Abstract
An international consortium released the first draft sequence of the human genome 10 years ago. Although the analysis of this data has suggested the genetic underpinnings of many diseases, we have not yet been able to fully quantify the relationship between genotype and phenotype. Thus, a major current effort of the scientific community focuses on evaluating individual predispositions to specific phenotypic traits given their genetic backgrounds. Many resources aim to identify and annotate the specific genes responsible for the observed phenotypes. Some of these use intra-species genetic variability as a means for better understanding this relationship. In addition, several online resources are now dedicated to collecting single nucleotide variants and other types of variants, and annotating their functional effects and associations with phenotypic traits. This information has enabled researchers to develop bioinformatics tools to analyze the rapidly increasing amount of newly extracted variation data and to predict the effect of uncharacterized variants. In this work, we review the most important developments in the field—the databases and bioinformatics tools that will be of utmost importance in our concerted effort to interpret the human variome.
Keywords: genomic variation, genome interpretation, genomic variant databases, gene prioritization, deleterious variants
INTRODUCTION
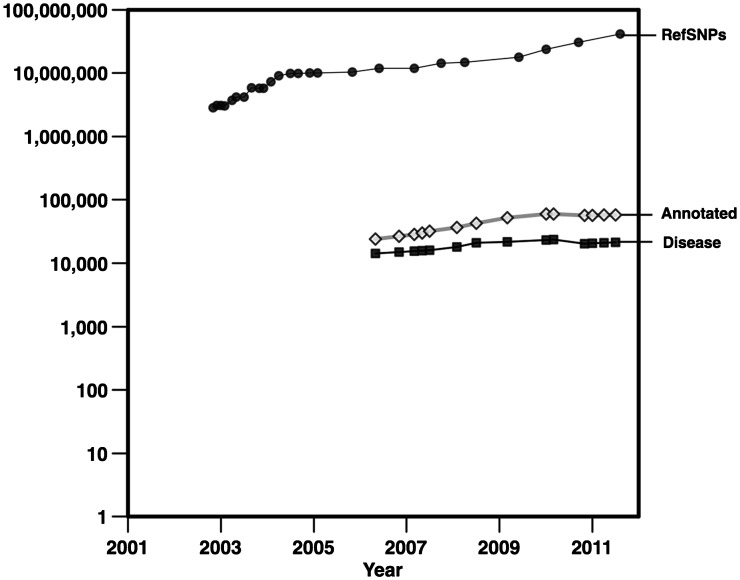
In 1990, the Human Genome Project was launched and, almost 14 years later, the complete sequence of the human genome (over 3 billion bp) was made available [1] at an estimated cost of $2.7 billion. Since then, genomic data has been collected at a continually increasing rate (Figure 1). The strategy for relating a genotype to a phenotype experimentally depends on the type of trait or disease being studied. Re-sequencing the associated gene in affected individuals versus a control population can elucidate variants causing Mendelian pathologies. For analyzing complex, multigenic diseases, sequencing all possible disease-associated regions is necessary.
Figure 1:
Growth in the number of genetic variations in dbSNP and SwissVar. RefSNPs shows the number of position-based clusters of variants from dbSNP [2]. Disease and Annotated show the numbers of disease-related and total annotated (either disease-related or neutral) nonsynonymous SNVs from the SwissVar database [3].
Although many types of genetic variations exist, the Single Nucleotide Variants (SNVs; mutations affecting exactly 1 nt in the genomic sequence) are most prevalent [4]. Many common Single Nucleotide Polymorphisms (SNPs; SNVs that substitute a nucleotide by another one and occur in at least 1% of the population) have been extensively evaluated for disease associations. The International HapMap Consortium was established in 2001 to detect the patterns in human DNA sequence variation and to determine their frequencies in different populations. One major goal of the effort was to enable the discovery of disease-related SNPs [5]. In 2007, using linkage data from HapMap, a genome-wide association study (GWAS) on 17 000 samples/∼500 K SNVs was performed to detect variants associated with seven common diseases [6]. To date, nearly 1000 GWAS, genotyping at least 100 K SNPs per individual, have been published [7]. Although these studies are important for identifying disease-associated variants, only a few thousand common SNPs have been significantly associated to specific phenotypic traits [8]. Moreover, a systematic analysis of lower frequency, rare SNVs was not possible with GWAS [9].
With the advent of modern high-throughput technology, the cost of sequencing whole genomes has continued to decrease to reach ∼$3000 today. These technological advances have also enabled the sequencing of individual genomes [10, 11] and the establishment of the 1000 Genomes Project Consortium. In the publication of its pilot results, the Consortium reported detecting over 16 million SNVs [4]—data that now needs to be analyzed for its association with various phenotypes. In addition, direct to consumer (DTC) companies are offering limited genotyping to provide insight into personal traits and disease risks [12]. It is expected that in the next few years, we will witness a second phase of the personal genomics wave, where SNP genotyping chips will be replaced by whole-genome sequencing. Despite more comprehensive databases and better methods for the analysis of genetic variants, the problem of genome interpretation is still far from being solved. Thus, the idea of a ‘$1000 genome, $1 000 000 genome interpretation’ was expressed by the president of the American College of Medical Genetics, Bruce Korf. In this review, we summarize the newly available genetic variation resources, methods for gene prioritization and algorithms for the prediction of variant effects for use in interpreting personal genomes.
DATABASES AND RESOURCES
As next-generation sequencing technologies continue to decrease in cost and increase in throughput, SNP chip-based genotyping will rapidly be replaced by whole-exome and whole-genome sequencing. Once an individual's genotype has been accurately determined, the first step in genotype interpretation is to identify each variant as known or novel, rare or common, and to determine if it has been previously associated with a disease. Identifying the structural and functional context of each variant is also critical for variant prioritization. A number of databases are freely available to aid the variant annotation process.
Databases for short length and structural variations
Several databases aid in the classification of variants as either known or novel, and rare or common (Table 1). The National Center for Biotechnology Information (NCBI) dbSNP database [2] is the largest source of short genetic variation data. dbSNP currently contains over 40 million, both common and rare human SNVs, short indels and microsatellites (Build 134, August 2011). Where available, the database also reports SNV clinical significance. The 1000 Genomes Project Consortium is a major contributor of novel variants to dbSNP, aiming to catalog 95% of human variants with an allele frequency of at least 1% in each of five major human population groups.
Table 1:
Databases and resources for personal genome interpretation
| Database | URL | Description | References |
|---|---|---|---|
| Short variations—SNVs, short indels | |||
| 1000 Genomes | http://www.1000genomes.org | Human short variants and inferred genotypes | [4] |
| dbSNP | http://www.ncbi.nlm.nih.gov/projects/SNP | Short variants from all species | [21] |
| HapMap | http://www.hapmap.org | Human short variants and population group haplotypes | [5] |
| Structural variations—structural rearrangements, CNVs, large indels | |||
| dbVar | http://www.ncbi.nlm.nih.gov/dbvar | Structural variants from all species | [13] |
| DGV | http://projects.tcag.ca/variation | Structural variants from healthy human controls | [14] |
| DGVa | http://www.ebi.ac.uk/dgva | Structural variants from all species | [13] |
| General variants associated with phenotypes | |||
| ClinVar | http://www.ncbi.nlm.nih.gov/clinvar | Human variant–disease associations (in development) | |
| HGMD | http://www.hgmd.org | Human variant–disease associations (inherited diseases) | [15] |
| OMIM | http://www.omim.org | Human variant–disease associations (includes extensive gene and phenotype descriptions) | [16] |
| SwissVar | http://swissvar.expasy.org | Human variant–disease associations (non-synonymous SNVs only) | [3] |
| GWAS and other association studies | |||
| dbGaP | http://www.ncbi.nlm.nih.gov/gap | Controlled access to individual genotype/phenotype data from association studies | [17] |
| EGA | http://www.ebi.ac.uk/ega | Controlled access to individual genotype/phenotype data from association studies | |
| GAD | http://geneticassociationdb.nih.gov | Mainly complex disease SNVs from association studies | [18] |
| NHGRI GWAS Catalog | http://www.genome.gov/gwastudies | Significant SNVs from GWAS | [7] |
| Cancer genes and variants | |||
| ICGC | http://www.icgc.org | Somatic variants from tumor sequencing projects | [19] |
| COSMIC | http://sanger.ac.uk/genetics/CGP/cosmic | Somatic variants from tumor sequencing and literature | [20] |
| Cancer Gene Census | http://sanger.ac.uk/genetics/CGP/Census | Comprehensive list of cancer-related genes | [21] |
| Cancer Gene Index | http://ncicb.nci.nih.gov/NCICB/projects/cgdcp | Comprehensive list of cancer-related genes, including gene–disease and gene–drug relationships | |
| TCGA | http://cancergenome.nih.gov | Somatic variants from tumor sequencing projects | [22] |
| Pharmacogenomic genes and variants | |||
| DrugBank | http://drugbank.ca | Drug properties and protein amino acid target sequences | [23] |
| PharmGKB | http://www.pharmgkb.org | Curated and text-mined variant–drug response associations | [24] |
| Crowdsourced genes and variants | |||
| Gene Wiki | http://en.wikipedia.org/wiki/Portal:Gene_Wiki | Human gene/protein annotations | [25] |
| SNPedia | http://www.snpedia.com | Human SNV–disease associations | |
| WikiGenes | https://www.wikigenes.org | Gene annotations from all species | [26] |
| Viewers of the structural and functional impact of variants | |||
| DMDM | http://bioinf.umbc.edu/dmdm | Aggregates human protein mutations at individual domain positions | [27] |
| LS-SNP/PDB | http://ls-snp.icm.jhu.edu/ls-snp-pdb | Variant/PDB structure viewer (includes multiple filters for selection of variants) | [28] |
| MutDB | http://mutdb.org | Variant/PDB structure viewer (includes SIFT predictions for nonsynonymous mutations) | [29] |
| SAAPdb | http://bioinf.org.uk/saap/db | Variant/PDB structure viewer (includes impact on physico-chemical and functional features) | [30] |
| StSNP | http://ilyinlab.org/StSNP | Variant/PDB structure viewer (includes physico-chemical impact for nonsynonymous mutations) | [31] |
| SNPeffect | http://snpeffect.vib.be | Variant/PDB structure viewer (includes predictions for variants to cause protein aggregation) | [32] |
| TopoSNP | http://gila-fw.bioengr.uic.edu/snp/toposnp | Variant/PDB structure viewer (includes location of variant on surface, in pocket or in core) | [33] |
The Consortium is expanding on the work of the International HapMap Project [5] to catalog genetic variation shared within and between members of various populations. So far, over 38 million variant sites have been identified within the framework of this effort (Phase 1 Low Coverage Data, May 2011). In addition, the Consortium data includes inferred genotypes for individual samples, useful for future association studies utilizing genotype imputation. The recently initiated UK10K Project (http://www.uk10k.org) will have even greater power to discover rare variants, identifying those with as little as 0.1% allele frequency. The project will conduct low coverage, whole-genome sequencing for 4000 healthy individuals, and whole-exome sequencing for 6000 individuals with a variety of extreme disease phenotypes to facilitate the discovery of rare variants associated with these diseases. Although more exhaustive in scope, the UK10K data is less accessible—access is managed by a consortium and requires acceptance of ‘terms and conditions’ to protect the privacy and interests of the study participants.
While databases like dbSNP and HapMap and projects like 1000 Genomes and UK10K focus primarily on short-length variants like missense, nonsense and short insertion and deletion mutations (indels), larger-scale structural rearrangements, copy number variants (CNVs) and large indels can also dramatically affect human phenotypes. NCBI's database of genomic structural variation (dbVar) [13] and the collaborative effort Database of Genomic Variants (DGV) [14] are two of the largest repositories for large-scale (typically >1 kb in length) structural variations. DGV only contains entries from healthy human controls, while dbVar contains entries from all species and includes variants with associated phenotypes. The DGV archive (DGVa) [13] is a new database maintained by the European Bioinformatics Institute that also contains structural variants from all species with associated phenotypes when available.
Genotype/phenotype annotation databases
Many specialized databases contain variant–disease associations that are commonly used to identify known deleterious mutations (Table 1). The Online Mendelian Inheritance in Man (OMIM) database [16] is a catalog of human genes and diseases. OMIM is manually curated and contains descriptions of over 13 000 genes and almost 7000 phenotypes (September 2011). Over 2600 genes in OMIM contain listings of specific allelic variants associated with disease. The SwissVar database [3] is another manually curated source of variant–phenotype association data. The database also includes a number of variant features, e.g. physico-chemical properties, affected functional features and conservation profiles for amino-acid changing variants in SwissProt proteins. SwissVar currently contains information on over 24 000 deleterious variants linked to over 3300 diseases (September 2011). The Human Gene Mutation Database (HGMD) [15] is a large collection of variants associated with human inherited diseases. HGMD is available in two versions: a free version for academic/nonprofit users, and a more regularly updated, paid professional version. The free version of HGMD contains associations of approximately 82 000 variants of all kinds to approximately 3000 diseases (September 2011). NCBI's ClinVar database, currently in development, aims to provide a freely available, comprehensive listing of variants associated with phenotypes along with links to regularly updated evidence for the associations. In addition to OMIM, SwissProt and HGMD, which list variants in all disease-associated genes, locus specific databases (LSDBs) report variants in a single gene, often related to a single disease. LSDBs are used as a source of variant data for both SwissProt and HGMD. A comprehensive list of links to LSDBs is provided on the Human Genome Variation Society website (http://www.hgvs.org/dblist/glsdb.html). The number of LSDBs and the quality of data contained therein has grown over recent years. However, at least one comprehensive study shows [34] that LSDBs would be better equipped to serve the research (and treatment) community with standardization of reported results and improvement of variant effect (and disease) annotation.
GWAS and other association study databases
As previously noted, large-scale GWAS have identified thousands of variants associated with disease. The National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies (NHGRI GWAS catalog) [35] conveniently lists significantly associated marker SNPs from these studies in a manually curated, online database. The catalog includes data on study designs, individual SNV P-values, odds ratios and links to the published studies. The Genetic Association Database (GAD) [18] predates the NHGRI GWAS catalog, and contains curated information on both positive and negative variant associations from GWAS and candidate gene association studies primarily from studies of common, complex diseases. In addition to the summary data in the GWAS catalog and GAD, the database of Genotype and Phenotype (dbGaP) [17] and the European Genome-phenome Archive (EGA) provide controlled access to individual-level genotype and phenotype data from many large-scale association studies.
Cancer gene and variant databases
Given the significance of somatic mutations in oncogenesis, several large-scale projects sequencing multiple cancer types have emerged including the Cancer Genome Atlas (TCGA) [22] and the Cancer Genome Project (CGP) (http://www.sanger.ac.uk/genetics/CGP). The International Cancer Genome Consortium (ICGC) [19] was developed to coordinate cancer sequencing projects around the world, including TCGA and the CGP, for over 50 different cancer types and subtypes. Data portals for ICGC and TCGA are available to retrieve open access variant data, and individual level, controlled access genotype data by application. Data from the CGP and curated mutations from the literature for a list of genes previously associated with cancer (the Cancer Gene Census list [21]) are available from COSMIC, the Catalog of Somatic Mutations in Cancer [20]. COSMIC (Release 54) currently contains data on over 177 000 mutations from almost 620 000 tumors. The NCI Cancer Gene Index is another comprehensive source of genes related to cancer, containing gene–disease and gene–drug relationships text-mined and manually validated from over 20 million MEDLINE abstracts.
Pharmacogenomic genes and databases
Specialized databases also now exist to link genes and genotypes with drug targets and drug response. The Pharmacogenomics Knowledgebase (PharmGKB) [36] contains both manually curated and automatically text-mined associations of human variations to drug response. The database includes information for variants in over 1500 genes related to approximately 375 drugs and almost 300 diseases (September 2011). DrugBank [23] is a more drug-focused resource containing structural, chemical and pharmacologic properties for over 6800 drugs (September 2011). DrugBank also contains the amino acid target sequences for individual drugs, enabling the identification of variants falling in drug binding sites.
Other resources and tools for personal genome interpretation
Crowdsourcing
Many annotation databases use automated searches followed by expert human curation to identify and validate variant–disease associations from literature. As the pace of association studies continues to increase, this process will become increasingly unsustainable. To address this problem, several databases have been developed to harness a crowdsourcing model for gene and variant annotation including Gene Wiki [25], WikiGenes [26] and SNPedia (http://www.SNPedia.com). While all of these resources include some information automatically extracted from public sources like PubMed, OMIM and dbSNP, the community contribution and curation could potentially provide more comprehensive and update-to-date information as new studies are published.
Viewers
A variant's structural/functional context is critical to determining its likelihood of disease involvement. Several web tools are available for viewing SNVs superimposed onto the corresponding Protein Data Bank (PDB) [37] protein structures. These tools highlight the changes in physico-chemical properties and display proximity to structural features like domain interfaces, binding sites, etc. LS-SNP/PDB [38], MutDB [29], SAAPdb [30], StSNP [31], SNPeffect [32] and TopoSNP [33] are just a few resources that provide this type of structural and functional annotation. By aggregating all SNPs and disease mutations from dbSNP, OMIM and SwissVar at the protein and domain levels, the Domain Mapping of Disease Mutations (DMDM) database [27] also adds the critical functional context provided by protein domains for variant characterization.
Epigenetics and transciptome Databases
Changes in gene regulation due to epigenetic mechanisms, other than variation in the DNA sequence, can also be disease associated. Local patterns of DNA methylation, chromatin structure and histone modification states, and nonprotein-coding RNAs (ncRNA), e.g. microRNAs, affect gene expression levels. Thus, genome-wide studies to comprehensively catalog the various structural and functional elements of the genome, as well as studies to map the epigenetic elements affecting gene expression levels, are now being undertaken. These will lead to a better understanding of genome complexity and gene regulation. The ENCyclopedia of DNA Elements Project (ENCODE) [39] includes studies to catalog the full human transcriptome including protein coding, noncoding and pseudogene transcripts, in addition to local chromatin states and methylation patterns. The National Institutes of Health Roadmap Epigenomics Mapping Consortium [40] was recently organized to map DNA methylation, histone modifications, chromatin accessibility and ncRNA transcripts in each human tissue and cell type. The ENCODE Project and NIH Roadmap Epigenome data are both available as annotation tracks viewable from the UCSC Genome Browser at http://genome.cse.ucsc.edu/encode and http://www.epigenomebrowser.org, respectively. Interesting reviews about large-scale epigenomics projects, association studies of epigenetic variation and computational epigenetics analysis have been recently published [41–43].
GENE PRIORITIZATION
Aberrations in normal gene function that result in the development of a disease define a ‘disease gene’. Proving a gene–disease relationship experimentally is expensive and time-consuming. Ranking candidate genes prior to experimental testing reduces the associated costs. Computational gene prioritization uses heterogeneous pieces of evidence to associate each gene with a given disease. Whereas experimental studies provide a lot of information, incorporation of other sources of evidence is necessary to narrow down the candidate search space. Gene prioritization techniques effectively translate heterogeneous experimental data into legible disease–gene associations.
Making sense of available data
A functional module, or molecular pathway, is generally defined as a series of interactions between molecules in the cell leading to a specific end point in cellular functionality. For the body to remain disease-free, all normally occurring processes, molecular interactions and pathways should function without major alterations. Moreover, since it is an oversimplification to view a single pathway as a discrete and independent entity, it is increasingly evident that different diseases, resulting from aberrations in different pathways, are also interdependent.
Identifying the pathways affected in the observed disease is a major challenge. A given pathway can be altered by gene expression changes, gene-product malformation, introduction of new pathway members, and/or environmental disruptions. Identification of gene–disease associations is complicated by gene pleiotropy, multigenic nature of most diseases, varied influence of environmental factors and overlying genome variation; i.e. any one source of information about a disease may or may not be sufficient to identify its specifics. Moreover, the available experimental data describing each of the biological concepts involved is itself very heterogeneous. Thus, using a combination of resources requires knowing how to meaningfully combine the extracted information (Table 2).
Table 2:
Available data sources and gene prioritization tools
A wide range of data sources can be used to formulate the associations and implications described herein. Existing tools try to take advantage of many of them. This table summarizes current web-available, state of the art gene prioritization methods. Note, some methods (e.g. PRINCE [68]) are not available online, but are downloadable for local installation. Letters in the Description column denote the data sources used: (E) Experimental observation: Linkage, association, pedigree, relevant texts and other data. (S) Sequence, structure, meta-data: Sequence conservation, exon number, coding region length, known structural domains and sequence motifs, chromosomal location, protein localization and other gene-centered information and predictions. (P) Pathway, protein–protein interaction, genetic linkage, expression: Disease–gene associations, pathways and gene–gene/protein–protein interactions/interaction predictions and gene expression data. (N) Non-human data: Information about related genes and phenotypes in other species. (O) Ontologies: Gene, disease, phenotype and anatomic ontologies. (M) Mutation associations and effects: information about existing mutations, their functional and structural effects and their association with diseases, predictions of functional or structural effects for the mutations in the gene in question. (L) Literature: mixed information of all types extracted from literature references (e.g. disease–gene correlation and nonontology based gene-function assignment).
The lines of evidence most robust in identifying genes as prime suspects for disease involvement are: GWAS or linkage analysis studies, similarity or linkage to and co-regulation/-expression/-localization with known disease genes, and participation in disease-associated pathways/compartments. Five notions commonly define these associations: (i) functional association: participation in a common pathway with other disease genes, (ii) cross-species association: orthologues generating similar phenotypes in other organisms, (iii) disease association: gene-product presence in disease pathways or associated cellular compartments and/or localization to affected tissues, (iv) mutant implication: candidate genes harboring functionally deleterious mutations in diseased individuals and (v) text implication: co-occurrence of gene and disease terms within scientific texts.
Functional association
Common pathway
Many gene prioritization tools use gene–gene (protein–protein) interaction and/or pathway information to prioritize candidates. Since diseases are results of pathway malfunction, disabling any of the pathway members may lead to similar phenotypes. Cancer-associated proteins, for example, are very strongly interconnected [69]. In general, genes responsible for generating similar diseases tend to participate in the same protein–protein interaction networks [70].
Regulatory information and genetic linkage
Co-regulated genes are often thought to be involved in the same molecular pathways [71]. However, co-regulated genes may reside in distinct pathways [72]. Moreover, co-expressed nonparalogous genes often demonstrate conservation of clustering across species [73], suggesting that co-expression clusters are evolutionarily advantageous and naturally selected [74]. Some researchers [75] argue that these clusters may represent groups of genes involved in high-level cooperation beyond the canonical description of cellular pathways. As such, deregulation of these clusters may manifest in disease. Note that whereas genetic linkage/co-regulation are valuable markers of disease association, they are not specific; i.e. a given disease-associated gene may be co-regulated with or linked to another disease-associated gene, where the two diseases are not necessarily identical.
Similar function
It is common to observe reduced/absent phenotypic effect in response to gene knockout/inactivation [76, 77]. This phenomenon is largely explained by functional compensation via partial interchangeability of paralogous genes. When functional compensation is insufficient, inactivation of any of the paralogs leads to same/similar disease. Thus, many tools use functional similarity to establish disease–gene association. Defining functional similarity is nontrivial. When utilizing ontologies, like GO [78], the question becomes a problem of assigning a ‘score’ to the similarity of two ontology nodes/subtrees. ‘Functional distance’ [79–82] calculations for any two genes within the context of the ontology suggest a well-defined way of annotating functional similarity.
Sequence/structure association
Sequence/structure homology is often used for transferring functional annotations from characterized genes/proteins to new ones [83]. Since functionally similar genes are likely to produce similar disease phenotypes, homology is also important in disease gene prioritization. Additionally, disease genes are distinctly associated with specific canonical features such as higher exon number, gene, protein and 3′-UTR length and distance to a neighboring gene, as well as lower sequence divergence from their orthologs [64, 84]. Proteins with signal peptides are also more commonly disease associated [64], whereas disordered proteins are often implicated in cancer [85].
Cross-species association
A high number of orthologs suggests essential genes prone to disease involvement. Orthologs also generally participate in similar molecular pathways across species, although different levels of function may be necessary for different organisms. Cross-species, tissue-specific phenotypic differences due to slightly varied sequences are thus useful for gene prioritization. Note that phenotype ontologies are necessary to facilitate [86] this comparison of organism phenotypes.
A correlation of co-expression of genes in different species is also a predictive measure for annotating disease genes [87, 88]. As discussed above, there is evidence for co-expression of genes that are not related in any known functional manner [89–91]. These co-expressed clusters may be evolutionarily advantageous [74, 75, 90], but are only evident as such if conserved throughout other species. Cross-species comparisons of protein co-expression are therefore useful for validating disease–gene co-expression associations (e.g. [87]).
Disease compartment association
Altered gene expression is expected in association with many common complex diseases [92]. Genes that are preferentially expressed in disease-affected tissues are likely candidates for disease association. Some proteins interact only in some tissues [93], so tissue specificity is important for finding the right protein–protein interaction networks. Similarly, disease-association with cellular pathways (e.g. ion channels) and compartments (e.g. plasma membrane) may indicate that pathway/compartment-specific gene-products are also disease associated.
Mutant implication
Every genetic disease is associated with some sort of mutation that alters normal functionality. Selection of candidates for further analysis is often based on observations of variants in diseased individuals, which are absent in healthy controls. Not all observed variants are deleterious. Most observed variations do not manifest phenotypically, and some are weakly deleterious or even beneficial. Many gene prioritization methods use mutation effect predictions to make their own inferences. Tools used to make these predictions are described in the ‘Genetic variant interpretation’ section of this article.
Text implication
Experimental findings of gene–disease associations are often reported in the literature. Some of the data is also machine accessible via various databases described above such as dbSNP [11], GAD [18] and COSMIC [20], or by depositing manually curated knowledge into databases like GeneRIF [94] and UniProt. However, a vast sea of data remains ‘hidden’ in the natural language text of scientific publications. Text mining tools have recently come of age [94–97], allowing for the identification of possible gene–gene and disease–gene correlations [98–100]. For example, the Information Hyperlinked Over Proteins, IHOP method [101] extracts gene/protein names in scientific texts and links these via pathology, phenotype, physiology and interaction information. Gene prioritization techniques also often rely on term co-occurrence statistics (e.g. PosMed [102], GeneDistiller [51]) and gene-function annotations (e.g. ENDEAVOR [103], PolySearch [61]).
The inputs, outputs and processing
Gene prioritization methods vary based on the inputs they require and the types of outputs they produce. For an excellent summary of methods and their inputs, see [48] and Table 3. Method inputs rely on two distinct notions: previously known information about the disease and the candidate search space. The disease information may include a list of genes known to be associated with the disease, the tissues and pathways it afflicts and any relevant keywords. The candidate search space may include the entire genome, or may be defined by the suspect genomic region, overexpression in the affected tissue, or other experimental results. Not surprisingly, the accuracy of the prioritization method often depends on the accuracy and specificity of the input data. Outputs of prioritization methods are generally limited to ranked lists of genes, often associated with test-performance values (e.g. P-values). Some methods only rank/order the top genes they select, while others manage the entire submitted list. Selected input and output requirements are important for a tool's acceptance by the biological community. A given method's ease of use often defines its utility as strictly as do its speed and accuracy.
Table 3:
Tools for the interpretation of single nucleotide variants
| Method | URL | Description | References |
|---|---|---|---|
| Methods for the prediction of stability change upon mutation | |||
| AutoMUTE | http://proteins.gmu.edu/automute/ | Delaunay tessellation and combined machine learning methods | [104] |
| CUPSAT | http://cupsat.tu-bs.de/ | Physics-based energy function | [105] |
| D-Mutant | http://sparks.informatics.iupui.edu/hzhou/ mutation.html | Statistical-based energy function | [106] |
| Fold-X | http://foldx.crg.es/ | Physics-based energy function | [107] |
| I-Mutant | http://gpcr2.biocomp.unibo.it/I-Mutant.htm | Sequence and Structure SVM-based method | [108] |
| PoPMuSiC | http://babylone.ulb.ac.be/popmusic | Statistical-based energy function optimized by ANN | [109] |
| Methods for the prediction of deleterious non-synonymous SNVs | |||
| PANTHER | http://www.pantherdb.org/ | Protein family HMM-based method | [110] |
| PhD-SNP | http://gpcr.biocomp.unibo.it/PhD-SNP.htm | Sequence and profile-based SVM method | [111] |
| PolyPhen | http://genetics.bwh.harvard.edu/pph | Decision Tree-based method | [112] |
| MutPred | http://mutdb.org/mutpred | Random forest approach including multiple scores | [113] |
| SIFT | http://sift-dna.org | Protein block alignment-based method | [114] |
| SNAP | http://rostlab.org/services/snap | Sequence profile-based neural network | [115] |
| SNPs3D | http://www.snps3d.org | Structure-based SVM predictor | [63] |
| SNPs&GO | http://snps-and-go.biocomp.unibo.it | Sequence profile and functional-based SVM | [116, 117] |
| Predictors of the impact of SNVs at DNA level | |||
| ANNOVAR | http://www.openbioinformatics.org/annovar | Scoring functional and evolutionary information | [118], |
| binCons | http://zoo.nhgri.nih.gov/binCons/index.cgi | Evolutive analysis with window-based approach | [119] |
| GERP | http://mendel.stanford.edu/SidowLab/downloads/gerp/ | Site-specific evolutive analysis | [120] |
| Gunby | http://pga.jgi-psf.org/gumby/ | Statistical-based evolutive analysis | [121] |
| Is-rSNP | http://www.genomics.csse.unimelb.edu.au/is-rSNP/ | Effect of variants in regulatory regions | [122] |
| MutationTaster | http://www.mutationtaster.org/ | Evolutionary conservation, splicing site changes and loss of protein features | [123] |
| PhastCONS | http://compgen.bscb.cornell.edu/phast | Phylogenetic HMM-based method | [124] |
| SCONE | http://genetics.bwh.harvard.edu/scone | Site-specific evolutive analysis | [125] |
| Skippy | http://research.nhgri.nih.gov/skippy | Predicts variants affecting splicing sites | [126] |
| VISTA | http://genome.lbl.gov/vista/index.shtm | Integrated approach including scores from different methods | [127] |
Finally, gene prioritization methods also differ in the algorithms they use to make sense of the data. Tools used include mathematical/statistical models and scoring methods (e.g. SUSPECTS [64], GeneProspector [52]), fuzzy logic (e.g. ToppGene [65, 66]), artificial learning devices (e.g. decision trees in PROSPECTR [62], neural networks in PosMed [102]), network/topology analysis approaches (e.g. DiseaseNet [127]), and vector/profile comparisons (e.g. CAESAR [44], MedSim [56]) among others. Most often, some combination of the above methods is used, but there is no single methodology that is objectively better than the others for the compilation of data from all sources.
GENETIC VARIANT INTERPRETATION
The interpretation of the functional impact of genetic variation is one of the most important tasks in personal genomics and personalized medicine [129]. Genomic variants have different effects depending on whether they occur in coding or noncoding regions. In coding regions, variants can change the amino acid sequence of the coded protein. In noncoding regions, they can affect transcription, splicing and binding. The recent 1000 Genomes Project Consortium work confirms that single nucleotide variants (SNVs) are the most common type of genetic variation [2]. Thus, understanding the functional effect of SNVs is one of the main goals of modern genetics/genomics studies [130]. Over the past 10 years, several methods have been developed to predict the impact of SNVs [131–133]. Here, we describe the information used by these algorithms, and present a selection of the most popular web-available tools for genome interpretation.
Computational approaches for genome interpretation
As noted, experimental studies to characterize the impact of SNVs are still expensive and time consuming. To partially overcome this limitation, several algorithms have been implemented to predict the effect of genetic variants (Table 3). All such methods take input information derived from sequence conservation, because it has been observed that functionally important regions of the genome tend to be more conserved through evolution than nonfunctional ones [134, 135]. The detection of functional and conserved sites depends on their locations in the genome. We currently have a better understanding of the relationship between the DNA sequence and function for coding regions than for noncoding ones [136]. Hence, the majority of methods have been designed to predict the effect of nonsynonymous SNVs (nsSNVs) and, only recently, a few supervised methods have been developed to evaluate the impact of genetic variants in noncoding regions.
Predicting the effect of nsSNVs
Methods for predicting the effect of nsSNVs estimate their probability of being disease-associated or functionally deleterious. The catalog of the relationships between molecular phenotypes and disease is far from complete. However, it is believed that the pathologic state results from amino acid substitutions affecting functionally critical residues and/or causing alterations in the structure of the folded protein, structural instability or protein aggregation [137]. Several methods have been developed for predicting the effects of amino acid substitutions. In particular, we describe two (not fully separable) classes: (i) predictors of nsSNV functional effects; i.e. modifying the catalytic site of an enzyme or affecting a residue involved in crucial interactions with partner molecules and (ii) those predicting the effect of nsSNVs on protein stability (Table 3).
Methods for the prediction of protein stability changes
Incorrect protein folding mechanisms and decreased stability are the major consequences of pathogenic nsSNVs [138], as they can cause a reduction in hydrophobic area, overpacking, backbone strain and/or loss of electrostatic interactions [139]. Although different thermodynamics measures can be used to assess the variation of stability upon mutation, one of the most common is the difference of the folding free energy change between the wild-type and mutated proteins (ΔΔG). Several methods have been developed to predict if a given nsSNV changes the stability of the protein structure. Some algorithms implement an energy function to compute the ΔΔG [106, 140–145], whereas others are based on machine-learning approaches [108, 146–148].
The methods relying on energy functions can be subdivided into (i) physics-based approaches that use a force field to describe the atomic interactions involved in the folding process [142–144] and (ii) statistical potential approaches that use an empirical energy function derived from the statistical analysis of the structural environment around the mutated site [106, 140, 141, 145]. More recently, an increasing amount of thermodynamic data, collected in web-available databases such as ProTherm [149], has allowed training machine learning methods to predict both the value and sign of the difference between the folding free energy of the wild-type and mutated proteins (ΔΔG). In 2010, the accuracy of the most popular web-available algorithms was assessed by reporting method performances on a set of thermodynamic data not included in the training set [150]. Although tested on data sets of different sizes, Dmutant [106], FoldX [107] and I-Mutant3.0 [147] scored the highest for predicting protein stability changes. This assessment showed that current methods for the prediction of stability changes due to nsSNVs reach a moderate level of accuracy (∼60%). Further improvements are therefore necessary to provide more reliable predictions.
Methods for the prediction of functional effects of nsSNVs
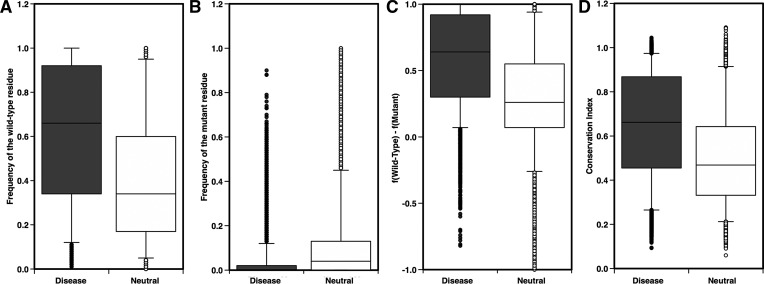
Efforts to design accurate algorithms for the prediction of functionally deleterious nsSNVs have resulted in a slew of available methods [130]. Considering evolution as the ultimate mutagenesis experiment, comparative sequence analysis is a powerful source of information taken into account by all the algorithms. A simple study performed on a dataset of nsSNVs extracted from SwissVar database [3] showed significant differences between the distribution of the frequencies of wild-type and mutant residues for the subsets of disease-related and neutral variants (Figure 2A and B). The median values for the frequencies of the wild-type residues in disease-related and neutral variants (0.66 and 0.34, respectively) confirm the idea that wild-type residues are more conserved in disease-associated nsSNVs. Analyzing the distributions of the frequencies of the mutant residues, it was shown that in ∼60% of the deleterious mutations, the mutant residue does not appear in any sequence of the multiple sequence alignment, whereas in ∼71% of the neutral mutations, the mutant residue appears at least once. In addition, the distribution of the difference between the frequencies of the wild-type and mutant residues, in Figure 2C, confirms the previous observations. Similar results were obtained when calculating the distributions of the conservation index as defined in Ref. [151] (Figure 2D).
Figure 2:
Distribution of the frequencies of wild-type (A) and mutant (B) residues, difference between the frequencies of wild-type and mutant residues (C) and Conservation Index [151] (D) for disease-related and neutral nsSNVs. Black and white bars show the distributions for disease-related and neutral nonsynonymous variants, respectively, for a set of 54 347 nsSNVs extracted from SwissVar database (October 2009). The data set was composed of 20 089 disease-related and 34 258 neutral mutations from 11 657 proteins. Sequence profiles were calculated from one run of the BLAST algorithm [152] over the UniRef90 database [153] and selecting only sequences with E-values lower than 10−9.
The discriminative power of evolutionary information is used in all prediction methods, although in different ways. For example, SIFT [114] uses blocks of conserved regions in proteins, PhD-SNP [111] and SNPs&GO [116] calculate the sequence profile by running the BLAST algorithm [152] over a database of sequences, PolyPhen [112] and SNAP [154] also include position-specific independent count (PSIC) scores, PANTHER [134] compares the sequence against a library of hidden Markov models from protein families and other methods perform the analysis of the DNA sequence by evaluating the selective pressure acting at the codon level [155, 156]. Predictors also use features from predicted or experimentally determined protein structures and available functional information [28, 112, 113, 117, 138, 154].
The algorithms for the detection of deleterious nsSNVs also differ in their training sets and underlying classification methodologies. Most are trained on an annotated set of nonsynonymous variants from OMIM, HGMD and/or SwissVar. Others, like SNAP, use mutations from the PMD database [157], collecting functional (as opposed to disease associated) data from mutagenesis experiments. In a recent work, the performance of a pool of methods was assessed on a curated set of nsSNVs [158]. This study showed that more sophisticated methods, such as MutPred [113] and SNAP [154] which include features from predicted structures and SNPs&GO [116] which uses functional information encoded in a Gene Ontology-based score, had the best performance.
With the increasing number of annotated nsSNVs, new algorithms can also be trained on a set of mutations associated to a specific class of diseases and/or proteins. Methods for the prediction of cancer-causing nsSNVs [159–161] are an interesting example, as is a new tool for predicting the effect of genetic variants in voltage-gated potassium channels [162].
Predicting the impact of genomic variants in noncoding regions
Until recently, the analysis of the effect of genetic variations strongly focused on those altering the protein sequence. The interpretation of genetic variants occurring in noncoding regions is also a challenging task. Although variants in noncoding regions may exhibit weaker effects than nsSNVs, it is evident that they constitute the majority of human genetic variations [4, 136], and are also likely to be disease-associated; i.e. ∼88% of weakly trait-associated variants from GWAS studies are noncoding [5]. Noncoding variants under purifying selection are five times more common than those in coding regions [163], and the detection of numerous regulatory variants with significant effect [15] has recently spurred interest in their computational annotation. Thus, a considerable number of methods are currently available to perform an evolutionary analysis of the nucleotide sequence to determine conserved regions across species. This approach, also applicable to protein sequences, is more complex for noncoding regions where there is no detectable conservation outside vertebrates [164]. The available algorithms for the detection of deleterious noncoding SNVs estimate the rate of evolution at the mutated position or consider a sliding window around the mutation site. Methods like binCons [119] and phastCons [124] implement a context dependent approach or a Hidden Markov model, in contrast to other algorithms such as GERP [120], SCONE [125] and Gumby [121] which calculate a position-specific score. This class of methods was also reviewed in a recent publication [165].
New approaches to predict the effect of mutations in noncoding regions focus specifically on genetic variations in regulatory regions and splicing sites. For example, Is-rSNP [122] uses a transcription factor position weight matrix and novel convolution methods to evaluate the statistical significance of the score. The RAVEN algorithm combines phylogenetic information and transcription factor binding site prediction to detect variations in candidate cis-regulatory elements [166]. Recently, a new method including features associated with the mutated site and its surrounding region and gene-based features has been used for the identification of functional, regulatory SNVs involved in monogenic and complex diseases [167]. SNVs affecting splicing sites and their surrounding regions can be evaluated using Skippy [126]. In Table 3, we listed a selection of methods for the prediction of the effect of SNVs.
Integrated tools for variant annotation
The steps for interpreting the net effects of variants from an individual genome or from a disease association study have previously been performed one at a time: filtering out common polymorphisms, identifying known deleterious mutations, functionally annotating and predicting the effects of novel variants and prioritizing variants for experimental follow-up. A number of integrated tools are now emerging to automate various portions of this pipeline including ANNOVAR [118], the Ensembl Variant Effect Predictor [168], GAMES [169], SeqAnt [170], Sequence Variant Analyzer (SVA) [171] and MutationTaster [123]. Frameworks for storing patient data along with associated analysis tools like i2b2 [172] and caBIG [173], and workflow management systems like Galaxy [174] and Taverna [175] that can be installed and run ‘on the cloud’, are also now available to automate and dramatically speed up variant annotation pipelines.
FUTURE OUTLOOK
Advances in high-throughput sequencing technology are generating a large amount of genetic variation data, thereby creating more complete models relating genotype to phenotype. The release of this information to publicly available databases has stimulated the development of several tools for genome interpretation. Although these methods have reached a promising level of accuracy, there are still many challenges to overcome before they will be directly applicable in a clinical setting. A number of recent studies address this concern [176, 177]. To make personal genome analysis a routine practice in the diagnosis and treatment of genetically determined phenotypes, the following challenges must be met: (i) defining standard and unified protocols for testing functional variation, (ii) designing integrated and publicly available resources of annotated genetic variants, (iii) developing holistic approaches to score the effect of multiple genetic variants, (iv) implementing user-friendly methods for the application of personal genomics in the health care context. The first challenge will require outlining easily reproducible experimental procedures necessary for data consistency. Curated data sets with standardized nomenclatures for the functional effects of genetic variants (easily parseable from the literature) will also be necessary. These resources will be useful for the development and benchmarking of new and more accurate methods for genome interpretation.
One of the most challenging aims for personal genomics will be the development of models able to capture the full complexity of the human genome. These models should take into account the linkage disequilibrium between different genomic regions and the possible effects of compensatory mutations. Bioinformatics will be particularly important for this challenge, enabling the design of heuristics to reduce the computational complexity of the problem. Since one of the primary goals of personal genomics is the development of computational methods for use in clinical diagnostics, an important issue is the usability of these tools. New clinical applications should be easily accessible, return useful and comprehensible results and perform their analyses in a reasonable run time. It will thus be crucial to adopt open access policies that, avoiding privacy/copyright issues, will allow the sharing of large sets of data and developed analysis tools. In particular these algorithms can be used to define a set of markers important for genetic counseling. In the near future we expect to have accurate disease-specific protocols for estimating the disease development and transmission risks inherent to a personal variome. These will be useful in the diagnosis of inherited disease, in preventative management and/or in family planning.
Recently, the Critical Assessment for Genome Interpretation (CAGI) experiments, assessing the accuracy of computational methods for genome interpretation over a blind set of data, and international meetings, e.g. ISMB SNP-SIG, AIMM and PSB, have drawn attention in the bioinformatics community to the challenges of the analysis of a personal genome. In the near future, these types of initiatives will be essential for organizing the necessarily interdisciplinary scientific environment for cracking the code of the human genome.
Key Points.
Vast amounts of variation data from genome sequencing studies need to be analyzed to understand its association with various phenotypes.
Well-curated databases, reliable tools for gene prioritization and accurate methods for predicting the impact of variants will be essential for the interpretation of personal genomes.
Standard and unified protocols for testing the functional impact of genetic variations are critical for their accurate annotation.
Experimental studies and computational models describing the gene/protein interaction networks and aiming at capturing the full complexity of the human genome will be key to leveraging personal genomic data for the precise diagnosis and effective treatment of disease.
FUNDING
The European Community through the Marie Curie International Outgoing Fellowship program (PIOF-GA-2009-237225 to E.C.); this work was supported by the National Institutes of Health (NIH) (1K22CA143148 to M.G.K.); Rutgers University, New Brunswick, School of Environmental and Biological Science (SEBS) start-up funds (to Y.B.); the Research Participation Program administered by Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between Department of Energy (DOE) and Food and Drug Administration (FDA) (to N.N.).
Acknowledgements
We acknowledge all the colleagues who contributed in the organization and panel discussions at the last PSB and ISMB SNP-SIG meetings. In particular, we would like to thank Can Alkan, Christopher Baker, Steven Brenner, Sean Mooney, John Moult, Pauline Ng, Burkhard Rost, Janita Thusberg and Mauno Vihinen. The mention of commercial products herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services. This is a contribution of the Food and Drug Administration and is not subject to copyright.
Biographies
Emidio Capriotti is a Marie Curie International Outgoing Fellow at the University of Balearic Islands (Spain). He was previously a postdoctoral researcher at the Department of Bioengineering, Stanford University. His main research lines include the development of algorithms for the prediction of protein and RNA three-dimensional structure and the effect of point protein mutations.
Nathan Nehrt is a bioinformatician and United States Food and Drug Administration Research Participation Program Fellow working with Maricel Kann at the University of Maryland, Baltimore County. His current research includes the classification of variants from human sequence data and the study of protein domain-based properties of human disease mutations.
Maricel Kann is an assistant professor at the University of Maryland, Baltimore County. Her research interests include classification of human variants, prediction of protein–protein interactions and the study of protein domains and their associations with disease. She is one of the leading experts in the area of translational Bioinformatics, and has chaired several international conferences.
Yana Bromberg is an assistant professor in the department of Biochemistry and Microbiology at Rutgers University, New Brunswick. Her research interests involve developing bioinformatics methods for the prediction/annotation of protein function and the analysis of genome variation across the full spectrum of life. Together with E.C., she chairs the annual SNP interest group meeting in the context of the ISMB/ECCB conference.
References
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mottaz A, David FP, Veuthey AL, et al. Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics. 2010;26:851–2. doi: 10.1093/bioinformatics/btq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 6.WTCC Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frazer KA, Murray SS, Schork NJ, et al. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 9.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 12.Ng PC, Murray SS, Levy S, et al. An agenda for personalized medicine. Nature. 2009;461:724–6. doi: 10.1038/461724a. [DOI] [PubMed] [Google Scholar]
- 13.Church DM, Lappalainen I, Sneddon TP, et al. Public data archives for genomic structural variation. Nat Genet. 2010;42:813–4. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Feuk L, Duggan GE, et al. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–14. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 15.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amberger J, Bocchini CA, Scott AF, et al. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–6. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker KG, Barnes KC, Bright TJ, et al. The genetic association database. Nat Genet. 2004;36:431–2. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 19.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–7. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 23.Knox C, Law V, Jewison T, et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39:D1035–41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–5. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huss JW, 3rd, Lindenbaum P, Martone M, et al. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38:D633–9. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann R. A wiki for the life sciences where authorship matters. Nat Genet. 2008;40:1047–51. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 27.Peterson TA, Adadey A, Santana-Cruz I, et al. DMDM: domain mapping of disease mutations. Bioinformatics. 2010;26:2458–9. doi: 10.1093/bioinformatics/btq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karchin R, Diekhans M, Kelly L, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–20. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 29.Singh A, Olowoyeye A, Baenziger PH, et al. MutDB: update on development of tools for the biochemical analysis of genetic variation. Nucleic Acids Res. 2008;36:D815–9. doi: 10.1093/nar/gkm659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurst JM, McMillan LE, Porter CT, et al. The SAAPdb web resource: a large-scale structural analysis of mutant proteins. Hum Mutat. 2009;30:616–24. doi: 10.1002/humu.20898. [DOI] [PubMed] [Google Scholar]
- 31.Uzun A, Leslin CM, Abyzov A, et al. Structure SNP (StSNP): a web server for mapping and modeling nsSNPs on protein structures with linkage to metabolic pathways. Nucleic Acids Res. 2007;35:W384–92. doi: 10.1093/nar/gkm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reumers J, Conde L, Medina I, et al. Joint annotation of coding and non-coding single nucleotide polymorphisms and mutations in the SNPeffect and PupaSuite databases. Nucleic Acids Res. 2008;36:D825–9. doi: 10.1093/nar/gkm979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stitziel NO, Binkowski TA, Tseng YY, et al. topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res. 2004;32:D520–2. doi: 10.1093/nar/gkh104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitropoulou C, Webb AJ, Mitropoulos K, et al. Locus-specific database domain and data content analysis: evolution and content maturation toward clinical use. Hum Mutat. 2010;31:1109–16. doi: 10.1002/humu.21332. [DOI] [PubMed] [Google Scholar]
- 35.Hindorff LA, Junkins HA, Hall PN, et al. A Catalog of Published Genome-Wide Association Studies. www.genome.gov/gwastudies (19 September 2011, date last accessed) [Google Scholar]
- 36.Gong L, Owen RP, Gor W, et al. PharmGKB: an integrated resource of pharmacogenomic data and knowledge. Curr Protoc Bioinformatics. 2008;Chapter 14:Unit14 17. doi: 10.1002/0471250953.bi1407s23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose PW, Beran B, Bi C, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan M, Diekhans M, Lien S, et al. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–2. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakyan VK, Down TA, Balding DJ, et al. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satterlee JS, Schubeler D, Ng HH. Tackling the epigenome: challenges and opportunities for collaboration. Nat Biotechnol. 2010;28:1039–44. doi: 10.1038/nbt1010-1039. [DOI] [PubMed] [Google Scholar]
- 43.Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1–10. doi: 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- 44.Gaulton KJ, Mohlke KL, Vision TJ. A computational system to select candidate genes for complex human traits. Bioinformatics. 2007;23:1132–40. doi: 10.1093/bioinformatics/btm001. [DOI] [PubMed] [Google Scholar]
- 45.Hutz JE, Kraja AT, McLeod HL, et al. CANDID: a flexible method for prioritizing candidate genes for complex human traits. Genet Epidemiol. 2008;32:779–90. doi: 10.1002/gepi.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erten S, Bebek G, Ewing RM, et al. DADA: Degree-Aware Algorithms for Network-Based Disease Gene Prioritization. BioData Min. 2011;4:19. doi: 10.1186/1756-0381-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, Chen Y, Sun F, et al. DomainRBF: a Bayesian regression approach to the prioritization of candidate domains for complex diseases. BMC Syst Biol. 2011;5:55. doi: 10.1186/1752-0509-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tranchevent L, Bonachela Capdevila F, Nitsch D, et al. A guide to web tools to prioritize candidate genes. Brief Bioinform. 2011;12:22–32. doi: 10.1093/bib/bbq007. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Iratxeta C, Bork P, Andrade-Navarro MA. Update of the G2D tool for prioritization of gene candidates to inherited diseases. Nucleic Acids Res. 2007;35:W212–6. doi: 10.1093/nar/gkm223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Iratxeta C, Wjst M, Bork P, et al. G2D: a tool for mining genes associated with disease. BMC Genet. 2005;6:45. doi: 10.1186/1471-2156-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seelow D, Schwarz JM, Schuelke M. GeneDistiller—distilling candidate genes from linkage intervals. PLoS One. 2008;3:e3874. doi: 10.1371/journal.pone.0003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu W, Wulf A, Liu T, et al. Gene Prospector: an evidence gateway for evaluating potential susceptibility genes and interacting risk factors for human diseases. BMC Bioinformatics. 2008;9:528. doi: 10.1186/1471-2105-9-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohler S, Bauer S, Horn D, et al. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82:949–58. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fontaine JF, Priller F, Barbosa-Silva A, et al. Genie: literature-based gene prioritization at multi genomic scale. Nucleic Acids Res. 2011;39:W455–61. doi: 10.1093/nar/gkr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.George RA, Liu JY, Feng LL, et al. Analysis of protein sequence and interaction data for candidate disease gene prediction. Nucleic Acids Res. 2006;34:e130. doi: 10.1093/nar/gkl707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlicker A, Lengauer T, Albrecht M. Improving disease gene prioritization using the semantic similarity of Gene Ontology terms. Bioinformatics. 2010;26:i561–7. doi: 10.1093/bioinformatics/btq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Driel MA, Bruggeman J, Vriend G, et al. A text-mining analysis of the human phenome. Eur J Hum Genet. 2006;14:535–42. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 58.Xiong Q, Qiu Y, Gu W. PGMapper: a web-based tool linking phenotype to genes. Bioinformatics. 2008;24:1011–3. doi: 10.1093/bioinformatics/btn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radivojac P, Peng K, Clark WT, et al. An integrated approach to inferring gene-disease associations in humans. Proteins. 2008;72:1030–7. doi: 10.1002/prot.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nitsch D, Tranchevent LC, Goncalves JP, et al. PINTA: a web server for network-based gene prioritization from expression data. Nucleic Acids Res. 2011;39:W334–8. doi: 10.1093/nar/gkr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng D, Knox C, Young N, et al. PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 2008;36:W399–405. doi: 10.1093/nar/gkn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adie EA, Adams RR, Evans KL, et al. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics. 2005;6:55. doi: 10.1186/1471-2105-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adie EA, Adams RR, Evans KL, et al. SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics. 2006;22:773–4. doi: 10.1093/bioinformatics/btk031. [DOI] [PubMed] [Google Scholar]
- 65.Chen J, Bardes EE, Aronow BJ, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Xu H, Aronow BJ, et al. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yandell M, Huff C, Hu H, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–42. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vanunu O, Magger O, Ruppin E, et al. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun J, Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11(Suppl 3):S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gandhi TK, Zhong J, Mathivanan S, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–93. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 71.van Noort V, Snel B, Huynen MA. Predicting gene function by conserved co-expression. Trends Genet. 2003;19:238–42. doi: 10.1016/S0168-9525(03)00056-8. [DOI] [PubMed] [Google Scholar]
- 72.Yu CL, Louie TM, Summers R, et al. Two distinct pathways for metabolism of theophylline and caffeine are coexpressed in Pseudomonas putida CBB5. J Bacteriol. 2009;191:4624–32. doi: 10.1128/JB.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hurst LD, Williams EJ, Pal C. Natural selection promotes the conservation of linkage of co-expressed genes. Trends Genet. 2002;18:604–6. doi: 10.1016/s0168-9525(02)02813-5. [DOI] [PubMed] [Google Scholar]
- 74.Elizondo LI, Jafar-Nejad P, Clewing JM, et al. Gene clusters, molecular evolution and disease: a speculation. Curr Genomics. 2009;10:64–75. doi: 10.2174/138920209787581271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dawkins R. The Selfish Gene. New York City: Oxford University Press; 1976. [Google Scholar]
- 76.Conant GC, Wagner A. Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc Biol Sci. 2004;271:89–96. doi: 10.1098/rspb.2003.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu Z, Steinmetz LM, Gu X, et al. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–6. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 78.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.del Pozo A, Pazos F, Valencia A. Defining functional distances over gene ontology. BMC Bioinformatics. 2008;9:50. doi: 10.1186/1471-2105-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lord PW, Stevens RD, Brass A, et al. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–83. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 81.Wang JZ, Du Z, Payattakool R, et al. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–81. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- 82.Nehrt NL, Clark WT, Radivojac P, et al. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput Biol. 2011;7:e1002073. doi: 10.1371/journal.pcbi.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Punta M, Ofran Y. The rough guide to in silico function prediction, or how to use sequence and structure information to predict protein function. PLoS Comput Biol. 2008;4:e1000160. doi: 10.1371/journal.pcbi.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lopez-Bigas N, Ouzounis CA. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32:3108–14. doi: 10.1093/nar/gkh605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iakoucheva LM, Brown CJ, Lawson JD, et al. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 86.Washington NL, Haendel MA, Mungall CJ, et al. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ala U, Piro RM, Grassi E, et al. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput Biol. 2008;4:e1000043. doi: 10.1371/journal.pcbi.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mootha VK, Lepage P, Miller K, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100:605–10. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukuoka Y, Inaoka H, Kohane IS. Inter-species differences of co-expression of neighboring genes in eukaryotic genomes. BMC Genomics. 2004;5:4. doi: 10.1186/1471-2164-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–8. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 91.Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang B-B, Wang J-G, Wang Y, et al. Gene Prioritization for Type 2 Diabetes in Tissue-specific Protein Interaction Networks. Syst Biol. 2009;10801131:319–28. [Google Scholar]
- 94.Mitchell JA, Aronson AR, Mork JG, et al. Gene indexing: characterization and analysis of NLM's GeneRIFs. AMIA Annu Symp Proc. 2003:460–4. [PMC free article] [PubMed] [Google Scholar]
- 95.Altman RB, Bergman CM, Blake J, et al. Text mining for biology–the way forward: opinions from leading scientists. Genome Biol. 2008;9(Suppl 2):S7. doi: 10.1186/gb-2008-9-s2-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blaschke C, Andrade MA, Ouzounis C, et al. Automatic extraction of biological information from scientific text: protein-protein interactions. Proc Int Conf Intell Syst Mol Biol. 1999:60–7. [PubMed] [Google Scholar]
- 97.Hirschman L, Yeh A, Blaschke C, et al. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Caporaso JG, Baumgartner WA, Jr, Randolph DA, et al. MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics. 2007;23:1862–5. doi: 10.1093/bioinformatics/btm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laurila JB, Naderi N, Witte R, et al. Algorithms and semantic infrastructure for mutation impact extraction and grounding. BMC Genomics. 2010;11(Suppl 4):S24. doi: 10.1186/1471-2164-11-S4-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mika S, Rost B. NLProt: extracting protein names and sequences from papers. Nucleic Acids Res. 2004;32:W634–7. doi: 10.1093/nar/gkh427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 102.Yoshida Y, Makita Y, Heida N, et al. PosMed (Positional Medline): prioritizing genes with an artificial neural network comprising medical documents to accelerate positional cloning. Nucleic Acids Res. 2009;37:W147–52. doi: 10.1093/nar/gkp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tranchevent LC, Barriot R, Yu S, et al. ENDEAVOUR update: a web resource for gene prioritization in multiple species. Nucleic Acids Res. 2008;36:W377–84. doi: 10.1093/nar/gkn325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Masso M, Vaisman II. Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics. 2008;24:2002–9. doi: 10.1093/bioinformatics/btn353. [DOI] [PubMed] [Google Scholar]
- 105.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–42. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–26. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schymkowitz J, Borg J, Stricher F, et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–10. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dehouck Y, Grosfils A, Folch B, et al. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009;25:2537–43. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- 110.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–34. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 112.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li B, Krishnan VG, Mort ME, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–50. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–8. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Calabrese R, Capriotti E, Fariselli P, et al. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–44. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 117.Capriotti E, Altman RB. Improving the prediction of disease-related variants using protein three-dimensional structure. BMC Bioinformatics. 2011;12(Suppl 4):S3. doi: 10.1186/1471-2105-12-S4-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Margulies EH, Blanchette M, Haussler D, et al. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–18. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cooper GM, Stone EA, Asimenos G, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prabhakar S, Poulin F, Shoukry M, et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–63. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Macintyre G, Bailey J, Haviv I, et al. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26:i524–30. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schwarz JM, Rodelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–76. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 124.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Asthana S, Roytberg M, Stamatoyannopoulos J, et al. Analysis of sequence conservation at nucleotide resolution. PLoS Comput Biol. 2007;3:e254. doi: 10.1371/journal.pcbi.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Woolfe A, Mullikin JC, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol. 2010;11:R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frazer KA, Pachter L, Poliakov A, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26:1057–63. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fernald GH, Capriotti E, Daneshjou R, et al. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27:1741–8. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cline MS, Karchin R. Using bioinformatics to predict the functional impact of SNVs. Bioinformatics. 2011;27:441–8. doi: 10.1093/bioinformatics/btq695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinform. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- 133.Tavtigian SV, Greenblatt MS, Lesueur F, et al. In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat. 2008;29:1327–36. doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci USA. 2004;101:15398–403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhu Q, Ge D, Maia JM, et al. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am J Hum Genet. 2011;88:458–68. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kumar S, Dudley JT, Filipski A, et al. Phylomedicine: an evolutionary telescope to explore and diagnose the universe of disease mutations. Trends Genet. 2011;27:377–86. doi: 10.1016/j.tig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thusberg J, Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum Mutat. 2009;30:703–14. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- 138.Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–73. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 139.Steward RE, MacArthur MW, Laskowski RA, et al. Molecular basis of inherited diseases: a structural perspective. Trends Genet. 2003;19:505–13. doi: 10.1016/S0168-9525(03)00195-1. [DOI] [PubMed] [Google Scholar]
- 140.Deutsch C, Krishnamoorthy B. Four-body scoring function for mutagenesis. Bioinformatics. 2007;23:3009–15. doi: 10.1093/bioinformatics/btm481. [DOI] [PubMed] [Google Scholar]
- 141.Gilis D, Rooman M. PoPMuSiC, an algorithm for predicting protein mutant stability changes: application to prion proteins. Protein Eng. 2000;13:849–56. doi: 10.1093/protein/13.12.849. [DOI] [PubMed] [Google Scholar]
- 142.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–87. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 143.Pitera JW, Kollman PA. Exhaustive mutagenesis in silico: multicoordinate free energy calculations on proteins and peptides. Proteins. 2000;41:385–97. [PubMed] [Google Scholar]
- 144.Prevost M, Wodak SJ, Tidor B, et al. Contribution of the hydrophobic effect to protein stability: analysis based on simulations of the Ile-96—Ala mutation in barnase. Proc Natl Acad Sci USA. 1991;88:10880–4. doi: 10.1073/pnas.88.23.10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou H, Zhou Y. Quantifying the effect of burial of amino acid residues on protein stability. Proteins. 2004;54:315–22. doi: 10.1002/prot.10584. [DOI] [PubMed] [Google Scholar]
- 146.Capriotti E, Fariselli P, Casadio R. A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 2004;20(Suppl 1):I63–8. doi: 10.1093/bioinformatics/bth928. [DOI] [PubMed] [Google Scholar]
- 147.Capriotti E, Fariselli P, Rossi I, et al. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics. 2008;9(Suppl 2):S6. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Capriotti E, Fariselli P, Calabrese R, et al. Predicting protein stability changes from sequences using support vector machines. Bioinformatics. 2005;21(Suppl 2):ii54–8. doi: 10.1093/bioinformatics/bti1109. [DOI] [PubMed] [Google Scholar]
- 149.Kumar MD, Bava KA, Gromiha MM, et al. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34:D204–6. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31:675–84. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 151.Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–12. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 152.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Suzek BE, Huang H, McGarvey P, et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–8. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 154.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–35. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arbiza L, Duchi S, Montaner D, et al. Selective pressures at a codon-level predict deleterious mutations in human disease genes. J Mol Biol. 2006;358:1390–404. doi: 10.1016/j.jmb.2006.02.067. [DOI] [PubMed] [Google Scholar]
- 156.Capriotti E, Arbiza L, Casadio R, et al. Use of estimated evolutionary strength at the codon level improves the prediction of disease-related protein mutations in humans. Hum Mutat. 2008;29:198–204. doi: 10.1002/humu.20628. [DOI] [PubMed] [Google Scholar]
- 157.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Res. 1999;27:355–7. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–68. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 159.Capriotti E, Altman RB. A new disease-specific machine learning approach for the prediction of cancer-causing missense variants. Genomics. 2011;98:310–7. doi: 10.1016/j.ygeno.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–7. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaminker JS, Zhang Y, Watanabe C, et al. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007;35:W595–8. doi: 10.1093/nar/gkm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stead LF, Wood IC, Westhead DR. KvSNP: accurately predicting the effect of genetic variants in voltage-gated potassium channels. Bioinformatics. 2011;27:2181–6. doi: 10.1093/bioinformatics/btr365. [DOI] [PubMed] [Google Scholar]
- 163.Davydov EV, Goode DL, Sirota M, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dehal P, Satou Y, Campbell RK, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–67. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 165.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–40. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 166.Andersen MC, Engstrom PG, Lithwick S, et al. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput Biol. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhao Y, Clark WT, Mort M, et al. Prediction of functional regulatory SNPs in monogenic and complex disease. Hum Mutat. 2011;32:1183–90. doi: 10.1002/humu.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McLaren W, Pritchard B, Rios D, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sana ME, Iascone M, Marchetti D, et al. GAMES identifies and annotates mutations in next-generation sequencing projects. Bioinformatics. 2011;27:9–13. doi: 10.1093/bioinformatics/btq603. [DOI] [PubMed] [Google Scholar]
- 170.Shetty AC, Athri P, Mondal K, et al. SeqAnt: a web service to rapidly identify and annotate DNA sequence variations. BMC Bioinformatics. 2010;11:471. doi: 10.1186/1471-2105-11-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ge D, Ruzzo EK, Shianna KV, et al. SVA: software for annotating and visualizing sequenced human genomes. Bioinformatics. 2011;27:1998–2000. doi: 10.1093/bioinformatics/btr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.caBIG Strategic Planning Workspace. The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Stud Health Technol Inform. 2007;129:330–4. [PubMed] [Google Scholar]
- 174.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hull D, Wolstencroft K, Stevens R, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–32. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dewey F, Chen R, Cordero S, et al. Phased whole genome genetic risk in a family quartet using a major allele reference sequence. PLOS Genet. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]