Master transcription factors play key roles in hematopoiesis by regulating cell fate decisions and terminal cell maturation. However, the mechanisms that integrate activity of transcriptional networks with cell signaling inputs are incompletely understood. In this study, Cantor and colleagues find that RUNX1 is tyrosine phosphorylated by Src family kinases (SFKs) and that this inhibits RUNX1 activity in megakaryocyte and CD8 T-lymphocyte differentiation. In addition, the authors demonstrate that RUNX1 directly binds to the nonreceptor tyrosine phosphatase Shp2 (PTPN11), which dephosphorylates RUNX1. Thus, this study delineates novel molecular mechanisms by which RUNX1 activity is regulated during hematopoietic cell differentiation.
Keywords: RUNX1, tyrosine phosphorylation, c-Src, Shp2, megakaryocyte, T cell
Abstract
Hematopoietic development occurs in complex microenvironments and is influenced by key signaling events. Yet how these pathways communicate with master hematopoietic transcription factors to coordinate differentiation remains incompletely understood. The transcription factor RUNX1 plays essential roles in definitive hematopoietic stem cell (HSC) ontogeny, HSC maintenance, megakaryocyte (Mk) maturation, and lymphocyte differentiation. It is also the most frequent target of genetic alterations in human leukemia. Here, we report that RUNX1 is phosphorylated by Src family kinases (SFKs) and that this occurs on multiple tyrosine residues located within its negative regulatory DNA-binding and autoinhibitory domains. Retroviral transduction, chemical inhibitor, and genetic studies demonstrate a negative regulatory role of tyrosine phosphorylation on RUNX1 activity in Mk and CD8 T-cell differentiation. We also demonstrate that the nonreceptor tyrosine phosphatase Shp2 binds directly to RUNX1 and contributes to its dephosphorylation. Last, we show that RUNX1 tyrosine phosphorylation correlates with reduced GATA1 and enhanced SWI/SNF interactions. These findings link SFK and Shp2 signaling pathways to the regulation of RUNX1 activity in hematopoiesis via control of RUNX1 multiprotein complex assembly.
Master transcription factors play key roles in hematopoiesis by regulating cell fate decisions and terminal cell maturation (Orkin and Zon 2008). Many of these factors act in combinatorial self-stabilizing networks that reinforce selected cell identity programs while repressing alternate lineage choices (Graf and Enver 2009). At the same time, hematopoiesis occurs in complex microenvironments and is influenced by external signaling events. Yet how cell signaling pathways communicate with the transcriptional networks to modulate differentiation remains incompletely understood.
The transcription factor RUNX1 (previously called AML-1 and CBFα2) is an essential regulator of definitive hematopoietic stem cell (HSC) ontogeny, HSC self-renewal, megakaryocyte (Mk) maturation, and lymphocyte differentiation. It is a central component of a dynamic combinatorial transcription factor network that includes GATA1/2, FLI1, TAL/SCL, and AP1 (Elagib et al. 2003; Huang et al. 2009; Wilson et al. 2010; Palii et al. 2011; Pencovich et al. 2011; Tijssen et al. 2011). RUNX1−/− mice die between embryonic day 11.5 (E11.5) and E12.5 with CNS hemorrhage and complete failure of all definitive hematopoiesis (Okuda et al. 1996; Wang et al. 1996). The latter phenotype is due to a block in the emergence of the first definitive HSCs from hemogenic endothelium in the aorto–gonadal–mesonephros (AGM) region (North et al. 1999; Mukouyama et al. 2000; Chen et al. 2009; Kissa and Herbomel 2010). Partial RUNX1 deficiency leads to premature HSC emergence from the AGM (Cai et al. 2000; North et al. 2002) and altered stem/progenitor cell balance in adult mice (Sun and Downing 2004; Jacob et al. 2010). Inducible RUNX1 loss also results in impaired Mk and lymphocyte differentiation as well as a mild myeloproliferative disorder (Ichikawa et al. 2004; Growney et al. 2005).
In humans, acquired RUNX1 deficiency is an early initiating event in up to 30% of all human leukemias (for review, see Speck and Gilliland 2002). RUNX1 mutations are also a poor prognostic indicator in de novo myelodysplastic syndrome (MDS) and myeloproliferative neoplasms (MPNs) (Nakao et al. 2004; Bejar et al. 2011; Vainchenker et al. 2011). Germline mutations leading to RUNX1 haploinsufficiency cause familial platelet disorder with propensity to develop AML (FPD/AML), an autosomal dominant syndrome characterized by thrombocytopenia, platelet dysfunction, and an ∼35% lifetime risk of developing MDS/AML (Song et al. 1999; Owen et al. 2008). Altered RUNX1 expression also predisposes to lymphoma in mice (Wotton et al. 2002; Kundu et al. 2005). Thus, tight regulation of RUNX1 activity levels is critical for normal hematopoiesis.
RUNX1 contains a number of autoinhibitory domains (IDs) that control its function. A negative regulatory DNA-binding (NRDB) domain inhibits DNA association. This is relieved when RUNX1 physically interacts with CBFβ and/or ETS family transcription factors (Ogawa et al. 1993; Goetz et al. 2000). Likewise, an ID located C-terminal to the transcriptional activation domain (AD) dampens transcriptional activity (Kanno et al. 1998). The mechanism that relieves this autoinhibition is unknown.
Tyrosine phosphorylation plays critical roles in cellular signaling events, particularly those controlling proliferation in response to cytokine, cell–cell, and cell–matrix interactions. Moreover, protein tyrosine kinases and phosphatases are frequently dysregulated in MPNs and hematologic malignancies. Although tyrosine phosphorylation is typically described in the context of membrane receptors and cytoplasmic proteins, transcription factors and other nuclear proteins can be functionally modified by tyrosine phosphorylation. In the present study, we show that RUNX1 is tyrosine phosphorylated on its NRDB and ID domains by Src family kinases (SFKs) and that this negatively regulates RUNX1 activity in megakaryocytic and T-lymphocyte differentiation. We also provide evidence that the nonreceptor tyrosine phosphatase Shp2 contributes to dynamic RUNX1 tyrosine dephosphorylation and that tyrosine phosphorylation alters RUNX1 multiprotein complex formation.
Results
RUNX1 is tyrosine phosphorylated in megakaryoblastic cells and primary thymocytes
To further understand RUNX1 regulatory mechanisms, we previously purified RUNX1-containing multiprotein complexes from nuclear extracts of 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced L8057 murine megakaryoblastic cells using metabolic biotin tagging and streptavidin affinity chromatography and identified associated proteins by mass spectrometry (Huang et al. 2009). In addition to known interacting proteins such as CBFβ, GATA1, GATA2, TAL1/SCL, FLI1, Sin3A, PRMT1, PML, CDK6, Smad2, and SWI/SNF components, we reproducibly and specifically obtained peptides corresponding to c-Src tyrosine kinase and the nonreceptor tyrosine phosphatase Shp2 (also called Ptpn11) (Supplemental Fig. S1; Supplemental Table S1). These interactions were validated in subsequent experiments (discussed further below; see Fig. 6, below; Supplemental Fig. S2).
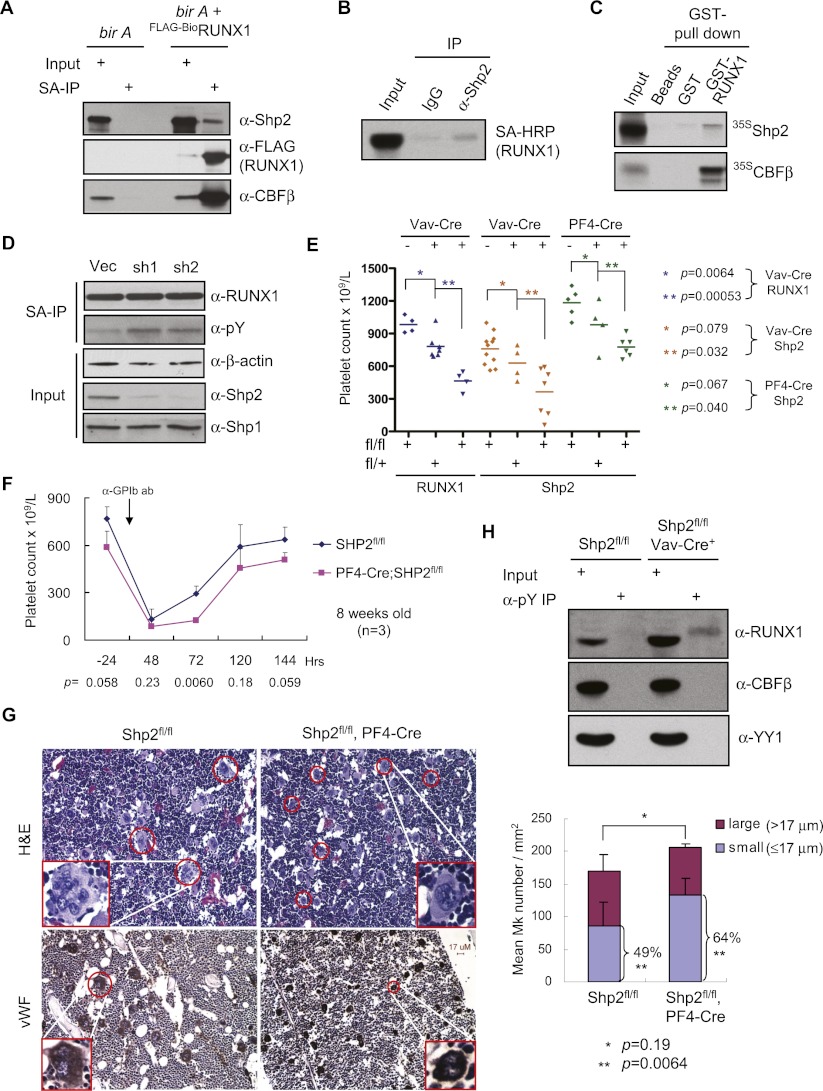
Figure 6.
Contribution of Shp2 to RUNX1 tyrosine dephosphorylation. (A) SA-IP of Flag-BioRUNX1 from TPA-induced L8057 cells. Western blots of the copurified material for Shp-2, RUNX1 (Flag), and CBFβ are shown; 1% of input is shown. (B) Co-IP of endogenous Shp2 and Flag-BioRUNX1 from TPA-induced L8057 cells. (IgG) Species-matched control antibody; 1.35% of input is shown. (C) GST pull-down of bacterially produced GST-RUNX1 and in vitro transcribed and translated [35S]-labeled Shp2 or CBF-β. Autoradiograms of the eluted material and 10% of the input are shown. (D) SA-IP and anti-pY Western blot of uninduced L8057 cells containing Flag-BioRUNX1 transduced with the empty vector or two independent lentiviral Shp2-targeting shRNA constructs. (E) Peripheral blood platelet counts of RUNX1fl/fl, Vav-Cre (7 wk old); Shp2fl/fl, Vav-Cre (6 wk old); and Shp2fl/fl, PF4-Cre (8 wk old) mice with different allele doses. Horizontal bars represent mean values. P-values are based on Student's one-tailed t-test. (F) Peripheral blood platelet counts of Shp2fl/fl or Shp2fl/fl, PF4-Cre 8-wk-old mice before and after anti-GPIbα antibody injection. n = 3 for both genotypes. P-values are indicated for each time point (Student's one-tailed t-test). (G, left) Hematoxylin/eosin stains of bone marrow from Shp2fl/fl or Shp2fl/fl, PF4-Cre mice obtained 72 h after anti-GPIbα antibody injection. Original magnification, 200×. Representative morphologically recognizable Mks are circled in red. (Insets) Enlarged representative Mks. (Bottom) von Willebrand factor (vWF) (Mk marker) in situ immunohistochemistry of bone marrow sections. (Right) Quantification of vWF+ cells per square millimeter is given. Small Mks are defined as having a maximum diameter of ≤17 μm (Long and Williams 1981). (H) RUNX1 tyrosine phosphorylation levels in thymocytes from 6-wk-old Shp2fl/fl, Vav-Cre or Shp2fl/fl littermates. Thymocyte extracts were incubated with anti-pY antibody-coupled beads. After washing, the bound beads were eluted using 100 mM phenyl phosphate. The eluates were examined by Western blot for RUNX1, CBFβ (negative control) and YY1 (negative control); 0.5% of the input is shown (50 μg of whole-cell lysate).
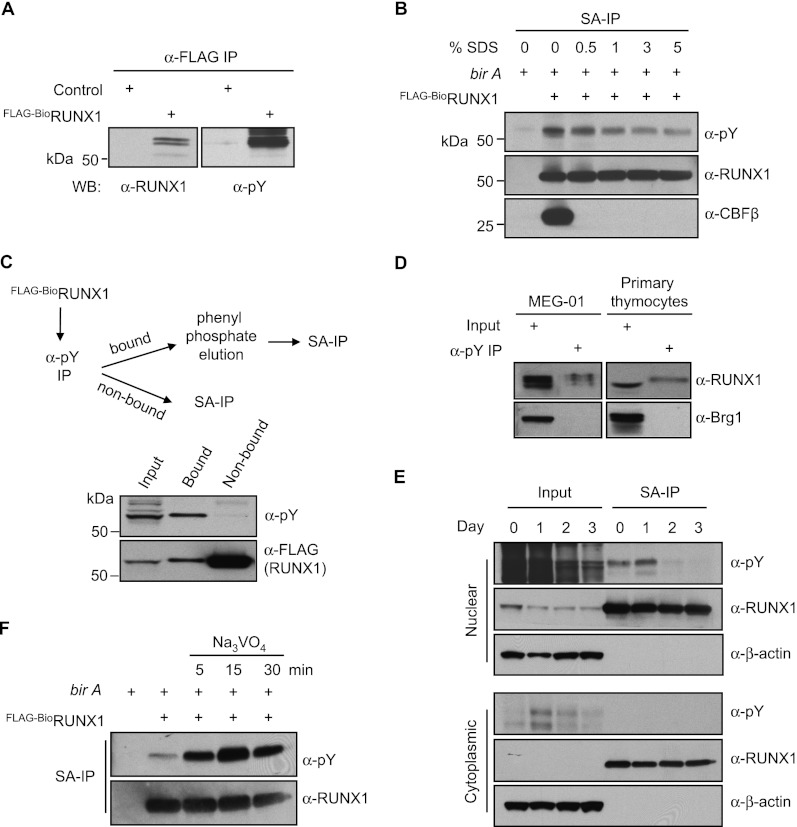
The physical association of RUNX1 with a tyrosine kinase and phosphatase led us to hypothesize that RUNX1 may be tyrosine phosphorylated itself. To test this, Flag-tagged and metabolically biotinylated RUNX1 (Flag-BioRUNX1) was immunoprecipitated from nuclear extracts of uninduced L8057 cells with an anti-Flag antibody and analyzed by Western blot using the pan anti-phosphotyrosine (pY) monoclonal antibody 4G10 (Millipore). This revealed bands migrating at the same molecular weight as Flag-BioRUNX1 (Fig. 1A). To exclude the possibility that these bands represent RUNX1-associated proteins, rather than RUNX1 itself, the experiment was repeated under denaturing conditions in which Flag-BioRUNX1 was streptavidin-precipitated in the presence of increasing concentrations of sodium dodecyl sulfate (SDS). As shown in Figure 1B, the anti-pY reactive band was retained with as much as 5% SDS, whereas the noncovalently bound protein CBFβ was completely dissociated with as little as 0.5% SDS. Quantitation using sequential anti-pY immunoprecipitation (IP) followed by streptavidin immunoprecipitation (SA-IP) revealed that up to ∼1%–10% of the nuclear Flag-BioRUNX1 is tyrosine phosphorylated in uninduced L8057 cells (Fig. 1C). Endogenous RUNX1 was also found to be tyrosine phosphorylated in human MEG-01 megakaryoblastic cells and primary murine thymocytes (Fig. 1D).
Figure 1.
RUNX1 tyrosine phosphorylation. (A) Anti-Flag IP from nuclear extracts of control (nontransfected) or Flag-BioRUNX1-expressing uninduced L8057 cells. Western blot of the purified material for RUNX1 (left) and phosphotyrosine (pY) (right) is shown. (B) SA-IP of nuclear extracts from uninduced L8057 cells expressing Flag-BioRUNX1 with and without birA (biotin ligase) in the presence of increasing concentrations of SDS. Western blot of the purified material is shown for pY, RUNX1, and CBFβ. (C) Quantitation of RUNX1 tyrosine phosphorylation levels. Nuclear extracts from uninduced L8057 cells containing Flag-BioRUNX1were incubated with α-pY antibody-conjugated agarose beads. After washing, the bound material was eluted with excess phenyl phosphate. SA-IP was performed on both bound and nonbound fractions and examined by anti-pY and Flag Western blot; 0.5% of the input is shown. (D) Tyrosine phosphorylation of endogenous RUNX1 from human MEG-01 cells and primary murine thymocytes. Nuclear extracts from MEG-01 cells or whole-cell lysates from primary thymocytes of 6-wk-old C57BL/6 mice were incubated with α-pY-bound beads. After washing, the bound material was eluted with 100 mM phenyl phosphate and examined by Western blot for RUNX1 and Brg1 (negative control). For MEG-01, 1.25% of the input is shown (25 μg of nuclear extract), and 0.5% input is shown for primary thymocytes (90 μg of whole-cell lysate). (E) Loss of RUNX1 tyrosine phosphorylation upon TPA-induced maturation of L8057 cells. SA-IP and α-pY Western blot of Flag-BioRUNX1 from nuclear extracts or cytoplasmic fractions of L8057 cells treated with 50 nM TPA for the indicated number of days; 1% of the input is shown. (F) Enhanced RUNX1 tyrosine phosphorylation with inhibition of tyrosine phosphatases. SA-IP and α-pY Western blot of Flag-BioRUNX1 from nuclear extracts of uninduced L8057 cells treated with 1.25 mM Na3VO4 for the indicated time.
RUNX1 tyrosine phosphorylation levels decrease during phorbol ester-induced L8057 megakaryoblastic cell maturation
We next examined whether RUNX1 tyrosine phosphorylation levels change during TPA-induced maturation of L8057 megakaryoblastic cells. This revealed a dramatic loss of RUNX1 tyrosine phosphorylation (Fig. 1E, top panel). By 3 d of treatment, a time when many of the cells are undergoing endomitosis and cytoplasmic maturation (Ishida et al. 1993), tyrosine phosphorylation levels are barely detectable. Cell fractionation studies indicate that tyrosine-phosphorylated RUNX1 localizes to the nuclear compartment and does not translocate to the cytoplasm upon TPA treatment (Fig. 1E, bottom panel). This suggests that loss of RUNX1 tyrosine phosphorylation is due to dephosphorylation. Consistent with this, brief treatment of the cells with the pan-tyrosine phosphatase inhibitor sodium orthovanadate (Na3VO4) markedly enhances RUNX1 tyrosine phosphorylation levels (Fig. 1F). Thus, RUNX1 tyrosine phosphorylation is dynamically regulated, and higher levels correlate with an immature cell state. Given that RUNX1 is required for normal Mk maturation (Ichikawa et al. 2004; Growney et al. 2005), this correlation suggests that tyrosine phosphorylation may inhibit RUNX1 function in megakaryopoiesis.
RUNX1 is phosphorylated by SFKs
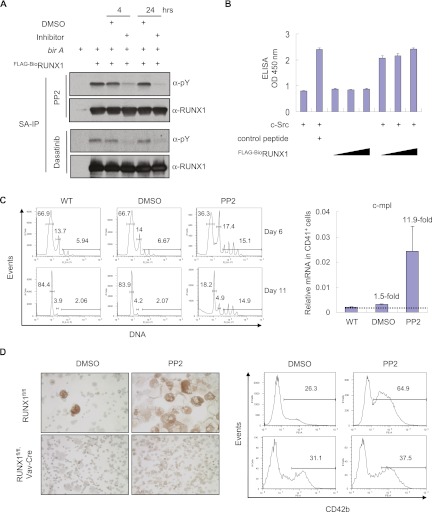
Inhibition of SFKs has previously been shown to markedly enhance megakaryopoiesis (Lannutti et al. 2005, 2006; Mazharian et al. 2011). In combination with the findings above, we hypothesized that SFKs may be responsible for RUNX1 tyrosine phosphorylation. To test this, uninduced L8057 cells containing Flag-BioRUNX1 were treated with the pan-SFK inhibitor PP2, and RUNX1 tyrosine phosphorylation levels were measured (Fig. 2A, top panel). In contrast to control cells treated with dimethyl sulfoxide (DMSO), the RUNX1 phosphotyrosine signal markedly diminished by 4 h and was nearly undetectable by 24 h. Similar findings were observed using Dasatinib, a clinically available SFK inhibitor (Fig. 2A, bottom panel). In vitro kinase assays using recombinant c-Src and Flag-BioRUNX1 purified from TPA-induced L8057 cells show a dose-dependent increase in RUNX1 tyrosine phosphorylation (Fig. 2B). Confocal immunofluorescence microscopy studies indicate partial overlapping localization patterns for c-Src and RUNX1 in L8057 cells (Supplemental Fig. S3). Collectively, these data indicated that c-Src and/or possibly additional SFKs are responsible for RUNX1 tyrosine phosphorylation in megakaryocytic cells.
Figure 2.
RUNX1 tyrosine phosphorylation by Src family kinases. (A) Inhibition of RUNX1 tyrosine phosphorylation with the SFK inhibitors PP2 or Dasatinib. SA-IP and α-pY Western blot of Flag-BioRUNX1 from nuclear extracts of uninduced L8057 cells treated with DMSO, 10 μM PP2, or 10 μM Dasatinib for 4 or 24 h. (B) c-Src in vitro phosphorylation of RUNX1. Purified recombinant c-Src was incubated with SA-purified Flag-Bio-RUNX1 from TPA-induced L8057 cells, washed with 3% SDS, and assayed by ELISA for pY. Control peptide is a known c-Src substrate, Tyr 160 (Cell Signaling). The data represent the mean of two measurements ± SEM. (C) SFK inhibition enhances megakaryopoiesis in fetal liver cultures. Flow cytometry plots for DNA content of whole murine E13.5 fetal liver cells cultured with 1% TPO conditioned medium with either no treatment (WT), DMSO, or 10 μM PP2 for 6 or 11 d. (Right panel) Quantitative RT–PCR (qRT–PCR) for c-mpl mRNA transcript levels from CD41+ flow-sorted cells from the same cultures on day 11. Levels are normalized to β-actin and represent the mean of two experiments ± SEM. (D, left) AChE stains of cytospun cells from E13.5 fetal liver cells of RUNX1fl/fl or RUNX1fl/fl, Vav-Cre mice (both also contain the Cre reporter allele Rosa26-flox-stopper-flox-EYFP) cultured with TPO for 12 d and with 10 μM PP2 or DMSO for 11 d. Mks stain orange/brown. (Right) CD42b flow cytometry plots of cells taken from day 6 of the culture. For the Runx1fl/fl, Vav-Cre (Rosa26-flox-stopper-flox-EYFP) mice, cells were first gated for EYFP expression.
In order to examine the contribution of RUNX1 to the enhancement of megakaryopoiesis by SFK inhibition, wild-type, RUNX1fl/fl or RUNX1fl/fl, Vav-Cre fetal liver cells from E13.5 murine embryos were cultured in thrombopoietin (TPO) and stem cell factor (SCF) with or without PP2. Consistent with earlier reports (Lannutti et al. 2005; Mazharian et al. 2011), PP2 treatment of wild-type or RUNX1fl/fl fetal liver cultures dramatically enhanced the number, size, ploidy, and percentage of CD42b+ (mature) Mks in the culture (Fig. 2C,D). This correlated with a marked (12-fold) increase in mRNA levels of the direct RUNX1 target gene c-mpl (Huang et al. 2009) in CD41+ flow-sorted cells (Fig. 2C, right panel). In contrast, treatment of RUNX1fl/fl, Vav-Cre fetal liver cells, which lack RUNX1, failed to increase the number of large, polyploid, acetylcholinesterase (AChE)-positive cells (Mks) or the percentage of CD42b+ cells in the culture (Fig. 2D). Thus, RUNX1 is required for the enhanced megakaryopoiesis observed with SFK inhibition.
RUNX1 is tyrosine phosphorylated in its NRDB and ID domains
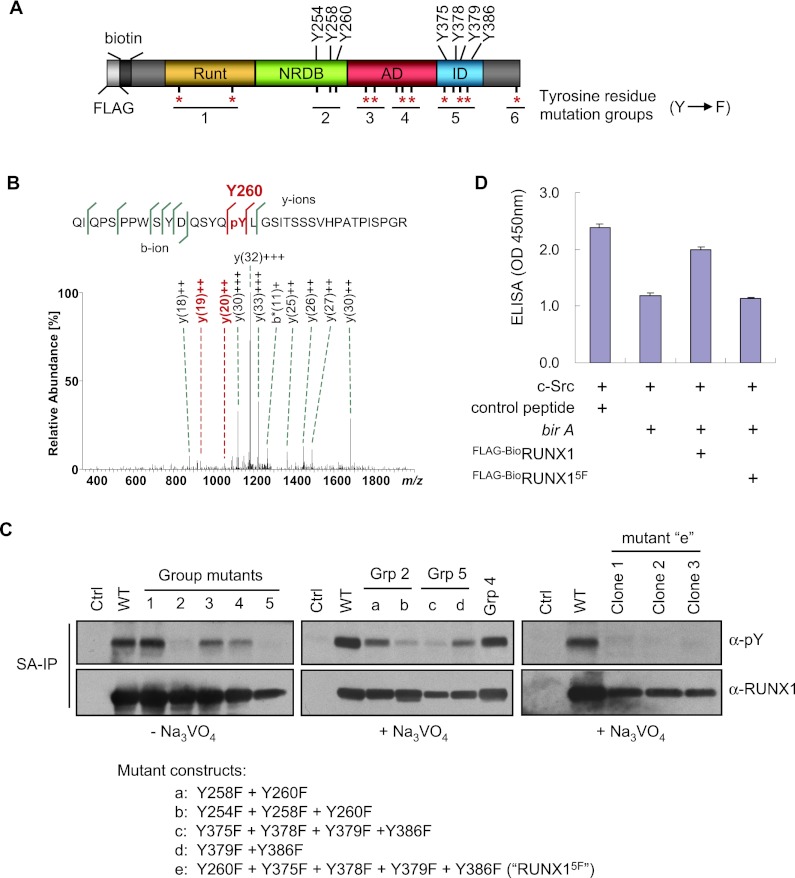
To examine the functional role of RUNX1 tyrosine phosphorylation more directly, we next mapped the phosphorylated tyrosine residues. Murine RUNX1 (isoform 3) contains 15 tyrosine residues, 10 of which are highly conserved among RUNX family members (RUNX1, RUNX 2, and RUNX 3) and across multiple species (Fig. 3A; Supplemental Fig. S4). Two complementary approaches were taken to identify the phosphorylated residues. In the first approach, Flag-BioRUNX1 was purified from uninduced Na3VO4-treated L8057 cells by SA affinity chromatography and examined by mass spectrometry for phosphorylation (Fig. 3B; Supplemental Fig. S5). This identified Tyr 260, located within the NRDB domain, as a phosphorylation target. Four peptides encompassing the three tyrosine residues within the AD and four tyrosine residues within the ID, respectively, were also found to be phosphorylated, but the low fragment ion intensities precluded unambiguous assignment of the phosphorylated residues within this region (Supplemental Fig. S5).
Figure 3.
Mapping of RUNX1 phosphorylated tyrosine residues. (A) Schematic diagram of RUNX1 (murine isoform 3) showing positions of its 15 tyrosine residues relative to functional domains. The most highly conserved tyrosine residues among species and different RUNX family members are indicated with a red asterisk (see Supplemental Fig. S4 for more details). The groups of tyrosine residues mutated to phenylalanine are indicated. (RUNT) rnt homology domain (DNA- and CBFβ-binding domain). (B) Fragment ion mass spectrum of the tryptic peptide QIQPSPPWSYDQSYQpYLGSITSSSVHPATPISPGR indicating phosphorylation of Tyr 260 (murine isoform 3 numbering). The y192+ and y202+ (both shown in red) localize the phosphorylation site to Y260. (C) SA-IP and α-pY Western blot of uninduced L8057 cells stably expressing Flag-BioRUNX1 group mutants (as shown in A). (Left panel) Cells without Na3VO4 treatment. (Middle and right panels) Cells were treated with 1.25 mM Na3VO4 for 15 min prior to SA-IP. (Right panel) Three independent clonal cell lines are shown. (D) c-Src in vitro kinase assay of Flag-BioRUNX1 and Flag-BioRUNX15F purified by streptavidin pull-down from TPA-induced L8057 cells. The data represent the mean of three measurements ± SEM. See the legend for Figure 2B for additional details.
As a second approach, L8057 cell lines were generated that stably express Flag-BioRUNX1 mutants in which tyrosine residues were replaced by phenylalanine. Six mutation groups were initially constructed based on the location of the tyrosine residues (Fig. 3A). The mutants were assayed for tyrosine phosphorylation by SA-IP followed by anti-pY Western blot (Fig. 3C, left panel). Mutation of both tyrosine residues within the runt domain (group 1) showed no effect on the RUNX1 tyrosine signal. The group containing Tyr 260 (group 2) showed significantly reduced pY levels, consistent with the mass spectrometry results. However, some residual signal was evident even with mutation of all three tyrosine residues within this group (Y254, Y258, and Y260), suggesting that additional residues are also phosphorylated. Group 3 and 4 mutants, involving residues within the AD, showed only minor reductions in signal. However, group 5, which contains the four tyrosine residues within the ID, showed significantly reduced tyrosine phosphorylation levels. Group 6, which corresponded to the C-terminal “VWRPY” Groucho repressor-interacting domain, showed no change (data not shown).
Additional mutants were generated to further dissect the phosphorylated tyrosine residues within these regions. These assays were performed in the presence of Na3VO4 to enhance assay sensitivity. As shown in Figure 3C (middle panel), mutation of Y258 and Y260 within group 2 reduced the total RUNX1 tyrosine phosphorylation level compared with wild-type RUNX1 or mutant group 4. However, addition of Y254 further reduced the levels. Likewise, mutation of Y379 and Y386 within group 5 reduced the overall tyrosine phosphorylation levels, but addition of Y375 and Y378 further decreased the levels. Last, we generated a mutant with phenylalanine substitution of Y260 and each of the four ID tyrosine residues (RUNX1Y260F, Y375F, Y378F, Y379F, Y386F mutant, hereafter referred to as “RUNX15F”). Three independent L8057 clones expressing this mutant showed RUNX1 tyrosine phosphorylation levels close to background (Fig. 3C, right panel). RUNX15F also failed to be phosphorylated by c-Src in vitro (Fig. 3D). These findings indicate that multiple RUNX1 tyrosine residues are phosphorylated under the conditions tested and that they reside predominantly within the NRDB and ID regions.
RUNX1 tyrosine phosphorylation negatively affects RUNX1 function in Mk maturation
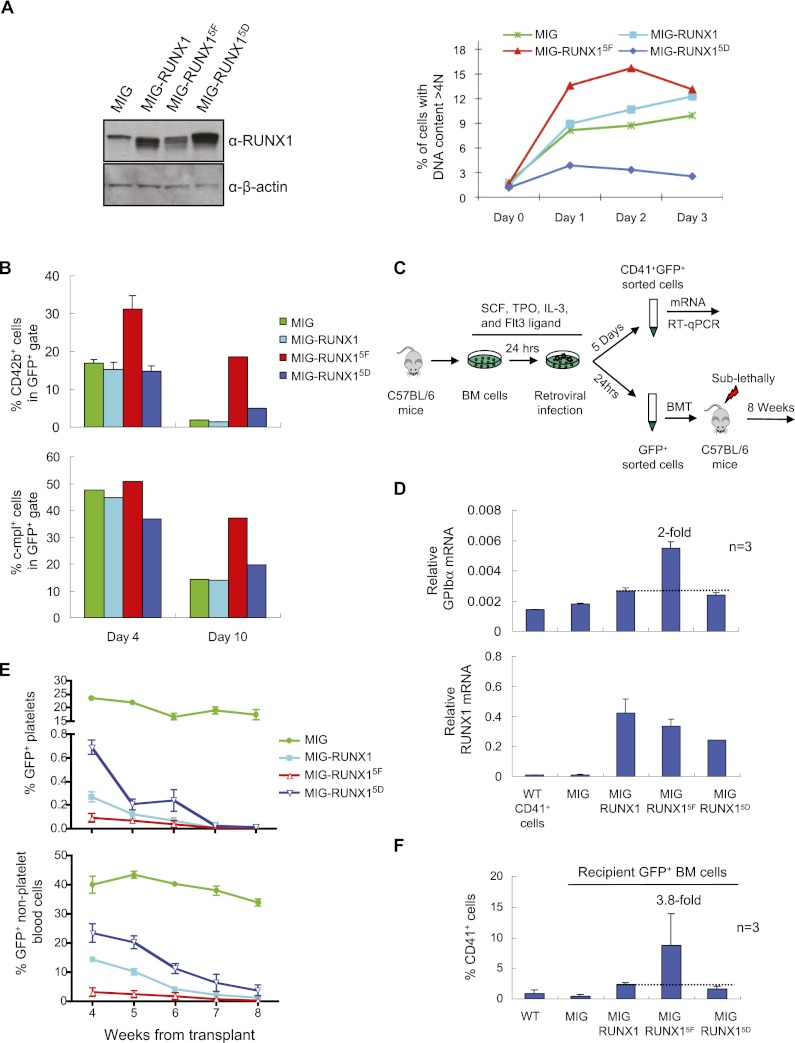
To test the functional significance of RUNX1 tyrosine phosphorylation, we began by retrovirally transducing L8057 cells with either wild-type RUNX1, the nonphosphorylatable RUNX15F mutant, or a mutant containing substitution of each of the five phosphorylated tyrosine residues with aspartic acid to mimic constitutive phosphorylation (RUNX1Y260D, Y375D, Y378D, Y379D, Y386D, hereafter referred to as “RUNX15D”). The retroviral vectors also express enhanced green fluorescence protein (GFP) as a bicistronic mRNA. After retroviral infection, GFP+ cells were flow-sorted and examined for RUNX1 protein levels. All of the constructs expressed RUNX1 at moderately higher levels than endogenous RUNX1, with RUNX15F somewhat lower than wild-type RUNX1 or RUNX15D (Fig. 4A, left panel). Despite lower levels, RUNX15F significantly enhanced TPA-induced polyploidization compared with wild-type RUNX1 (Fig. 4A, right panel). RUNX15D inhibited polyploidization, indicating dominant-negative activity. Retroviral expression of RUNX15F in primary murine fetal liver cells also significantly increased the percentage of CD42b- and c-mpl-positive cells compared with wild-type RUNX1 and RUNX15D after culturing in TPO (Fig. 4B). To dissect the functional role of individual tyrosine residues, additional RUNX1 mutants were constructed containing phenylalanine substitution of Y375, Y378, Y379, and Y386 (“Runx14F”) or substitution of only Y379 and Y386 (“Runx12F”). Both of these mutants also led to increased Mk maturation marker expression compared with wild-type RUNX1 when overexpressed in fetal liver-derived Mks (Supplemental Fig. S6). This suggests that Y379 and Y386 are key residues involved in RUNX1 regulation by tyrosine phosphorylation.
Figure 4.
Inhibitory role of tyrosine phosphorylation on RUNX1 function in megakaryopoiesis. (A) Retroviral expression of RUNX1 phosphorylation mutants in L8057 cells. (Left) RUNX1 Western blot of GFP+-sorted L8057 cells retrovirally transduced with the empty vector (MIG), wild-type RUNX1 (MIG-RUNX1), RUNX1Y260F, Y375F, Y378F, Y379F, Y386F (MIG-RUNX15F), or RUNX1Y260D, Y375D, Y378D, Y379D, Y386D (MIG-RUNX15D). (Right) DNA ploidy analysis of the cells induced with 50 nM TPA for the indicated time. The percentage of cells with DNA content >4N is shown. (B) Flow cytometry analysis of GFP+ cells for CD42b and c-mpl expression of E13.5 murine fetal liver cells transduced with each of the retroviral constructs and placed in liquid culture with TPO for either 4 or 10 d. (C) Schematic diagram of retroviral transduction experiments of bone marrow from C57BL/6 wild-type mice. (D) qRT–PCR analysis for GPIbα and RUNX1 mRNA transcripts in GFP+CD41+ or CD41+ wild-type cells flow-sorted after 5 d of liquid culture with TPO. Measurements were normalized to β-actin mRNA levels and represent the mean of three experiments ± SEM. Fold change relative to wild-type RUNX1 is indicated. (E) Loss of engraftment with overexpression of RUNX1 and RUNX1 tyrosine phosphorylation mutants. The percentage of GFP+ platelets (top) or all other GFP+ peripheral blood cells (bottom) in recipient mice at the indicated time from transplant is shown. An equal number of GFP+ cells were injected into each mouse at the start of the transplant. (F) Flow cytometry analysis of GFP+ bone marrow cells for CD41 expression in recipient mice 8 wk following transplant. Measurements represent the mean of three experiments ± SEM. The fold change relative to wild-type RUNX1 is indicated.
To examine the role of RUNX1 tyrosine phosphorylation in a more physiologic setting, whole bone marrow cells from wild-type C57BL/6 mice were transduced with the RUNX15F, RUNX15D, or control retroviral constructs. Some of the cells were transferred to liquid culture containing TPO. The remaining cells were sorted for GFP expression and transplanted into sublethally irradiated wild-type recipient mice (Fig. 4C). For the liquid cultures, GFP+ CD41+ cells were flow-sorted on day 5 of culture. RUNX15F produced GPIbα (CD42b) mRNA levels that were about twofold higher than wild-type RUNX1, despite being expressed at lower levels (Fig. 4D). The RUNX15D construct generated slightly lower levels compared with wild type.
Analysis of the transplanted mice was limited by marked loss of engraftment associated with RUNX1 overexpression, as has been previously reported for human RUNX1b (Tsuzuki et al. 2007) and RUNX1c (Challen and Goodell 2010). Interestingly, the impaired engraftment, as measured by loss of peripheral blood GFP+ cells, was significantly worse for the bone marrow donor cells transduced with RUNX15F compared with wild-type RUNX1 (Fig. 4E). The RUNX15D construct was not as severe as wild-type RUNX1, although it still suppressed. Although the number of GFP+ cells in the bone marrow was quite low at 8 wk following transplant, the mean percentage of GFP+CD41+ cells was 3.8-fold higher in the mice receiving RUNX15F compared with wild-type RUNX1 transduced cells (Fig. 4F). The RUNX15D recipient mice had a slightly lower percentage of GFP+CD41+ cells.
RUNX1 tyrosine phosphorylation negatively affects RUNX1 function in CD8 T-cell development
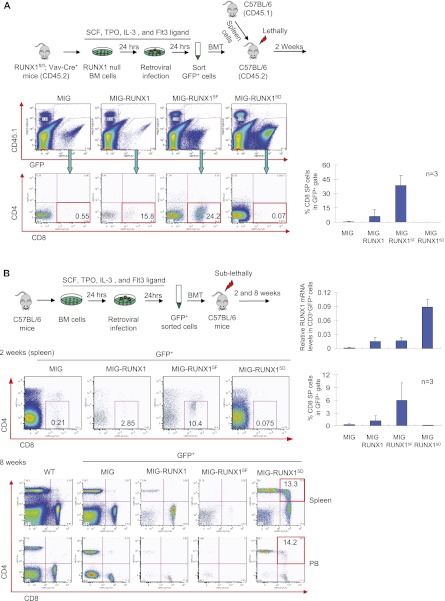
To test the functional significance of RUNX1 tyrosine phosphorylation in T-cell differentiation, whole bone marrow cells from RUNX1fl/fl, Vav-Cre donor mice (CD45.2) were retrovirally transduced with the RUNX1 expression constructs described above and transplanted into lethally irradiated wild-type recipient mice (CD45.2), along with CD45.1 supporting splenocytes (Fig. 5A). After 2 wk, GFP+, CD45.1− spleen cells from the recipient mice were examined by flow cytometry for CD4 and CD8 expression. Consistent with earlier reports (Hayashi et al. 2000, 2001), mice that received the RUNX1-null bone marrow cells transduced with the empty vector had low levels of CD8 single-positive (SP) cells (0.41% ± 0.28%), whereas those receiving wild-type RUNX1 construct had rescue of CD8 SP cell differentiation (5.94% ± 6.98%). Importantly, the RUNX15F mutant construct consistently produced more CD8 SP cells (38.5% ± 10.6%) compared with wild type. The RUNX15D construct failed to rescue CD8 SP cell development (0.09% ± 0.02%).
Figure 5.
Inhibitory role of tyrosine phosphorylation on RUNX1 function in CD8 T-cell differentiation. (A) Rescue assay of CD8 T-cell differentiation. (Top) Schematic diagram of retroviral transduction rescue experiments of bone marrow from Runx1fl/fl, Vav-Cre mice. (Bottom left) Representative flow cytometric plots for GFP, CD45.1, CD4, and CD8 of spleen cells from recipient mice 2 wk following transplantation. (Right) Quantitation of flow cytometry data (n = 3; error bars represent mean ± SEM). (B) Dominant effects of RUNX1 tyrosine phosphorylation mutants on T-cell differentiation. (Top left) Schematic diagram of retroviral transduction experiments of bone marrow from wild-type C57BL/6 mice. (Bottom) Representative flow cytometry plots for CD4 and CD8 of GFP+ cells from spleen or peripheral blood (PB) at 2 or 8 wk following transplantation. (Top right) qRT–PCR analysis for RUNX1 mRNA levels from CD3+GFP+-sorted splenocytes at 2 wk. (Bottom right) Quantitation of the flow cytometry data at 2 wk (n = 3; mean ± SEM).
We also examined potential dominant-negative activity of tyrosine phosphorylated RUNX1 on T-cell differentiation (Fig. 5B). For these experiments, bone marrow cells from C57BL/6 wild-type mice were retrovirally transduced and injected into sublethally irradiated wild-type recipient mice. After 2 wk, the RUNX15F construct again gave rise to considerably more GFP+ CD8 SP cells than wild-type RUNX1 (6.0% ± 4.0% vs. 1.2% ± 1.2%) in the spleen, despite being expressed at equivalent levels. RUNX15D was similar to the empty vector. After 8 wk, mice receiving the empty vector transduced cells had splenic CD4 to CD8 ratios of ∼2–2.5, and few CD4 CD8 double-positive (DP) cells. Overexpression of wild-type RUNX1 and RUNX15F led to a decrease in the CD4 to CD8 SP ratio (CD4:CD8 ratio ∼0.1–0.3). In contrast, RUNX15D failed to skew the population toward CD8 SP cells (CD4:CD8 ratio 3.2) and instead led to a marked increase of CD4 CD8 DP cells, with cells expressing a continuum of CD4 and CD8 levels. Similar findings were observed in peripheral blood mononuclear cells. This suggests that overexpression of RUNX15D blocked T-cell maturation and/or derepressed CD4 expression. Thus, as in Mk maturation, tyrosine phosphorylation negatively affects RUNX1 activity in T-cell CD8 SP cell differentiation and acts in a dominant fashion.
Shp2 contributes to RUNX1 dephosphorylation in Mks and thymocytes
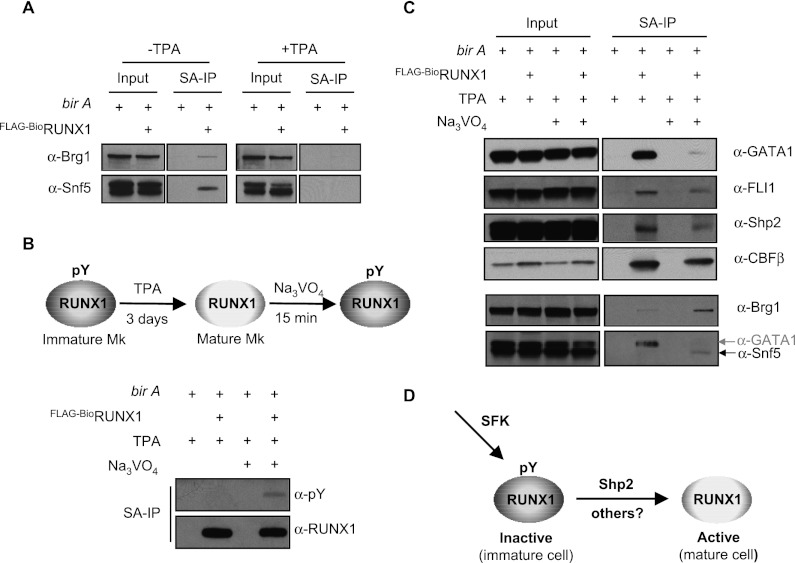
Given our identification of Shp2 in RUNX1-containing complexes, we next investigated whether Shp2 contributes to RUNX1 dephosphorylation. Indirect immunofluorescence microscopy demonstrated partial nuclear Shp2 localization in TPA-induced L8057 cells and primary fetal liver-derived murine Mks (Supplemental Fig. S7). This is consistent with earlier reports of partial Shp2 nuclear localization (Yuan et al. 2003; Xu et al. 2005). We next validated the interaction between RUNX1 and Shp2 by independent SA pull-down experiments from TPA-induced L8057 cells containing Flag-BioRUNX1, reverse coimmunoprecipitation (co-IP) assays of endogenous Shp2 and Flag-BioRUNX1, and GST pull-down assays of recombinant RUNX1 and Shp2 (Fig. 6A–C).
To test whether Shp2 contributes to RUNX1 dephosphorylation, lentiviral shRNA knockdown was performed in L8057 cells, and RUNX1 tyrosine phosphorylation levels were measured. As shown in Figure 6D, two independent shRNA constructs significantly reduced Shp2 protein levels but did not affect the related protein Shp1. This led to an increase in RUNX1 tyrosine phosphorylation levels compared with the empty vector, consistent with a contributory role of Shp2 in RUNX1 dephosphorylation.
To examine the role of Shp2 in megakaryopoiesis in vivo, we generated Shp2fl/fl, Vav-Cre and Shp2fl/fl, PF4-Cre (Mk-specific deletion) conditional knockout mice. Similar to RUNX1fl/fl Vav-Cre mice, these mice are significantly thrombocytopenic compared with non-Cre transgenic littermates (Fig. 6E). Heterozygous animals do not have statistically significant platelet count differences compared with wild type. Following transient immune-mediated thrombocytopenia, Shp2fl/fl, PF4-Cre mice had a significant delay in platelet recovery compared with control Shp2fl/fl mice at 72 h following antibody injection (Fig. 6F). Many of the Mks from Shp2fl/fl, PF4-Cre mouse bone marrow have a smaller size, reduced cytoplasm, and less lobulated nuclei than controls and partially resemble the micromegakaryocytes seen with RUNX1 deficiency (Fig. 6G; Ichikawa et al. 2004; Growney et al. 2005).
Prior work has also shown that both RUNX1 and Shp2 deficiency lead to impaired T-cell differentiation at the DN3 to DN4 stage using Lck-directed conditional allele excision (Taniuchi et al. 2002; Nguyen et al. 2006). In order to determine the role of Shp2 on RUNX1 tyrosine dephosphorylation in vivo, we examined RUNX1 tyrosine phosphorylation levels in thymocytes from Shp2fl/fl, Vav-Cre 6-wk-old mice. As shown in Figure 6H, loss of Shp2 led to a marked increase in RUNX1 tyrosine phosphorylation levels in primary thymocytes. We conclude that Shp2 contributes to RUNX1 tyrosine dephosphorylation in Mks and thymocytes.
RUNX1 tyrosine phosphorylation alters key protein–protein interactions
We next explored the mechanisms whereby tyrosine phosphorylation inhibits RUNX1 function in megakaryopoiesis. Cell fractionation studies show that tyrosine phosphorylated RUNX1 properly localizes to the cell nucleus (Fig. 1E), and confocal immunofluorescence microscopy shows no significant change in bulk RUNX1 subcellular localization after treatment of cells with PP2 or Na3VO4 (Supplemental Fig. S3).
RUNX1 makes a number of important protein–protein interactions that modulate its function. We and others have shown that RUNX1 synergistically interacts with the transcription factors GATA1 and Fli1 during terminal Mk maturation (Elagib and Goldfarb 2007; Huang et al. 2009). RUNX1 also physically and functionally associates with the SWI/SNF chromatin remodeling complex (Bakshi et al. 2010; Yu et al. 2012), and we observe that this interaction diminishes during TPA-induced L8057 cell maturation (Fig. 7A). In order to determine whether RUNX1 tyrosine phosphorylation correlates with altered interactions with these factors, TPA-induced L8057 cells containing Flag-BioRUNX1 were treated with or without Na3VO4 for 15 min. As shown in Figure 7B, addition of Na3VO4 restores RUNX1 tyrosine phosphorylation in TPA-treated cells. This correlates with a dramatic decrease in GATA1 and slight decreases in Fli1, Shp2, and CBFβ binding (Fig. 7C). In contrast, interactions with the SWI/SNF core components Brg1 and Snf5 are significantly enhanced. Thus, tyrosine phosphorylation may inhibit RUNX1 function in part by altering key protein–protein interactions.
Figure 7.
Correlation of altered RUNX1 protein–protein interactions with tyrosine phosphorylation. (A) Loss of interaction between RUNX1 and SWI/SNF core components Brg1 and Snf5 during TPA-induced L8057 maturation. SA-IP of Flag-BioRUNX1 and Western blots for Brg1 and Snf5 are shown; 5% of the input is shown. (B) Restoration of RUNX1 tyrosine phosphorylation in TPA-induced L8057 cells treated with Na3VO4. SA-IP of Flag-BioRUNX1 in L8057 cells induced for 3 d with 50 nM TPA and treated for 15 min with DMSO or 1.25 mM Na3VO4. Western blot for pY and RUNX1 is shown. (C) Changes in RUNX1 protein–protein interactions associated with RUNX1 tyrosine phosphorylation restoration. SA-IP of Flag-BioRUNX1 in L8057 cells induced for 3 d with 50 nM TPA and treated for 15 min with DMSO or 1.25 mM Na3VO4. Western blots for GATA-1, FLI1, Shp2, CBFβ, Snf5, and Brg1 are shown; 0.5% of input is shown. (D) Schematic model for regulation of RUNX1 activity via SFKs and Shp2.
Discussion
In this study, we uncovered a direct regulatory role of SFK-mediated tyrosine phosphorylation on RUNX1 activity in Mk and T-cell differentiation. Our data support an inhibitory function in these lineages, which may be in part due to altered RUNX1 protein–protein interactions. Moreover, we show that Runx1 tyrosine phosphorylation is dynamically regulated by tyrosine phosphatases such as Shp2.
At least six SFKs, including Fyn, Lyn, Fgr, Hck, Src, and Yes, are expressed in primary Mks (Lannutti et al. 2003). Our in vitro data suggest that Src itself is involved in RUNX1 tyrosine phosphorylation. However, our data do not exclude contributions of additional SFKs. Fyn and Lyn are activated upon TPO receptor signaling (Lannutti et al. 2003), and Lyn-null mice have markedly enhanced megakaryopoiesis (Lannutti et al. 2006), making these potential additional candidates. In T cells, the SFK family member Lck positively regulates CD4 lineage development, while Lck deficiency directs commitment to the CD8 lineage (Zamoyska et al. 2003). Our data showing enhanced CD8 SP cell differentiation with the nonphosphorylatable RUNX1 mutant (RUNX15F) makes Lck a potential candidate SFK involved in RUNX1 tyrosine phosphorylation in the T-cell lineage.
Shp2 is the first characterized proto-oncogenic tyrosine phosphatase. It was previously shown to dephosphorylate the transcription factor HOXA10 within the nuclear compartment (Lindsey et al. 2007). In the current study, we identified Shp2 as a component of RUNX1 multiprotein complexes in nuclear extracts from megakaryocytic cells and provided evidence that it plays a role in RUNX1 tyrosine dephosphorylation in cellular assays and in vivo. Genetic deletion of Shp2 in Mks alters Mk development and reduces peripheral blood platelet number, similar to RUNX deficiency. Likewise, both Shp2 and RUNX1 loss lead to blocked T-cell differentiation at the same maturational stage (Taniuchi et al. 2002; Nguyen et al. 2006). Conversely, oncogenic (activating) Shp2 mutations lead to a CD8 SP T-cell lymphoproliferative disorder in mice (Mohi et al. 2005). These effects mirror our findings of enhanced CD8 T-cell development with the nonphosphorylatable RUNX15F construct and impaired CD8 SP T-cell development with the phosphomimmetic RUNX15D (Fig. 5). Collectively, our findings are consistent with an SFK–RUNX1–Shp2 regulatory axis in megakaryopoiesis and CD8 T-cell development (Fig. 7D). In this model, SFKs maintain RUNX1 in an inactive state. Shp2 and perhaps additional nonreceptor tyrosine phosphatases then activate RUNX1 by removing the phosphate groups and modulating RUNX1 protein–protein interactions.
Despite the importance of activating Shp2 mutations in human MPNs and leukemia, the key physiologic Shp2 substrates remains unclear. In this study, we identified Shp2 in a nonbiased screen of RUNX1-associated factors and showed that it binds directly to RUNX1. Moreover, we demonstrated that loss of Shp2 leads to increased RUNX1 tyrosine phosphorylation levels in vivo. We propose that RUNX1 is an important physiologic Shp2 substrate.
Recently, Goh et al. (2010) reported that Src phosphorylates the RUNX family member RUNX3 in human gastrointestinal cell lines. Consistent with our findings, they show that tyrosine phosphorylation inhibits RUNX3 activity. However, the mechanisms may be different. In their cell system, tyrosine phosphorylation results in cytoplasmic sequestration of RUNX3, whereas we found normal nuclear localization of tyrosine phosphorylated RUNX1 in megakaryocytic cells. It is possible that SFK-mediated phosphorylation of RUNX family transcription factors has evolved as a general means to negatively modulate their function, but by different mechanisms dependent on family member and/or cell type.
A number of RUNX1 mutant molecules were used in our study. While we cannot exclude the possibility that the mutations have nonspecific effects through protein misfolding, this seems unlikely since (1) mutants with the phenylalanine substitution (RUNX15F) produced even greater CD8 T-lymphocyte and Mk differentiation activity than the wild-type molecule, and the aspartic acid substitutions (RUNX15D) had the opposite effect; (2) we observed similar effects of RUNX15F on Mk development when we mutated only two residues of RUNX1 (RUNX12F) (Supplemental Fig. S6); (3) the inhibitor studies, which avoid the use of mutant proteins, produced parallel results; and (4) the results produced by the mutants mirror effects seen with Shp2 loss-of-function and gain-of-function mutations.
Only up to ∼1%–10% of the nuclear pool of RUNX1 molecules is tyrosine phosphorylated under the conditions we tested. Although this is in the same general range as observed for tyrosine phosphorylated cytoplasmic proteins, it raises the question of how modification of this small subset of RUNX1 molecules leads to dominant effects when assayed in whole-cell or animal studies? It is possible that there is only a small fraction of the RUNX1 pool that is normally functional within the nucleus. Stein and colleagues (Zaidi et al. 2004) showed that a subpopulation of RUNX1 and RUNX2 molecules are normally targeted to the nuclear matrix and that this is essential for their activity. Interestingly, the nuclear matrix attachment site resides within the transactivation domain and is flanked by the phosphoryated tyrosine residues that we identified. Thus, the effects of tyrosine phosphorylation may be confined to the pool that gets targeted to the nuclear matrix. Further studies will be required to investigate this possibility.
SFKs are typically activated upon integrin and cytokine receptor signaling and are influenced by cell–cell and cell–matrix contacts. Mk maturation is spatially compartmentalized within the bone marrow. Mk progenitors leave the osteoblastic niche and migrate in an immature state to vascular sinusoids, where they make physical contacts with vascular sinusoidal endothelial cells. It is in this location that terminal Mk maturation occurs, allowing proplatelets formed by the Mks direct access to the vascular space. T-cell development is also spatially compartmentalized within the thymus and is influenced by cell–cell and cell–matrix interactions. Thus, it is possible that the actions of SFKs on RUNX1 help coordinate spatial cues with Mk and T-cell differentiation.
RUNX1 deficiency is an early initiating event in human leukemia and MDS and typically results from haploinsufficiency or generation of dominant-negative-acting molecules. Yet, in many of these cases, the wild-type allele is left intact. Our results suggest that SFK inhibition may be a useful means to enhance functional activity of the residual wild-type RUNX1 protein. A number of SFK inhibitors are now clinically available, including Dasatanib, which we have shown to inhibit RUNX1 tyrosine phosphorylation (Fig. 2). Future studies will be aimed at investigating the clinical utility of SFK inhibitors in the treatment of RUNX1 deficiency disorders.
In summary, our data demonstrate direct tyrosine phosphorylation of RUNX1 and its functional role in Mk/T-lymphocyte differentiation. Moreover, we uncovered a RUNX1-centered regulatory axis involving SFK and Shp2 cell signaling. These findings help connect key cell signaling pathways to master hematopoietic transcription factor control in complex microenvironments.
Materials and methods
Materials
All chemicals were purchased from Sigma unless specified otherwise. See Supplemental Table S2 for sources of all antibodies.
Plasmid construction
The cDNA encoding murine RUNX1 (isoform3, gift from Nancy Speck) was cloned into the vector MSCV-IRES-GFP vector for retroviral expression experiments. Phosphorylation point mutants were generated using the Qiagen site-directed mutagenesis kit following the manufacturer's instructions. The associated online primer design software (Qiagen) was used to design primers. Shp2 shRNAs constructs were designed to target murine Shp2 using RNAi Codex (http:// http://cancan.cshl.edu/cgi-bin/Codex/Codex.cgi). See the Supplemental Material for hairpin sequences.
Cell culture and transfection
Cells were cultured in 5% CO2 at 37°C. Culture medium was supplemented with 100 U/mL penicillin/streptomycin (Pen/Strep) and 2 mM L-glutamine. MEG-01 and L8057 cells were cultured as previously described (Ishida et al. 1993) and induced to differentiate with 50 nM TPA (Sigma). Generation and culturing of L8057 cells stably expressing Flag-BioRUNX1 are as previously described (Huang et al. 2009). PLAT-E cells and primary murine fetal liver cells were cultured in Dulbecco's modified Eagle's high-glucose medium (DMEM) supplemented with 10% fetal calf serum (FCS). Murine bone marrow cells were cultured in Isocove's modified Dulbecco's medium (IMDM) supplemented with 10% FCS. PLAT-E cells were transfected using FuGene 6 reagent (Roche) according to the manufacturer's instructions.
Mice
Generation of RUNX1fl/fl (Growney et al. 2005) and Shp2fl/fl (Zhu et al. 2011) mice has been previously described. These were bred to PF4-Cre (Tiedt et al. 2007), Vav-Cre (Stadtfeld and Graf 2005), and/or Rosa26-flox-stopper-flox-EYFP (Srinivas et al. 2001) mice. All experiments involving mice were approved by the Animal Care and Use Committee at Children's Hospital Boston.
RUNX1 multiprotein complex purification and proteomic analysis
Methods for Flag and SA purification of Flag-BioRUNX1-containing multiprotein complexes and identification of associated proteins by microcapillary liquid chromatography and tandem mass spectrometry of tryptic peptides have been previously described (Huang et al. 2009). See the Supplemental Material for details.
Protein–protein interaction experiments
Several different protein pull-down assays were performed in this study: (1) Endogenous Shp2 pull-down assays: These were performed using an anti-Shp2 antibody (Santa Cruz Biotechnology, C-18) and the Pierce co-IP kit following the manual instructions. (2) Small-scale SA purification of Flag-BioRUNX1: This was performed as previously described (Huang et al. 2009). (3) GST pull-down experiments: GST or GST-RUNX1 were produced in bacteria and measured as previously described (Huang et al. 2009). In vitro translation of Shp2 or CBFβ incorporated with 35S-methionine was performed using the TNT-Coupled Reticulocyte Lysate systems (Promega) following the manufacturer's instructions. One microgram of GST or GST fusion proteins and 15 uL of 35S-Shp2 or 35S-CBFβ were incubated with glutathione sepharose 4B beads (GE Healthcare) in HEMGT-150 buffer rotated top-to-end overnight at 4°C. The beads were washed four times for 15 min each with HEMGT-150 buffer, boiled in SDS sample buffer, and loaded onto a SDS-PAGE gel. The gels were then stained with colloidal Coomassie blue, dried in a gel drier (Bio-Rad), and exposed to Kodak BioMax MS film. (4) Purification of biotinylated RUNX1 under denaturing conditions: Nuclear extracts from L8057 cells stably expressing Flag-BioRUNX1 (or mutants) were incubated with SA-coupled agarose beads for 1 h at room temperature (small scale) or 2 h at room temperature (large scale) with up to 5% of SDS. For all SA-IP assays following the initial titration, 3% SDS was used. SA-agarose beads were then washed four times for 15 min each with the same percentage of SDS used in the IP. Flag-BioRUNX1 was eluted by heating for 5 min at 100°C. One millimolar Na3VO4 and 1 mM NaF were added to the lysis and IP buffers to prevent loss of tyrosine phosphorylation during this procedure and the following procedure. (5) Sequential IP for purification of tyrosine phosphorylated RUNX1: An anti-phosphotyrosine purification kit was purchased from Millipore. Enrichment of phosphotyrosine proteins was performed following the manual instructions. Tyrosine phosphorylated Flag-BioRUNX1 was purified from eluted phosphotyrosine proteins using SA-agarose beads in the IP buffer containing 3% SDS. For MEG-01 and primary thymocyte experiments, 10 mg of nuclear extract protein (MEG-01) cells or whole-cell lysates (thymocytes from 6-wk-old C57BL/6 mice using 1% NP-40 whole-cell lysis buffer) were incubated with anti-pY antibody-conjugated agarose beads (Millipore) overnight at 4°C. After washing in IP buffer four times for 15 min each, the bound proteins were eluted by adding 100 mM phenyl phosphate and concentrated by tricholoroacetic acid (TCA) precipitation. Enriched material was boiled with SDS loading buffer for SDS-PAGE and Western blot. For the experiments examining RUNX1 tyrosine phosphorylation in thymocytes from the Shp2fl/fl and Shp2fl/fl, Vav-Cre mice, 17 mg of whole-cell lysate protein was used for the anti-pY IP.
Retroviral infection of primary Mks
Retroviral particle production and cell infection followed standard procedures. See the Supplemental Material for details.
Bone marrow transplantation
Bone marrow was harvested from 8- to 12 wk-old RUNX1fl/fl, Vav-Cre+ or wild-type C57BL/6 mice by crushing femurs, tibias, ileum, and spine with IMDM medium containing 2% FCS and 2% penicillin/streptomycin and passing through a 100-μm cell strainer. Single-cell suspensions of whole bone marrow were retrovirally infected as described above for fetal liver Mks, except that a cocktail of recombinant cytokines (50 ng/mL TPO, 100 ng/mL SCF, 20 ng/mL IL-3, 50 ng/mL Flt3 ligand) and concentrated retroviral supernatants were used. Twenty-four hours following infection, GFP+ cells were sorted by flow cytometry and washed with phosphate-buffered saline (PBS) twice, and the same number of cells were injected (4.3 × 105 or 5.6 × 105 cells, depending on the experiment) per animal via the retro-orbital venous plexus of 8- to 10-wk-old lethally (10 Gy divided into two fractions) or sublethally (7.2 Gy) irradiated recipient C57BL/6 mice. In the experiments using lethal irradiation, 2 million CD45.1 spleen cells from wild-type donor mice were coinjected as supporting cells. Lethally irradiated mice were housed in sterilized cages and fed with autoclaved food and sulfamethoxazole-treated water. Peripheral blood counts were obtained weekly beginning 4 wk following transplantation using a Drew HemaVet 950FS animal automated blood cell analyzer. Mice were sacrificed at 2 or 8 wk after transplantation for spleen and bone marrow hematopoietic analysis, respectively.
Transient immune-mediated thrombocytopenia
Two micrograms per gram body weight of anti-GPIbα antibody (Emfret) or normal IgG was injected through the mouse retro-orbital venous plexus. Peripheral blood platelet counts were obtained 24 h prior to injection and every 24 h after injection up to 144 h. Additional injected mice were euthanized for bone marrow and spleen histologic and immunochemistry analysis at 72 h after injection.
Manipulating RUNX1 tyrosine phosphorylation by PP2 or Na3VO4
L8057 cells expressing Flag-BioRUNX1 were incubated with 10 μM PP2 or Dasatanib for 4 or 24 h, or 1.25 mM Na3VO4 for 15 min or as indicated in the figures. Control cells were incubated with equivalent volumes of DMSO. Flag-BioRUNX1 was purified by SA affinity chromatography in the presence of 3% SDS as described in Protein–Protein Interaction Experiments. The anti-phosphotyrosine antibody 4G10 (Millipore) was used to detect tyrosine phosphorylated proteins in the Western blot assays.
In vitro kinase assays
L8057 cells stably expressing Flag-BioRUNX1 or Flag-BioRUNX15F were induced with 50 nM TPA for 3 d to generate non-tyrosine phosphorylated RUNX1. SA pull-down was performed from crude nuclear extracts as described above. SA beads containing the bound material were treated with recombinant c-Src using an in vitro kinase kit (Cell Signaling) following the manual instructions. After the reaction, the beads were washed with 3% SDS IP buffer four times for 15 min each to remove c-Src and other noncovalently bound proteins from the beads. The beads were then washed briefly with IP buffer and PBS three times each to remove any residual SDS. The ELISA product was read at 450 and 650 nm.
Analysis of RUNX1 tyrosine phosphorylation by mass spectrometry
L8057 cells containing Flag-BioRUNX1 were pretreated with 1.25 mM Na3VO4 for 15 min. SA-IP was performed from crude nuclear extract in the presence of 3% SDS. Flag-BioRUNX1 was eluted by heat denaturation and separated by SDS-PAGE. After staining with Colloidal Coomassie blue, bands corresponding to Flag-BioRUNX1 were cut into ∼1-mm3 pieces and digested with sequencing-grade trypsin (Promega) at a concentration of 12.5 ng/uL in 100 mM ammonium bicarbonate overnight at 37°C. The peptides were extracted with 100 mM ammonium bicarbonate and acetonitrile and then lyophilized. See the Supplemental Material for details of mass spectrometry analysis.
Flow cytometry
Standard flow cytometry procedures were followed. See the Supplemental Material for details.
Immunofluorescence staining and confocal microscopy
L8057 cells were cytospun after treatment with 50 nM TPA for 5 d, with 10 μM PP2 for 20 h, or with 1.25 mM Na3VO4 for 15 min. DMSO-treated and nontreated cells served as control. The cells were blocked with 5% goat serum overnight at 4°C. Anti-RUNX1 (1:200 dilution; Abcam) and anti-c-Src (1:100 dilution; Santa Cruz Biotechnology) antibodies were incubated at room temperature for 1 h After washing with PBS, the cells were incubated with anti-rabbit (Alexa 594) and anti-mouse (Alexa 488; 1:5000 dilution) for 1 h at room temperature, washed with PBS, and stained with DAPI. Immunofluorescence images were obtained using an LSM700 confocal microscope and analyzed with ImageJ software.
Quantitative RT–PCR (qRT–PCR)
Total mRNA was extracted from the cells using the Qiagen micro- or mini-RNA extraction kit following the manual instructions. Reverse transcription was performed using Bio-Rad Reverse Transcription kit, and quantitative PCR was performed in the presence of SYBR Green dye (Invitrogen) using a MyIQ real-time PCR instrument (Bio-Rad). See the Supplemental Material for PCR primer sequences.
Histology and immunohistochemistry staining
Seventy-two hours after anti-GPIbα antibody injection, Shp2fl/fl or Shp2fl/fl, PF4-Cre mouse femurs were fixed in 4% paraformaldehyde (PFA). Paraffin-embedded slides were prepared and hematoxylin and eosin staining was performed at the Dana-Farber/Harvard Cancer Center Research Pathology Core facility. A standard immunohistochemistry protocol was followed for von Willebrand factor (vWF) staining. In brief, antigens were retrieved by incubating the sections in 20 μg/mL proteinase K solution in TE buffer for 30 min at 37°C. Antigens were bound using a 1/500 dilution of vWF antibody (Dako), followed by detection using ABC-HRP and DAB (Vector Laboratories) according to the manufacturer's protocols. Nine photomicrographs of each sample were taken randomly. vWF-positive Mks were counted as small immature (≤17 μm in diameter) or large mature (>17 μm in diameter) Mks (Long and Williams 1981).
Statistical analysis
All data are expressed as the mean ± standard error of the mean (SEM). Statistical significance was determined using the Student's t-test (one-tailed). Differences were considered significant when the P-value is <0.05.
Acknowledgments
We thank Benjamin Neel, Rebecca Chan, Alan Friedman, Gordon Chan, Huafeng Xie, Xi Wang, and Zhe Li for helpful advice, and Nancy Speck, Yotis Senis, Martha Sola-Visner, and Wei Tong for sharing reagents and/or unpublished data. We also thank Gary Gilliland for providing RUNX1fl/fl mice, Thomas Graf for Vav-Cre mice, and Radek Skoda for PF4-Cre mice. This work was supported by NIH grants R01 HL082952 (to A.B.C.) and R01 HL096125 (to G.-S.F.) and a grant from the Gabrielle's Angel Foundation (to A.B.C.). H.H., H.S., and A.B.C. designed the experiments. H.H. carried out all experiments, except as otherwise stated for the other contributors. A.J.W. performed vWF immunohistochemistry experiments. Z.W. contributed to mass spectrometry analysis of RUNX1 phosphorylation. Y.S. performed Shp2 immunofluorescence studies. T.B.M. contributed to RUNX1 multiprotein complex purification and characterization. H.H.Z. constructed Shp2 conditional knockout mice. G-S.F. contributed Shp2 conditional knockout mice and assisted with interpretation of data. H.S. performed mass spectrometry analysis of RUNX1 tyrosine phosphorylation. H.H. and A.B.C. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.192054.112.
References
- Bakshi R, Hassan MQ, Pratap J, Lian JB, Montecino MA, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS 2010. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J Cell Physiol 225: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, et al. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, Dzierzak E 2000. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13: 423–431 [DOI] [PubMed] [Google Scholar]
- Challen GA, Goodell MA 2010. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol 38: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457: 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib KE, Goldfarb AN 2007. Regulation of RUNX1 transcriptional function by GATA-1. Crit Rev Eukaryot Gene Expr 17: 271–280 [DOI] [PubMed] [Google Scholar]
- Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN 2003. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101: 4333–4341 [DOI] [PubMed] [Google Scholar]
- Goetz TL, Gu TL, Speck NA, Graves BJ 2000. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol Cell Biol 20: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH, Jang JW, Li YH, Chi XZ, Lee KS, Wee H, et al. 2010. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J Biol Chem 285: 10122–10129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T 2009. Forcing cells to change lineages. Nature 462: 587–594 [DOI] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, et al. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Natsume W, Watanabe T, Abe N, Iwai N, Okada H, Ito Y, Asano M, Iwakura Y, Habu S, et al. 2000. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J Immunol 165: 6816–6824 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Abe N, Watanabe T, Obinata M, Ito M, Sato T, Habu S, Satake M 2001. Overexpression of AML1 transcription factor drives thymocytes into the CD8 single-positive lineage. J Immunol 167: 4957–4965 [DOI] [PubMed] [Google Scholar]
- Huang H, Yu M, Akie TE, Moran TB, Woo AJ, Tu N, Waldon Z, Lin YY, Steen H, Cantor AB 2009. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Mol Cell Biol 29: 4103–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Hirai H, et al. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med 10: 299–304 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Levin J, Baker G, Stenberg PE, Yamada Y, Sasaki H, Inoue T 1993. Biological and biochemical characteristics of murine megakaryoblastic cell line L8057. Exp Hematol 21: 289–298 [PubMed] [Google Scholar]
- Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, Littman DR, Asou N, Ito Y 2010. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood 115: 1610–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y 1998. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol 18: 2444–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115 [DOI] [PubMed] [Google Scholar]
- Kundu M, Compton S, Garrett-Beal L, Stacy T, Starost MF, Eckhaus M, Speck NA, Liu PP 2005. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood 106: 3621–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannutti BJ, Shim MH, Blake N, Reems JA, Drachman JG 2003. Identification and activation of Src family kinases in primary megakaryocytes. Exp Hematol 31: 1268–1274 [DOI] [PubMed] [Google Scholar]
- Lannutti BJ, Blake N, Gandhi MJ, Reems JA, Drachman JG 2005. Induction of polyploidization in leukemic cell lines and primary bone marrow by Src kinase inhibitor SU6656. Blood 105: 3875–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannutti BJ, Minear J, Blake N, Drachman JG 2006. Increased megakaryocytopoiesis in Lyn-deficient mice. Oncogene 25: 3316–3324 [DOI] [PubMed] [Google Scholar]
- Lindsey S, Huang W, Wang H, Horvath E, Zhu C, Eklund EA 2007. Activation of SHP2 protein-tyrosine phosphatase increases HoxA10-induced repression of the genes encoding gp91(PHOX) and p67(PHOX). J Biol Chem 282: 2237–2249 [DOI] [PubMed] [Google Scholar]
- Long MW, Williams N 1981. Immature megakaryocytes in the mouse: Morphology and quantitation by acetylcholinesterase staining. Blood 58: 1032–1039 [PubMed] [Google Scholar]
- Mazharian A, Ghevaert C, Zhang L, Massberg S, Watson SP 2011. Dasatinib enhances megakaryocyte differentiation but inhibits platelet formation. Blood 117: 5198–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohi MG, Williams IR, Dearolf CR, Chan G, Kutok JL, Cohen S, Morgan K, Boulton C, Shigematsu H, Keilhack H, et al. 2005. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell 7: 179–191 [DOI] [PubMed] [Google Scholar]
- Mukouyama Y, Chiba N, Hara T, Okada H, Ito Y, Kanamaru R, Miyajima A, Satake M, Watanabe T 2000. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta–gonad–mesonephros region. Dev Biol 220: 27–36 [DOI] [PubMed] [Google Scholar]
- Nakao M, Horiike S, Fukushima-Nakase Y, Nishimura M, Fujita Y, Taniwaki M, Okuda T 2004. Novel loss-of-function mutations of the haematopoiesis-related transcription factor, acute myeloid leukaemia 1/runt-related transcription factor 1, detected in acute myeloblastic leukaemia and myelodysplastic syndrome. Br J Haematol 125: 709–719 [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Ke Y, Zhang EE, Feng GS 2006. Conditional deletion of Shp2 tyrosine phosphatase in thymocytes suppresses both pre-TCR and TCR signals. J Immunol 177: 5990–5996 [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126: 2563–2575 [DOI] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA 2002. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16: 661–672 [DOI] [PubMed] [Google Scholar]
- Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K 1993. Molecular cloning and characterization of PEBP2 β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 α. Virology 194: 314–331 [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330 [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI 2008. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen CJ, Toze CL, Koochin A, Forrest DL, Smith CA, Stevens JM, Jackson SC, Poon MC, Sinclair GD, Leber B, et al. 2008. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy (FPD/AML). Blood 112: 4639–4645 [DOI] [PubMed] [Google Scholar]
- Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, et al. 2011. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J 30: 494–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencovich N, Jaschek R, Tanay A, Groner Y 2011. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood 117: e1–e14 doi: 10.1182/blood-2010-07-295113 [DOI] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, et al. 1999. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 23: 166–175 [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG 2002. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer 2: 502–513 [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4 doi: 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Graf T 2005. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development 132: 203–213 [DOI] [PubMed] [Google Scholar]
- Sun W, Downing JR 2004. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 104: 3565–3572 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111: 621–633 [DOI] [PubMed] [Google Scholar]
- Tiedt R, Schomber T, Hao-Shen H, Skoda RC 2007. Pf4-Cre transgenic mice allow generating lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood 109: 1503–1506 [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK et al. 2011. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell 20: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki S, Hong D, Gupta R, Matsuo K, Seto M, Enver T 2007. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Med 4: e172 doi: 10.1371/journal.pmed.0040172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA 2011. New mutations and pathogenesis of myeloproliferative neoplasms. Blood 118: 1723–1735 [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci 93: 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7: 532–544 [DOI] [PubMed] [Google Scholar]
- Wotton S, Stewart M, Blyth K, Vaillant F, Kilbey A, Neil JC, Cameron ER 2002. Proviral insertion indicates a dominant oncogenic role for Runx1/AML-1 in T-cell lymphoma. Cancer Res 62: 7181–7185 [PubMed] [Google Scholar]
- Xu R, Yu Y, Zheng S, Zhao X, Dong Q, He Z, Liang Y, Lu Q, Fang Y, Gan X, et al. 2005. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood 106: 3142–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, et al. 2012. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell 45: 330–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Yu WM, Yuan Z, Haudenschild CC, Qu CK 2003. Role of SHP-2 tyrosine phosphatase in the DNA damage-induced cell death response. J Biol Chem 278: 15208–15216 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS 2004. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J 23: 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoyska R, Basson A, Filby A, Legname G, Lovatt M, Seddon B 2003. The influence of the src-family kinases, Lck and Fyn, on T cell differentiation, survival and activation. Immunol Rev 191: 107–118 [DOI] [PubMed] [Google Scholar]
- Zhu HH, Ji K, Alderson N, He Z, Li S, Liu W, Zhang DE, Li L, Feng GS 2011. Kit–Shp2–Kit signaling acts to maintain a functional hematopoietic stem and progenitor cell pool. Blood 117: 5350–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]