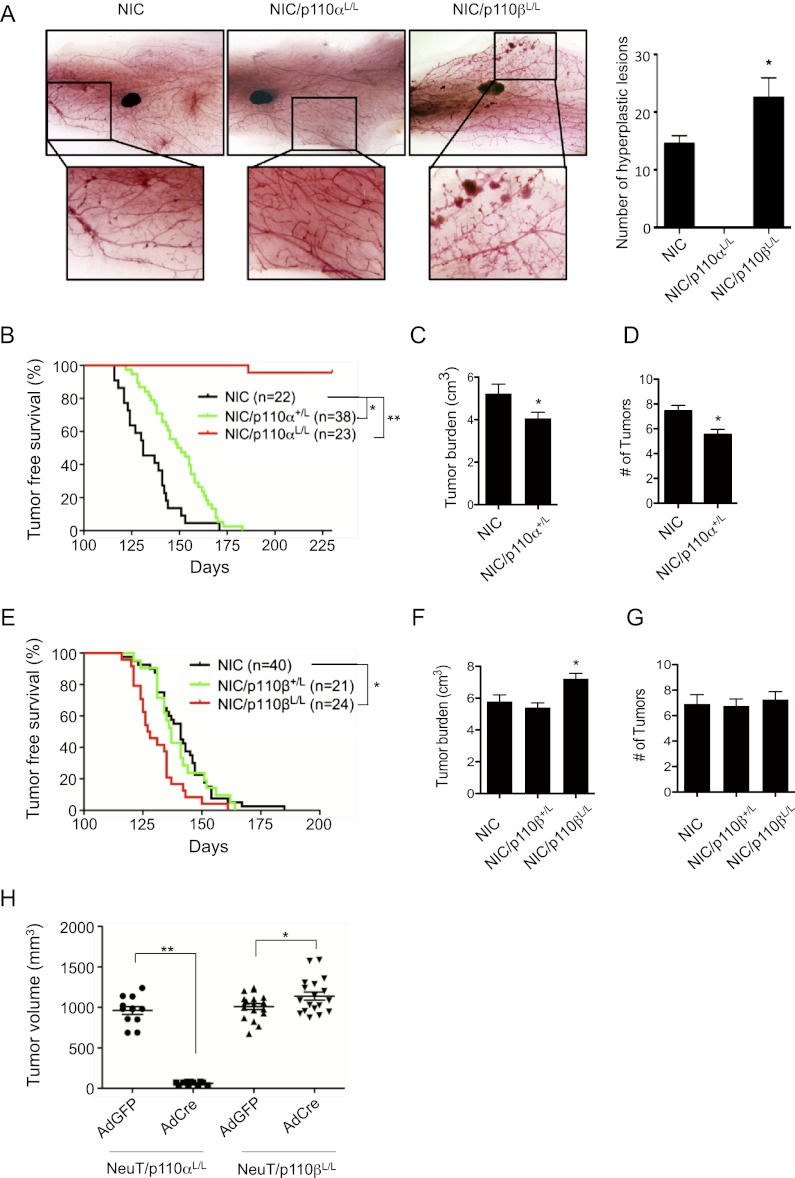
To examine the roles of p110α and p110β in a more disease-relevant model of breast tumors, we introduced floxed p110α or p110β alleles into a well-characterized HER2/Neu model—MMTV-Neu-IRES-Cre (NIC)—that coexpresses activated Neu and Cre recombinase (the latter via an IRES) under the control of a single MMTV promoter (Ursini-Siegel et al. 2008). This approach eliminates the need for an additional MMTV-Cre transgene and circumvents the stochastic nature of the expression of Neu and deletion of p110 that occurs when expression of the relevant genes are driven by separate promoters. Interestingly, whole-mount analysis of mammary glands from mice prior to palpation of tumor lesions revealed that, while the mammary glands of NIC females display hyperplastic lesions, NIC/p110αL/L female mice exhibit normal ductal outgrowth (Fig. 3A). It is intriguing that, while dramatically impaired mammary gland development is seen in MMTV-Cre/p110αL/L mice, NIC/p110αL/L mice had normal ductal outgrowth. Similarly, lobulo–alveolar budding is substantially increased in the p110β-deficient mammary glands in MMTV-Cre/p110βL/L mice but not in the NIC/p110βL/L mammary glands. We do not have a clear explanation for these observations, but one possibility is that ablation of p110 isoforms in mammary glands is mediated by different MMTV-Cre lines that fire their MMTV promoters at different stages of development and/or within different epithelial cell populations. The MMTV-Cre (Wagner et al. 1997) line expresses Cre during the embryonic stage before or at the beginning of the formation of the rudimentary ductal structure, which might result in more complete deletion of p110α or p110β and more significant developmental phenotypes. The MMTV promoter in NIC mice is active in luminal mammary epithelial cells after birth (Ursini-Siegel et al. 2008). Thus, the phenotypes seen in NIC/p110αL/L and NIC/p110βL/L could be related to luminal expression of Cre in the NIC population. Alternatively, the different phenotypes in the two models may reflect at least some degree of compensatory signaling arising from ErbB2 action in mammary epithelial cells.
Figure 3.
Effects of p110α or p110β ablation on HER2/Neu-driven mammary tumorigenesis. (A) Representative whole mounts of mammary glands from mice carrying MMTV promoter-driven NIC (n = 9), NIC/p110αL/L (n = 6), and NIC/p110βL/L (n = 10) at 10–12 wk of age before any formation of palpable tumor lesions. Bar graph shows the number of hyperplastic lesions in mammary glands of the indicated genotype (n = 5 each). (*) P < 0.05. (B) Kaplan-Meier curve of tumor-free survival of mouse cohorts of the indicated genotypes. T50NIC = 131 d, T50NIC/p110α+/L = 150 d. (*) P < 0.001; (**) P < 10 × 10−17 (log-rank test). (C) Average tumor burden per mouse (±SEM) for each cohort of mice at study endpoint (6 wk post-palpation of the first tumor in each mouse). (*) P < 0.05 (Student's t-test). (D) Average number of tumors per animal (±SEM) at study endpoint as described in C. (*) P < 0.005 (Student's t-test). (E) Kaplan-Meier curve of tumor-free survival of mouse cohorts of the indicated genotypes. T50NIC = 141 d, T50NIC/p110β+/L = 137 d, T50NIC/p110βL/L = 127.5 d. (*) P < 0.002 (log-rank test). (F) Average tumor burden per mouse (±SEM) for each cohort of mice at study endpoint as described in C. (*) P < 0.05 (Student's t-test). (G) Average number of tumors per animal (±SEM) at study endpoint (as described in C). (H) Volumes of tumor transplants at day 28 post-injection of MMTV-NeuT/p110αL/L (n = 11) or MMTV-NeuT/p110βL/L (n = 18) MMECs treated in vitro with AdCreGFP or AdGFP. (*) P < 0.05; (**) P < 0.0001 (Student's t-test).
Analyses of Ki67 expression in ducts and lobules demonstrated that Neu induced robust cell proliferation in mammary glands of NIC mice at the preneoplastic or early neoplastic stage (Supplemental Fig. 3A). While the Ki67-positive cells were significantly reduced in the mammary glands of NIC/p110αL/L mice, the number of proliferating cells significantly increased in NIC/p110βL/L mice (Supplemental Fig. 3A), thus providing the cellular mechanism by which p110α is important for Neu-induced neoplastic transformation, and p110β plays a negative role in Neu-mediated transforming activity.
We next followed cohorts of virgin females with NIC, NIC/p110αL/L, and NIC/p110βL/L genotypes to assess the roles of PI3K isoforms in HER2/Neu-initiated tumor development. NIC mice develop palpable tumors at 131 d of age, with each mouse carrying multiple tumors (Fig. 3B-D). Although all NIC/p110αL/+ mice developed tumors, the time to tumor onset was significantly extended to 150 d (P < 0.001), and tumor burden and occurrence were also significantly reduced (Fig. 3B–D; Supplemental Fig. 3B). Notably, ablating both copies of p110α virtually blocked tumor formation, with only one mouse in the cohort of 23 NIC/p110αL/L mice developing a single tumor over an observation period of 230 d (Fig. 3B). This solitary HER2/Neu tumor, which developed when the mouse was 186 d old, appears to have truly escaped p110α dependence, as Western blotting confirms the loss of p110α expression in the tumor (Supplemental Fig. 3C). Further studies following NIC/p110αL/L mice for longer times will be necessary to determine whether more p110α-independent HER2/Neu tumors occur and to elucidate the underlying mechanisms for such escapees. In contrast, p110β deletion resulted in a more rapid HER2/Neu mammary tumor occurrence and an increased tumor burden but did not alter the number of tumors per mouse (Fig. 3E–G; Supplemental Fig. 3D). These results are consistent with our findings in the MT tumor model described above.
The p110α isoform is required for the growth of established tumors driven by HER2/Neu
Because of the coupled expression of HER2/Neu and Cre, the NIC/p110 model limits our analysis to determination of whether a PI3K isoform is required for the initiation of HER2/Neu tumor development. To ascertain whether HER2/Neu tumors are dependent on PI3K for maintaining tumor cells in their transformed state, we crossed floxed p110 mice with another HER2/Neu mammary tumor model, MMTV-NeuT (Lucchini et al. 1992). Established NeuT-driven tumors carrying floxed p110 alleles were isolated, and dissociated tumor cells were treated with AdCre to delete p110. These cells were then transplanted into the mammary fat pads of athymic mice. Consistent with the results from the NIC model, deletion of p110α blocked the growth of NeuT-driven tumor transplants. Loss of p110β slightly but significantly increased tumor volume (Fig. 3H). Taken together, these data suggest that HER2/Neu-driven tumor cells are critically dependent on the p110α isoform of PI3K to maintain their transformed state, while p110β may negatively regulate HER2/Neu transforming activity.
The effects of p110α or p110β loss on oncogenic MT- or HER2/Neu-driven PI3K signaling in primary mammary tumor cells
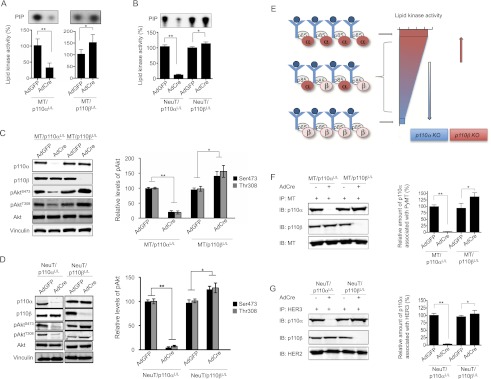
To explore the potential mechanism underlying these surprising, opposite effects of p110α or p110β loss on MT- and HER2/Neu-mediated tumorigenesis, we performed biochemical analyses on tumor-derived MMECs to determine how the PI3K activity associated with oncogenic MT or HER2/Neu is affected by the absence of p110α or p110β. We isolated MMECs from MT- or NeuT-driven tumors carrying floxed p110α or p110β alleles, treated them with AdCreGFP or AdGFP as a control, and isolated GFP-expressing cells by FACS. To study the effect of ablating p110 isoforms on MT- or NeuT-associated PI3K activity, cells were starved to minimize confounding signals from the activation of other receptors. While MT binds to the p85/p110 complex directly (Schaffhausen and Roberts 2009), HER3 recruits PI3K to an activated HER2/Neu (Supplemental Fig. 4A). We confirmed that treatment of NeuT tumor cells with a HER2 inhibitor, lapatinib, abolished formation of the HER3/PI3K complex and PI3K activities associated with HER3, as well as phosphorylation of AKT, in cells (Supplemental Fig. 4B–D). In addition, equal amounts of MT and HER3 were found in the immunoprecipitates from the various cell lysates (Supplemental Fig. 4E,F). We therefore measured PI3K activity in MT or HER3 immunoprecipitates in vitro and found robust lipid kinase activity associated with MT or HER3 in control cells treated with AdGFP. While deletion of p110α significantly reduced 3′ phosphoinositide synthesis, depletion of p110β increased MT- or HER3-associated PI3K activity (Fig. 4A,B). In parallel, greatly reduced pAkt levels were seen in p110α-deficient cells derived from MT or NeuT tumors, and increased levels of pAkt were observed in p110β-ablated cells (Fig. 4C,D). These data suggest that p110α is the major PI3K isoform mediating oncogenic MT- and HER2/Neu-driven signaling and that p110β plays a negative regulatory role in modulating the strength of PI3K signaling downstream from activated receptors.
Figure 4.
Effects of p110α or p110β ablation on MT- or HER2/Neu-mediated PI3K signaling. (A) Lipid kinase activity associated with MT immunoprecipitates from MMTV-MT/p110αL/L or MMTV-MT/p110βL/L tumor-derived cells treated in vitro with AdCreGFP or AdGFP as indicated. The bars represent relative lipid kinase activity ± SD (n = 7). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (B) Lipid kinase activity associated with HER3 immunoprecipitates from MMTV-NeuT/p110αL/L or MMTV-NeuT/p110βL/L tumor-derived cells treated in vitro with AdCre or AdGFP. The bars represent relative lipid kinase activity ± SD (n = 12). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (C) Western blots for p110α, p110β, pAktS473, pAktT308, and vinculin (as a loading control) in cell lysates prepared from MMTV-MT tumor cells carrying p110αL/L or p110βL/L. Cells were treated in vitro with AdGFP or AdCre and starved without growth factors before being subjected to lysis. The bar graph shows the relative levels of pAkt (±SD) (n = 7 for each). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (D) Western blots for p110α, p110β, pAktS473, pAktT308, and vinculin (as a loading control) in cell lysates prepared from MMTV-NeuT tumor cells carrying p110αL/L or p110βL/L. All cells were treated in vitro with AdGFP or AdCre and starved without growth factors before being subjected to lysis. The bar graph shows the relative levels of pAkt (±SD) (n = 9 for each). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (E) Schematic diagram illustrating a competition model in which the competitive occupancy by p110α and p110β—the two PI3K isoforms with low and high lipid kinase activities, respectively—determines the overall PI3K activity associated with ligand-bound, activated RTKs. Ablation of p110α is expected to result in greatly reduced PI3K activity. Conversely, loss of p110β liberates binding sites on receptors, which can then be occupied by the more active p110α, consequently increasing signal output. (F) Representative Western blots of MT immunoprecipitates of the indicated lysates are shown. The bar graph represents the relative amount of p110α associated with MT immunoprecipitates in the indicated lysates (n = 7, ±SD). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (G) Representative Western blots of HER3 immunoprecipitates of the indicated lysates are shown. The bar graph represents the relative amount of p110α associated with HER3 immunoprecipitates in the indicated lysates (n = 12, ±SD). (*) P < 0.05; (**) P < 0.005 (Student's t-test).
Given previous reports that p110α has much higher specific lipid kinase activity than p110β (Beeton et al. 2000; Knight et al. 2006; Zhao et al. 2006), one would predict that the level of receptor-associated lipid kinase activity should reach a maximum level if all RTKs were bound with p110α. Conversely, if most RTKs were associated with p110β isoform, the PI3K activity would be much reduced (Fig. 4E). We therefore hypothesize that the occupancy of receptors by the less active p110β titrates the levels of the more active p110α that can associate with receptors and thus modulates the overall lipid kinase activity and the amount of PIP3 production in response to RTK activation (Fig. 4E). Thus, loss of p110β liberates binding sites on activated receptors that can then be occupied by the more active p110α, consequently increasing signal output (Fig. 4E). To test this hypothesis, we assessed the amount of p110α bound to MT or HER2/HER3 complexes in the absence of p110β. Indeed, the abundance of p110α associated with MT or HER3 was increased in cells lacking p110β (Fig. 4F,G).
To further corroborate the suppressive effects of p110β, we stably introduced MT or NeuT into wild-type mouse embryonic fibroblasts (MEFs), p110β knockout (βKO) MEFs, βKO MEFs with exogenous expression of p110β (βKO +β), and βKO MEFs with exogenous expression of kinase-dead p110β-K805R (βKO +KR) (Supplemental Fig. 4G). These cells were starved to emphasize the MT- or NeuT-driven pAKT and eliminate other confounding growth factor signals. Our results show that, while ablation of p110β results in increased pAKT levels in MEFs expressing MT or NeuT, overexpression of either wild-type (βKO +β) or kinase-dead (βKO +KR) versions of p110β markedly reduced pAKT levels in these cells (Supplemental Fig. 4G). We further show that the reduced pAKT levels are associated with increased binding of p110β to MT or HER3 in cells expressing wild-type or KR p110β (Supplemental Fig. 4G). Together, these data are consistent with our model that the effect of p110β loss on MT- or HER2/Neu-driven tumorigenesis may be due to removal of the less active p110β from competition with the more active p110α for binding sites on the oncogenic MT or HER2/Neu receptor. This results in increased PI3K signaling and in turn leads to increased oncogenic transformation.
Interestingly, however, the effect of increased p110α binding in the absence of p110β is relatively more subtle in the HER2/Neu model than in the MT model. Our hypothesis predicts that, as the number of binding sites for PI3K on the activated receptor increases, the effect of p110β ablation will decrease (see Supplemental Fig. 4H for a schematic diagram explaining the model). Specifically, our model suggests that, where there are relatively small numbers of activated receptors capable of binding p85, we would expect to see an increase in receptor-associated PI3K activity and that the magnitude of the increase would be determined by the relative numbers of p110α and p110β and the relative difference in their specific PI3K activities (Supplemental Fig. 4H, left panel). At the other extreme, if there are an infinite number of active receptors that “soak up” all of the p110 molecules, leaving no free p110α to compensate when p110β is ablated, the result of p110β ablation is reduced PI3K activity. This circumstance would be similar to the situation presented by expression of a kinase-dead p110β (Ciraolo et al. 2008) or inactivation of p110β by inhibitors (see below) and would be predicted to give a modest decrease in receptor-associated PI3K activity (Supplemental Fig. 4H, right panel). For most receptors with intermediate abundance, the effect of ablation of p110β would be expected to result in changes varying from a modest increase to a modest decrease in receptor-associated PI3K activity (Supplemental Fig. 4H, middle panel). This model predicts the relatively large effects seen in the MT model, since there are a very limited number of MT molecules per cell that are capable of binding PI3K (Bolen et al. 1987; Morrison et al. 1988), while the transgenic MMTV-HER2/Neu cells that overexpress active HER2/Neu receptors (Siegel et al. 1999) would be expected to exhibit more modest effects upon p110β ablation.
The effects of p110α or p110β loss on growth factor signaling in normal primary mammary epithelial cells
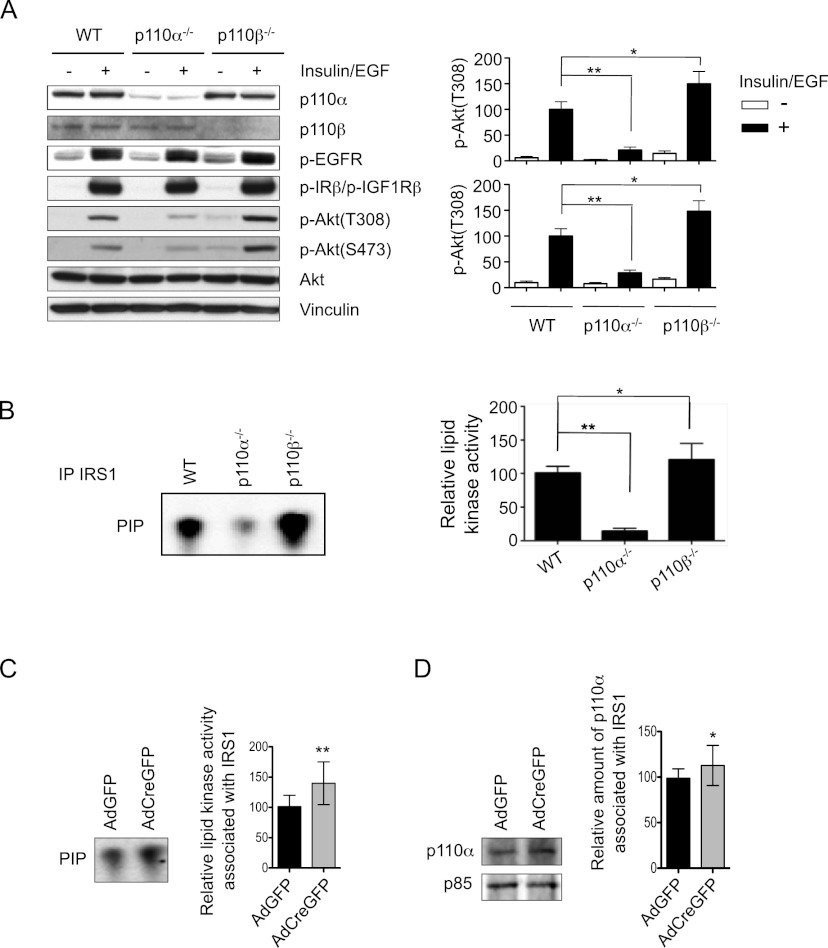
Given the competition model that we proposed for p110 isoforms in oncogenic receptor-mediated signaling and our initial observation of the effects of p110α or p110β loss in normal mammary gland development, we hypothesized that these isoforms might also play similar roles in mediating certain growth factor signaling in normal MMECs. To test this hypothesis, we isolated MMECs from mammary glands of wild-type and floxed p110α or p110β mice and treated these cells with AdCre to ablate p110α or p110β. To determine the effects of p110α or p110β ablation on signaling in response to growth factor stimulation and subsequent RTK activation, these MMECs were starved and then stimulated with insulin and EGF, the main growth factors in MMEC culture medium. Western blot analyses showed robust phosphorylation of endogenous EGFR and IR/IGF1R in all MMECs in response to insulin/EGF stimulation (Fig. 5A). As expected, pAkt levels (Fig. 5A) as well as the amount of lipid kinase activity that can be immunoprecipitated with IRS1 (Fig. 5B) in response to insulin/EGF were greatly reduced in cells ablated for p110α, suggesting that p110α is the major PI3K isoform mediating growth factor signaling in MMECs. In contrast, pAkt levels were increased in cells ablated for p110β (Fig. 5A), and increased lipid kinase activity could be immunoprecipitated with IRS1 in p110β-null cells (Fig. 5B). To further test this intriguing finding, we carried out additional experiments with MMECs isolated from floxed p110β mice and treated them with AdCreGFP or AdGFP. Cells were starved and then stimulated with insulin before being subjected to immunoprecipitation with IRS. Consistently, both the lipid kinase activity and the amount of p110α associated with IRS were significantly increased (Fig. 5C,D). Consistent with our competition model of PI3K isoforms in RTK-mediated signaling described above (Fig. 4E), these results identify p110α as the primary PI3K isoform in normal MMECs responding to insulin and EGF stimulation and support a regulatory role for p110β in fine-tuning the strength of PI3K signaling downstream from these growth factors. Our model presupposes that the levels of p110α and p110β are roughly comparable in MMECs. Using recombinant proteins as standards, we determined that the ratio of p110α:p110β in MMECs is ∼60:40 (Supplemental Fig. 5) These data thus provide potential mechanistic insights for the impairment of mammary gland development in the absence of p110α and the moderately hypermorphic mammary gland phenotype observed in the absence of p110β.
Figure 5.
Effects of p110α or p110β ablation on growth factor signaling in MMECs. (A) Western blot analyses of the phosphorylation levels of EGFR, IRβ/IGF1Rβ, and Akt in AdCre-treated wild-type (WT), p110αL/L, or p110βL/L MMECs in response to insulin and EFG stimulation after ablation of p110α or p110β. The bar graphs represent the mean ± SD of pAkt(Thr308) and pAkt(Ser473) (n = 4). (*) P < 0.05; (**) P < 0.005 (Student's t-test). (B) Relative lipid kinase activity associated with IRS1 immunoprecipitates in AdCre-treated wild-type (WT), p110αL/L, or p110βL/L MMECs in the presence of insulin and EGF. The bar graph represents the mean ± SD of the relative lipid kinase activity (n = 6). (*) P ≤ 0.05; (**) P < 0.005 (Student's t-test). (C) Relative lipid kinase activity associated with IRS1 immunoprecipitates of MMECs isolated from p110βL/L mice and treated in vitro with AdGFP and AdCreGFP as indicated. Cells were starved for 4 h before insulin stimulation. Bars represent mean ± SD (n = 12). (**) P < 0.005. (D) Relative amounts of p110α associated with IRS1 immunoprecipitates in MMECs isolated from p110βL/L mice and treated in vitro with AdGFP and AdCreGFP as indicated. Cells were starved for 4 h before insulin stimulation. Bars represent mean ± SD (n = 14). (*) P < 0.05
The effects of selective pharmacological inhibition of p110α or p110β on oncogenic receptor-driven PI3K signaling and tumorigenesis
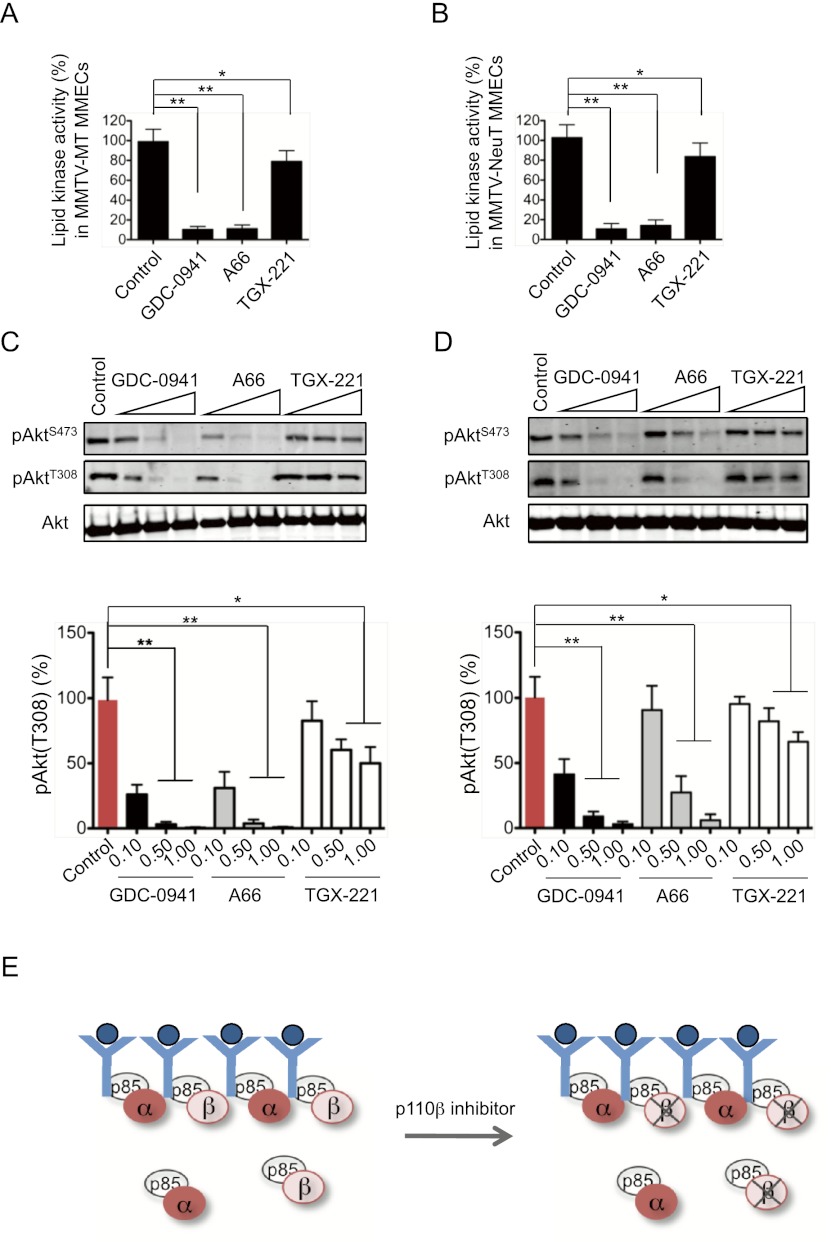
Since clinical efforts targeting PI3K in cancer involve inhibition of kinase activities rather than ablation of kinase proteins, it is important to examine the effects of specific pharmacological inactivation of the p110α or p110β isoform on oncogenic receptor-driven PI3K signaling and tumorigenesis. In particular, we wanted to determine whether there is a difference between p110β inhibition as opposed to p110β deletion on oncogenic signals driven by MT or HER2/Neu. To investigate this, we isolated primary mammary tumor cells from tumor-bearing MMTV-MT or MMTV-NeuT mice and subjected these cells to inhibitor studies using GDC-0941, a pan-PI3K inhibitor (Raynaud et al. 2009); A66, a p110α-selective inhibitor (Sun et al. 2010); and TGX-221, a p110β-selective inhibitor (Jackson et al. 2005). We first determined that the amounts of p110α or p110β activity associated with the MT/p85 or NeuT/HER3/p85 complexes were not altered when cells were treated with these PI3K inhibitors (Supplemental Fig. 6A,B). This implies that p110α or p110β remains associated with the receptor complexes despite being inactivated. As expected, both pan-PI3K and p110α-selective inhibitors dramatically reduced lipid kinase activity and pAkt in MT and NeuT tumor cells (Fig. 6A–D). Our model predicts that there would be no compensatory effects upon using p110β inhibitors, since the inactivated p110β is still able to compete with p110α for binding sites on oncogenes or receptors (Fig. 6E). Indeed, TGX-221 treatment reduced both lipid kinase activity and pAkt levels in these cells, albeit to a much lesser extent than did GDC-0941 or A66 (Fig. 6A–D). To confirm the full inhibitory activity of TGX-221 against p110β, we tested TGX-221 in p110α knockout cells and showed that it fully blocked Akt activation (Supplemental Fig. 6C).
Figure 6.
Effects of PI3K inhibitors on lipid kinase activity and pAkt abundance. (A) Lipid kinase activity associated with MT immunoprecipitates in MT tumor-derived MMECs treated with various PI3K inhibitors (GDC-0941, 0.1 μM; A66, 0.1 μM; and TGX-221, 0.25 μM) as indicated following growth factor starvation. The bars represent relative lipid kinase activity ± SD (n = 6). (*) P < 0.05; (**) P < 0.001 (Student's t-test). (B) Lipid kinase activity associated with HER3 immunoprecipitates in NeuT tumor-derived MMECs treated with various PI3K inhibitors (GDC-0941, 0.1 μM; A66, 0.1 μM; and TGX-221, 0.25 μM) as indicated. The bar graph represents relative lipid kinase activity ± SD (n = 6). (*) P < 0.05; (**) P < 0.001 (Student's t-test). (C) Western blot analyses of pAkt in MMTV-MT tumor-derived cells treated with various PI3K inhibitors (in micromolar) as indicated following growth factor starvation. The bar graphs show the relative amounts of pAkt(T308) ± SD (n = 6). (*) P < 0.01; (**) P < 0.001 (Student's t-test). (D) Western blot analyses of pAkt in MMTV-NeuT tumor-derived cells treated with various PI3K inhibitors (in micromolar) as indicated. The bar graphs show the relative amounts of pAkt(T308) ± SD (n = 6). (*) P < 0.01; (**) P < 0.001 (Student's t-test). (E) Schematic diagram illustrating that p110β remains associated with the receptor complex in the presence of a p110β inhibitor, thus blocking the increased p110α binding and increased signal output observed in the p110β knockout scenario.
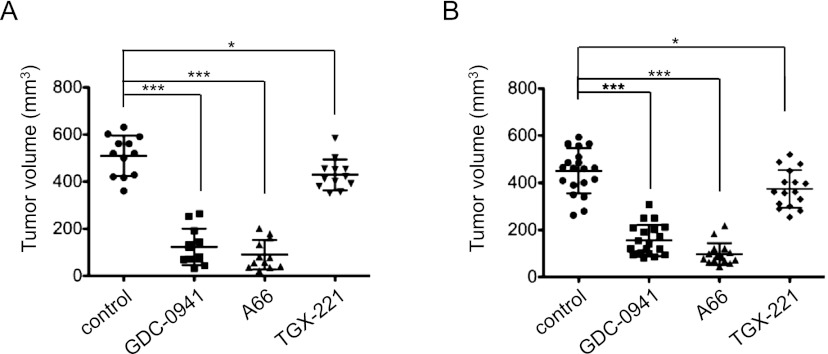
To further assess isoform-specific pharmacological inhibition of PI3K in vivo, we performed mammary fat pad transplantation of athymic mice with primary tumors arising from transgenic mice with MMTV-MT and MMTV-NeuT genotypes. Transplanted tumor-bearing mice were treated with vehicle, GDC-0941, A66, or TGX-221. All three inhibitors affected the growth of both MT and NeuT tumors in a manner consistent with their effects in vitro, with TGX-221 producing only a small reduction, while both the pan-PI3K and p110α inhibitors dramatically impaired tumor growth (Fig. 7A,B).
Figure 7.
Effects of PI3K inhibitors on the growth of orthotopic tumor transplants derived from MMTV-MT or MMTV-NeuT tumors. (A) Tumor volumes of MMTV-MT tumor transplants treated with various PI3K inhibitors as indicated (n = 12 per group). (*) P < 0.01; (***) P < 0.0001 (Student's t-test). (B) Tumor volumes of MMTV-NeuT tumor transplants treated with PI3K inhibitors as indicated (control, n = 20; GDC-0941, n = 20; A66, n = 16; and TGX221, n = 16). (*) P < 0.01; (***) P < 0.0001 (Student's t-test).
Discussion
The key role of PI3K as an effector of most oncogenic RTKs has been known for some time, and clinical trials are currently under way to test the effectiveness of pan-PI3K inhibitors in tumors featuring RTK activation. However, given the development of second-generation PI3K inhibitors targeting specific isoforms, it is important to determine the individual roles of the two ubiquitously expressed isoforms of PI3K, p110α and p110β, in tumors driven by activated RTKs. Using both genetic and pharmacological approaches, we demonstrate for the first time that p110α is the primary PI3K isoform responsible for oncogenic RTK signaling and tumorigenesis in mammary epithelium. Our data are consistent with a model in which the high specific lipid kinase activity of p110α isoform underlies its functional importance.
The most intriguing and surprising finding was the observation of significant enhancement of both mammary ductal side branching and mammary tumorigensis driven by MT or HER2/Neu upon genetic ablation of p110β. Detailed biochemical analyses revealed that p110β down-modulates PI3K activity and Akt phosphorylation in primary mammary tumor cells isolated from MT- and HER2/Neu-driven tumors. A potential mechanism underlying this novel negative role of p110β in RTK signaling comprises a competition model in which the less active p110β competes with the more active p110α for receptor-binding sites, thereby down-tuning the level of lipid kinase activity associated with receptors. This model is supported by both genetic and pharmacological studies and provides one mechanism underlying the divergent roles of p110α and p110β in mammary gland tumorigenesis.
Notably, our model makes a striking prediction in the context of heterozygous loss of p110α: The loss of 50% of p110α should increase the ratio of p110β to p110α, reduce the amount of p110α available for binding to activated oncogenes or receptors, and significantly reduce the oncogenic transformation of MT or HER2/Neu in mammary glands (Figs. 2, 3). This is most obvious in the case of MT transformation, where heterozygous loss of p110α severely impairs MT-mediated tumorigenesis. Our model predicts that additional loss of a single copy of p110β should restore the ratio of p110α/p110β and hence restore MT-mediated transforming activity. This is exactly what is observed with doubly heterozygous deletion of p110α/p110β on MT-induced tumor formation (Supplemental Fig. 7).
It has long been known that the level of PIP3 is tightly regulated by the activities of two opposing enzymes, PI3K and PTEN lipid phosphatase, which act as “on/off” switches, respectively. Our current study suggests a potential novel second mechanism by which the overall level of PI3K activity can be fine-tuned through the relative occupancy of binding sites on activated RTKs by two PI3K isoforms with different lipid kinase activities. While PTEN negatively regulates PIP3 levels via dephosphorylation, our data show that p110β can modulate the generation of PIP3 by reducing the total lipid kinase activity bound to activated receptors, thus allowing the abundance of the critical second cellular messenger PIP3 to be precisely regulated by these two complementary mechanisms.
In addition, however, p110β plays a necessary and positive role in certain tumors driven by PTEN loss (Jia et al. 2008; Torbett et al. 2008; Wee et al. 2008). Similar genetic behavior has previously been noted for the gene encoding the p85 adapter protein (Mauvais-Jarvis et al. 2002; Ueki et al. 2002, 2003; Taniguchi et al. 2006); p85 has a dual role as a negative regulator of the ground-state activity of p110 catalytic kinase subunits while also functioning to dock the kinases to activating receptors. In the case of p110β, its negative effect lies at least in part in its role as a weak kinase competing with the more potent p110α isoform for binding sites on RTKs. Its positive function in certain tumors arising from PTEN loss may arise from a requirement for p110β in mediating signals downstream from particular activating inputs such as certain GPCRs or integrins (Hirsch et al. 2000; Guillermet-Guibert et al. 2008; Jia et al. 2008; Ciraolo et al. 2010; Martin et al. 2010). Thus, it has been hypothesized that, in at least some PTEN loss-driven tumors, the signal activating PI3K comes from a GPCR. There is also a second potential mechanism underlying p110β's positive role in PTEN-null tumors: It has been proposed that p110β is responsible for the basal level of lipid kinase activity observed in cells (Knight et al. 2006; Dbouk et al. 2010). In some PTEN-null tumors, the signal amplified by PTEN loss may arise from this baseline activity (Jia et al. 2008). In fact, a recent study that revealed the structure of the p110β/p85 complex in addition elucidated a distinctive mode of regulation of p110β by p85 that contributes to the unique properties of p110β (Dbouk et al. 2010; Vogt 2011; Zhang et al. 2011). Since PTEN loss simply removes a “brake” from the PI3K pathway, the key to understanding the roles of the p110 isoforms in transformation induced by loss of PTEN lies in determining the genetic/tissue-specific signaling inputs activating the PI3K pathway in each context
To date, the array of PI3K inhibitors that are in preclinical and clinical development consists largely of nonisoform selective (pan) inhibitors, and patients with HER2-positive breast cancer are candidates for such PI3K-targeted therapy. However, isoform-selective molecules are emerging in the clinic that, because of their increased specificity, offer considerable promise of both efficacy and safety advantages over pan-PI3K inhibitors. Our isoform-specific inhibitor study suggests that, while there are no tumor-stimulating effects of inhibiting p110β, p110β will generally not be an effective target in treating oncogenic RTK-dependent tumors. Notably, we demonstrate that a selective p110α inhibitor under development for clinical use effectively blocks both oncogenic HER2-driven signaling and tumor growth, suggesting the utility of p110α-specific inhibitors either alone or likely in combination with HER2-targeted therapies such as trastuzumab as therapeutic agents for HER2-positive disease.
Materials and methods
Animals
MMTV-Cre (Wagner et al. 1997), p110αL/L (Zhao et al. 2006), and p110βL/L (Jia et al. 2008) mice were backcrossed 10 generations to the FVB/N background before being subjected to this study. MMTV-MT (Guy et al. 1992) and NIC (Ursini-Siegel et al. 2008) were in the FVB/N background. MMTV-NeuT mice were in the Balb/c background (Lucchini et al. 1992). NCrNu female mice (Taconic) were used for orthotopic transplantation. All animals were housed and treated in accordance with protocols approved by the Institutional Animal Care and Use Committees of Dana-Farber Cancer Institute and Harvard Medical School.
Biochemical analyses
MMECs prepared from wild-type mammary glands or tumors were starved either 4 h or overnight in DMEM/F12 medium (Invitrogen) before being lysed for biochemical analyses in 1% nonidet P-40 lysis buffer containing a protease inhibitor cocktail (Roche). After centrifugation, cleared lysates were separated on SDS-PAGE gels and transferred to nitrocellulose membranes. Western blot assays were performed with primary antibodies against MT (Pallas et al. 1986), phospho-AktS473, phospho-AktT308, Akt, p110α, p110β, p-EGFR, and p-IRβ/p-IGF1Rβ (all from Cell Signaling Technology), and pan-p85, HER2, HER3, and IRS1 (all from Millipore). Immunoprecipitations were set up as follows: Lysates containing 1 mg of total protein from MT tumors were incubated with 10 μg of MT antibody (rat monoclonal antibody, GeneTex), and lysates containing 500 μg of total protein from NeuT tumors were incubated with 5 μg of antibodies to HER2 or HER3. Lysates containing 300 μg of total protein from wild-type mammary glands were incubated with 2 μg of anti-IRS1. All immunoprecipitations were carried out overnight, and protein A/G sepharose (30 μL of 50% bead slurry) (Santa Cruz Biotechnology) was added for 2 h. Beads were then collected by centrifugation, followed by three washes with lysis buffer. The resulting immunoprecipitates were either subjected to Western blot analysis or used in a lipid kinase assay.
Mammary gland whole mounts and quantitation of mammary gland branching
The fourth set of mammary glands was harvested and mounted on glass slides, fixed in an ethanol/glacial acetic acid mixture, rehydrated, and stained with carmine red followed by dehydration and a final clearing in toluene as described previously (Wiseman et al. 2003). Whole mounts of the fourth set of mammary glands from 12-wk-old MMTV-Cre and MMTV-Cre/p110β animals were photographed using a Nikon SMZ-U microscope (2× magnification), and ductal branch points were marked and counted by hand. Five fields were counted per sample.
Mammary epithelial cell preparation, culture, adenovirus infection, and primary transplantation assays
Mammary glands obtained from virgin wild-type or transgenic females were digested in medium containing 160 U/mL collagenase, 1.25 U/mL dispase, 100 U/mL hyaluroindase, 100 μg/mL gentamicin, and 100 U/mL penicillin/streptomycin for 4 h at 37°C followed by multiple spin-break centrifugations in basal medium. The collected MMECs were plated in growth medium containing insulin and EGF, subjected to two rounds of infection with either AdCreGFP or AdGFP (GeneTransfer Vector Core), and FACS-sorted 48 h post-infection. For transplantation assays, cultured primary MMECs were trypsinized to obtain single cells. For mammary duct outgrowth, MMECs (1 × 105 cells) were injected into the cleared inguinal mammary glands of 3-wk-old recipient mice as described (Deome et al. 1959). The transplanted glands were excised from mice 8 wk post-transplantation and stained with carmine red (Sigma) for whole-mount analysis. For tumor growth, cells (2 × 106 MT or NeuT tumor cells) were resuspended in phosphate-buffered saline (PBS) containing 25% Matrigel (BD Biosciences) and injected into the inguinal mammary fat pads of nude mice.
Tumorigenesis studies
Nulliparous mice from different cohorts were subjected to regular palpation, and tumor onset was determined upon palpation of the first tumor lesion in each individual mouse. To determine the overall tumor burden of an individual MT transgenic animal, all mammary glands of each mouse were collected, and their cumulative wet weight was measured. The tumor growth was determined by the difference between the weight of a tumor-bearing mammary gland and the weight of a normal mammary gland. For the NIC model, the total number of tumors as well as their cumulative volume were determined for each animal at the study endpoint, which was 6 wk after detection of the first palpable tumor. Tumor volumes were measured twice a week with calipers and calculated by the following formula: (4/3π) × (length/2) × (width/2)2 (Wapnir et al. 1996).
Quantification of premalignant lesions
Whole mounts of the fourth set of mammary glands of 14-wk-old NIC, NIC/p110αL/L, and NIC/p110βL/L animals were photographed using a Nikon SMZ-U microscope (2× magnification), and premalignant, hyperplastic nodules were marked and counted.
Immunohistochemical staining for Ki67 in mammary gland tissue
The fourth pair of mammary glands of NIC, NIC/p110αL/L, and NIC/p110βL/L animals was isolated and mounted on glass slides before fixation in 10% buffered formalin. Sections were then stained according to routine protocol. In brief, sections were dewaxed and rehydrated, followed by antigen retrieval by boiling in citrate buffer. Ki67 antibody (Vector Laboratories) was added overnight, and positive nuclei were detected by diaminobenziidine (Sigma). Sections were then counterstained with Mayer's hematoxylin.
Lipid kinase assay
Lipid kinase assays were performed as previously described (Zhao et al. 2006). Briefly, MT or HER3 immunoprecipitates of freshly prepared cell lysates were subjected to an in vitro kinase assay using phosphatidylinositol (PI) (Avanti Polar Lipids) as a substrate. The phosphorylated lipids were resolved by thin-layer chromatography (TLC) overnight. TLC plates were then exposed on a storage phosphor screen (Molecular Dynamics), which was scanned using a STORM imaging machine. The radioactive product of the reaction, phosphatidylinositol-3-phosphate (PIP), was quantified by ImageQuant software.
Inhibitor studies in vitro
Primary MT tumor- or NeuT tumor-derived MMECs were treated with the pan-PI3K inhibitor GDC-0941 (Axon MedChem), the p110α-specific inhibitor A66 (Selleck), and p110β-specific inhibitor TGX221 (Cayman Chemical) at various concentrations as indicated in the text in vitro. To inhibit HER2-mediated phosphorylation of HER3, we used lapatinib at the indicated doses on primary NeuT tumor-derived MMECs that were starved overnight. We examined lipid kinase activity, pAkt levels, and interaction with PI3K isoforms by immunoprecipitations of untreated versus lapatinib-treated cells.
Inhibitor studies in vivo
For in vivo studies, 2 × 106 MT or NeuT MMECs were resuspended in 30% Matrigel and subcutaneously injected into NCrNu recipient animals. Treatment was started the following day and continued once daily for 25 d. GDC-0941 was administered by gavage at 125 mg/kg in 10% DMSO, 5% Tween20, and 85% water. A66 was administered by intraperitoneal injection at 100 mg/kg in 10% DMSO, 5% Tween20, and 85% water. TGX221 was injected adjacent to the tumor site at 0.2 mg per injection in 10% DMSO and 90% water.
Acknowledgments
We thank Dr. Linda K. Clayton for critical reading and suggestions. We thank Dr. Roderick Bronson and the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core for histopathological analyses. This work was supported by NIH grants CA030002 (to T.M.R.), CA050661 (to T.M.R.), CA134502 (to J.J.Z.), P50CA089393-08S1 (to J.J.Z.), and CA148164-01 (to T.M.R. and J.J.Z.); Stand Up to Cancer Dream Team Translational Research Grant; a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209) (T.M.R. and J.J.Z.); the Department of Defense (BC051565 to J.J.Z.); the V Foundation (J.J.Z.); the Claudia Adams Barr Program (J.J.Z.); CIHR (MOP 89751 to W.M.); Terry Fox Team Grant (NCIC 02002 to W.J.M.); a CRC Chair in Molecular Oncology (W.M.); and a DOD studentship (W81XWH-09-01-0079 to T.R.). In compliance with Harvard Medical School guidelines, we disclose that T.M.R. and J.J.Z. are consultants for Novartis Pharmaceuticals, Inc.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.191973.112.
References
- Beeton CA, Chance EM, Foukas LC, Shepherd PR 2000. Comparison of the kinetic properties of the lipid- and protein-kinase activities of the p110α and p110β catalytic subunits of class-Ia phosphoinositide 3-kinases. Biochem J 350: 353–359 [PMC free article] [PubMed] [Google Scholar]
- Bolen JB, DeSeau V, O'Shaughnessy J, Amini S 1987. Analysis of middle tumor antigen and pp60c-src interactions in polyomavirus-transformed rat cells. J Virol 61: 3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, Costa C, Azzolino O, Gonella C, Rubinetto C, Wu H, et al. 2008. Phosphoinositide 3-kinase p110β activity: Key role in metabolism and mammary gland cancer but not development. Sci Signal 1: ra3 doi: 10.1126/scisignal.1161577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciraolo E, Morello F, Hobbs RM, Wolf F, Marone R, Iezzi M, Lu X, Mengozzi G, Altruda F, Sorba G, et al. 2010. Essential role of the p110β subunit of phosphoinositide 3-OH kinase in male fertility. Mol Biol Cell 21: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk HA, Pang H, Fiser A, Backer JM 2010. A biochemical mechanism for the oncogenic potential of the p110β catalytic subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci 107: 19897–19902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ Jr, Bern HA, Blair PB 1959. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res 19: 515–520 [PubMed] [Google Scholar]
- Engelman JA 2009. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer 9: 550–562 [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7: 606–619 [DOI] [PubMed] [Google Scholar]
- Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B 2006. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature 441: 366–370 [DOI] [PubMed] [Google Scholar]
- Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, Salpekar A, Pearce W, Meek S, Millan J, Cutillas PR, et al. 2008. Angiogenesis selectively requires the p110α isoform of PI3K to control endothelial cell migration. Nature 453: 662–666 [DOI] [PubMed] [Google Scholar]
- Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K, Vanhaesebroeck B 2008. The p110β isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110γ. Proc Natl Acad Sci 105: 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J 2007. Binding of ras to phosphoinositide 3-kinase p110α is required for ras-driven tumorigenesis in mice. Cell 129: 957–968 [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol 12: 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP 2000. Central role for G protein-coupled phosphoinositide 3-kinase γ in inflammation. Science 287: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Ichaso N, Dilworth SM 2001. Cell transformation by the middle T-antigen of polyoma virus. Oncogene 20: 7908–7916 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, et al. 2005. PI 3-kinase p110β: A new target for antithrombotic therapy. Nat Med 11: 507–514 [DOI] [PubMed] [Google Scholar]
- Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, et al. 2008. Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature 454: 776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, et al. 2006. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell 125: 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163: 2113–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng H, Roberts TM, Zhao JJ 2009. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8: 627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini F, Sacco MG, Hu N, Villa A, Brown J, Cesano L, Mangiarini L, Rindi G, Kindl S, Sessa F, et al. 1992. Early and multifocal tumors in breast, salivary, harderian and epididymal tissues developed in MMTY-Neu transgenic mice. Cancer Lett 64: 203–209 [DOI] [PubMed] [Google Scholar]
- Martin V, Guillermet-Guibert J, Chicanne G, Cabou C, Jandrot-Perrus M, Plantavid M, Vanhaesebroeck B, Payrastre B, Gratacap MP 2010. Deletion of the p110β isoform of phosphoinositide 3-kinase in platelets reveals its central role in Akt activation and thrombus formation in vitro and in vivo. Blood 115: 2008–2013 [DOI] [PubMed] [Google Scholar]
- Mauvais-Jarvis F, Ueki K, Fruman DA, Hirshman MF, Sakamoto K, Goodyear LJ, Iannacone M, Accili D, Cantley LC, Kahn CR 2002. Reduced expression of the murine p85α subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J Clin Invest 109: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK, Browning PJ, White MF, Roberts TM 1988. Tyrosine phosphorylations in vivo associated with v-fms transformation. Mol Cell Biol 8: 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Schley C, Mahoney M, Harlow E, Schaffhausen BS, Roberts TM 1986. Polyomavirus small t antigen: Overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J Virol 60: 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R 2004. Human cancer, PTEN and the PI-3 kinase pathway. Semin Cell Dev Biol 15: 171–176 [DOI] [PubMed] [Google Scholar]
- Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, Folkes A, Gowan S, De Haven Brandon A, Di Stefano F, et al. 2009. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: From PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther 8: 1725–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen BS, Roberts TM 2009. Lessons from polyoma middle T antigen on signaling and transformation: A DNA tumor virus contribution to the war on cancer. Virology 384: 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Ryan ED, Cardiff RD, Muller WJ 1999. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: Implications for human breast cancer. EMBO J 18: 2149–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopasakis VR, Liu P, Suzuki R, Kondo T, Winnay J, Tran TT, Asano T, Smyth G, Sajan MP, Farese RV, et al. 2010. Specific roles of the p110α isoform of phosphatidylinsositol 3-kinase in hepatic insulin signaling and metabolic regulation. Cell Metab 11: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK 2010. Cancer-derived mutations in the regulatory subunit p85α of phosphoinositide 3-kinase function through the catalytic subunit p110α. Proc Natl Acad Sci 107: 15547–15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Tran TT, Kondo T, Luo J, Ueki K, Cantley LC, Kahn CR 2006. Phosphoinositide 3-kinase regulatory subunit p85α suppresses insulin action via positive regulation of PTEN. Proc Natl Acad Sci 103: 12093–12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torbett NE, Luna-Moran A, Knight ZA, Houk A, Moasser M, Weiss W, Shokat KM, Stokoe D 2008. A chemical screen in diverse breast cancer cell lines reveals genetic enhancers and suppressors of sensitivity to PI3K isoform-selective inhibition. Biochem J 415: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Yballe CM, Brachmann SM, Vicent D, Watt JM, Kahn CR, Cantley LC 2002. Increased insulin sensitivity in mice lacking p85β subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci 99: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Fruman DA, Yballe CM, Fasshauer M, Klein J, Asano T, Cantley LC, Kahn CR 2003. Positive and negative roles of p85α and p85β regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J Biol Chem 278: 48453–48466 [DOI] [PubMed] [Google Scholar]
- Ursini-Siegel J, Hardy WR, Zuo D, Lam SH, Sanguin-Gendreau V, Cardiff RD, Pawson T, Muller WJ 2008. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. EMBO J 27: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B 2010. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 11: 329–341 [DOI] [PubMed] [Google Scholar]
- Vogt PK 2011. PI3K p110β: More tightly controlled or constitutively active? Mol Cell 41: 499–501 [DOI] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L 1997. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res 25: 4323–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapnir IL, Wartenberg DE, Greco RS 1996. Three dimensional staging of breast cancer. Breast Cancer Res Treat 41: 15–19 [DOI] [PubMed] [Google Scholar]
- Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C 2008. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci 105: 13057–13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM 1985. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature 315: 239–242 [DOI] [PubMed] [Google Scholar]
- Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ, Soloway P, Itohara S, Werb Z 2003. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol 162: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, Stephens LR, Williams RL 2011. Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell 41: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM 2006. The p110α isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc Natl Acad Sci 103: 16296–16300 [DOI] [PMC free article] [PubMed] [Google Scholar]