MicroRNAs (miRNAs) are emerging as cooperating factors that promote the activity of oncogenes in tumor formation and disease progression. Yet, the identification of miRNA targets that cooperate with tumor drivers remains challenging. In this study, Cohen and colleagues use a genetic strategy in Drosophila to identify EGFR-cooperating factors in tumorigenesis. The authors show that the growth regulatory miRNA bantam and its target, Socs36E, are cooperating factors in EGFR-driven tumorigenesis and metastasis in a Drosophila model of epithelial transformation. In addition, the human ortholog of SOCS36E, SOCS5, behaves as a candidate tumor suppressor in cellular transformation in cooperation with EGFR/RAS pathway activation. Thus, this study provides novel insight into the role of SOCS5 in cell transformation.
Keywords: microRNA, cell proliferation, apoptosis, growth control, cancer
Abstract
MicroRNAs (miRNAs) are emerging as cooperating factors that promote the activity of oncogenes in tumor formation and disease progression. This poses the challenge of identifying the miRNA targets responsible for these interactions. In this study, we identify the growth regulatory miRNA bantam and its target, Socs36E, as cooperating factors in EGFR-driven tumorigenesis and metastasis in a Drosophila model of epithelial transformation. bantam promotes growth by limiting expression of Socs36E, which functions as a negative growth regulator. Socs36E has only a modest effect on growth on its own, but behaves as a tumor suppressor in combination with EGFR activation. The human ortholog of SOCS36E, SOCS5, behaves as a candidate tumor suppressor in cellular transformation in cooperation with EGFR/RAS pathway activation.
Cancer genomes accumulate a wide variety of mutations. Cancer genome sequence data provide an unparalleled depth of information about the changes that occur as a result of selective pressure during disease progression. At the same time, they pose a challenge in distinguishing “driver” mutations that are causally linked to disease from “passenger” mutations that may be present, but with limited impact on disease (Stratton 2011). Activating mutations in EGFRs and downstream effectors in the PI3K/AKT and MAPK signaling pathways have been recognized as driver mutations in human cancer (Paez et al. 2004; Stephens et al. 2004; Citri and Yarden 2006; Sibilia et al. 2007). Understanding these modifications has a major impact in guiding therapeutic choice and in understanding the basis for clinical resistance to targeted therapies. However, much remains to be learned about other factors that may cooperate with important driver mutations to promote disease progression (Greenman et al. 2007; Wood et al. 2007)
Drosophila provides a genetically tractable model in which to study oncogene cooperation because the signaling pathways involved in tumor formation and metastasis are well conserved from flies to humans (for review, see Miles et al. 2011; Rudrapatna et al. 2012). For example, a model based on expression of an oncogenic form of Ras was found to be insufficient to direct tumor formation and metastasis without additional mutations (Brumby and Richardson 2003; Pagliarini and Xu 2003).
In this study, we identify the bantam microRNA (miRNA) and its target, Socs36E, as cooperating factors in EGFR-driven tumorigenesis and metastasis in a Drosophila tumor model. bantam has been implicated in the control of cell proliferation and apoptosis during tissue growth (Hipfner et al. 2002; Brennecke et al. 2003) and is a key effector of the growth-promoting activity of the Hippo signal transduction pathway (Nolo et al. 2006; Thompson and Cohen 2006). bantam has also recently been identified as a target of the EGFR pathway that links EGFR and Hippo pathway signaling in growth control (Herranz et al. 2012). We identified Socs36E as a target of bantam in growth control. Under normal conditions, Socs36E functions as a negative growth regulator with a modest effect on tissue growth rates. However, under conditions of elevated EGFR pathway activity, SOCS36E and its human ortholog, SOCS5, behave as tumor suppressors.
Results
A Drosophila model for EGFR-induced epithelial transformation
Carcinomas originating in epithelial tissues are among the most common human cancers. To search for cooperating oncogenes in an epithelial context, we made use of the Drosophila wing imaginal disc to screen for genes cooperating with EGFR to promote tumor formation. We made use of the TARGET system, in which GAL4 is used to direct spatially defined target expression, and the temperature-sensitive GAL80 protein (Gal80ts) permits temporal control of GAL4 activity (McGuire et al. 2003). A strain was generated containing Gal80ts under the control of the ubiquitously expressed tubulin promoter (tubulin-Gal80ts), combined with apterous-Gal4 (apG4), UAS-EGFR to permit EGFR overexpression, and UAS-GFP to mark the GAL4-expressing cells. As illustrated in Figure 1, GAL80 is active at 18°C and represses GAL4. These animals were viable and fertile, and wing discs did not show any growth or patterning abnormalities (Fig. 1A). At 29°C, GAL80 is inactivated, allowing GAL4-dependent expression of UAS transgenes. Larvae transferred to 29°C showed overgrowth of the wing disc in the dorsal compartment where apG4 was expressed (Fig 1B, green). This strain was designed to allow the introduction of different genetic lesions into the EGFR overexpression background with a single genetic cross.
Figure 1.
A simple genetic model for conditional activation of the EGFR pathway. The strain for temporal and spatial regulation of EGFR expression consists of Gal80ts expressed in all cells under control of the tubulin promoter together with apterous-Gal4, UAS-EGFR, and UAS-GFP. (A) At 18°C, GAL80ts is active and represses GAL4 activity. The UAS transgenes are not expressed. (B) At 29°C, GAL80ts is inactive, allowing apterous-Gal4 to drive UAS-EGFR and UAS-GFP in the dorsal compartment of the wing imaginal disc. Use of the GAL80 system to limit GAL4 activity makes it possible to maintain this genetic combination as a viable fertile strain, which facilitates its use in screening.
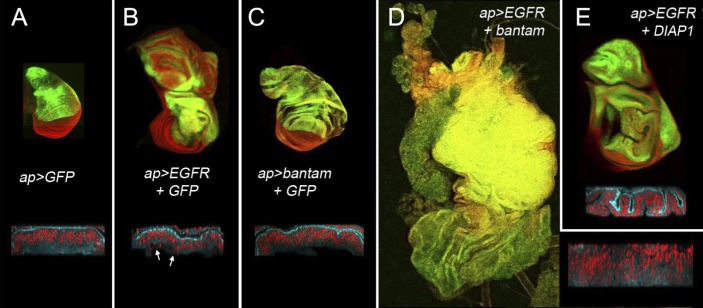
The wing disc is a pseudostratified epithelium. Cells are densely packed, with apically located junctional complexes. Nuclei appear to be organized into “layers” below the junctions. This arrangement can be visualized by the apical concentration of DE-cadherin (Fig. 2A, blue in the bottom panel) and using DAPI to label nuclei (Fig. 2A, red in the bottom panel). Despite being thrown into folds as a consequence of overgrowth (Fig. 2B), EGFR-expressing discs retained normal apico–basal polarity and epithelial organization (Fig. 2B). However, dying cells were observed dropping out of the epithelial layer (Fig. 2B, arrows). Apoptosis is commonly observed under conditions of high EGFR/RAS pathway activity in the imaginal discs (Karim and Rubin 1998; Hipfner and Cohen 2003).
Figure 2.
A Drosophila model of epithelial–mesenchymal transformation. (A–E) Confocal micrographs of third instar wing imaginal discs of the following genotypes: apG4:UAS-GFP (A); apG4:UAS-EGFR, UAS-GFP (B); apG4:UAS-bantam, UAS-GFP (C); apG4:UAS-EGFR, UAS- bantam (D); and apG4:UAS-EGFR, UAS-DIAP1 (E). Nuclei (red) were labeled with DAPI. GFP is shown in green. Insets show optical cross-sections of other discs of the indicated genotypes labeled with antibody to DE-cadherin (blue) and DAPI (red). Arrows in B indicate dying cells dropping out of the disc epithelium.
bantam miRNA has previously been implicated in the control of cell proliferation in Drosophila (Hipfner et al. 2002; Brennecke et al. 2003). Under conditions comparable with those used for EGFR expression, bantam overexpression produced overgrowth of the wing disc (Fig. 2C). The overgrowing disc retained normal epithelial organization. Apoptosis was not observed because bantam acts in part through down-regulation of the proapoptotic gene hid (Brennecke et al. 2003).
Synergistic interaction between bantam and EGFR
To our surprise, coexpression of bantam together with EGFR resulted in massive overgrowth and produced tissue with a strikingly different appearance (Fig. 2D). Epithelial organization was compromised, reflected by the loss of apically concentrated DE-cadherin (Fig. 2D, inset). The tissue appeared to grow as a disordered mass of cells. In this respect, the disc resembled tissue undergoing neoplastic transformation that can result from loss of apico–basal polarity (Bilder 2004).
To ask whether the interaction with bantam could be explained by suppression of EGFR-induced apoptosis, we coexpressed EGFR with the Drosophila inhibitor of apoptosis DIAP1. This suppressed EGFR-induced apoptosis, reflected by improved epithelial order, the absence of dying cells (Fig. 2E), and a reduction in the number of cell expressing activated Caspase 3 (Supplemental Fig. S1). It also led to some increase in the amount of tissue growth but did not lead to loss of epithelial integrity (Fig. 2E). Coexpression of the apoptosis inhibitor p35 produced similar results (Supplemental Fig. S1). The cooperative effect of bantam with EGFR suggests that bantam may have additional targets affecting growth and tissue organization that act in conjunction with its known anti-apoptotic targets.
Identification of Socs36E as a bantam target
To identify bantam targets that might cooperate with EGFR in this context, we made use of a biochemical method based on immunopurification of miRNA-containing ribonucleoprotein complexes. Antibodies to HA-tagged Ago1 were used to immunoprecipitate miRNPs from Drosophila S2 cells, where bantam is abundant (Hong et al. 2009). Socs36E mRNA was enriched by Ago immunoprecipitation (Ago-IP) (P < 0.001) and has one potential bantam target site in its 3′ untranslated region (UTR) (Fig. 3A). Socs36E had previously been identified genetically as a negative regulator of the JAK–STAT and EGFR pathways (Callus and Mathey-Prevot 2002; Almudi et al. 2009), making it a promising candidate for further analysis.
Figure 3.
Socs36E is a bantam target. (A) Diagram depicting the RA and RB isoforms of Socs36E mRNA. Orange denotes the 5′ noncoding exon, and blue denotes coding exons. Red arrows denote the 3′ UTRs of the two isoforms. Green indicates the position of the predicted bantam target site (not to scale). Predicted pairing between bantam and this site is shown below. Note that the site is atypical in having an extended region of 15 residues pairing to the 5′ end of the miRNA without the typical central bulge, as well as in having a number of G:U base pairs (red). A mutant version of the Socs36E 3′ UTR was generated with the pairing region deleted. (B) Histogram showing luciferase assays testing the functionality of the bantam site in the Socs36E 3′ UTR. S2 cells were transfected to express the luciferase reporter with or without bantam for wild-type or mutant Socs36E 3′ UTR (∼600-base-pair [bp] fragment). A bantam sensor with two perfect sites provided a positive control, and the empty luciferase vector provided a negative control. The difference between wild-type and mutant 3′ UTRs was statistically significant (P = 0.001, Student's t-test). Data represent mean ± SD from three independent experiments. (C–E) Confocal micrographs of the wing pouch region of early third instar wing discs labeled to visualize SOCS36E protein by antibody staining (green; shown separately in gray at right). Gal4 protein is shown in red.
To test whether bantam can directly regulate Socs36E, we constructed luciferase reporters with a fragment of the 3′ UTR containing the predicted bantam target site (Fig. 3A). Overexpression of bantam in S2 cells reduced the expression of the cotransfected Socs36E reporter by 44% (Fig. 3B, [*] P = 0.005 compared with control). This down-regulation was largely eliminated when the predicted bantam site was deleted (Fig. 3B, P = 0.001 compared with the intact reporter). The mutant reporter was not significantly different from the empty vector control (Fig. 3B), whereas the positive control reporter containing two perfect bantam sites was down-regulated by 58% (Fig. 3B, sensor, P = 0.001 compared with control), slightly more effective than the Socs36E reporter.
Next, we asked whether bantam regulates endogenous Socs36E expression in vivo. SOCS36E protein is uniformly expressed in the wing disc during the rapid proliferative phase in the early third larval instar (Fig. 3C). Overexpression of bantam under apG4 control resulted in reduced SOCS36E levels (Fig. 3D). Reciprocally, depletion of bantam by expression of a UAS-bantam-sponge (Becam et al. 2011) led to an increase in SOCS36E protein levels (Fig. 3E). The expression levels of control proteins were unaffected by these treatments. Taken together, these results suggest that bantam acts directly to limit SOCS36E expression in vivo.
SOCS36E is a negative growth regulator
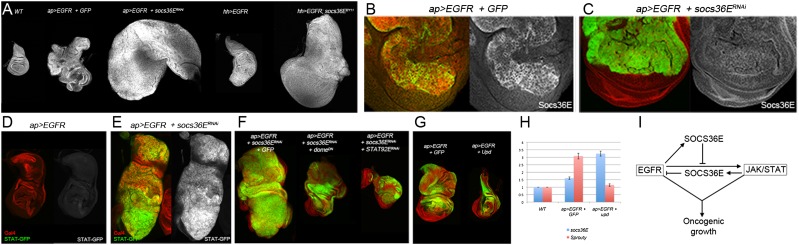
To examine the role of Socs36E in tissue growth, we made use of a null allele, Socs36EEY11, generated by imprecise excision of a P-element inserted at the Socs36E locus (Almudi et al. 2009). Socs36EEY11 mutant flies showed increased wing size (Fig. 4A, P < 0.001). Consistent with this adult phenotype, localized depletion of Socs36E by expression of a UAS-Socs36ERNAi transgene resulted in increased growth in the wing disc (Fig. 4B, P < 0.001; Supplemental Fig. S2). In both scenarios, the effect of removing Socs36E activity produced relatively modest overgrowth. Reciprocally, overexpression of Socs36E reduced growth (Fig. 4B, P < 0.001). Thus, SOCS36E appears to function as a negative growth regulator.
Figure 4.
Socs36E is a negative growth regulator. (A) Cuticle preparations of adult wings from w- Canton S control flies and Socs36EEY11 homozygous mutants. Socs36EEY11 is a null allele generated by imprecise excision of a P-element inserted at the Socs36E locus (Almudi et al. 2009). The increase in wing size was ∼10% and was statistically significant (P < 0.001; n = 11 wings for each genotype). (B) Histogram plotting the average ratio of the area of the dorsal (D) and ventral (V) compartments of the wing imaginal disc (mean ± SD). D compartments were labeled by apG4 expression. The area outlining the wing pouch from the fold to the DV boundary was measured for both compartments, and the ratio was calculated. Details and representative images are provided in Supplemental Figure S1. Reduction of the D/V ratio in discs overexpressing Socs36E was statistically significant compared with the GFP control (P < 0.001, Student's t-test). Discs expressing Socs36ERNAi showed an ∼10% increase in the D/V ratio (P < 0.001). (C) Histogram plotting the average wing area from flies expressing GFP, the bantam-sponge together with GFP, or the bantam-sponge together with Socs36ERNAi under MS1096-Gal4 control.
Does regulation of Socs36E contribute to bantam function in vivo? Depletion of bantam by expression of the bantam-sponge reduced wing size. This effect was partially offset by coexpression of the UAS-Socs36ERNAi transgene in place of the UAS-GFP control (Fig. 4C, [*] P < 0.001). This suggests that limiting the increase in Socs36E levels by expression of an RNAi transgene reduced the severity of the bantam phenotype. Thus, bantam promotes tissue growth by down-regulating a negative growth regulator, Socs36E.
EGFR up-regulates Socs36E to maintain tissue homeostasis
To ask whether the synergistic effect of bantam with EGFR was due to down-regulation of Socs36E, we used the UAS-RNAi transgene to deplete Socs36E in EGFR-overexpressing discs. This combination produced massive overgrowth (Fig. 5A) far in excess of the effects of Socs36E depletion on its own (Fig. 4; Supplemental Fig. S2A) and comparable with that obtained by coexpression of bantam with EGFR (Fig. 1). Overgrowth also resulted in clones of cells coexpressing the UAS-RNAi transgene together with EGFR (Supplemental Fig. S2B). Synergistic overgrowth was also observed when Socs36E was depleted in discs expressing the oncogenic mutant form of Rasv12 (Supplemental Fig. S2C). As a genetically independent test of the role of Socs36E, we overexpressed EGFR in a Socs36E-null mutant background. This also led to massive overgrowth compared with EGFR expression in an otherwise normal genetic background (Fig. 5A, right).
Figure 5.
Reduction in Socs36E levels potentiates the effects of EGFR leading to EMT. (A) Low-magnification images of wing discs labeled with DAPI. From left, apG4 control (WT); apG4 UAS-EGFR, UAS-GFP; and apG4 UAS-EGFR, UAS-Socs36ERNAi. The two discs at right show hedgehog-Gal4 driving UAS-EGFR in an otherwise normal background and in a Socs36E-null mutant background. hhG4 was used for this experiment to bypass the need to make a recombinant chromosome carrying apG4 and the Socs36E allele, which are close together on Chromosome II. (B) apG4 UAS-EGFR, UAS-GFP wing disc labeled with GFP (green) and anti-SOCS36E (red, shown separately at right). (C) apG4 UAS-EGFR, UAS-Socs36ERNAi wing disc labeled with GFP (green) and anti-SOCS36E (red). (D,E) Discs carrying a STAT-GFP reporter transgene labeled with anti-Gal4 (red) and anti-GFP (green). The GFP channel is shown separately. (F) Effect of blocking JAK/STAT signaling on the synergy between EGFR and SOCS36E. (Red) DAPI; (green) GFP. (Left) apG4 UAS-EGFR, UAS-Socs36ERNAi + UAS-GFP. (Middle) apG4 UAS-EGFR, UAS-Socs36ERNAi + UAS-DomeDN. (Right) apG4 UAS-EGFR, UAS-Socs36ERNAi + UAS-Stat92ERNAi. (G) Effect of coexpression of Upd on EGFR-induced growth. (Red) DAPI; (green) GFP. (Left) apG4 UAS-EGFR + UAS-GFP. (Right) apG4 UAS-EGFR + UAS-Upd. (H) Relative levels of Socs36E and sprouty mRNAs measured by quantitative real-time RT–PCR. RNA samples were isolated from wing imaginal discs of the indicated genotypes. (WT) apG4 control. A single experiment is shown. Error bars indicate technical replicates. Comparable results were obtained in three independent biological replicates. (I) Model of the relationship between EGFR, SOCS36E, and JAK/STAT pathway activity.
To explore the basis for this interaction, we examined Socs36E levels in discs overexpressing EGFR. Socs36E mRNA and protein levels were increased by EGFR (Fig. 5B,H). Coexpression of the Socs36E RNAi transgene together with EGFR restored SOCS36E protein to near-endogenous levels (Fig. 5C). These findings suggest that EGFR induces expression of a negative growth regulator, SOCS36E, which serves to limit the ability of EGFR to drive tissue growth. Massive overgrowth resulted when the Socs36E “brake” was disengaged.
How might SOCS36E act to limit EGFR-induced growth? Drosophila Socs36E has been shown to antagonize EGFR activity (Callus and Mathey-Prevot 2002; Almudi et al. 2009). Interestingly, human SOCS5, an ortholog of SOCS36E, is induced by EGF stimulation and can feed back to down-regulate EGFR levels (Kario et al. 2005; Nicholson et al. 2005). We therefore considered the possibility that SOCS36E-mediated negative feedback might limit the magnitude of signaling via EGFR effector pathways. However, we observed that further increasing EGFR activity (using two copies of the UAS-EGFR transgene) was not sufficient to cause loss of epithelial integrity, although it did drive a further increase in growth. This contrasts with the loss of epithelial integrity when EGFR was expressed in the Socs36E mutant background or in combination with the Socs36ERNAi transgene. Although Socs36E depletion can increase EGFR pathway activity (Supplemental Fig. S3A,B), the synergistic effect of Socs36E depletion with EGFR cannot be explained solely by increased EGFR activity.
SOCS36E has also been reported to negatively regulate JAK/STAT activity (Callus and Mathey-Prevot 2002; Rawlings et al. 2004a; Baeg et al. 2005). We observed a low level of activity of a STAT-GFP reporter transgene (Bach et al. 2007) in discs expressing EGFR alone, but this reporter was strongly induced in discs simultaneously depleted of Socs36E (Fig. 5D,E). Depletion of Socs36E alone under apG4 control in an otherwise normal growth context had little effect on STAT-GFP reporter levels (Supplemental Fig. S3C). Thus, SOCS36E limits the capacity of EGFR to induce JAK/STAT activity (Fig. 5I).
In this context, it is interesting that Socs36E is a transcriptional target for regulation by JAK/STAT signaling (Karsten et al. 2002). The STAT-GFP reporter is based on the promoter fragment from the Socs36E locus (Bach et al. 2007). Hence, up-regulation of STAT-GFP is likely to reflect expression of the endogenous Socs36E locus as a consequence of Socs36E depletion under conditions of high EGFR activity.
JAK/STAT signaling has been implicated in tumor growth and invasion induced by activated Rasv12 in Drosophila (Wu et al. 2010). To test whether JAK/STAT contributed to the overgrowth presented in the UAS-EGFR, UAS-Socs36ERNAi combination, we reduced JAK/STAT pathway activity by coexpressing a dominant-negative form of the receptor (DomeDN) or expressing an RNAi transgene directed against STAT92E (STAT92ERNAi). Down-regulation of JAK/STAT pathway activity by either of these two means greatly reduced the magnitude of the overgrowth caused by the EGFR Socs36ERNAi combination (Fig. 5F). Hence, the observed activation of the JAK/STAT pathway (Fig. 5E) is required for the massive overgrowth in the UAS-EGFR, UAS-Socs36ERNAi combination.
In the Rasv12 model, loss of scribble was found to activate JNK signaling, leading to expression of the ligand Unpaired (Upd) (Wu et al. 2010). Upd expression in the eye imaginal disc under eyeless-Gal4 control was sufficient to synergize with Rasv12 in tumor formation (Wu et al. 2010). It was therefore surprising to find that coexpression of Upd had the opposite effect, reducing EGFR-induced growth of the wing disc (Fig. 5G). To explore the basis for this, we examined expression levels of Socs36E and sprouty mRNA. sprouty is a transcriptional target of the EGFR signaling pathway. EGFR expression alone increased both Socs36E and sprouty mRNA levels (Fig. 5H). Interestingly, Upd coexpression led to a further increase in Socs36E mRNA but unexpectedly caused a reduction in sprouty mRNA levels. This suggests that increased activation of the JAK/STAT pathway by Upd led to a net decrease in EGFR activity. The observed increase in Socs36E levels is likely to contribute to this, but we do not exclude other routes by which JAK/STAT activity could feed back on EGFR pathway activity.
These findings suggest a model in which SOCS36E plays a key role mediating a regulatory feedback mechanism linking EGFR and JAK/STAT pathway activity (Fig. 5I). In agreement with the results reported in Wu et al. (2010), we saw that EGFR and JAK/STAT cooperate in oncogenic transformation. Our findings suggest that the EGFR and JAK/STAT pathways are both able to induce Socs36E expression. SOCS36E, in turn, serves to limit activity of both pathways. Under conditions of EGFR activation, SOCS36E serves as a brake on JAK/STAT activity. Loss of this brake contributes to the massive growth observed when EGFR and JAK/STAT are up-regulated together.
SOCS36E behaves as a tumor suppressor under conditions of elevated EGFR activity
Depletion of Socs36E led to loss of apico–basal polarity and loss of epithelial organization in EGFR-expressing discs, visualized by loss of apically localized DE-cadherin and Dlg, compared with discs expressing EGFR alone or controls (Fig. 6A). These morphological changes were accompanied by elevated expression of Matrix Metalloprotease-1 and snail mRNAs (Fig. 6B), molecular markers of tumor invasiveness and epithelial–mesenchymal transition (EMT) (Egeblad and Werb 2002; Barrallo-Gimeno and Nieto 2005; Peinado et al. 2007).
Figure 6.
Depletion of Socs36E leads to malignant transformation of tissue overexpressing EGFR. (A) Optical cross-sections of third instar wing discs labeled with DAPI to show the nuclei (blue), anti-DE-cadherin (red), and Dlg (green). (B) Histogram showing mRNA levels of mmp1 and snail measured by quantitative real time PCR. RNA was prepared from discs of the indicated genotypes. Data were normalized to rp49. The data show mean ± SD from three technical replicates of a representative experiment. Comparable results were obtained in independent experiments. (C) Two images of the same host fly implanted with fragments of an apG4 UAS-EGFR, UAS-GFP, UAS-Socs36ERNAi wing imaginal disc at 1 d and 15 d after implantation of the GFP-labeled disc fragment (white arrowhead). (D) Summary of the results of disc fragment injection experiments. (E) Examples of GFP-positive metastases found in the eye (left) and invading the gut (right).
These observations prompted us to explore the potential of EGFR-expressing tissue with reduced Socs36E activity to form tumors. Transplantation of imaginal disc fragments into the abdomens of adult hosts provides an in vivo assay system for tumor formation and metastasis (Caussinus and Gonzalez 2005). Disc fragments expressing EGFR and GFP initially grew quickly but regressed without producing large tumors or affecting host viability. In contrast, fragments of discs coexpressing EGFR and GFP together with the Socs36ERNAi transgene grew rapidly to kill the host flies (Fig. 6C,D). In 65% of animals with tumors, GFP-expressing cells were also found growing at sites distant from the implanted disc fragment. Figure 6E shows examples of GFP-labeled metastasis in the eye and invading the gut (additional examples are in Supplemental Fig. S4). This prompted us to look for evidence of metastasis in the donor larvae prior to disc fragment transplantation. We observed GFP-labeled wing disc tissue invading the nerve cord of the brain, as well as scattered GFP-positive cells that appear to have migrated away from the main mass of the tissue (Supplemental Fig. S4D). These observations suggest that SOCS36E is a critical element in a homeostatic mechanism to prevent tumor formation in a context of EGFR up-regulation.
Human SOCS5 behaves as a candidate tumor suppressor in an EGFR/RAS-dependent cell transformation assay
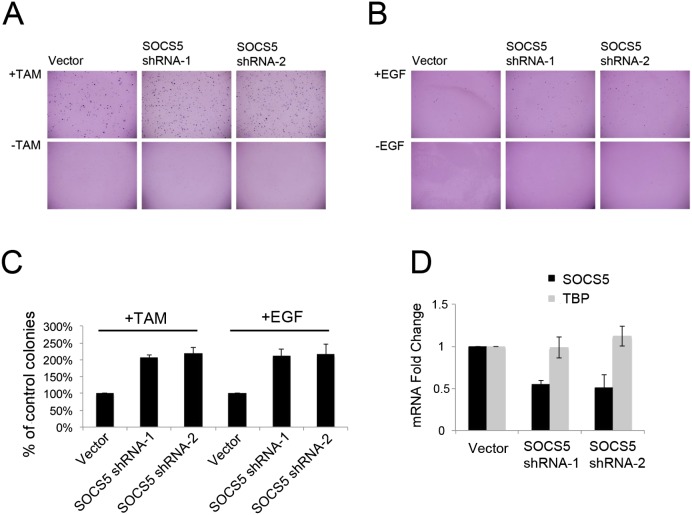
The identification of SOCS36E as a cofactor that promoted oncogenic transformation and metastasis in the fly model prompted us to ask whether SOCS5, the closest mammalian ortholog, might have a similar role. We made use of an in vitro assay for transformation of primary human BJ fibroblast cells based on anchorage-independent colony growth in soft agar (Voorhoeve and Agami 2003). The cells were transduced with retroviral constructs to direct expression of (1) hTERT, which encodes the catalytic subunit of the telomerase to allow replicative immortality; (2) inducible oncogenic H-RasV12 fused to the ligand-binding domain of the estrogen receptor (ER-H-RasV12) to activate Ras signaling upon addition of 4-hydroxy-tamoxifen (De Vita et al. 2005); (3) shRNAs to deplete p53 and p16 to overcome RasV12-induced growth arrest; and (4) SV40 small T antigen, which allows anchorage-independent growth in soft agar (Hahn et al. 1999, 2002). As expected, activation of ER-H-RasV12 by addition of 4-hydroxy-tamoxifen promoted colony formation in soft agar (Fig. 7A). To test the effects of depleting SOCS5 in this assay, we made shRNA vectors targeting two independent sites in SOCS5. Both shRNAs were effective in depleting SOCS5 mRNA to ∼50% of control levels upon transduction into BJ cells (Fig. 7D, measured by quantitative real time PCR, P < 0.05 for each shRNA). Depletion of SOCS5 potentiated the effects of RasV12 activation and led to an approximately twofold increase in the number of colonies compared with RasV12 induction alone (Fig. 7C, P = 0.002 for shRNA-1, P = 0.006 for shRNA-2).
Figure 7.
Reduced SOCS5 expression enhanced EGFR/MAPK-induced cellular transformation (A,B) Representative images showing colonies of BJ/ET-RASV12-ERTAM cells grown in soft agar with the indicated shRNA treatments: empty vector control and two independent shRNAs targeting SOCS5 (shRNA-1 and shRNA-2). (A) Cells were treated with 10−7 M 4-hydroxy-tamoxifen to activate RasV12 (+TAM, top panels) or left untreated (−TAM, bottom panel). (B) Cells were treated with 0.1 μg/mL EGF (+EGF, top panel) or left untreated (−EGF, bottom panel). (C) Colony formation in soft agar produced by the indicated treatments. Data were normalized to the empty shRNA vector control and are presented as a percentage of control colony number. Data represent mean ± SD from three independent retroviral transduction experiments. Student's t-test was used to assess differences between SOCS5 shRNA treatments and empty vector control. P = 0.002 for SOCS5 shRNA-1; P = 0.006 for SOCS5 shRNA-2. (D) Quantitative real-time PCR to measure the efficacy of shRNA-mediated depletion of SOCS5 (Student's t-test, P = 0.004). mRNA fold change is shown relative to empty vector control for three independent retroviral transduction experiments. Data were normalized to GAPDH. TBP was used as a negative control.
We also tested the ability of SOCS5 depletion to promote colony formation in cells stimulated with EGF ligand to activate the EGFR/MAPK pathway in place of RasV12. Use of EGF ligand produced fewer and smaller colonies in the transformation assay (Fig. 7B), but the effects of ligand activation were again potentiated by shRNA-mediated depletion of SOCS5 (Fig. 7C, P = 0.009 for shRNA-1, P = 0.02 for shRNA-2). Depletion of SOCS5 alone was not sufficient to promote colony formation, suggesting that it acts cooperatively with EGFR pathway activation in cell transformation. Although SOCS5 has been linked to EGFR turnover in mammalian cells (Kario et al. 2005; Nicholson et al. 2005), cooperation with activated RAS suggests that, as in the fly model, SOCS5 may act by a mechanism distinct from direct modulation of EGFR to promote cellular transformation.
Discussion
Oncogene cooperation in a Drosophila model for EMT, tumor formation, and metastasis
Loss of epithelial integrity is an important characteristic that distinguishes benign tissue overgrowth from invasive, metastatic carcinoma. Although activation of the EGFR pathway is sufficient to drive proliferation, concomitant induction of apoptosis limits the proliferative capacity of the tissue. Suppression of apoptosis enhanced EGFR-driven growth but did not lead to loss of epithelial integrity. Our findings show that coexpression of the bantam miRNA or depletion of the bantam target Socs36E caused the EGFR-expressing tissue to undergo EMT: loss of epithelial integrity and up-regulation of the EMT markers snail and mmp1. These features are typically associated with transformation and invasiveness of epithelial cancers (for review, see Thiery et al. 2009).
We were intrigued by the finding that Socs36E had a limited capacity as a negative growth regulator under normal conditions but proved to be an important regulator under conditions in which the EGFR pathway was hyperactivated. This relationship also holds true for SOCS5 and the EGFR pathway in human cell transformation.
Although SOCS proteins can serve as a brake on EGFR activity, we presented evidence that SOCS36E is also likely to act on other pathways that contribute to tissue homeostasis. In addition to their roles as regulators of receptor tyrosine kinases (RTKs), SOCS proteins have been shown to down-regulate the JAK/STAT pathway (for review, see Alexander 2002; Rawlings et al. 2004b). There is a body of evidence suggesting that EGFR signaling acts by various means to promote STAT activation in cancer cells (Quesnelle et al. 2007). Here we presented evidence for an incoherent feed-forward regulatory motif in which EGFR acts positively to promote STAT activity while also acting negatively via SOCS36E proteins to limit STAT activity (Fig. 5I). Failure to limit EGFR-induced STAT activation results in loss of growth control and epithelial integrity in the Drosophila model and in cellular transformation in the human cell model. Incoherent feed-forward relationships of this sort allow for regulatory interactions to be self-limiting (for review, see Herranz and Cohen 2010).
Oncogenic miRNAs and tumor-suppressive targets
Identification of the tumor-suppressive activity of Socs36E followed from its identification as a target of a growth regulatory miRNA. Although bantam miRNA is not obviously conserved in mammals, it acts through a conserved target. This prompted us to explore human miRNAs with seed sequences similar to bantam. Human miR-450b-3p and bantam share six out of seven residues in the seed sequence (Ibanez-Ventoso et al. 2008). SOCS5 is not predicted to be a target of miR-450b-3p by computational target prediction programs, as most require near-perfect seed matching (e.g., TargetScanHuman 5.2). However, a potential target site was identified using RNAHybrid (Supplemental Fig. S5; Rehmsmeier et al. 2004). The site has characteristics of a 3′ compensatory site, in which extensive 3′ pairing compensates for an imperfect seed match (Brennecke et al. 2005). Although little is known about miR-450b-3p function to date, this observation raises the possibility of a conserved functional relationship that may merit investigation.
The functional similarity between Drosophila SOCS36E and human SOCS5 in tumor suppression suggests that miRNA target identification could be a powerful tool in uncovering novel cancer genes, regardless of evolutionary conservation of the miRNA sequences. Interestingly, the mammalian SOCS5 contains predicted binding sites for several other miRNAs, including miR-19 and miR-9. Both miR-19 and miR-9 are up-regulated in breast cancer cells. miR-19 has been shown to repress PTEN mRNA (Mu et al. 2009; Olive et al. 2009) and affect drug sensitivity (Liang et al. 2011). miR-9 has been shown to repress DE-cadherin and promote metastasis and tumor invasion (Ma et al. 2010). It is conceivable that both miRNAs might target SOCS5 and thereby contribute to tumor formation. Aberrant expression of miRNAs could lead to less robust regulation of signaling levels or dynamics and promote oncogenesis.
Materials and methods
Drosophila strains
The stocks used are described in the following references: MS1096-Gal4 (Capdevila and Guerrero 1994), ap-Gal4 (Calleja et al. 1996), UAS-EGFR (Buff et al. 1998), UAS-Socs36E (Callus and Mathey-Prevot 2002), UAS-bantam-sponge (Becam et al. 2011), UAS-socs36ERNAi (stock 51821, Vienna Drosophila RNAi Center), UAS-bantam-GFP (Brennecke et al. 2003), UAS-upd (obtained from Pernille Rørth), UAS-domeDN (UAS-domeΔCYT) (Brown et al. 2001), and UAS-DIAP1; Dlg-GFP (Fly Trap ID: YC0005).
Regulated overgrowth using the Gal4/Gal80ts system
For experiments involving use of the Gal4/Gal80 system to direct tissue overgrowth, embryos were collected from crosses of the indicated genotypes for 24 h at 18°C and allowed to develop for 5 d at 18°C to maintain the Gal80-dependent repression of Gal4 until the larvae reached early third instar. Larvae were then transferred to 29°C to induce Gal4 activity and raised for 3 d at 29°C before being processed for immunostaining. The EGFR-overexpressing larvae do not pupate and can continue to grow for up to 5 d.
Immunostaining
Rat and rabbit polyclonal antibodies to the SOCS36E protein were provided by F. Serras and used at 1:300 dilution. Other primary antibodies were rabbit anti-Gal4 (Santa Cruz Biotechnology), rat anti-DE-cadherin (Developmental Studies Hybridoma Bank), and mouse anti-dpERK (Cell Signaling Technology).
Imaginal disc transplantation
Wing imaginal discs were removed from third instar larvae into PBS and cut into fragments using electrolytically sharpened tungsten needles. Fragments were implanted into the abdomens of virgin female yw flies as described (Caussinus and Gonzalez 2005). Implantation was performed using sharpened glass capillaries, and implanted females were kept at 29°C.
Luciferase reporter assays
Drosophila S2 cells were cotransfected with the following plasmids: a tubulin-driven bantam miRNA construct, a tubulin-driven Renilla luciferase, and a tubulin-driven firefly luciferase reporter fused with the intact Socs36E 3′ UTR sequences or a mutant version with pairing region deleted as shown in Figure 1A. bantam sensor luciferase reporter was used as a positive control (Brennecke et al. 2003). Dual luciferase assays were performed 72 h after transfection following the manufacturer's instructions (Promega). Standard deviations were from three independent transfection experiments.
shRNA plasmid construction
The following primers were used: SOCS5 shRNA-1 Forward (GATCCGGAGTCCACACACAGATTGATTACTCGAGTAATCAATCTGTGTGTGGACTTTTTTA) and Reverse (AGCTTAAAAAAGTCCACACACAGATTGATTACTCGAGTAATCAATCTGTGTGTGGACTCCG), and SOCS5 shRNA-2 Forward (GATCCGGTGCAATTCCACAAGCTAATTGCTCGAGCAATTAGCTTGTGGAATTGCATTTTTA) and Reverse (AGCTTAAAAATGCAATTCCACAAGCTAATTGCTCGAGCAATTAGCTTGTGGAATTGCACCG). shRNA sequences were from the RNAi consortium (Sigma). Oligos were annealed and cloned to pRetrosuper-Blast YFP vector for retrovirus production (Voorhoeve and Agami 2003).
Mammalian cell culture
Cells were grown in DMEM containing 10% FCS. Human primary BJ fibroblasts were genetically modified with retrovirally transduced p53 shRNA, p16INK4A shRNA, small T antigen, inducible RasV12-ERTAM, and hTERT as described (Voorhoeve and Agami 2003). Retrovirus was made by calcium-phosphate transduction of Eco-Pack 2 (Clontech) and harvested 20 h later. BJ cells were selected with blasticidin-selective medium 48 h after transduction for 4–6 d.
Soft agar colony formation assay
Cells were resuspended in DMEM containing 0.4% low-melting agarose (Sigma, type VII) and 10% FCS and seeded onto a coating of 1% low-melting agarose in DMEM containing 10% FCS in six-well culture dishes. After 2–3 wk of growth (2 wk for tamoxifen treatment and 3 wk for EGF treatment), plates were stained using an MTT assay and photographed under identical settings, and colony formation was scored using Matlab 2001b.
Real-time quantitative PCR
RNA was extracted from BJ cells by Trizon (Invitrogen), treated with DNase I (Promega), and performed using oligo-dT based reverse transcription of the SuperScript III system (Invitrogen). Real-time quantification was performed using SYBR Green reagents on an ABI 7500 fast real-time PCR platform (Applied Biosystems).
Acknowledgments
We thank S.F. Lim for technical assistance, and Florenci Serras and Marina Ruiz-Romero for reagents and their invaluable help in teaching HH imaginal disc transplantation.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.192021.112.
References
- Alexander WS 2002. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol 2: 410–416 [DOI] [PubMed] [Google Scholar]
- Almudi I, Stocker H, Hafen E, Corominas M, Serras F 2009. SOCS36E specifically interferes with Sevenless signaling during Drosophila eye development. Dev Biol 326: 212–223 [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH 2007. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns 7: 323–331 [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N 2005. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev 19: 1861–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA 2005. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 132: 3151–3161 [DOI] [PubMed] [Google Scholar]
- Becam I, Rafel N, Hong X, Cohen SM, Milan M 2011. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development 138: 3781–3789 [DOI] [PubMed] [Google Scholar]
- Bilder D 2004. Epithelial polarity and proliferation control: Links from the Drosophila neoplastic tumor suppressors. Genes Dev 18: 1909–1925 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the pro-apoptotic gene hid in Drosophila. Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85 doi: 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC 2001. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol 11: 1700–1705 [DOI] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE 2003. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J 22: 5769–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buff E, Carmena A, Gisselbrecht S, Jimenez F, Michelson AM 1998. Signalling by the Drosophila epidermal growth factor receptor is required for the specification and diversification of embryonic muscle progenitors. Development 125: 2075–2086 [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G 1996. Visualization of gene expression in living adult Drosophila. Science 274: 252–255 [DOI] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B 2002. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene 21: 4812–4821 [DOI] [PubMed] [Google Scholar]
- Capdevila J, Guerrero I 1994. Targeted expression of the signalling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J 13: 4459–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E, Gonzalez C 2005. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet 37: 1125–1129 [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y 2006. EGF–ERBB signalling: Towards the systems level. Nat Rev Mol Cell Biol 7: 505–516 [DOI] [PubMed] [Google Scholar]
- De Vita G, Bauer L, da Costa VM, De Felice M, Baratta MG, De Menna M, Di Lauro R 2005. Dose-dependent inhibition of thyroid differentiation by RAS oncogenes. Methods Enzymol 19: 76–89 [DOI] [PubMed] [Google Scholar]
- Egeblad M, Werb Z 2002. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2: 161–174 [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA 1999. Creation of human tumour cells with defined genetic elements. Nature 400: 464–468 [DOI] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 22: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Cohen SM 2010. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev 24: 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Hong X, Cohen SM 2012. Mutual repression by bantam miRNA and capicua links the EGFR/MAPK and hippo pathways in growth control. Curr Biol 22: 651–657 [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Cohen SM 2003. The Drosophila sterile-20 kinase slik controls cell proliferation and apoptosis during imaginal disc development. PLoS Biol 1: E35 doi: 10.1371/journal.pbio.0000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner DR, Weigmann K, Cohen SM 2002. The bantam gene regulates Drosophila growth. Genetics 161: 1527–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Hammell M, Ambros V, Cohen SM 2009. Immunopurification of Ago1 miRNPs selects for a distinct class of microRNA targets. Proc Natl Acad Sci 106: 15085–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Vora M, Driscoll M 2008. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 3: e2818 doi: 10.1371/journal.pone.0002818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Rubin GM 1998. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125: 1–9 [DOI] [PubMed] [Google Scholar]
- Kario E, Marmor MD, Adamsky K, Citri A, Amit I, Amariglio N, Rechavi G, Yarden Y 2005. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J Biol Chem 280: 7038–7048 [DOI] [PubMed] [Google Scholar]
- Karsten P, Hader S, Zeidler MP 2002. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev 117: 343–346 [DOI] [PubMed] [Google Scholar]
- Liang Z, Li Y, Huang K, Wagar N, Shim H 2011. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res 28: 3091–3100 [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. 2010. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768 [DOI] [PubMed] [Google Scholar]
- Miles WO, Dyson NJ, Walker JA 2011. Modeling tumor invasion and metastasis in Drosophila. Dis Model Mech 4: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A 2009. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23: 2806–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson SE, Metcalf D, Sprigg NS, Columbus R, Walker F, Silva A, Cary D, Willson TA, Zhang JG, Hilton DJ, et al. 2005. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc Natl Acad Sci 102: 2328–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G 2006. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol 16: 1895–1904 [DOI] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L 2009. miR-19 is a key oncogenic component of mir-17-92. Genes Dev 23: 2839–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T 2003. A genetic screen in Drosophila for metastatic behavior. Science 302: 1227–1231 [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A 2007. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Quesnelle KM, Boehm AL, Grandis JR 2007. STAT-mediated EGFR signaling in cancer. J Cell Biochem 102: 311–319 [DOI] [PubMed] [Google Scholar]
- Rawlings JS, Rennebeck G, Harrison SM, Xi R, Harrison DA 2004a. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol 5: 38 doi: 10.1186/1471-2121-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA 2004b. The JAK/STAT signaling pathway. J Cell Sci 117: 1281–1283 [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrapatna VA, Cagan RL, Das TK 2012. Drosophila cancer models. Dev Dyn 241: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M 2007. The epidermal growth factor receptor: From development to tumorigenesis. Differentiation 75: 770–787 [DOI] [PubMed] [Google Scholar]
- Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O'Meara S, Smith R, Parker A, et al. 2004. Lung cancer: Intragenic ERBB2 kinase mutations in tumours. Nature 431: 525–526 [DOI] [PubMed] [Google Scholar]
- Stratton MR 2011. Exploring the genomes of cancer cells: Progress and promise. Science 331: 1553–1558 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial–mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM 2006. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126: 767–774 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, Agami R 2003. The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4: 311–319 [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Wu M, Pastor-Pareja JC, Xu T 2010. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature 463: 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]