Alternative RNA processing plays important roles in such diverse processes as cell differentiation and tissue development and is mediated by cell type-specific combinations of transacting factors binding to cis-acting RNA elements. In this study, Darnell and colleagues provide the first genome-wide biochemical description of targets of the neuronal/brain isoform of the polypyrimidine tract-binding protein Ptbp2. By using high-quality HITS-CLIP mapping studies and Ptbp2-null mice, the authors found that the major action of Ptbp2 is to inhibit adult-specific alternative exons by binding pyrimidine-rich sequences upstream of and/or within them. Thus, this study provides novel insight into the role of Ptbp2 in early development and of RNA regulation in stem cell and early neuronal development.
Keywords: HITS-CLIP, polypyrimidine tract-binding protein, RNA-binding map, alternative splicing, brain development, neuronal progenitor
Abstract
Two polypyrimidine tract RNA-binding proteins (PTBs), one near-ubiquitously expressed (Ptbp1) and another highly tissue-restricted (Ptbp2), regulate RNA in interrelated but incompletely understood ways. Ptbp1, a splicing regulator, is replaced in the brain and differentiated neuronal cell lines by Ptbp2. To define the roles of Ptbp2 in the nervous system, we generated two independent Ptbp2-null strains, unexpectedly revealing that Ptbp2 is expressed in neuronal progenitors and is essential for postnatal survival. A HITS-CLIP (high-throughput sequencing cross-linking immunoprecipitation)-generated map of reproducible Ptbp2–RNA interactions in the developing mouse neocortex, combined with results from splicing-sensitive microarrays, demonstrated that the major action of Ptbp2 is to inhibit adult-specific alternative exons by binding pyrimidine-rich sequences upstream of and/or within them. These regulated exons are present in mRNAs encoding proteins associated with control of cell fate, proliferation, and the actin cytoskeleton, suggesting a role for Ptbp2 in neurogenesis. Indeed, neuronal progenitors in the Ptbp2-null brain exhibited an aberrant polarity and were associated with regions of premature neurogenesis and reduced progenitor pools. Thus, Ptbp2 inhibition of a discrete set of adult neuronal exons underlies early brain development prior to neuronal differentiation and is essential for postnatal survival.
Alternative pre-mRNA processing is a major mechanism contributing to the diversification of genetic output, allowing nearly every primary transcript (Pan et al. 2008; Wang et al. 2008) to generate multiple alternative mRNAs. Through alternative mRNA splicing and polyadenylation, mRNA isoforms can be generated that differ in their protein-coding and noncoding sequences and thereby encode biochemically diverse polypeptides and RNAs subject to different post-transcriptional controls. Alternative RNA processing plays important roles in such diverse processes as cell differentiation and tissue development (Chen and Manley 2009; Nilsen and Graveley 2010; Kalsotra and Cooper 2011) and is increasingly recognized to be linked to human diseases (Licatalosi and Darnell 2006, 2010; Cooper et al. 2009).
The biologic diversity of alternative splicing is regulated in large part by tissue-restricted controls thought to be mediated by cell type-specific combinations of trans-acting factors binding to cis-acting RNA elements. Recent computational efforts to elucidate regulatory codes associated with alternative mRNA expression have defined combinations of motifs and regulatory factors that are prime candidates to regulate alternative RNA processing in different tissues (Sugnet et al. 2006; Castle et al. 2008; Barash et al. 2010; Zhang et al. 2010). However, such approaches to understanding tissue and developmental stage-specific RNA regulation require confirmatory biochemical and genetic evidence in relevant biologic contexts (Licatalosi and Darnell 2010).
The polypyrimidine tract-binding (PTB) proteins are an important family of RNA-binding proteins thought to be involved in cell-specific alternative splicing. Polypyrimidine tract-binding protein (PTB, hnRNPI, or Ptbp1) is a widely expressed and multifunctional RNA-binding protein with roles in mRNA stability, transport, translation, and pre-mRNA processing, including the repression of exons through binding to pyrimidine-rich sequences (Sawicka et al. 2008). Ptbp1 is able to antagonize the binding or activity of core splicing factors and interfere with either cross-exon or cross-intron interactions between components of the general splicing machinery (Wagner and Garcia-Blanco 2001; Spellman et al. 2005; Llorian et al. 2010). Recent global analyses of Ptbp1 function in HeLa cells have demonstrated that Ptbp1 binding within and upstream of alternative exons is associated with exon silencing and have identified a smaller number of Ptbp1-dependent exons in which Ptbp1 binding to sequences downstream from the regulated exon is associated with alternative exon inclusion (Llorian et al. 2010). These findings agree with observations originally made with the Nova family of splicing regulators and subsequently several other tissue-restricted RNA-binding proteins, including RbFox2, Mbnl1, Tia1/2, hnRNPC, hnRNPL, and TDP-43 (Licatalosi et al. 2008; Zhang et al. 2008; Licatalosi and Darnell 2010; Tollervey et al. 2011; Witten and Ule 2011), in which the position of protein binding determines the outcome of alternative splicing.
Ptbp1 is present in most mammalian tissues but is replaced in some cell types by paralogous proteins, including ROD1 in hematopoietic cells and Ptbp2 (originally termed brPTB [Polydorides et al. 2000] or nPTB [Markovtsov et al. 2000]) in the nervous system and testis. While Ptbp1 has been proposed to be a negative regulator of neuron-specific exons in nonneural tissues, the functions of Ptbp2 in the brain are not clear. For example, PTB-binding CU-rich motifs are enriched upstream of alternative exons that are repressed in nonneuronal tissues, which is consistent with a role for Ptbp1 in splicing repression but also raises the question as to whether Ptbp2 also has the same splicing activity in the nervous system (Castle et al. 2008). In neuroblastoma and HeLa cells, Ptbp1 and Ptbp2 regulate the alternative splicing of distinct sets of RNA targets, suggesting that the roles of Ptbp1/2 proteins may differ in different cellular contexts (Boutz et al. 2007; Spellman et al. 2007). Cell type-specific expression of PTB proteins can be regulated by tissue-restricted microRNAs (miRNAs) such as miR-124 (Makeyev et al. 2007) and by cross-regulation, as Ptbp1 binds to Ptbp2 pre-mRNA to introduce a premature termination codon and trigger degradation of Ptbp2 mRNA by the nonsense-mediated decay pathway (Boutz et al. 2007; Makeyev et al. 2007; Spellman et al. 2007). In cultured cells, knockdown of Ptbp1 expression by siRNAs or overexpression of miR-124 leads to increased Ptbp2 levels and subsequent alterations in pre-mRNA splicing that parallel those observed in cell culture models of neuronal differentiation (Boutz et al. 2007; Makeyev et al. 2007). Together, these studies identified a regulatory switch from Ptbp1 to Ptbp2 expression associated with neuronal differentiation in cell culture. The keys to understanding the function of Ptbp2 in the nervous system are the identification of its native RNA targets and a genetic model to investigate Ptbp2 function in vivo.
Here we present the results of a multifaceted approach to understanding Ptbp2 function in the mouse brain. We show that Ptbp2 is expressed in neuronal progenitors as well as in differentiated neurons and some astrocytes and is essential for postnatal survival and the proper splicing of hundreds of mRNAs in the brain. Using high-throughput sequencing cross-linking immunoprecipitation (HITS-CLIP) methods (Ule et al. 2003; Licatalosi et al. 2008; Darnell 2010; Licatalosi and Darnell 2010), we generated a transcriptome-wide map of direct Ptbp2–RNA interactions in vivo. These experiments revealed that the major action of Ptbp2 is to bind primary transcripts upstream of alternate exons to inhibit splicing of adult exons in the developing mouse brain. These target RNAs predicted previously unrecognized roles for Ptbp2 in suppressing developmentally regulated splicing events controlling the neural stem cell cycle and neuronal differentiation in mitotic neural progenitor cells.
Results
Ptbp2 is an essential protein that is broadly expressed in the nervous system
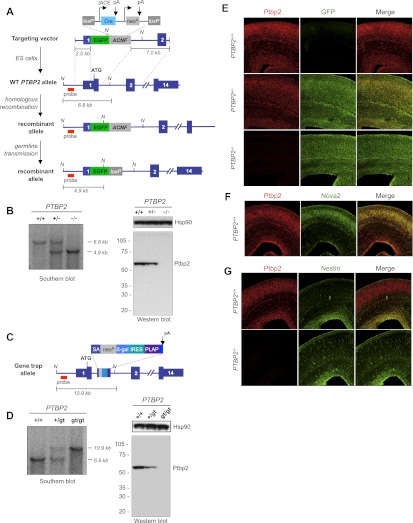
To generate a mouse model to study Ptbp2 function in vivo, Ptbp2 was targeted for homologous recombination in embryonic stem (ES) cells by replacing a 99-nucleotide (nt) DNA fragment (encompassing the translation start codon and first exon–intron junction) with DNA encoding EGFP and the self-excising ACNF cassette (Fig. 1A; Bunting et al. 1999). Animals homozygous for the recombinant allele (Ptbp2−/−) lacked Ptbp2, as demonstrated by Western blot analysis (Fig. 1B, right panel). Although grossly indistinguishable from wild-type (Ptbp2+/+) and heterozygous (Ptbp2+/−) littermates, Ptbp2-null (Ptbp2−/−) mice died soon after birth or within 10–15 min of delivery by cesarean section at embryonic day 18.5 (E18.5). Postnatal lethality was also observed in Ptbp2-null animals generated from ES cells bearing a gene trap cassette insertion in Ptbp2 intron 1 that abrogates expression of Ptbp2 (Ptbp2gt/gt) (Fig. 1C,D, right panel).
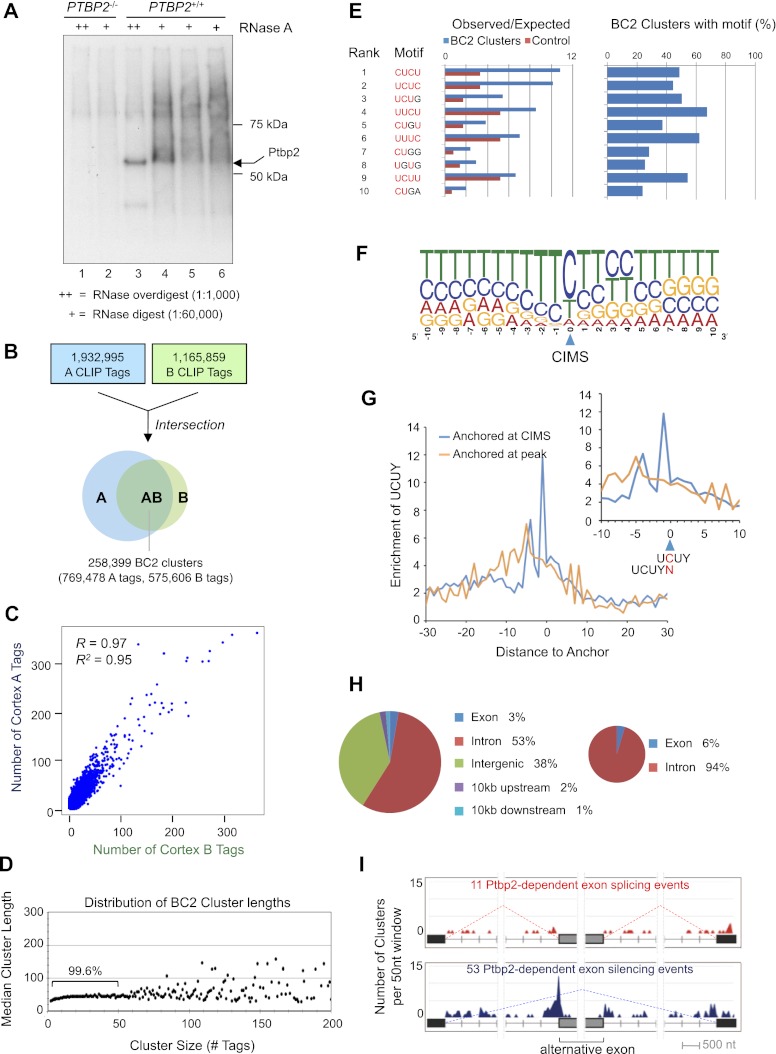
Figure 1.
Ptbp2 is broadly expressed in the developing mouse brain, including neuronal precursors. (A) Gene targeting strategy to generate Ptbp2-null mice, including schematic of the targeting vector, wild-type (WT) allele, and recombinant allele before and after Cre-mediated recombination at loxP sites in the male germline. (B) Southern blot confirmation of genotypes (position of probe indicated) and Western blot analysis of animals derived from the gene targeting strategy (whole brain E18.5) with polyclonal anti-Ptbp2 antibody raised against full-length protein (Polydorides et al. 2000) and anti-Hsp90. The position of the Southern blot probe is indicated, as are NcoI sites (N) used to digest genomic tail DNA. (C) Schematic of the gene trap Ptbp2 allele from ES cell line NPX210, bearing an intronic cassette consisting of a splice acceptor (SA), neomycin resistance (neoR) and β-galactosidase (β-gal) genes, an internal ribosome entry site (IRES), and coding sequence for placental alkaline phosphatase (PLAP). (D) Southern blot and Western blot analysis of animals derived from gene trap ES cells, as in B. (E) Sections of Ptbp2 wild-type (+/+), heterozygous (+/−), and Ptbp2 knockout (KO [−/−]) E18.5 brain are stained with antibodies to Ptbp2 and GFP. Ptpb2 is nuclear, while GFP is cytoplasmic, and cells with red nuclei and green cytoplasm are evident in heterozygotes. (F) Costaining of Ptbp2 and Nova2 in wild-type E18.5 brains. (G) Costaining of Ptbp2 and Nestin in wild-type and knockout E18.5 brains.
Immunohistochemical analyses demonstrated that Ptbp2 is broadly expressed in the embryonic and postnatal mouse brain (Fig. 1E–G; Supplemental Fig. S1); coincident staining of Ptbp2 and GFP (“knocked in” to the Ptbp2 locus) confirmed the specificity of Ptbp2 staining. The specificity of the Ptbp2 and GFP antibodies was also confirmed by the absence of appreciable immunofluorescence signal in Ptbp2−/− and wild-type (Ptbp2+/+) samples, respectively (Fig. 1E,G; Supplemental Fig. S1B,D,H). In the brain, Ptbp2 expression was observed in the nuclei of diverse cell types, including Nova2-positive cortical neurons (Fig. 1F), periventricular nestin-positive progenitor cells (Fig. 1G; Supplemental Fig. S1D,F,H), and, to a lesser extent, glutamine synthetase (GS)-positive astrocytes in the hippocampus, cerebellum, and cortex (Supplemental Fig. S1G). Ptbp2 expression was also observed in dorsal root ganglion of the spinal cord (Supplemental Fig. S1E) and in proliferating progenitor cells in the embryonic brain and eye (Supplemental Fig. S1C). Together, these observations indicate that Ptbp2 is expressed in neurons and astrocytes in the central and peripheral nervous systems in embryonic and postnatal mice, with robust expression in proliferating progenitors and post-mitotic neurons.
Ptbp2 is required for alternative mRNA regulation in the mouse brain
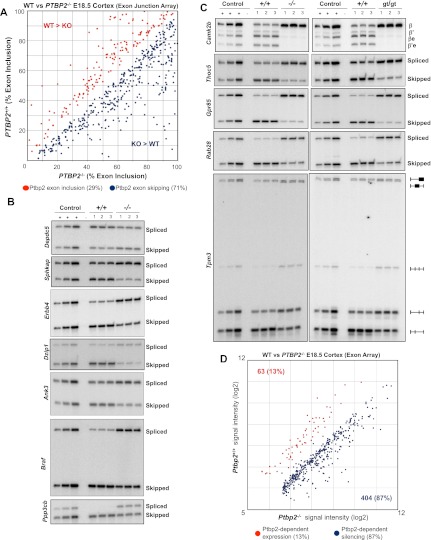
To determine whether Ptbp2 deletion affects the expression of alternatively spliced mRNAs in the developing brain, RNA from Ptbp2+/+ and Ptbp2−/− embryos was used to probe exon junction splicing-sensitive microarrays. ASPIRE3 analysis, previously used to identify reciprocal changes in the levels of alternatively spliced isoforms (Ule et al. 2005b; Licatalosi et al. 2008), identified 521 exons with significant splicing level differences in Ptbp2 knockout compared with wild-type brains (P < 0.01, ΔI Rank > 5) (Fig. 2A). These included changes in the expression of entire exons (cassette-type, mutually exclusive, and 5′- or 3′-terminal exons) or exon length due to alternative 5′ or 3′ splice site selection. These findings were validated by semiquantitative RT–PCR analysis of RNA from three pairs of Ptbp2+/+ and Ptbp2−/− littermates, confirming expression level changes for 36 of 36 candidate exons tested (Figs. 2, 6 [below]; Supplemental Table 1A; Supplemental Fig. S2). Notably, the majority of Ptbp2 action was to repress alternative splicing; i.e., there was increased expression (inclusion) of alternate exons in Ptbp2−/− brains in nearly three-quarters of instances (371 of 521, 71%). This was in contrast to the activity of Nova2 in the E18.5 mouse neocortex, which enhances the expression of approximately as many exons as it represses (47% repressed, 53% enhanced; n = 586 exons from age-matched E18.5 wild-type vs. Nova2-null cortices) (Supplemental Fig. S2N). Strikingly, some cases of Ptbp2-dependent splicing inhibition revealed major effects on exon inclusion; for example, from <50% in wild-type mice to near-maximal inclusion levels in Ptbp2 knockout neocortices (lower quadrant of Fig. 2A).
Figure 2.
Ptbp2-dependent alternative splicing in the brain. (A) Relative exon inclusion levels in wild-type (WT) and Ptbp2 knockout (KO) brains (ASPIRE3, P < 0.01, ΔIrank > 5). Red dots indicate alternative exons with higher inclusion in wild-type compared with knockout brains, and blue dots represent exons with increased inclusion in knockout compared with wild-type brains. (B) RT–PCR validation of seven ASPIRE candidates (additional candidates in are shown in the Supplemental Figures). In all cases, the control sample consists of an equal-parts mixture of wild-type and knockout cDNA amplified at different cycle numbers (see the Materials and Methods). (C) Comparison of alternative splicing in both Ptbp2-null strains (−/−, gene targeting; gt/gt, gene trap) (for additional examples, see Fig. 6). (D) Probeset signal intensities from MoEx 1.0ST analysis of wild-type and Ptbp2-null brains.
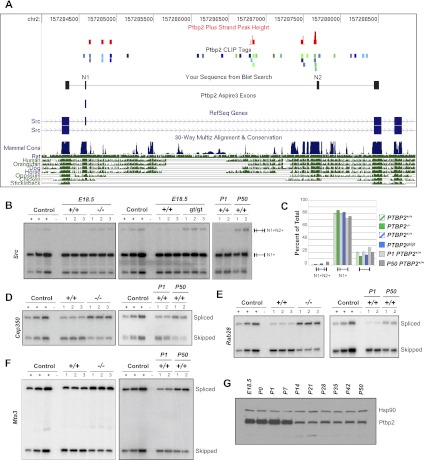
Figure 6.
Ptbp2 inhibits developmentally regulated adult-enriched alternative exons. (A) Distribution of Ptbp2 binding in a region of the c-src gene containing the alternative N1 and N2 exons. Also shown is a BLAT search result following Sanger sequencing of the N1+N2+ isoform purified from acrylamide gels shown in B. (B) RT–PCR analysis of c-src splicing in both Ptbp2-null strains (and wild-type littermates, left and center panels) and in neonatal (P1) and adult (P50) wild-type brains (right panel). Control lanes correspond to cDNA mixtures amplified at different cycle numbers (see the Materials and Methods). (C) Quantification of c-src RT–PCR products from B. (D–F) Analysis of Ptbp2-regulated exons in Cep350, Rab28, and Mta3 in wild-type and Ptbp2-null E18.5 brains (left) and wild-type neonatal and adult brains (right). (G) Western blot analysis of Ptbp2 and Hsp90 in wild-type brains at the indicated embryonic and postnatal days.
To independently examine Ptbp2-dependent mRNA variants, we also analyzed RNA from the second Ptbp2−/− strain (Ptbp2gt/gt), focusing on target exons that vary considerably in their sensitivity to Ptbp2 deletion. Importantly, neocortices from both Ptbp2-null strains (Ptbp2−/− and Ptbp2gt/gt) had nearly identical changes in expression of alternative mRNA variants compared with wild-type controls (Figs. 2C, 6B,C [below]). These data further validate the results of the splicing microarrays and are consistent with the shared postnatal lethal phenotype of both Ptbp2−/− and Ptbp2gt/gt mice.
Comparative analysis of RNA expression in Ptbp2+/+ and Ptbp2−/− E18.5 neocortices using a second microarray platform (Affymetrix MoEx 1.0ST exon arrays) also showed that a major consequence of Ptbp2 deletion is increased expression of alternatively spliced exons. An “exon-level” analysis (Clark et al. 2007) identified changes in the expression of 467 probe selection regions or “exons” (≥1.5-fold, P < 0.005) (Fig. 2D). The majority of these (404, 87%) showed an increase in alternative exon expression in Ptbp2-null embryonic brain, consistent with a role for Ptbp2 in repressing the steady-state level of specific alternatively spliced mRNAs. A transcript-level analysis showed that very few genes display significant differences in overall transcript abundance in Ptbp2−/− brains (Supplemental Table 1B,C). Two transcripts down-regulated in Ptbp2−/−, Snap25 and Dzip1, contain exons that are misspliced in Ptbp2−/− brains (see Figs. 5B, 2B, respectively). In these two cases, loss of Ptbp2 leads to inclusion of exons that introduce premature termination codons that are likely to cause nonsense-mediated mRNA decay, underscoring the conclusion that, in general, Ptbp2 does not appear to have direct, splicing-independent roles in regulating mRNA stability in the embryonic brain. Together, two different microarray platforms indicate that a major function of Ptbp2 in the embryonic mouse brain is to negatively regulate expression of specific alternative exons.
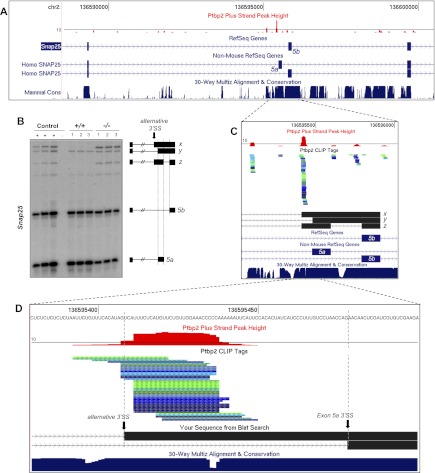
Figure 5.
Ptbp2 represses aberrant alternative 3′ splice site selection. Ptbp2 silences alternative 3′ splice site utilization in Snap25 pre-mRNA. (A) Distribution of Ptbp2 CLIP tags in a region of Snap25 containing the alternative 5a and 5b exons. CLIP tag density is represented by a histogram where the black line indicates a CLIP tag density of 10 overlapping tags. (B) RT–PCR analysis of Snap25 5a/5b splicing in wild-type and Ptbp2-null brains. (C) Zoom-in of a region of Snap25 showing Ptbp2 CLIP tags and splice site utilization and intron retention identified from PCR products x, y, and z. (D) Zoom-in of C showing the position of the Ptbp2 CLIP cluster relative to the alternative 3′ splice site up-regulated in Ptbp2-null brains.
Mapping biologically reproducible Ptbp2–RNA interactions in the mouse brain
Splicing microarrays delineate Ptbp2-sensitive transcripts but do not address whether these targets might be directly or indirectly regulated (Licatalosi and Darnell 2010). To identify direct Ptbp2 RNA targets in the mouse neocortex, a transcriptome-wide library of Ptbp2–RNA interactions was generated by HITS-CLIP (Ule et al. 2003, 2005a; Licatalosi et al. 2008). After UV irradiation of mouse neocortices, Ptbp2–RNA complexes were evident at or above (with high or low RNase concentration, respectively) the size of Ptbp2 (Fig. 3A), and complexes were isolated from two E18.5 littermates and processed in parallel (cortex A and cortex B) (Fig. 3B; Supplemental Fig. S3A). The specificity of the anti-Ptbp2 antibody used for CLIP was confirmed by the failure to precipitate radiolabeled RNA from UV cross-linked Ptbp2-null brains (Fig. 3A, lanes 1,2). Purified RNA from each biologic replicate neocortex CLIP sample was separated into four equal portions prior to CLIP cDNA library construction, and each was sequenced individually to generate technical replicates (Supplemental Fig. S3A). The resulting reads (ranging from 1.4 × 107 to 2.1 × 107 per replicate) were filtered and aligned to the mouse genome (Bowtie; mm9). Unambiguously mapped Ptbp2 CLIP tags were further collapsed to remove potential PCR duplicates, resulting in 1.9 × 106 neocortex A and 1.2 × 106 neocortex B unique tags, respectively (Supplemental Fig. Table S2A). These unique tags were pooled to identify Ptbp2–RNA interactions that were biologically reproducible, represented as 258,399 clusters (termed BC2, for biologic complexity = 2) containing overlapping CLIP tags from A and B neocortices (769,478 A tags [39.8% of total A] and 575,606 B tags [49.4% of total B]) (Fig. 3B; Supplemental Fig. S3A). The biologic reproducibility was assessed by comparing the number of A and B tags present in each cluster, which were highly reproducible between two different mouse brains (R2 = 0.95) (Fig. 3C). These observations allowed an approximation of the Ptbp2 footprint on RNA of ∼50 nt, as this was the length of the vast majority of Ptbp2 clusters, and this length plateaued with increasing numbers of tags/cluster (Fig. 3D).
Figure 3.
Ptbp2 HITS-CLIP. (A) Autoradiogram of nitrocellulose membrane containing protein–RNA complexes purified from Ptbp2-null (lanes 1,2) and wild-type (lanes 3–6) E18.5 mouse brains treated with a high (++) or low (+) concentration RNase A. (B) Summary of CLIP tag filtering and clustering to identify BC2 clusters. (C) Analysis of cortex A and B CLIP tag counts in each BC2 cluster. (D) Comparison of the length and numbers of CLIP tags for each BC2 cluster. (E) Top 10 tetramers present in BC2 sequences as determined by differences between the observed and expected frequency in BC2 compared with the shuffled control sequences. (F) CIMS-based identification of UCUY as a consensus binding site for Ptbp2. (G) Enrichment of UCUY near CIMS. (H) Genomic distribution of BC2 clusters (Ptbp2-binding events). (I) Distribution of BC2 clusters near alternative exons that are misspliced in Ptbp2-null brains. The total number (Y-axis) of BC2 clusters centered in each 50-nt window relative to alternative and flanking constitutive splice sites is shown for exons that exhibit increased skipping (top panel) and splicing (bottom panel) in Ptbp2 knockout compared with wild-type brains. Only BC2 clusters residing within the regions specified in the figure are displayed.
We used multiple measures to define CLIP tag clusters with higher stringency. As one measure, we performed a meta-analysis to rank clusters based on a χ2 score that reflects their intra- and interexperiment reproducibility, as detailed previously (Darnell et al. 2011). Using an empirical threshold (χ2 score ≥ 20), we obtained 19,767 high-confidence robust clusters, among which 18,998 clusters have tags identified in five or more technical replicates. We also searched CLIP tag cluster peaks in annotated genes that are statistically higher than one would expect from uniform random distributions (see the Materials and Methods for details; Xue et al. 2009). Among the robust clusters, about half (9972, 50.2%) are from 2842 annotated genes, and a vast majority (9861 or 98.9%) of these genic clusters have a peak height that was significantly higher than a uniform background distribution (P < 0.01, with Bonferroni correction; see the Materials and Methods for details). When we compared clusters in genic and deep intergenic regions (>10 kb from annotated genes), we found almost all deep intergenic clusters (94.7%) at least partially overlap with repetitive elements (LINE elements in particular; 64.5%), while genic clusters have a much lower proportion of repetitive clusters (46.7%). Taken together, our data demonstrate that Ptbp2 binds to thousands of annotated transcripts in mouse brains as well as a large number of unannotated regions.
An important asset of HITS-CLIP is that the method is able to very precisely identify RNA–protein interaction sites. To identify sequence features most commonly associated with Ptbp2–RNA interactions, nucleotide motifs (4–8 nt long) were counted in reproducible Ptbp2 HITS-CLIP clusters and, as a control, in a set of shuffled control sequences with the same nucleotide content. The most overrepresented features associated were CU- and UCU-rich sequences (Fig. 3E; Supplemental Fig. S3B), in agreement with previously defined sequence elements necessary for RNA binding and regulation by PTB proteins (Chan and Black 1997; Perez et al. 1997a; Reid et al. 2009b). Importantly, each of the top four ranking tetramers (CUCU, UCUC, UCUG, and UUCU) can be bound by each of the RNA recognition motifs of Ptpb1 (RRM1, RRM2, RRM3, and RRM4 bind YCU, CU[N]N, YCUNN, and YCN, respectively) (Oberstrass et al. 2005). Interestingly, G residues were relatively well tolerated within the pyrimidine-rich binding motif, but A residues were not. Together, these results indicate that the Ptbp2 CLIP clusters demarcate pyrimidine-rich sites of biologically reproducible Ptbp2–RNA interactions occurring in vivo.
To further confirm these motif patterns, we performed a cross-linking-induced mutation site (CIMS) analysis, which we previously demonstrated can determine protein–RNA interaction sites with single-nucleotide resolution (Zhang and Darnell 2011). We realigned reads with high sequencing quality scores using novoalign to identify reverse transcriptase errors at sites of RNA–protein cross-linking. As reported previously, we found that deletions, but not substitutions and insertions, are uniquely enriched in CLIP tags (Supplemental Table S2B), suggesting that deletions characteristically marked cross-link sites. Overall, one or more deletions were found in 7.4% unique CLIP tags, a rate comparable with that observed in Ago mRNA CLIP tags (∼8%) but lower than that observed in Nova CLIP tags (∼15%), implying that different amino acid–nucleotide cross-link adducts can result in variations in reverse transcriptase errors. We then clustered deletions according to their genomic coordinates and identified 1169 reproducible CIMS (false discovery rate [FDR] < 0.001). Examination of sequence composition anchored at CIMS clearly identified the UCUY motif (Fig. 3F), consistent with the high-affinity consensus of the Ptbp1 protein, as defined by in vitro RNA selection (Perez et al. 1997b). This motif is sharply enriched in specific positions around the CIMS, compared with flanking sequences (Fig. 3G). Importantly, the cross-link site is most frequently mapped to the cytidine of the UCUY element, which argues against presumed cross-linking bias to uridine in some published studies (Meisenheimer et al. 2000; Urlaub et al. 2002).
Ptbp2 negatively regulates splicing by binding upstream of and within alternative exons
Fifty-six percent of the BC2 clusters consisting of a minimum of 10 tags (13,647 of 24,529) mapped to annotated RefSeq genes (3362 RefSeq identifications), with 94% of these in introns (Fig. 3H). The prevalence of Ptbp2 binding to intronic sequences is consistent with a role for Ptbp2 in pre-mRNA processing and addresses concerns that CLIP would be unable to detect pre-mRNA due to the relative abundance of mature mRNAs in steady-state cells (Reid et al. 2009). Given this relative abundance, our HITS-CLIP and immunofluorescence data (Fig. 1; Supplemental Fig. S1) together demonstrate that the vast majority of Ptbp2 in the embryonic neocortex is in the nucleus.
To further explore the relationship between Ptbp2 binding and pre-mRNA splicing, we examined BC2 clusters near alternative exons identified from the splicing junction array analysis, restricting our analysis to exons subject to cassette-type regulation. Sixty-four Ptbp2-dependent cassette-type splicing events were identified (with ΔI > 0.1) in which at least one biologically reproducible Ptbp2 CLIP cluster was present within the entire local sequence region extending from the upstream to the downstream constitutive exons that flank the Ptbp2-sensitive exon (11 Ptbp2-dependent splicing events and 53 Ptbp2-dependent skipping). The coordinates of Ptbp2 CLIP-binding sites were compiled onto a single master transcript to view Ptbp2-binding patterns associated with multiple Ptbp2-dependent exons simultaneously. The region containing the greatest number of CLIP tags (Supplemental Fig. S3C) and BC2 clusters (Fig. 3I) extended from −250 to +50 relative to the 3′ splice acceptor site of regulated cassette exons that Ptbp2 silences in the brain. No single region could be identified that showed a marked preference for Ptbp2 binding in cases where exons are included in a Ptbp2-dependent manner, either in this analysis or with a normalized complexity map (Supplemental Fig. S3D; Licatalosi et al. 2008).
The presence of one or more high-density BC2 clusters within and/or the immediately preceding a silenced exon is a common feature of exons repressed by Ptbp2 (for representative examples, see Supplemental Fig. S3E–H). In some cases, BC2 clusters proximal to alternative exons are the densest CLIP tag clusters in the entire transcript and can reveal new Ptbp2-regulated exons not evident by our microarray analyses. For example, 60% of the Ptbp2 CLIP tags present in BC2 clusters mapping to Stam2 (signal transducing adaptor molecule 2) pre-mRNA reside in a single cluster immediately upstream of an exon that is alternatively spliced in different mouse tissues (Fig. 4A,B; Curlin et al. 2006), newly predicting a role for Ptbp2 its regulation. RT–PCR analysis of Ptbp2+/+ and Ptbp2−/− littermates confirmed that Ptbp2 is required for silencing of this exon in the embryonic neocortex (Fig. 4C).
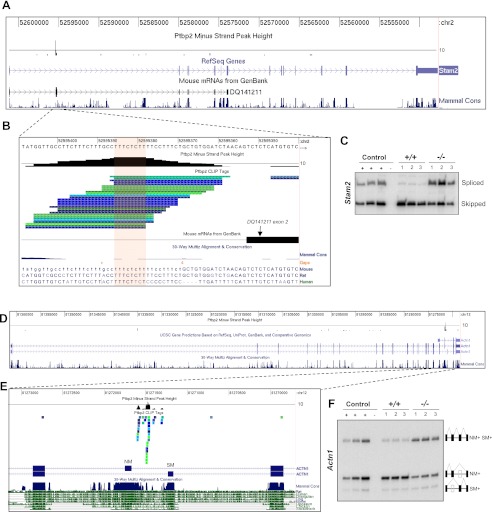
Figure 4.
Ptbp2 binding upstream of alternative exons silences exon inclusion in vivo. Examples of high-density BC2 CLIP tag clusters predicting Ptbp2 action to repress exon inclusion. (A) Distribution of Ptbp2 CLIP tags to the Stam2 gene. Tag density is represented by a histogram where the black line indicates a CLIP tag density of 10 overlapping tags. (B) Zoom-in view of a high-density Ptbp2 CLIP cluster immediately upstream of the Ptbp2-dependent Stam2 alternative exon. (C) RT–PCR confirmation of aberrant (increased) splicing of Stam2 exon 2 in Ptbp2-null brains compared with wild-type littermate controls. (D) Distribution of Ptbp2 CLIP tags to the Actn1 gene. (E) Zoom-in view of Ptbp2 binding in the intron flanking the alternative NM and SM exons. (F) RT–PCR confirmation of a role for Ptbp2 in the regulation of Actn1 NM/SM splicing.
The Ptbp2-binding map was also informative in cases where adjacent exons were spliced in a coordinated manner. For example, analysis of Ptbp2 binding to Dnm1 pre-mRNA revealed that the most dense BC2 cluster in the transcript was located upstream of an exon that is spliced in a mutually exclusive and Ptbp2-dependent manner with the preceding exon (Supplemental Figs. S2E, S4A,B). Similarly, coordinately regulated alternative exons in Tpm3 pre-mRNA sensitive to Ptbp2 deletion were associated with Ptbp2 BC2 clusters upstream of the Ptbp2-repressed exon (Supplemental Fig. S4C).
Control of mutually exclusive splicing by PTB proteins has been extensively studied in the regulation of the SM (smooth muscle) and NM (nonmuscle) exons in α-actinin 1 (Actn1) pre-mRNA, for which in vitro splicing and protein–RNA-binding assays have defined sites of Ptbp1–RNA interaction involved in SM/NM splicing regulation. Analysis of Ptbp2 CLIP tag distribution on Actn1 transcripts revealed that the greatest density of CLIP tags corresponded precisely to the regions determined to be high-affinity binding sites for Ptbp1 and necessary for Ptbp1-mediated splicing of Actn1 NM/SM exons in reporter constructs (Figs. 4D,E; Matlin et al. 2007). RT–PCR analysis of Ptbp2+/+ and Ptbp2−/− neocortexes confirmed that loss of Ptbp2 was associated with decreased expression of Actn1 alternative mRNAs containing the NM exon alone (from 87% of Actn1 mRNAs in wild-type to 48% in Ptbp2-null neocortexes) and an increase in Actn1 mRNAs containing both the NM and SM exons (from 7% of Actn1 mRNAs in wild-type to ∼44% in Ptbp2-null neocortexes) (Fig. 4F). Thus, the primary action of Ptbp2 binding to Actn1 pre-mRNA in the embryonic brain is to maintain expression of the NM isoform by repressing an Actn1 isoform containing both the NM and SM exons, and this likely alters the EF-hand domain and calcium sensitivity of Actn1 (Kremerskothen et al. 2002).
High CLIP tag density upstream of coordinately regulated exons revealed new aspects of Ptbp2 regulation. For example, multiple BC2 clusters mapped to evolutionarily conserved sequences flanking the 5a and 5b alternative exons in Snap25 pre-mRNA (Fig. 5A), and RT–PCR analysis of Snap25 mRNA in wild-type and Ptbp2 knockout mice confirmed a role for Ptbp2 in exon 5a/5b alternative splicing (Fig. 5B). However, we also noticed unanticipated high-molecular-weight RT–PCR products that became clearly evident in Ptbp2−/− neocortexes (Fig. 5B, labeled x and z), which resulted from the use of a novel 3′ splice site positioned 70 nt upstream of the annotated exon 5a 3′ splice site (Fig. 5C,D). Whereas RT–PCR products x and y may represent Snap25 pre-mRNA splicing intermediates (with retained introns), product z corresponds to spliced Snap25 mRNA containing the extended 5a exon (due to use of the upstream 3′ splice site) together with exon 5b, an isoform predicted to contain a premature termination codon. Again, these transcripts may be degraded by the nonsense-mediated decay pathway, consistent with the reduced Snap25 mRNA abundance predicted by microarray analysis (Supplemental Table S1B) and confirmed by RT–PCR (Fig. 5B). An additional example of Ptbp2 binding to prevent aberrant alternative 3′ splice site use is illustrated in Sympk pre-mRNA (Supplemental Fig. S5), supporting a role for Ptbp2 in maintaining splicing fidelity.
We also observed that Ptbp2 loss in the embryonic brain resulted in increased splicing of exons previously reported to be developmentally regulated or “adult-enriched.” Examples include aberrant splicing of adult alternative exons in c-src (Figs. 6A–C), Actn1, Snap25, Camk2b, and Dbn1 mRNAs (Pyper and Bolen 1990; Shirao et al. 1990; Brocke et al. 1995; Kremerskothen et al. 2002; Bark et al. 2004). To assess whether other Ptbp2 target exons are also spliced in a developmentally regulated manner, we compared expression of alternatively spliced target exons in wild-type neonatal (postnatal day 1 [P1]) and adult (P50) brains. This analysis identified a host of Ptbp2 target exons in the embryonic neocortex that are normally up-regulated during postnatal brain development. Of 31 splicing events analyzed, 28 showed developmental regulation in wild-type brains, and of these, 22 Ptbp2-regulated exons were increased in abundance in adult versus neonatal brains, including exons in Rab28, Mta3, Cep350, Sympk, Ppp3cb, Tpm3, Sphkap, Gpr85, Sorbs2, Dnm1, Pum2, Camk2g, and Dnm1 pre-mRNAs (Fig. 6D–F; Supplemental Fig. S6). These results are paralleled by finding that Ptbp2 levels decrease in postnatal mouse brains, beginning at a time of intense synaptogenesis (P14), with lower levels then maintained through young adulthood (Fig. 6G). These observations are consistent with the conclusion that Ptbp2 regulates the relative levels of expressed alternative mRNA isoforms in the neocortex through negative regulation of developmentally controlled exons.
Ptbp2 deletion impairs neural stem cell polarity and proliferation
To further explore the biologic functions of Ptbp2-mediated RNA regulation in the brain, we performed a gene ontology analysis using a list of 249 genes bearing Ptbp2-sensitive alternative exons identified from the splicing junction array (those with ΔI > 0.10, ΔI Rank > 5, P < 0.01). Among the recurring gene ontology terms enriched among Ptbp2-sensitive genes were plasma membrane, signaling, morphogenesis, cytoskeleton, and adhesion (Supplemental Tables S3A–D). Among the KEGG pathways associated with Ptbp2-regulated genes were multiple signaling pathways (including Vegf, Mapk, ErbB, and calcium signaling), many of which overlap with one another and impact on cell proliferation (Supplemental Fig. S7; Supplemental Table S3D).
The observations that Ptbp2 suppressed adult exon usage in development while coordinately regulating splicing in transcripts encoding factors that regulate cytoskeletal remodeling and cell proliferation led us to investigate whether Ptbp2 deletion affects cell proliferation in vivo. Since we observed strong staining of Ptbp2 in the mitotic ventricular zone of the embryonic brain (Fig. 1; Supplemental Fig. S1), we performed short-term BrdU-labeling experiments to label cell populations in S phase in E14.5 embryos. Immunohistochemical analysis of BrdU staining revealed mislocation of S-phase cells in Ptbp2-null brains, as evidenced by small ectopic BrdU clusters in the lateral wall of the ventricular zone (Fig. 7A,B). These ectopic clusters consisted of small numbers of cells (not all BrdU-positive cells were mislocalized), were not observed in wild-type or Ptbp2+/− brains, and were reproducibly observed in three of three biologic replicate Ptbp2−/− brains. Ectopic clusters often associated with mislocalized M-phase cells (identified by PH3 staining) residing away from the wall of the lateral ventricle, where stem cell-like neural progenitor cells undergo mitosis (Fig. 7B). In the Ptbp2−/− brain, clusters of aberrant cell division in the ventricular zone of the cortex and ganglionic eminence (GE) were associated with expression of Dcx, a marker of immature neurons (Fig. 7C,D, respectively), indicating ectopic neurogenesis. Importantly, ectopic Dcx-positive cells were Pax6-negative (Fig. 7E), indicating that in addition to aberrant neural stem cell polarity, Ptbp2 loss was associated with a reduction of the neural progenitor pool (Pax6-positive cells) and premature neurogenesis (Dcx-positive cells). Together, these analyses reveal a role for Ptbp2 in the proper regulation of neural progenitor proliferation and differentiation.
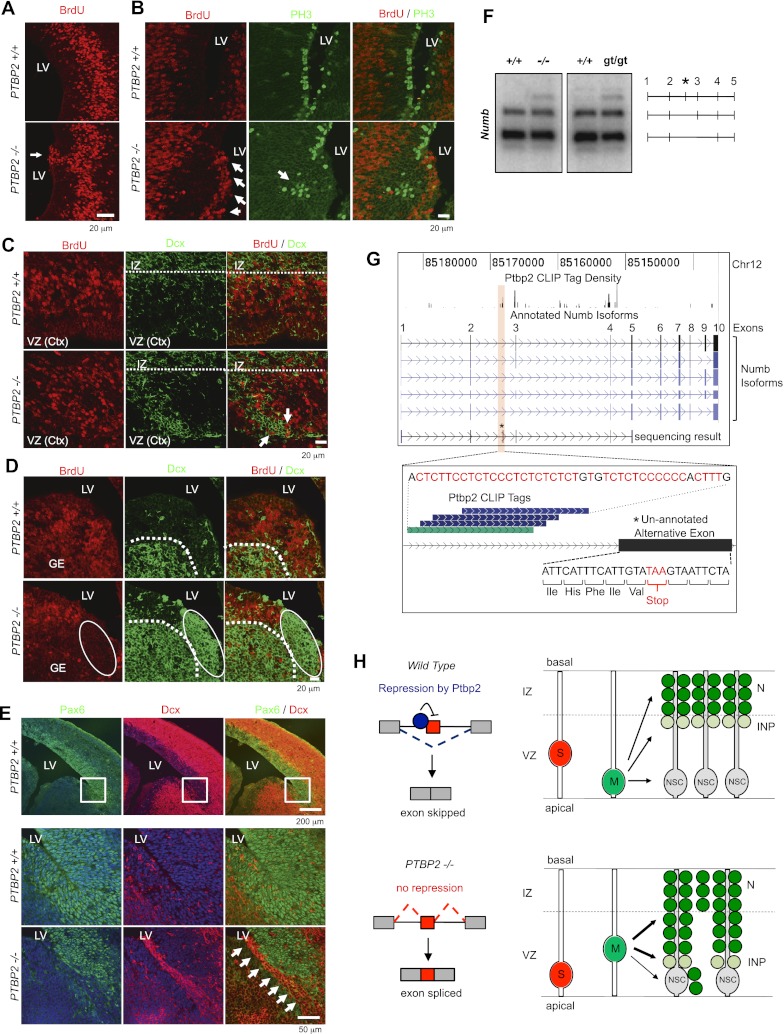
Figure 7.
Ptbp2 deletion results in aberrant neural stem cell polarity and premature neurogenesis. Immunohistochemical analysis of E14.5 wild-type (+/+) and Ptbp2-null (−/−) brains using BrdU (A), BrdU and PH3 (B), BrdU and Dcx (C,D), and Pax6 and Dcx (E) antibodies. (LV) Lateral ventricle; (IZ) intermediate zone; (VZ) ventricular zone; (GE) ganglionic eminence. (F) RT–PCR analysis of Numb pre-mRNA splicing in wild-type (+/+) and Ptbp2-null (−/−, gt/gt) brains to monitor splicing of an alternative exon (exon 3, using primers corresponding to exons 1 [forward] and 5 [reverse]) indicates expression of aberrantly processed Numb mRNA in both Ptbp2-null strains at E18.5. The identity of RT–PCR products is shown on the right, including an unannotated isoform (top band) containing an additional exon (asterisk, confirmed by Sanger sequencing). (G, top panel) Genome browser view of Ptbp2 CLIP tag density along the Numb gene, including annotated alternative mRNA isoforms and the unannotated isoform identified by sequencing RT–PCR products. (Bottom panel) Zoom-in view of a 79-nt region of Numb showing the unannotated Ptbp2-repressed alternative exon and a BC2 CLIP cluster centered ∼20 nt upstream of the 3′ splice site. (H) Model summarizing a role for Ptbp2 in exon repression and the consequences to neural stem cell polarity and cell cycle. The results of a HITS-CLIP analysis of Ptbp2 targets and the morphology of Ptbp2−/− mice suggest a model in which Ptbp2 normally acts on a regulatory cascade in neural progenitors cycling between S and M phase and differentiating into neural stem cells (NSCs) and indirect neural pathway (INP) progenitors in the ventricular zone (VZ). (Top panel) Ptbp2's immediate action is primarily to repress inclusion of alternative exons; in aggregate, these mediate changes in proteins (cell cycle, cell cytoskeleton, and cell fate) that inhibit aberrant cell cycle entry in periventricular progenitors. For example, Ptbp2 may inhibit “activating” exons in transcripts encoding cell cycle proteins, restrict cytoskeletal proteins to those that align the mitotic spindle orthogonal to the ventricular surface, and inhibit the function of regulators of basal cell division. (Bottom panel) In Ptbp2−/− mice brains, neural progenitors show increased cell cycle entry in the ventricular zone (A–D) and increased indirect neural pathway progenitors (E) that may result from skewed mitotic spindle orientation and/or actions on regulators of basal cell cycle.
Discussion
The functions of RNA-binding proteins have traditionally been studied using reductionist approaches, including the analysis of reporter constructs in vitro and in transfected cells. While mechanistically powerful, these studies do not yield a systems-biologic, global understanding of RNA-binding protein function in different physiologic and cellular contexts. Validation of the roles of specific RNA-binding proteins in RNA regulation in vivo requires biochemical and genetic systems to identify direct RNA–protein interactions and assess their functional significance in their native cellular context (Licatalosi and Darnell 2010). Here we used such a multidisciplinary approach to show that Ptbp2 is an essential RNA-binding protein that functions as a negative regulator of alternative exon splicing in the embryonic mouse brain, regulating transcripts encoding cell cycle and cytoskeletal proteins and culminating in the control of the genesis of neuronal precursors.
Ptbp2 regulates splicing of brain transcripts and is essential for postnatal survival
We generated Ptbp2-null mice using two independent approaches: a conventional gene targeting strategy, replacing an essential portion of the Ptbp2 gene with EGFP by homologous recombination (generating Ptbp2−/−), and using ES cells bearing an intronic gene trap, which prevents expression of full-length Ptbp2 mRNA while yielding readouts of Ptbp2 gene activity (EGFP and β-galactosidase/PLAP, Ptbp2gt/gt). Importantly, Ptbp2-null mice generated from both approaches exhibited neonatal lethality and nearly identical splicing changes in the target exons analyzed (these findings could not be attributed to compensatory changes in Ptbp1, whose levels were unchanged in Ptbp2-null brain Western blot analysis) (Figs. 2C, 6B,C; data not shown). For example, both Ptbp2-null lines had identical levels of Camk2b and c-src alternative exon inclusion, even though these transcripts vary considerably in their sensitivity to Ptbp2 deletion (Figs. 2C, 6B, respectively). Thus, both Ptbp2 deletion strains are useful tools to study Ptbp2 function during embryogenesis, and each contains different marker/reporter genes to facilitate monitoring of Ptbp2 expression, post-transcriptional control, and the properties of Ptbp2-expressing cells (Leighton et al. 2001).
Coordinated changes in the expression of alternative mRNAs significantly contribute to the diversification and fine-tuning of genetic output during cell differentiation (Kalsotra and Cooper 2011). Regulatory roles for PTB proteins have previously been described in a variety of experimental model systems. These include roles in the regulation of pre-mRNA processing (splicing and 3′ end formation) and mRNA localization, stability, and translation. Although recombinant Ptbp2 has been shown to stabilize one mRNA in testis extract (Xu and Hecht 2007), microarray analysis of Ptbp2−/− brains showed very few changes in the mRNA steady-state level in Ptbp2-null brains (other than Ptbp2 itself, only 12 mRNAs showed significant changes, ranging from 1.5-fold to 2.21-fold; P < 0.01, six up-regulated and six down-regulated). Among these, two of the genes encode RNA-binding proteins: Nanos2 and Brunol6. Two other pre-mRNAs (Dzip1 and Snap25) were confirmed by RT–PCR to be aberrantly spliced in Ptbp2 knockout brains to generate mRNA with a premature termination codon, thus likely targeting the transcripts for degradation by the nonsense-mediated mRNA decay pathway. While we do not exclude the possibility that Ptbp2 directly regulates the stability of the remaining 10 transcripts in a splicing-independent manner, we did not detect robust CLIP clusters in their 5′ and 3′ untranslated regions (UTRs) (data not shown), raising the possibility that these are indirect effects.
In contrast to the transcript-level analysis, we found an important role for Ptbp2 in shaping the transcriptome of the embryonic brain through alternative splicing. Ptbp2 binds predominantly to introns, where it acts on multiple transcripts to repress the expression of developmentally regulated alternative exons. Exon-level analysis performed with conventional and exon junction microarrays and ASPIRE3 identified hundreds of genes with differential expressions of exonic sequences in Ptbp2−/− brains. Notably, the vast majority (87% and 71%, respectively) exhibited increased splicing in the absence of Ptbp2, consistent with a major role for Ptbp2 as a repressor of exon splicing. Careful dissection of the actions of PTB proteins (primarily Ptbp1) in splicing assays has previously revealed a plethora of different actions that can result in splicing repression. Ptbp1 can interfere with U2AF65 binding to polypyrimidine tracts (Lin and Patton 1995; Singh et al. 1995; Spellman and Smith 2006), multimerize and loop RNA to block splicing factor access and create “zones of silencing” (Wagner and Garcia-Blanco 2001), or bind stem–loop IV of U1 snRNA directly (Sharma et al. 2011) to repress alternative exon splicing of the c-src N1 exon. Here we show that Ptbp2 directly binds c-src pre-mRNA in the mouse brain (Fig. 6A) and that deletion of Ptbp2 is associated with a small increase in N1 exon inclusion as well as increased expression of a developmentally restricted isoform of c-src mRNA containing both the N1 and N2 exons (Fig. 6B). Experiments in cell culture have suggested that Ptbp1 and Ptbp2 regulate splicing of overlapping and distinct sets of mRNAs and that Ptbp2 may be a weaker repressor of exon splicing than Ptbp1 (Spellman et al. 2005; Boutz et al. 2007; Makeyev et al. 2007). Bioinformatic approaches have identified an enrichment of UC-rich motifs upstream of alternative exons spliced in the brain and an anti-correlation between Ptbp1 expression and the level of alternative exon inclusion (Castle et al. 2008). While these observations are consistent with a role for Ptbp1 in splicing repression, they do not answer how Ptbp2 acts in the nervous system, where it replaces Ptbp1 and is speculated to be a less potent silencer of alternative exon splicing. Our analysis of Ptbp2 in mouse neocortexes demonstrates that Ptbp2 loss results in robust increases in exon inclusion—in some cases, near maximal levels. While we cannot exclude the possibility that some of the splicing changes in Ptbp2-null brains are mediated by indirect effects of Ptbp2 deletion, we would conclude that transcripts in which we see splicing changes and direct Ptbp2 binding in the region of the alternative exons are likely direct targets, consistent with our prior experience with Nova-dependent splicing maps (Licatalosi et al. 2008; Licatalosi and Darnell 2010). Moreover, we did not observe significant changes in the expression of genes encoding splicing, transcription, and chromatin regulatory factors in Ptbp2-null brains, nor did we observe these processes to be associated with gene ontology terms enriched among Ptbp2-dependent RNAs.
HITS-CLIP provided a means to identify the in vivo targets of Ptbp2 in an unbiased yet comprehensive manner. Consistent with the high degree of similarity between the RNA-binding domains of Ptbp1 and Ptbp2, reproducible Ptbp2-binding sites (BC2 HITS-CLIP clusters) were enriched in UCU-rich sequences. The HITS-CLIP map also illustrates functional similarities between Ptbp1 and Ptbp2. For example, Ptbp2 CLIP tags mapping to Actn1 pre-mRNA precisely overlap previously determined Ptbp1-binding sites associated with regulation of NM/SM exon splicing (Matlin et al. 2007). These results suggest that differences in Ptbp1/2-binding maps and biology likely reflect the action of cell-specific accessory factors rather than different binding preferences of the two proteins. Comparative in vitro studies using fragments of the c-src gene have suggested that Ptbp2 may not be an effective inhibitor of exon splicing. We show here that this claim was premature, since Ptbp2 does inhibit c-src splicing by direct binding to pyrimidine-rich sequences associated with exon N2, an exon whose splicing is always associated with inclusion of exon N1. Although the change in c-src splicing is small in Ptbp2-null compared with wild-type neocortexes, it was observed in all biologic replicates of both Ptbp2-null strains and was similar in magnitude to that observed in wild-type brains during postnatal development. We cannot exclude the possibility that a subset of neurons exhibits large changes in c-src splicing and that the magnitude of the splicing change during development is masked by analyses of whole cortexes.
The intersection of HITS-CLIP data with exons misspliced in knockout mice here parallels results observed with the neuron-specific RNA-binding protein Nova2 (Licatalosi et al. 2008), validating biochemical (Dredge et al. 2005) and bioinformatic approaches (Ule et al. 2006) and, in both cases, predicting position-dependent actions on alternative splicing, in which the position of binding in pre-mRNA determines the outcome of alternative exon inclusion or exclusion. Subsequent studies of Ptbp1 in tissue culture cells (Xue et al. 2009), in conjunction with reanalysis of these data (Llorian et al. 2010), as well as subsequent studies of HITS-CLIP maps of Fox1/2 (Zhang et al. 2008; Yeo et al. 2009), hnRNP C (Konig et al. 2010), MBNL1 (Du et al. 2010), TIA1/2 (Wang et al. 2010), and TDP-43 (Tollervey et al. 2011; for reviews, see Corrionero and Valcarcel 2009; Licatalosi and Darnell 2010; Witten and Ule 2011), reveal that such position-dependent actions are a general rule governing alternative splicing regulation to which we found Ptbp2 also adheres.
The genome-wide nature of HITS-CLIP allows exploration of new Ptbp2 actions. Annotation of Ptbp2 CLIP tags showed a surprising number of BC2 sequences (∼40%–50% of the most robust clusters) that map to unique genomic loci that do not overlap with annotated genes. The nature and functional significance of these interactions is currently being investigated, as is the functional significance, if any, of Ptbp2–RNA interactions observed in RefSeq 5′ and 3′ UTRs, although the latter comprise very minor fractions of RefSeq-mapped BC2 sequences (0.2% and 2.7%, respectively). Moreover, while recent global analyses of Ptbp1 function in HeLa cells have uncovered unexpected roles for Ptbp1 as an enhancer of splicing in a small cohort of exons, a consensus binding pattern associated with Ptpb2-dependent exon inclusion was less clear in our data. This may be due to the small number of exons included in this analysis, the lack of a general binding profile common to each target, or an indirect role in such regulation. Interestingly, we also found examples in which Ptbp2-binding sites mapped precisely to alternative 3′ splice sites misregulated in Ptbp2-null brains, further highlighting the resolution and power of the integrative analyses performed here.
Ptbp2 function in neuronal differentiation and development
Ptbp2 levels are high in early development, consistent with a role to inhibit adult exons and maintain fidelity of both neuronal progenitors and migrating neurons. Ptbp2 levels decrease with postnatal age (Fig. 6G), coincident with the alleviation of Ptbp2-mediated silencing of developmentally regulated exons and suggesting that tuning of the relative levels and activities of multiple trans-acting splicing factors that include Ptbp2 has a net action to regulate neuronal development. Reduced Ptbp2 protein level was not associated with detectable accumulation of the Ptbp2 splice variant associated with Ptbp2 post-transcriptional control (data not shown) and was modestly reduced during postnatal brain development, consistent with multiple layers of Ptbp2 post-transcriptional control, as previously suggested (Boutz et al. 2007). The decrease in Ptbp2 protein begins at approximately P14, a time of intense synaptogenesis and plasticity, and these levels are roughly maintained thereafter (at least until P50, the latest age we examined). This suggests that reduction of Ptbp2 action, and concomitant options to regulate adult exons, may be an important component of neuronal plasticity.
A convergent set of data have suggested a role for Ptbp2 in early brain development. In a number of cell culture models, induction of neural differentiation is associated with a miR-124-dependent switch from Ptbp1 to Ptbp2 expression and significant changes in alternative mRNA biogenesis. These observations, together with the finding that Ptbp2 is expressed in mature neurons (Boutz et al. 2007) and the absence of miR-124 in ventricular zone neural precursors, have suggested that the induction of progenitors into post-mitotic neurons is accompanied by a switch from Ptbp1 to Ptbp2 expression (Boutz et al. 2007), mediated by expression of miR-124 (Makeyev et al. 2007). Here, we clarify the role of Ptbp2 in neuronal precursors by analyzing Ptbp2-null mice, showing that Ptbp2 is expressed in neural progenitors and regulates splicing events associated with the regulation of neural stem cell polarity and suppression of premature neurogenesis.
In the rodent brain, the nucleus of neuronal progenitor cells resides at the apical surface of the ventricle during mitosis and in a more basal position during S phase. Although the molecular details that underlie nuclear transit in neuronal progenitors, known as interkinetic nuclear migration (INM), are not well understood, experimental evidence indicates a dependence on calcium signaling, cell junctions, and remodeling of the cytoskeletal architecture (Taverna and Huttner 2010), all of which are gene ontology terms overrepresented among the top Ptbp2 target mRNAs identified by our splicing microarray analysis. Transit of the nucleus during INM is associated with cell fate determination and is believed to control the exposure of nuclei to neurogenic or proliferative cues (Murciano et al. 2002; Baye and Link 2008; Del Bene et al. 2008). The loss of Ptbp2 protein from the ventricular zone in Ptbp2−/− brains was associated with reduced numbers of Pax6-positive cells and increased Dcx-positive cells, suggesting a skewing of the mitotic cycle and neurogenic pathway, perhaps consistent with an indirect pathway of neuronal differentiation. The irregularity of these cell clusters suggests that near-threshold phenomena are being breached. Such clusters may represent abnormal division and fate (neurogenesis) of ventricular neural stem cells, in which case the role of Ptbp2 would be to suppress cell fate decisions; for example, through modulation of cell signaling pathways (Supplemental Table S3D; Supplemental Fig. S7). Alternatively, cell orientation may be disturbed such that the balance between differentiation of dividing neural stem cells (ventricular) and dividing intermediate neural precursors (normally located away from the ventricular surface) is disturbed. Such scenarios may emerge from the disturbance of the fine temporal regulation of the splicing of Ptbp2 alternative exon targets encoding mitotic and neurogenic factors.
Presumably, such a role for Ptbp2 would relate to actions on specific mRNA targets, which might ultimately be identifiable with HITS-CLIP analysis specifically in neuronal progenitor populations. The list of RNA targets identified here (admittedly only an indirect approximation, given that targets were identified from whole neocortexes at E18.5) provides some clues. For example, differentiation of neuronal progenitors is coupled to orientation of the plane of cell division, which in turn is controlled by atypical PKC (Prkci) and its action on basal determinants of cell division (Numb, Brat [Trim3], prospero [Prox1], and others) (Knoblich 2010). Interestingly, Ptbp2−/− mice show defects in both cell fate and cell division pathways, including Erbb and Numb mRNA processing (Figs. 2B, 7F). We explored a role for Ptbp2 in Numb pre-mRNA splicing based on the presence of Ptbp2 CLIP tags near an alternative exon encoding part of the phosphotyrosine-binding domain important for Numb localization and interaction with signaling pathway components (exon 3 in Fig. 7G; Dho et al. 1999). RT–PCR analysis confirmed that expression of Numb alternative mRNAs is altered in both Ptbp2-null strains and identified an unannotated alternative exon containing an in-frame stop codon that is specifically expressed in Ptbp2-null brains (Fig. 7F,G). This observation indicates that Ptbp2-dependent splicing may function to regulate Numb protein levels in specific cell types; for example, through nonsense-mediated decay. We propose that a net action of Ptbp2 in neuronal progenitors is to suppress splicing of alternative exons, including developmentally regulated “adult” exons in transcripts encoding key determinants of cell fate, acting in aggregate to inhibit cell division, maintain cell polarity, and mediate the temporal control of neurogenesis (Fig. 7H).
Materials and methods
Generation of Ptbp2-null mice
A PTBP2 targeting construct was generated to replace part of exon 1 (including the translation initiation codon) and intron 1 with an EGFP-ACNF cassette. After ES cell selection, recombinant clones were identified by Southern blot and inserted into blastocysts, and chimeric mice were propagated after autoexcision of the ACNF cassette. A second line was generated from ES cell line NPX210 (BayGenomics), and gene trap mice were again confirmed by Southern blot.
Immunohistochemistry
Immunofluorescence microscopy for Ptbp2, GFP, Nestin, Nova, GS, BrdU, PH3, Dcx, Pax6, Hsp90 (BD Transduction Laboratories), and BrdU labeling was performed as described (Yano et al. 2010).
HITS-CLIP and data analysis
HITS-CLIP was performed on UV-irradiated tissue from E18.5 mice as described (Ule et al. 2005a; Licatalosi et al. 2008), using an anti-Ptbp2 polyclonal antibody (Polydorides et al. 2000) for immunoprecipitation. CLIP tags were sequenced on an Illumina GAIIX at the Rockefeller University Genomics Resource Center. Reads were mapped to the mouse genome (mm9) with Bowtie and collapsed to identify unique tags (Darnell et al. 2011). Significant CLIP tag cluster peaks were identified as described in the Supplemental Material, and CIMS analysis was performed as previously described (Zhang and Darnell 2011).
Microarray analysis
Microarray analyses were performed using Affymetrix Exon Junction (MJAY) and MoEx 1.0ST arrays, and data analysis was performed using ASPIRE as previously described (Licatalosi et al. 2008).
RT–PCR validation
RT–PCR was performed on RNA from mouse neocortical samples as described (Licatalosi et al. 2008), under conditions where product amplification was determined to be linear, with results quantified by PhosphorImager analysis using Quantity One (Bio-Rad). In all cases, control lanes 1–3 contain an equal mixture of wild-type and knockout cDNA amplified at n − 1, n, and n + 1 cycles as described in the Supplemental Material. Lane 4 corresponds to PCR performed at n + 1 cycles using an equal mixture of wild-type and knockout samples processed in the absence of reverse transcriptase (RT− control)
Acknowledgments
We thank members of the Darnell laboratory for advice and suggestions throughout the course of this work, and Scott Dewell for help with Illumina data analysis. We thank Jernej Ule and Melis Kayikci for running our exon array data on the ASPIRE3 program. We are grateful to the NIH for support to C.Z. (K99GM95713) and R.B.D. (NS40955 and NS34389). R.B.D. is an HHMI Investigator.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.191338.112.
References
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Bark C, Bellinger FP, Kaushal A, Mathews JR, Partridge LD, Wilson MC 2004. Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J Neurosci 24: 8796–8805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA 2008. Nuclear migration during retinal development. Brain Res 1192: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares MJ, Black DL 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocke L, Srinivasan M, Schulman H 1995. Developmental and regional expression of multifunctional Ca2+/calmodulin-dependent protein kinase isoforms in rat brain. J Neurosci 15: 6797–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR 1999. Targeting genes for self-excision in the germ line. Genes Dev 13: 1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40: 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RC, Black DL 1997. The polypyrimidine tract binding protein binds upstream of neural cell- specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol 17: 4667–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE 2007. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol 8: R64 doi: 10.1186/gb-2007-8-4-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, Valcarcel J 2009. RNA processing: Redrawing the map of charted territory. Mol Cell 36: 918–919 [DOI] [PubMed] [Google Scholar]
- Curlin M, Lucic V, Gajovic S 2006. Splice variant of mouse Stam2 mRNA in nervous and muscle tissue contains additional exon with stop codon within region coding for VHS domain. Croat Med J 47: 16–24 [PMC free article] [PubMed] [Google Scholar]
- Darnell RB 2010. HITS-CLIP: Panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H 2008. Regulation of neurogenesis by interkinetic nuclear migration through an apical–basal notch gradient. Cell 134: 1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho SE, French MB, Woods SA, McGlade CJ 1999. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem 274: 33097–33104 [DOI] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB 2005. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J 24: 1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. 2010. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol 17: 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA 2011. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA 2010. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremerskothen J, Teber I, Wendholt D, Liedtke T, Bockers TM, Barnekow A 2002. Brain-specific splicing of α-actinin 1 (ACTN1) mRNA. Biochem Biophys Res Commun 295: 678–681 [DOI] [PubMed] [Google Scholar]
- Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M 2001. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410: 174–179 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2006. Splicing regulation in neurologic disease. Neuron 52: 93–101 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Patton JG 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1: 234–245 [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T 2007. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol 20: 7463–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Southby J, Gooding C, Smith CW 2007. Repression of α-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA 13: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenheimer KM, Meisenheimer PL, Koch TH 2000. Nucleoprotein photo-cross-linking using halopyrimidine-substituted RNAs. Methods Enzymol 318: 88–104 [DOI] [PubMed] [Google Scholar]
- Murciano A, Zamora J, Lopez-Sanchez J, Frade JM 2002. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci 21: 285–300 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. 2005. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science 309: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Perez I, McAfee JG, Patton JG 1997a. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 36: 11881–11890 [DOI] [PubMed] [Google Scholar]
- Perez I, Lin CH, McAfee JG, Patton JG 1997b. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778 [PMC free article] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci 97: 6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyper JM, Bolen JB 1990. Identification of a novel neuronal C-SRC exon expressed in human brain. Mol Cell Biol 10: 2035–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DC, Chang BL, Gunderson SI, Alpert L, Thompson WA, Fairbrother WG 2009. Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence. RNA 15: 2385–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE 2008. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem Soc Trans 36: 641–647 [DOI] [PubMed] [Google Scholar]
- Sharma S, Maris C, Allain FH, Black DL 2011. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol Cell 41: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirao T, Kojima N, Terada S, Obata K 1990. Expression of three drebrin isoforms in the developing nervous system. Neurosci Res Suppl 13: S106–S111 doi: 10.1016/0921-8696(90)90039-6 [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176 [DOI] [PubMed] [Google Scholar]
- Spellman R, Smith CW 2006. Novel modes of splicing repression by PTB. Trends Biochem Sci 31: 73–76 [DOI] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW 2005. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans 33: 457–460 [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW 2007. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27: 420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. 2006. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol 2: e4 doi: 10.1371/journal.pcbi.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Huttner WB 2010. Neural progenitor nuclei IN motion. Neuron 67: 906–914 [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB 2005a. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods 37: 376–386 [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. 2005b. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37: 844–852 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Urlaub H, Hartmuth K, Luhrmann R 2002. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods 26: 170–181 [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol 21: 3281–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J 2010. iCLIP predicts the dual splicing effects of TIA–RNA interactions. PLoS Biol 8: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J 2011. Understanding splicing regulation through RNA splicing maps. Trends Genet 27: 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hecht NB 2007. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3′UTR. Biol Reprod 76: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB 2010. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66: 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 29: 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB 2010. Integrative modeling defines the nova splicing-regulatory network and its combinatorial controls. Science 329: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]