Abstract
Honeybees produce silken cocoons containing four related fibrous proteins. High levels of each of the honeybee silk proteins can be produced recombinantly by fermentation in E. coli. In this study we have used electrospinning to fabricate a single recombinant honeybee silk protein, AmelF3, into nanofibres of around 200 nm diameter. Infrared spectroscopy found that the molecular structure of the nanofibres was predominantly coiled coil, essentially the same as native honeybee silk. Mats of the honeybee nanofibres were treated with methanol or by water annealing, which increased their ß-sheet content and rendered them water-insensitive. The insoluble mats were degraded by protease on a time scale of hours to days. The protease gradually released proteins from the solid state and these were subsequently rapidly degraded into small peptides without the accumulation of partial degradation products. Cell culture assays demonstrated that the mats allowed survival, attachment and proliferation of fibroblasts. These results indicate that honeybee silk proteins meet many prerequisites for use as a biomaterial.
Keywords: Silk, Electrospinning, nanofibres, coiled coil, Apis mellifera
1. Introduction
The field of tissue engineering has invested considerable efforts into development of scaffolds that mimic the structure and the biological functions of the extracellular matrix (ECM). The ECM is principally constructed from nano-scale fibers composed of the proteins collagen, elastin, fibronectin and laminin, as well as proteoglycans, glycosaminoglycans and hyaluronic acid. Much of the information guiding cells to their correct location, cell anchorage and cell proliferation is encoded in the protein sequences of the ECM. The native proteins of the ECM have therefore attracted much attention as tissue engineering scaffold materials. Collagen in particular has been of interest because it is non-toxic, produces a minimal immune response, promotes attachment and proliferation of a range of cells, is resorbed in the body and can be fabricated into different forms (reviewed in [1]). However, collagen that is regenerated from natural sources is variable in composition and behavior, and requires chemical treatments to generate suitable mechanical properties. In order to achieve high levels of reproducibility and control, the research community has developed alternative artificial scaffolds such as poly(ethylene glycol), poly lactic acid, poly caprolactone, polyphophates, ceramics and metals. Although these materials provide suitable bulk and mechanical properties for a range of tissue engineering applications (reviewed in [2]), generally they lack chemical cues to promote good cell attachment and proliferation. The most widespread current strategy to improve cell attachment is the generation of composite scaffolds where naturally sourced ECM proteins, usually collagen, are used to coat scaffolds generated from synthetic materials.
A possible alternative material for construction of tissue scaffolds is the silk of honeybees. Honeybees spin silk threads that they use to structurally reinforce their soft waxen hives. The threads are strong and highly extensible (132 MPa breaking stress; 204% breaking strain [3]). The silks are composed of four small (approximately 32 KDa), non-repetitive proteins that do not require post-translational modification [4]. The proteins can be expressed recombinantly in E. coli at high yield [5] and potentially at large scale. Circular dichroism and dynamic light scattering studies of recombinant honeybee silk proteins show that they readily self-assemble into their native coiled coil structure [6–8]. Solutions of recombinant proteins can be fabricated into fibers with mechanical properties approaching those of the native silk [3]. Although the honeybee silk proteins are particularly rich in alanine (29–33%), they contain a wide variety of other amino acids, including 20–25% charged amino acids (Arg, Asp, Glu and Lys) and substantial levels (4–8%) of leucine, asparagine, glutamine, threonine and valine [4]. The combination of a consistent protein production method, good mechanical properties, and the chemical functionalities arising from the complex amino acid composition make honeybee silk unlike any existing biomaterial and of considerable interest. Furthermore, the proteins have low constraint on their primary sequence [9], and therefore offer the potential to be engineered to encode signaling sequences from the ECM that direct specific cell adhesion.
Electrospinning is a technique that generates nanoscale fibers from charged solutions of concentrated polymers. As the topology of electrospun fibers resembles parts of the ECM, electrospun fibers of synthetic polymers and regenerated natural polymers have been thoroughly explored as scaffolds for tissue engineering applications (reviewed by [10]). In this work we electrospun nanoscale fibres of recombinant honeybee silk protein, and systematically describe their structure, cell response and in vitro enzyme degradation, in order to determine the suitability of electrospun honeybee silk as a scaffold biomaterial.
2. Materials and methods
2.1. Preparation of honeybee silk proteins
Honeybee silk protein AmelF3 was expressed into the inclusion bodies of E. coli as described by Weisman et al. [5]. Inclusion bodies were purified using BugBuster Master Mix (Novagen) according to the manufacturer's protocol and then the silk protein was solubilised in 3% sodium dodecyl sulfate (SDS) with 2 h incubation at 60°C. Detergent was extracted by addition of KCl (300 mM final concentration) causing precipitation of potassium dodecyl sulfate (KDS). KDS precipitate was removed by centrifugation at 16,000 g for 5 min and the solution was dialyzed against 20% PEG 8000 until the protein concentration reached 12.5%. Silk/polyethylene oxide (PEO) blends were prepared by adding the required amount of 5.0% (wt/vol) PEO (900,000 MW, Sigma) into the silk solution and gently stirring for 20 min.
2.2. Electrospinning
The silk/PEO solution was delivered through a 16G stainless steel capillary maintained at high electric potential at a flow rate of 5 – 10 μl/min using a Sage syringe pump (Thermo Scientific). The collector was a grounded aluminum foil placed on a 10 cm diameter aluminum plate. Silk was collected directly on the aluminum foil except for the cell study, where it was collected on 9×9 mm2 glass cover slips that were placed on top of the usual aluminum foil. The applied voltage, solution flow rate and distance from capillary to collector for each solution are described in Table 1. Electrospinning was conducted at room temperature (20–22°C) and humidity levels of 16–17%.
Table 1.
Conditions for electrospinning recombinant honeybee silk protein solutions.
| Additive | Silk:PEO ratio | Fiber diameter (nm) | Voltage (kV) | Distance (cm) | Flow rate (ml/min) | Fiber quality |
|---|---|---|---|---|---|---|
| 1% PEO | 12.5 : 1 | 200 | 9.8 | 15 | 0.015 - 0.008 | Uniform diameter |
| 0.7% PEO | 18.7 : 1 | 200 | 10 | 15 | 0.015 - 0.008 | Uniform diameter |
| 0.5% PEO | 25.0 : 1 | 200 | 9 – 12 | 15 – 20 | 0.02 – 0.008 | Fibers uneven |
| 0.4% PEO | 31.3 : 1 | <100 | 9 – 12 | 15 – 20 | 0.02 – 0.008 | Fibers beaded |
| None | - | - | 10 – 20 | 10 – 20 | 0.02 | No fibers obtained |
2.3. Microscopy
Electrospun fibers were analyzed by scanning electron microscopy (SEM) and polarized optical microscopy. For SEM imaging, the silk mats were sputter-coated with platinum/palladium using a 208 HR Sputter Coater (Cressington) and examined using a Zeiss Ultra 55 FESEM at 5 kV. Polarized optical microscopy of mats was performed on a phase contrast microscope with a TCS SP2 scanner (Leica Microsystems).
2.4. Fourier Transform Infrared spectroscopy (FTIR)
FTIR analysis of the electrospun mats were performed with a Jasco FT/IR-6200 Spectrometer, equipped with a deuterated triglycine sulfate detector and a multiple reflection, horizontal MIRacle ATR attachment (Ge crystal, Pike Tech). The instrument was continuously purged with nitrogen gas to remove atmospheric water vapor. Each measurement incorporated 128 scans (wavenumber from 600 to 4000 cm−1) that were Fourier transformed using a Genzel-Happ apodization function to yield spectra with a nominal resolution of 4 cm−1. Secondary structure peak assignments were as described in [11–13]: absorption bands in the frequency range of 1610~1625cm−1 represent β-sheet structure; absorption bands around the frequencies of 1650 cm−1, 1640 cm−1, and 1630 cm−1 were ascribed to the coiled-coil fingerprint spectrum, and peaks above 1660 cm−1 were ascribed to β-turns.
2.5. Cell culture
Green fluorescent protein (GFP)-expressing rabbit corneal fibroblasts (rCFs) were generated after isolating primary stromal fibroblasts cells from excised rabbit corneas (Pel-Freeze) as previously described [14,15] and cultured in rabbit fibroblast media (rFB media) consisting of Dulbecco's modified Eagle's medium, 10% fetal bovine serum, 1% penicillin-streptomycin-fungizone, and maintained at 5% CO2. The previously described lentivirus system [16] was used to generate the GFP-expressing line of rCFs (GFP-rCFs). The rCFs were transduced at multiplicity of infection (MOI) of 1, with 5 mL of virus-containing supernatant (100,000 virus particles/mL) added to 5×105 rCFs. An additional 5 mL of rFB media and protamine sulfate (6 mg/mL) was added to enhance infection. The cells were incubated for 3 h with the lentivirus then washed twice with PBS before adding rFB media. Fluorescence microscopy and fluorescence-activated cell sorting analysis was used to evaluate the efficiency of GFP transduction. Stable GFP-rCFs were determined by the presence of fluorescence in cells over multiple cell passages. For seeding, GFP-rCFs were grown to confluence, detached from their substrates using 0.05% Trypsin (Gibco), and then re-plated at passage 10–12 (P10–12) onto prepared electrospun honeybee mats as described below.
For cell adhesion and proliferation assays the electrospun mats on 9×9 mm2 glass cover-slips were treated with methanol for 1 hr to make them water-insensitive, air dried for 2 h then placed in low binding 24-well plates (Ultra Low Cluster Plate, Corning). The mats were sterilized by submerging in 70% ethanol for 20 min. The FTIR spectrum of mats after ethanol treatment was the same as before treatment indicating that the ethanol did not affect silk protein structure. Ethanol was aspirated and the fibers were air-dried for 30 min then soaked in deionised water for 12 h to leach out the PEO. The mats were finally soaked in rFB media for 2 hr before cell seeding. GFP-rabbit corneal fibroblasts (P10–12) were seeded on the mats at low density (5 × 103 cells/well). Cells and mats were incubated for 7 days at 37°C and 5% CO2 with the media replaced with fresh media on days 1, 3, 5 and 7.
The cell number and coverage on the mats was measured in quadruplicate at days 1, 3 and 7 after staining cells with AlamarBlue as follows: the media was replaced with 500 μL of fresh media and 50 μL of AlamarBlue and the sample incubated for 4 h at 37°C and 5% CO2 then fluorescence emission at 610 during exposure to 570 nm light was measured using a microplate reader (Molecular Devices Corp) and analyzed with Softmax Pro 5. Cell number was calculated from a standard curve generated from the fluorescence emission of 0, 2,000, 5,000, 10,000, 20,000 or 40,000 cells per well in an equivalent 24-well plate. The cell coverage on the mats was estimated using ImageJ on six representative pictures for each sample by quantifying the amount of green fluorescence coming from the cells in comparison to the amount of surrounding black.
2.6. In vitro proteolytic degradation
Biodegradability of electrospun mats was assessed in vitro by incubation with α-chymotrypsin (TLCK-treated, activity 54 U/mg protein; Sigma-Aldrich) and trypsin (sequencing grade modified trypsin, activity 313 U/mg protein; Promega). α-chymotrypsin cleaves protein amide bonds at the carboxyl group of accessible F,Y,W,M or L residues (but not before P) and trypsin cleaves at the carboxyl group of accessible K or R residues (but not before P), therefore the proteases are predicted to cleave the honeybee AmelF3 protein up to 27 or 36 times, respectively, to generate single amino acids or peptides up to 73 amino acids in length.
Samples of silk mat were immersed in methanol for 10 min to make them water insensitive, and then weighed with a microbalance (CAHN C-31). Sample weights varied between 142 and 318 μg. The samples were washed in deionized water overnight, sterilized by treatment in 70% ethanol for 20 min, rinsed in sterile PBS, then incubated in freshly prepared solutions of α-chymotrypsin (1.3 U/ml) in 24-well plates at 37 °C for 1, 3, 8, 24, 48 or 96 h. During longer incubations, 1.3 U/ml fresh enzyme was added every 24 hrs. Mat fragments remaining after 24 hrs incubation with α-chymotrypsin were stained with commassie blue protein dye, collected by centrifugation and subsequently digested with trypsin. Control mats were immersed in PBS buffer without enzyme. At each time point, solutions were collected and heated at 80°C for 10 min to denature any remaining protease. Where possible, samples were air dried for 2 days and weighed. The amount of protein in solution was determined using the QuantiPro bicinchoninic acid (BCA) protein assay (Sigma-Aldrich), which detects peptides containing three or more residues. As 5% of the residues of AmelF3 protein (16 amino acids of the total 320) are predicted to be cleaved into fragments of less than 3 residues, the BCA assay results are expected to reflect an underestimation of the total peptide amount in α-chymotrypsin digested solutions of up to 5%. Nu-PAGE 12% Bis-Tris SDS polyacrylamide gel electrophoresis (SDS-PAGE; Invitrogen) was used to separate and estimate the molecular weight of degradation products. Liquid chromatography / mass spectrometry (LC/MS) experiments on peptide solutions were carried out on an Agilent LC/MSD Trap XCT instrument and analyzed by Agilent SpectrumMill software.
3. Results and Discussion
3.1. Electrospinning parameters and characterization of electrospun fibers
Solutions containing only honeybee silk protein (>95% purity as estimated by SDS-PAGE) at 12.5% (w/v) did not electrospin at any of the voltage and distance to collector combinations tested. Although the protein solution was able to maintain a stable droplet at the capillary tip, no jet was observed. Higher concentrations of AmelF3 solutions gelled and therefore could not be electrospun. When PEO was blended into the silk solution at concentrations below 0.5% (w/v) PEO, fibers could be electrospun. However, the fibers were non-uniform and frequently exhibited beading at all voltage and distance combinations tested (Figure 1A). When PEO was included in the silk solution at 0.67% or 1.0%, uniform fibers could be electrospun under conditions described in Table 1.
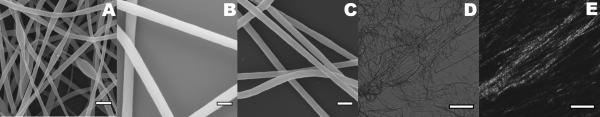
Figure 1.
Honeybee silk electrospun fibers. Scanning electron microscopy images of fibers from 12.5% AmelF3 protein solutions containing 0.4% (A), 0.67% (B) and 1% (C) PEO. Scale bar: 500 nm. Bright field (D) and polarized (E) light microscopy images of the same area of electrospun fibers, demonstrating fiber birefringence under polarized light. Scale bar: 100 μm.
The morphologies of individual fibers and fibrous mats were examined by SEM. The fibers spun from 12.5% silk protein in 1.0% and 0.67% PEO blends were uniformly cylindrical and approximately 200 nm in diameter (Figure 1B and 1C). The molecular alignment within the fibers was examined by polarized light microscopy. When imaged under cross-polarized light, the fibers were birefringent, indicating that a substantial degree of molecular orientation had occurred during the spinning process (Figure 1D and 1E). Birefringence was lost by post-spinning treatments and therefore it is likely that the feature was attributable to alignment of the PEO, which was washed out of the fibers in the post-spinning treatments, rather than alignment of the silk protein. Molecular alignment of PEO, visualized by birefringence, has previously been reported to occur during electrospinning [17].
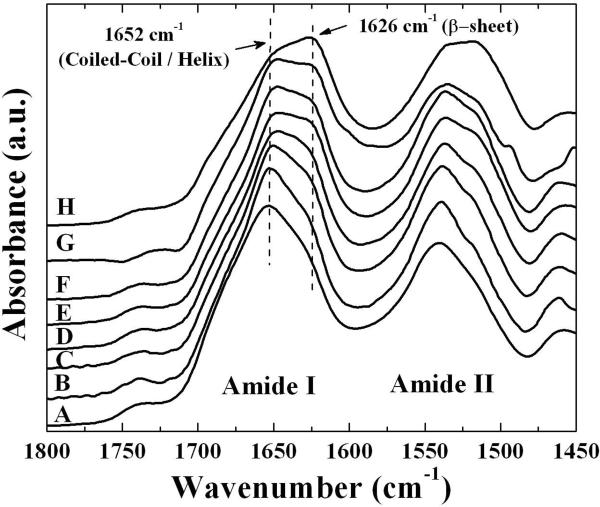
The protein structure of the electrospun fibers was analyzed by infrared spectroscopy. The FTIR spectrum of the untreated fibers showed a dominant feature at 1652 cm−1 with a shoulder at 1626 cm−1 in the amide I region (Figure 2A). This spectrum is very similar to the spectra observed for films cast from recombinant honeybee silk and for native silk [5], and is consistent with the silk proteins having a predominantly coiled coil molecular structure with a lower proportion of ß-sheet,.
Figure 2.
Fourier transformed infrared (FTIR) spectra of the amide I and II regions of electrospun honeybee silk before and after treatments to make the fibers water insensitive. As spun fibers (A), after water annealing for 7 min (B), 15 min (C), 30 min (D), 60 min (E), 120 min (F), overnight (G); and after methanol treatment (H). Assignments of bands to structures are described in the text.
3.2. Structural changes induced by post-spinning treatments of fibers
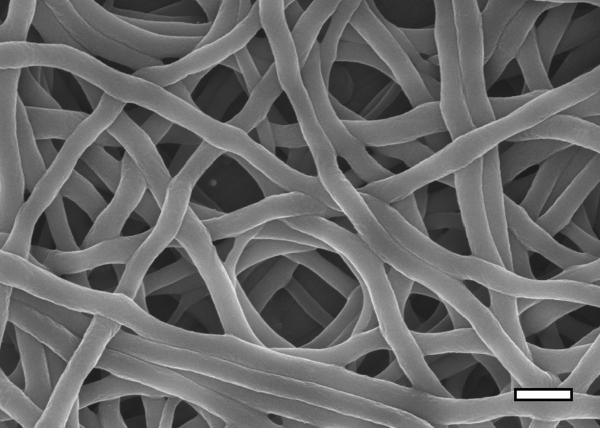
Two post-spinning treatments were employed to render the honeybee electrospun fibers insoluble: water annealing and methanol treatment. Both methods are commonly used to induce ß-sheet formation in reconstituted silkworm silk and result in water insensitive material [18–22]. Methanol treatment resulted in the surface of fibers becoming rougher (Figure 3). Similar effects seen for silkworm silk were attributed to phase separation between the silk proteins and PEO during extraction of the PEO in the methanol bath [23]. PEO was observed washing out from the honeybee silk fibers and forming opaque droplets during the methanol treatment, so it is likely that the roughness of the fibers relates to this process. FTIR analysis demonstrated that the molecular structure of the fibers changed as the material became insoluble, with both methanol and water annealing treatments strengthening a spectral peak at 1626 cm–1, indicating an increase in ß-sheet content in the silk proteins (Figures 2B–H). Water annealing led to a gradual increase in ß-sheet content over at least 16 hrs (Figure 2B–G) whereas methanol treatment had a rapid greater impact on the ß-sheet content (Figure 2H). Apparently the ß-sheet content of the electrospun honeybee silk nanofibres can be directly controlled by varying post-spinning treatments, allowing structural modification of this material for different biomedical applications.
Figure 3.
Scanning electron microscopy image of electrospun honeybee silk after methanol treatment to make fibers water-insensitive. Scale bar: 500 nm.
3.3. Cell response to honeybee nanofibres
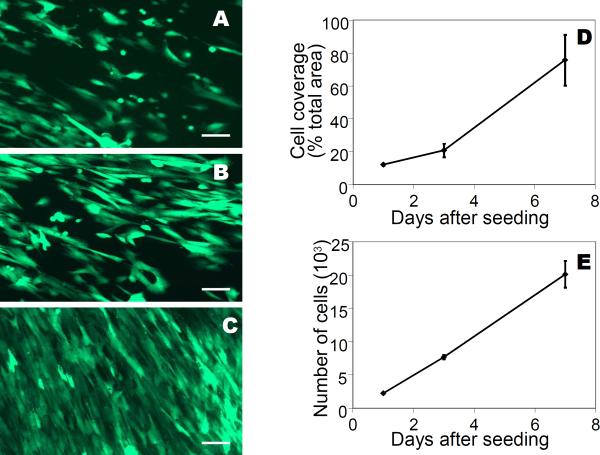
Fibroblasts are a standard cell type to assess compatibility in preliminary screening of biomaterials. Analysis is simplified when the fibroblasts are modified to express green fluorescent protein (GFP) to enable imaging through non-transparent mats using fluorescence microscopy. We therefore used GFP expressing-fibroblasts in order to evaluate cell adherence and proliferation on the electrospun honeybee silk fiber mats. Microscopic analysis of the electrospun honeybee silk mats showed that cells adhered and spread at day 1, were proliferating at day 3 and reached confluence by day 7 (Figure 4A–C). The absolute cell surface area on the mats, calculated from the surface green fluorescence, increased around six times from 12% of the total available area on day one to 75.6% on day 7 (Figure 4D). The number of cells on the mats, determined by an AlamarBlue assay, increased nine-fold from 2.2 × 103 on day 1 to 20 × 103 on day 7 (Figure 4E). These measurements reflect the fact that the cells were confluent toward the end of the experiment with comparatively less space to spread than at the beginning of the experiment. The results show that cells are able to survive, adhere and proliferate on honeybee electrospun mats.
Figure 4.
Adherence and proliferation of cells on electrospun honeybee silk mats. Fluorescent images of GFP expressing-fibroblasts grown on electrospun fiber mats after 1 day (A), 3 days (B), and 7 days (C). Scale bar: 100 microns. Cell number and coverage on the mats (D). Error bars are standard deviations of the mean.
The efficient cell adhesion and proliferation on honeybee silk mats may be attributable to the surface chemistry of the silk. The AmelF3 protein contains 12% acidic residues and 11% basic residues (Sutherland et al., 2006), which is comparable to the 9% acidic residues and 8% basic residues of collagen, a major component of the ECM. The assembly of collagen triple-helices sequesters a third of residues, all glycine, in the core, raising the proportion of charged residues on the fibre surface to around 25%. Similarly, coiled coil assembly sequesters a hydrophobic and alanine-rich core, leaving the fibre surface rich in charged residues, so with a surface chemistry close to that of collagen.
3. 4. Biodegradability
The biodegradability of the electrospun honeybee silk materials in vitro was assessed by α-chymotrypsin and trypsin degradation. Both proteases are predicted to cleave the protein multiple times. However when silkworm silk proteins are degraded with α-chymotrypsin, cleavage sites within the ß-sheet crystals in silkworm silk are less accessible to the enzyme and therefore there is a discrepancy between predicted and observed cleavage events [24–26]. Given the ß-sheet structures present in the honeybee silk mats after post-spinning treatments, it was unclear whether all sites would be available to this enzyme and therefore α-chymotrypsin digests were complemented with subsequent trypsin digests.
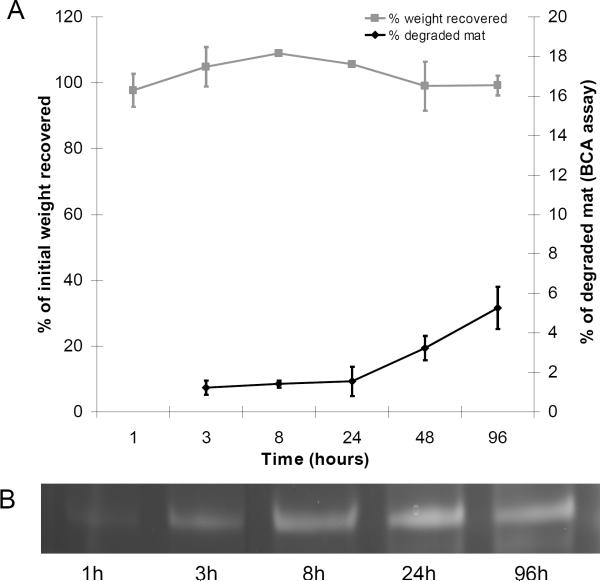
Control mats incubated for 4 days in buffer without enzyme did not lose statistically significant mass (Figure 6), indicating minimal solubilisation of the mats in the buffer solution over the time of the experiment. Analysis of the buffer solution by BCA protein concentration assay and SDS-PAGE protein gel indicated low-level accumulation of full-length silk protein in solution, equivalent to losing about 1% of the mass of the mat per day. No silk protein degradation products were found in the buffer solution. These results indicate that the mats were stable over the course of the biodegradability experiment in the absence of protease.
Figure 6.
Stability of control electrospun mats in buffer. No significant change in mat weight was detected during the incubation period (A). SDS-PAGE showing accumulation of small quantities of full-length AmelF3 protein in the buffer (B).
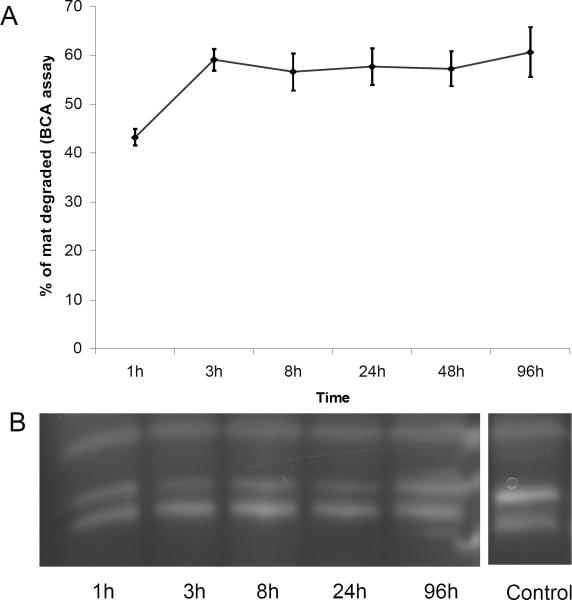
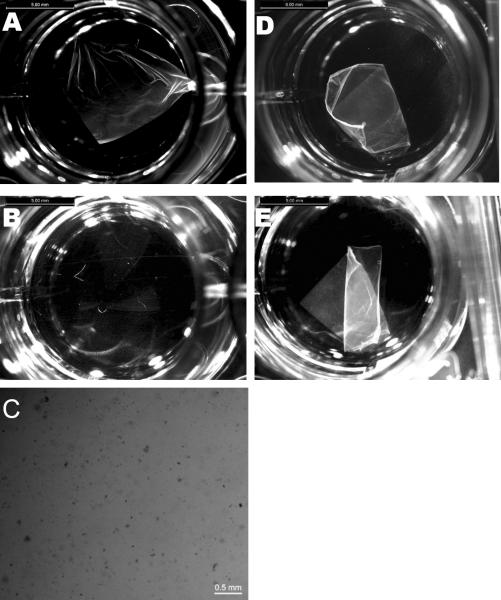
The mats incubated with α-chymotrypsin changed rapidly in the first 24 hrs, becoming thinner and more transparent than the initial samples (Figure 5). The remaining material lost mechanical integrity and broke into smaller pieces when subjected to shaking. The concentration of protein in the supernatant indicated that over 60% of the mat was digested within the first 8 hrs (Figure 7). No further visual change in the mats or increase in the peptide concentration of the supernatant was observed after the initial 24 hrs despite the addition of further enzyme, suggesting that all the accessible α-chymotrypsin sites on the mats had been cleaved. SDS PAGE analysis, which detects peptides larger than ~3 kDa, did not detect any peptide fragments in the digest solution, suggesting that after initial protease cleavage of the protein in the solid substrate the protein is rapidly digested at multiple sites (Figure 7). LC/MS analysis of the digest solution after fifteen minutes incubation identified 12 peptides that matched predicted chymotryptic peptides of the AmelF3 sequence (46% sequence coverage; Supplementary Figure 1a). The LC/MSD trap data was not quantitative and therefore the ratios of the peptides could not be determined. However, the identified peptides were distributed through the protein sequence (44% of the predicted coiled coil region, 52% of the N-terminal region and 49% of C-terminal region,) confirming that after an initial cleavage event the entire AmelF3 protein was rapidly further digested. The stability of parts of the honeybee silk mats towards chymotrypsin digestion may indicate that local regions of the mats have crystalline or ordered structure that limits protease access and inhibits the initial chymotrypsin cleavage.
Figure 5.
Bright field microscopy of electrospun honeybee silk mats. Mat before adding α-chymotrypsin (A), and same mat after 24 h incubation in α-chymotrypsin (B). Commassie Blue stained fragments of mat after α-chymotrypsin digestion (C). Control mat in PBS at t=0 (D), and same mat after 24 h (E). Scale bars: 2 mm.
Figure 7.
Susceptibility of electrospun mats to α-chymotrypsin digestion. The percent of mat weight change determined from the increase in protein concentration in the digest solution (A). SDS-PAGE analysis of proteins present in the digest solution showing only chymotrypsin bands (B, lane XX). Lane YY is a pure α-chymotrypsin solution.
The remnant mat fragments that were resistant to α-chymotrypsin digestion were stained with Commassie Blue. Microscopy revealed that the fragments remaining were clumps of up to 100 μm diameter (Figure 5). The Commassie stained fragments disappeared completely after incubation with trypsin. Analysis of the trypsin digest solution by LC/MS identified 10 tryptic peptide matches to AmelF3 (34% sequence coverage; Supplementary Figure 1b). As with the chymotrypsin digested peptides, the identified peptides were distributed through the protein sequence (38% of the predicted coiled coil region, 30% of N-terminal region, and 20% of C- terminal region) suggesting complete protein digestion. These results indicate that honeybee silk is readily biodegradable and that the expected degradation products are small peptides.
4. Conclusions
Recombinant honeybee silk proteins can be produced at high levels by controlled fermentation in E. coli and therefore avoid issues around reproducibility and contamination associated with similar naturally sourced materials. Honeybee silk protein AmelF3 can be electrospun into mats of uniform fibers of around 200 nm diameter, and the protein structure of the fibres can be controlled by post-spinning treatments. When seeded at low levels, fibroblast cells survived, attached well and proliferated on the fibers to reach confluence within seven days. Unlike silkworm silk, honeybee silk was degraded by proteases into small peptides without the accumulation of partial degradation products. These positive results justify more detailed exploration of use of honeybee silk as a tissue engineering scaffold.
Supplementary Material
Acknowledgements
We thank Sri Sriskantha for protein preparation. We also thank CSIRO, the AFOSR (Award Nos. FA9550-09-1-0332 and FA9550-10-1-0172), and the NIH P41 Tissue Engineering Resource Center for funding sections of this work. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of funding agencies.
References
- [1].Friess W. Collagen-Biomaterial For Drug Delivery. Eur J Pharm Biopharm. 1998;45:113–136. doi: 10.1016/s0939-6411(98)00017-4. [DOI] [PubMed] [Google Scholar]
- [2].Blit PH, Shen YH, Ernsting MJ, Woodhouse KA, Santerre JP. Bioactivation of porous polyurethane scaffolds using fluorinated RGD surface modifiers. J Biomed Mat Res. 2010;94A:1226–1230. doi: 10.1002/jbm.a.32804. [DOI] [PubMed] [Google Scholar]
- [3].Hepburn HR, Chandler HD, Davidoff MR. Extensometric properties of insect fibroins: the green lacewing cross-ß, honeybeeα -helical and greater waxmoth parallel-ß conformations. Insect Biochem. 1979;9:69–77. [Google Scholar]
- [4].Sutherland TD, Campbell PM, Weisman S, Trueman HE, Sriskantha A, Wanjura WJ, et al. A highly divergent gene cluster in honeybees encodes a novel silk family. Genome Res. 2006;16:1414–21. doi: 10.1101/gr.5052606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weisman S, Haritos VS, Church JS, Huson MG, Mudie ST, Rodgers AJW, et al. Honeybee silk: Recombinant protein production, assembly and fiber spinning. Biomaterials. 2010;31:2695–2700. doi: 10.1016/j.biomaterials.2009.12.021. [DOI] [PubMed] [Google Scholar]
- [6].Sutherland TD, Church JS, Hu X, Huson MG, Kaplan DL, Weisman S. Single honeybee silk protein mimics properties of multi-protein silk. PLoS ONE. 2011;6:e16489. doi: 10.1371/journal.pone.0016489. doi:10.1371/journal.pone.0016489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rudall KM. Silk and other cocoon proteins. In: Florkin M, Mason HS, editors. Comparative biochemistry. Vol. 4. Academic Press; New York: 1962. pp. 397–433. [Google Scholar]
- [8].Atkins EDT. A four-strand coiled-coil model for some insect fibrous proteins. J Mol Biol. 1967;24:139–41. [Google Scholar]
- [9].Sutherland TD, Weisman S, Trueman TE, Sriskantha A, Trueman JWH, Haritos VS. Conservation of essential design features in coiled-coil silks. Molec Biol Evol. 2007;24:2424–2432. doi: 10.1093/molbev/msm171. [DOI] [PubMed] [Google Scholar]
- [10].Zhang X, Reagan MR, Kaplan DL. Electrospun silk biomaterial scaffolds for regenerative medicine. Adv Drug Deliv Rev. 2009;61:988–1006. doi: 10.1016/j.addr.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hu X, Kaplan D, Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39:6161–6170. [Google Scholar]
- [12].Hu X, Kaplan D, Cebe P. Dynamic protein-water relationships during beta-sheet formation. Macromolecules. 2008;41:3939–3948. [Google Scholar]
- [13].Hu X, Lu Q, Kaplan DL, Cebe P. Microphase separation controlled beta-sheet crystallization kinetics in fibrous proteins. Macromolecules. 2009;42:2079–2087. [Google Scholar]
- [14].Chan KY, Haschke RH. Isolation and culture of corneal cells and their interactionswith dissociated trigeminal neurons. Exp Eye Res. 1982;35:137–156. doi: 10.1016/s0014-4835(82)80062-6. [DOI] [PubMed] [Google Scholar]
- [15].Chan KY, Patton DL, Cosgrove YT. Time-lapse videomicroscopic study of in vitro wound closure in rabbit corneal cells. Invest Ophth Vis Sci. 1989;30:2488–2498. [PubMed] [Google Scholar]
- [16].Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, et al. A lentivirus-based systemto functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNAinterference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- [17].Jaeger R, Schonherr H, Vancso GJ. Chain packing in electro-spun poly(ethylene oxide) visualized by atomic force microscopy. Macromolecules. 1996;29:7634–7636. [Google Scholar]
- [18].Ishida M, Asakura T, Yokoi M, Saito H. Solvent- and mechanical-treatment-induced conformational transition of silk fibroins studied by high-resolution solid-state 13C NMR spectroscopy. Macromolecules. 1990;23:88–94. [Google Scholar]
- [19].Tsukada M, Gotoh Y, Nagura M, Minoura N, Kasai N, Freddi G. Structual changes of silk fibroin membranes induced by immursion in methanol aqueous solutions. J Polym Sci Part B: Polym Phys. 1994;32:961–968. [Google Scholar]
- [20].Wilson D, Valluzzi R, Kaplan DL. Conformational transitions in model silk peptides. Biophys J. 2000;78:2690–2701. doi: 10.1016/S0006-3495(00)76813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jin HJ, Park J, Karageorgiou V, Kim UJ, Valluzzi R, Cebe P, et al. Water-insoluble silk films with reduced beta-sheet content. Adv Funct Mater. 2005;15:1241–1247. [Google Scholar]
- [22].Lu Q, Hu X, Wang XQ, Kluge JA, Lu SZ, Cebe P, et al. Water-insoluble silk films with silk I structure. Acta Biomater. 2010;6:1380–1387. doi: 10.1016/j.actbio.2009.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jin H-J, Fridrikh SV, Rutledge GC, Kaplan DL. Electrospinning Bombyx mori silk with poly(ethylene oxide) Biomacromolecules. 2002;3:1233–1239. doi: 10.1021/bm025581u. [DOI] [PubMed] [Google Scholar]
- [24].Arai T, Freddi G, Innocenti R, Tsukada M. Biodegradation of Bombyx mori silk fibroin fibers and films. J Appl Polym Sci. 2004;91:2383–90. [Google Scholar]
- [25].Lotz B, Gonthier-Vassal A, Brack A, Magoshi J. Twisted single crystals of Bombyx mori silk fibroin and related model polypeptides with beta structure. A correlation with the twist of the beta sheets in globular proteins. J Mol Biol. 1982;156:345–57. doi: 10.1016/0022-2836(82)90333-3. [DOI] [PubMed] [Google Scholar]
- [26].Li M, Ogiso M, Minoura N. Enzymatic degradation behavior of porous silk fibroin sheets. Biomaterials. 2003;24:357–65. doi: 10.1016/s0142-9612(02)00326-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.