Abstract
The explosion of biomedical data, both on the genomic and proteomic side as well as clinical data, will require complex integration and analysis to provide new molecular variables to better understand the molecular basis of phenotype. Currently, much data exist in silos and is not analyzed in frameworks where all data are brought to bear in the development of biomarkers and novel functional targets. This is beginning to change. Network biology approaches, which emphasize the interactions between genes, proteins and metabolites provide a framework for data integration such that genome, proteome, metabolome and other -omics data can be jointly analyzed to understand and predict disease phenotypes. In this review, recent advances in network biology approaches and results are identified. A common theme is the potential for network analysis to provide multiplexed and functionally connected biomarkers for analyzing the molecular basis of disease, thus changing our approaches to analyzing and modeling genome- and proteome-wide data.
Keywords: network biology, bioinformatics
INTRODUCTION
Network biology is an emerging field that attempts to integrate -omics data of various and seemingly disparate types into a biologically meaningful framework suitable for joint analysis. Biological regulation is a complex process, and the effects of single genes and proteins—while potent in the context of model systems and model organisms—often are diffuse in the context of a complex background of genetic variation typical in populations layered on a complex set of environmental stimuli. Redundancies of function, driving cooperation or competition between different genes and proteins, are hallmarks of population fitness and robust response to the environment. Network biology approaches take the logical step beyond both single gene and pathway analysis, attempting to detect and ultimately model the complex multi-dimensional interactions of cells, organs and organisms. In this review, we highlight several areas where significant progress has been made in the last 12–18 months in using network biology to analyze disease phenotypes. These examples expand and extend network frameworks to integrate multiple types of biological data to enable deeper mechanistic and medical insight. In particular integration of gene expression and protein–protein interaction (PPI) data remains a potent theme. We note progress in coupling disease-gene association data with network analysis methods and we review explicit modeling of biochemical networks to understand disease and ultimately predict therapeutic response.
INTEGRATING DIVERSE DATA SETS AND IMPLICATIONS FOR TRANSLATIONAL SCIENCE
In recent years, high-throughput methodologies, such as the yeast two-hybrid assays (Y2H) [1–3] and co-immunoprecipitation followed by mass spectrometry (AP/MS) [4, 5], have been widely used to identify physical PPIs for a wide range of organisms. Yeast two-hybrid assays provide a high-throughout assessment of binding for two domains or molecular entities and can be carried out on a genomic scale. AP/MS focuses on the identification of complexes through engineering of expressed ‘baits’ and cellular pullout of ‘prey’ interactors, which are identified by mass spectrometry. These methods have evolved such that assessment of sensitivity and specificity of the discovered interactions and development of sophisticated databases of the interactions is now commonplace. In addition to these physical measures of network connections, ‘genetic’ interactions, which identify cause and effect relationships without specific knowledge of whether the interactions are direct, have been mapped for many organisms [6, 7]. Although these databases of network interactions currently have not attained full ‘coverage’, e.g. many interactions are likely to be missing (false negatives) and calculating the rate of false positives is challenging, such technologies have revolutionized our understanding of biological function as a network of interactions, enabling analyses on a systems scale rather than at the level of individual genes or proteins. It is clear that many prevalent diseases such as cancer, diabetes and heart disease are not solely caused by the action of single genes, but rather by alterations in the functioning of a complex web of networks and pathways. Meanwhile, genome-wide measurements of multiple organisms and individuals made at the genome, transcriptome, metabolome and proteome level present new opportunities for both data integration and potential translation of findings to medical practice. We are now in a position to integrate these multiple types of systems level -omics data sets through various models of network biology. Such workflows will lead to discovery of new insights to understand these complex diseases and the system perturbations accompanying disease phenotype. As tools such as next-generation sequencing and mass-spectrometry instruments for analyzing the genome, proteome and metabolome keep improving and new types of -omics data emerge; developing computational frameworks for the integration of these different layers of data presents a challenge. Although pipelines to integrate large and diverse data sets and narrow them down to connected pathways that have prognostic value are emerging (Figure 1) (adapted from Ref. [8]) applying them productively in the context of clinical decision making has not yet been realized. Nevertheless, these methods are productive in identifying disease-associated pathways or biomarkers. Although bench scientists and clinicians recognize the need to translate these methods and knowledge to the bedside, the interdisciplinary nature of these studies and the lack of easy to use tools are impeding progress. Ideally, the goal of translational science with respect to diverse -omics data sets is to allow a scientist/clinician with limited computational skills to connect multiple layers of patient specific information to arrive at a more informed prognosis and, by identification of patient-specific molecular information, permit the design of optimal treatment choices, at least in the context of clinical trials if not in patient care. Although these ultimate goals are desirable, connecting even the simpler variables of diagnosis, prognosis or prediction of response to integrated -omics data is challenging. To overcome these challenges, we suggest the translational bioinformatics community consider the following as potential opportunities:
Utilize existing models of biology [e.g. protein–protein and genetic interaction networks, Gene Ontology (GO), known pathways] to integrate diverse -omics data using emerging novel tools providing multiplexed network biomarkers of disease. In many cases integration of all available data sets can be a starting point driving research.
Based on network biomarkers discovered from initial studies, develop and apply methods to segregate individual patients or patient classes with respect to clinical phenotypes or disease outcomes.
Develop tools and methods to visualize patient and disease-specific data from multiple sources enabling visualization of the disease states of patients.
Refine network biology approaches and network biomarkers by comparison of multiple patient cohorts validating the relationship of the network modules to phenotype.
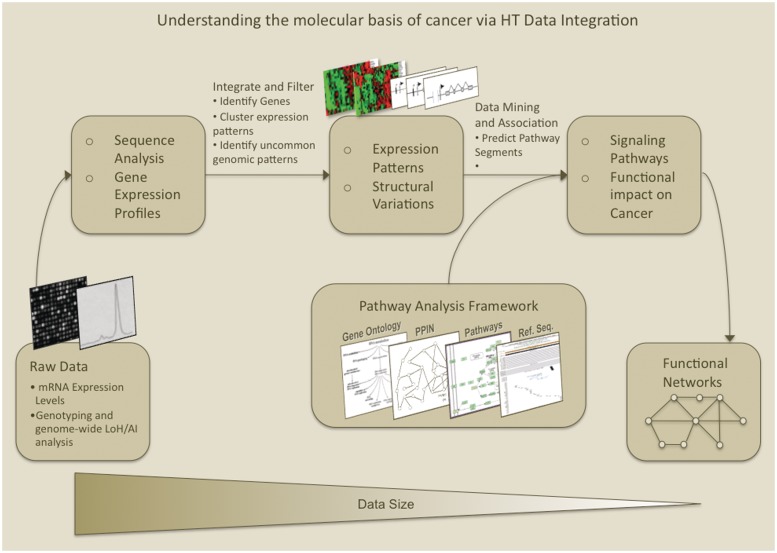
Figure 1:
Workflow for high-throughput data integration to help understand the molecular basis of cancer. An integrative -omics signaling network identification process workflow that begins with processing tissue-specific data (instrument outputs) is shown. Microarray data is normalized to make comparisons of expression levels and transformed to select genes for further analysis. Genome-wide genotyping signals are analyzed to identify regions (and hence regional genes) for both tumor and normal tissue (or non-cancerous cells). Next, genomic regions with significant aberrations are merged with their corresponding microarray probes to create expression profiles. In this analysis step, expression profiles are used to calculate Pearson's coexpression correlations among gene pairs. These results are fed into the Pathway Analysis Framework. Integrating gene–gene coexpression values, annotations from GO, known signaling pathways, protein sequence information, PPI networks and protein subcellular co-localization data, pathways are predicted and filtered. Significant pathway subnetworks are merged to form signaling networks connecting genes of interest. The networks and genomic alterations identified are put together to create a descriptive functional network, creating a molecular basis for the cancer studied. This type of workflow, which we utilized, can be applied to using integrative systems biology approaches to study cancer and other pathologies [8].
These general approaches are outlined in the examples that follow, providing a model for scientific discovery and validation in the new era of network biology.
SYSTEMS BIOLOGY OF DIFFERENTIAL GENE EXPRESSION: INTEGRATING TRANSCRIPTOMIC AND INTERACTOMIC DATA
Whole genome expression data, measured in terms of the mRNA transcripts present in a given sample (i.e. the transcriptome), has been valuable in characterizing cellular perturbations [9]. Identification of genes or gene sets that are differentially expressed in various disease states have enabled discovery of novel biomarkers for such clinical tasks as diagnosis and prognosis, as well as identifying potential targets for therapeutic intervention [10]. To this end, interpretation of differential gene expression from a systems perspective has the potential to shed light into the molecular mechanisms of complex diseases.
Systems biology approaches to differential expression analysis can be roughly classified into three categories: (i) signature-based, (ii) pathway-based and (iii) network-based analysis of differential gene expression. Signature-based approaches construct genome-wide expression signatures for diseases or drugs by comprehensively integrating multiple case-control data sets [11]. They then use these signatures to identify similarities between pairs of diseases, pairs of drugs [12] or disease and drug pairs [13] based on the similarity of expression signatures under the respective conditions (e.g. in samples with the disease or in samples under treatment). Subsequently, they predict new indications for drugs based on patterns of similarity between different drugs [13].
Pathway-based approaches mainly focus on relatively well-characterized cellular pathways and aim to systematically identify pathways that are enriched in products of differentially expressed genes. Among these, gene-set enrichment analysis (GSEA) has been quite popular in the last few years [14]. GSEA takes as input a set of genes (e.g. genes that code for proteins in a particular pathway) and aims to assess the overall rank of the genes in the set among all genes in the gene expression data set in terms of their differential expression in the disease of interest. If the genes coding for proteins in a pathway rank significantly higher compared to other genes in the entire genome, then the pathway is considered dysregulated in the disease. GSEA has been applied to the identification of dysregulated pathways in a large number of diseases and phenotypes, including breast cancer and obesity, among others. Following GSEA, many other methods have been developed for pathway-based differential expression analysis with improved statistical procedures [15].
While being quite useful, pathway-based analysis of differential gene expression has important limitations. In particular, pathway-based approaches restrict the functional relationships among genes (and their products) to well-characterized (and well-studied) pathways. Therefore, these approaches are generally not able to characterize the differential expression of relatively less studied genes, discover novel functional links among genes, or identify disease-specific crosstalk between different pathways. Furthermore, these methods rely on the assumption that genes coding for proteins in an ‘active’ pathway should show an evident correlation in their expression levels, which may not be necessarily true. PPI networks (i.e. the interactome) offer an invaluable resource in this regard. Since PPI networks are derived from high-throughput interaction data (e.g. Y2H and AP/MS), as well as comprehensive mining of other biological data sources (e.g. phylogenetic profiles, structural similarities, common citations), they contain potential functional links that are not captured from the perspective of canonical pathways that are characterized in detail [16]. Furthermore, since PPI networks provide a comprehensive map of functional interactions in the cell, they are also useful for global analyses that take into account network topology. To this end, network-based analysis of differential gene expression can effectively discover multiple interacting markers for disease and help generate novel hypotheses related to mechanisms of disease.
A commonly considered problem in network-based analysis of differential gene expression is the identification of sub-networks of the human PPI network that are significantly dysregulated with respect to a disease of interest. Here, the term sub-network refers to a group of proteins that are functionally linked to each other through PPIs (i.e. they induce a connected sub-graph of the PPI network). The input to the dysregulated sub-network discovery problem is genome-wide case–control expression data for a specific disease and a network of PPIs. The objective is to discover sub-networks of the PPI network that exhibit collective dysregulation with respect to the disease. Two key methodological challenges in this regard are (i) development of a scoring scheme to assess the collective dysregulation of multiple interacting genes and (ii) development of efficient computational algorithms to search for sub-networks with significant scores.
Common approaches to sub-network scoring differ from each other in terms of the order in which they integrate individual genes. A common approach is to first score the differential expression of each gene individually using a standard statistical test (e.g. t-test), and subsequently compute sub-network scores as an aggregation of these individual differential expression scores [17, 18]. While these methods are useful in identifying functionally related genes that are differentially expressed with respect to disease (‘active’ functional modules), they provide limited systems level insights since they assess the differential expression of functionally related genes individually. In other words, these methods cannot capture patterns of coordinated dysregulation at the level of individual samples.
An alternate strategy for scoring sub-networks is to first integrate expression levels of genes in a sub-network to construct a representative expression profile for the sub-network, and subsequently assess the ability of this representative profile in discriminating disease and control samples. The discriminative ability of an expression profile is often quantified in terms of its mutual information with phenotype, i.e. the reduction in the uncertainty of a sample's phenotype upon observation of the sub-network's expression profile for that sample [19–24]. These information-theoretic methods differ from each other in how they compute expression profiles for a sub-network. In a seminal paper, Chuang et al. [19] used ‘sub-network activity’ as the expression profile of a sub-network, which is defined as the aggregate expression of the genes in the sub-network. As illustrated in Figure 2, the concept of sub-network activity is also applied to integration of proteomic, transcriptomic and interactomic data to identify transcriptionally dysregulated sub-networks concentrated around post-translationally dysregulated proteins in colon cancer [20]. While quite useful, this additive scheme captures the coordination between the dysregulation of interacting gene products to a limited extent as interacting genes may not be additive in their functions. Observing that coordinated changes in the mRNA-level expression of interacting proteins can exhibit combinatorial patterns as well, Chowdhury et al. [21] quantized gene expression data and represented the expression profile of a sub-network as a multi-dimensional random variable that represents the combination of expression states of the genes in the sub-network. As a stronger information-theoretic measure of coordinated dysregulation, Anastassiou [22] proposed synergy, which is defined as the difference between the overall mutual information of sub-network state and the mutual information of sub-states of sub-network state.
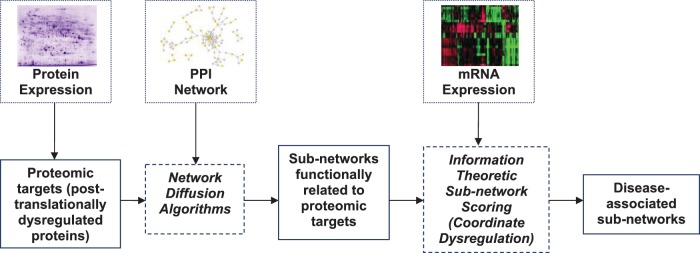
Figure 2:
A data integration framework for using disparate -omic data sets together to identify functional sub-networks in complex phenotypes. Data/experimental procedures are shown on the upper panel, inferred information shown in solid boxes on the lower panel, computational algorithms are shown in dashed boxes on the lower panel are shown by solid lines pipeline for identification of disease-associated sub-networks. This framework was used to identify PPI sub-networks dysregulated in late-stage colorectal cancer, revealing novel targets that are dysregulated at the post-translational level, but were not captured by untargeted proteomic analysis (45).
Besides mutual information, several alternate measures for assessing sub-network dysregulation have been recently proposed. These measures include the density of dysregulated genes in a subset of disease samples [25], the number of disease samples that can be distinguished from control samples by at least one gene in the sub-network [26], the linear separation between disease and control samples in the multi-dimensional space induced by expression profiles of the genes in the sub-network [27], and the ability of a decision tree constructed from the sub-network in discriminating disease and control samples [28].
Since the sub-network space of the human PPI network is of exponential size, searching for sub-networks with significant dysregulation is a challenging computational problem. In order to tackle these challenges, greedy heuristics [19, 26], branch-and-bound algorithms [21, 25], and randomized search algorithms [27], were proposed. Sub-networks identified by these algorithms were used as features for classification in various applications. It was repeatedly shown by several studies that such network-based classifiers outperform traditional gene expression based classifiers in predicting metastasis of breast [19, 25] and colorectal cancers [20, 21], response to chemotherapy [27], and progression of glioma [28].
An alternate approach to assessment of network-level dysregulation is to infer disease-specific networks by identifying interactions that are dysregulated in disease. As examples of this approach, Watkinson et al. [23] identified pairs of genes with synergistic differential expression in prostate cancer by clustering samples represented as points in the two-dimensional space induced by the expression levels of the pairs of genes and correlating this clustering with disease state. Similarly, Mani et al. [24] identified dysregulated interactions in B-cell lymphomas by constructing B-cell specific-networks and scoring the interactions in these networks using mutual information.
Overall, we expect continued advances in developing network models of disease driven by analysis of genome-wide expression data; these data are fertile ground for applying emerging graph theoretical algorithms to -omics data.
INTEGRATING GENOME-WIDE ASSOCIATION DATA AND NETWORK BIOLOGY TO UNDERSTAND DISEASE
Characterization of disease-associated variation in human genomes is an important step towards enhancing our understanding of the cellular mechanisms that drive complex diseases, with many potential applications in personalized medicine. In the last decade, genome-wide linkage and association studies (GWAS) based on comparison of healthy and affected populations have been quite useful in identifying genetic variants, particularly single nucleotide polymorphisms (SNPs) and more recently copy number variants (CNVs) that are potentially linked with disease [29]. However, many limitations of GWAS are being increasingly pronounced. These limitations, which pose significant challenges to effective identification of genes associated with complex diseases and their use in clinical applications, include the following:
The number of loci being monitored is in the order of millions while the number of patients is often limited to several thousands; therefore multiple hypothesis testing poses restrictions on evaluation of significance, leading to many candidate loci with moderate P-values [30].
Susceptible loci identified by GWAS so far generally account for a limited fraction of the genotypic variation in patient populations [31].
Predictive models based on identified loci have modest success in classifying phenotype (risk assessment) and therefore are of limited practical use [32, 33].
Many of the SNPs identified by GWAS do not have clear functional implications that provide insights into the mechanistic bases of disease; however, they might indeed have key regulatory roles [34].
Recognition of these limitations lead to concerns about the significant cost of the studies and a growing perception that the benefits to society are not yet matching the investment and thus that improved approaches are needed. This trend has not gone unrecognized by bioinformaticians, and explanations for the missing heritability have focused on rare variants [35] and interactions (epistasis) [36] as likely culprits.
Multi-gene analysis is commonly proposed as a logical next step to understanding the relationship of gene and disease [31]. The question is how to go about it without exponentially increasing the number of tests required? A promising direction in this regard is the development of multi-gene frameworks by incorporating biological data from other sources to confine the search space for combinations of variants to be tested [37]. This approach potentially has many favorable consequences, including reduction of the number of hypotheses (gene combinations) to be tested [38] and a context for gaining insights into the functional bases of genetic interactions [39, 40]. Consequently, computational methods are rapidly being developed to integrate GWA data with disparate -omics data sets for prioritizing and identifying combinations of genes that are most likely to be functionally associated with the disease of interest. Many methods utilize available pathway and annotation information to identify pathways that are significantly enriched in disease-associated genes [41, 42]. Recently, more sophisticated methods are also developed to assess disease association of pathways by directly integrating genotypes of the genes that take part in the pathway (as opposed to performing enrichment analysis based on association scores of individual genes) [43]. PPI networks also offer an invaluable resource in this regard, since they provide functional information in a network context and they can be obtained at a large scale via high-throughput screening [23].
Computational methods for identifying epistatic interactions using PPI data are still in relative infancy [31]. However, in the context of similar applications, use of PPI data has demonstrated great success in enhancing the outcome of GWAS [44]. These applications include prioritization of candidate disease genes and identification of disease-gene enriched sub-networks of the human PPI network.
Network-based prioritization of candidate disease genes
Methods for candidate gene prioritization are based on empirical evidence suggesting that products of genes that are implicated in clinically similar diseases are clustered together into ‘hot spots’ in PPI networks [45]. Motivated by these observations, many methods have been developed to search the PPI networks for interacting partners of known disease genes to narrow down the set of candidate genes implicated by GWAS. In one of the pioneering studies, Lage et al. [46] score candidate disease genes based on the association of their interacting partners with diseases clinically similar to the disease of interest. While such methods prove quite useful, they do not utilize knowledge of PPI networks to their full potential. In particular, they do not consider interactions among proteins that are not coded by candidate genes, which might also be useful in understanding indirect functional relationships between candidate genes and genes implicated in clinically similar diseases.
Information-flow based approaches to disease-gene prioritization are grounded on the notion that products of genes that have an important role in a disease are expected to exhibit significant network crosstalk to each other in terms of the aggregate strength of paths that connect the corresponding proteins. These methods, which include random walk with restarts [47] and network propagation [48], take as input the disease association scores of individual proteins (e.g. association with a disease clinically similar to the disease of interest). Subsequently, they propagate this information across the PPI network to compute network-based disease association scores for all proteins in the network. Finally, using these network-based association scores, they rank the proteins coded by genes in the implicated linkage interval for the disease of interest (Figure 3).
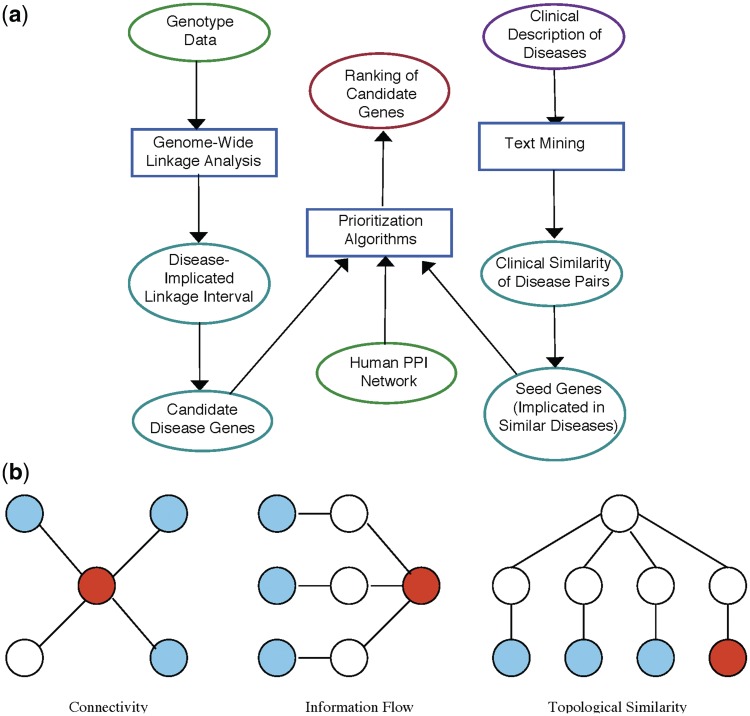
Figure 3:
Network-based prioritization of candidate disease genes. (A) Flow chart for network-based prioritization algorithms: -omic data are shown by green ellipses (top left), clinical data are shown by purple ellipses (top right), intermediary data are shown by cyan ellipses (left and right bottom two ellipses), computational algorithms and statistical analyses are shown by boxes, overall outcome of the framework is shown by a red ellipse (top middle). (B) Key principles employed by prioritization algorithms: Each panel shows part of a hypothetical PPI network, blue nodes (light grey) represent products of seed genes, red nodes (dark grey) represent products of candidate genes. Connectivity-based algorithms rank candidate genes based on their products direct interactions with product's of seed genes; information-flow based algorithms rank candidate genes based on themultiplicity of network paths between their products and products of seed genes; topological similarity based algorithms rank candidate genes based on the similarity of their products' location in the PPI network to that of the products of candidate genes.
Algorithms for candidate gene prioritization are often evaluated via leave-one-out cross-validation studies using data from Online Mendelian Inheritance in Man (OMIM). This database provides previously identified disease–gene associations for hundreds of human diseases and thousands of genes. In order to assess the performance of a prioritization algorithm, each disease–gene association in the database is considered. First, the association between the gene (named the target gene) and the disease (named the disease of interest) is removed from the database. Then a virtual linkage interval is constructed by selecting a number of genes in chromosomal proximity of the target gene as candidate genes. Subsequently, using a human PPI network and the clinical similarity of the disease of interest to other diseases, these candidate genes are ranked by the algorithm being tested. The final ranking of the target gene is then used as an indicator of the performance of the algorithm.
Systematic experimental studies show that, information-flow based approaches, which take into account the multiplicity of network paths between candidate genes and genes involved in clinically similar diseases, drastically improve the accuracy of network-based disease-gene prioritization, as compared to methods that only consider direct interactions [44]. However, these methods tend to favor highly connected gene products, since such proteins are likely to receive more flow [49]. While many disease-associated genes have many known interactions, loosely connected gene products are also of great interest for generating novel information, since such genes are likely to be less studied. Motivated by these considerations, network algorithms that use topological similarity instead of network connectivity are also proposed. These methods assess the potential disease association of each candidate gene based on the notion that proteins with similar roles in disease may be located similarly in terms of their proximity to other proteins in the network [50].
Identification of disease-gene enriched subnetworks
Since GWAS generally return many genetic variants that are moderately associated with disease. Aggregation of these moderate association scores within a functional context may reveal groups of functionally related proteins with significant aggregate association score. Based on this hypothesis, Jia et al. [51] integrate GWAS results for breast and pancreatic cancer using PPI data. They convert SNP markers in a GWAS data set to their associated genes, with the P-value of the genes being a function of the SNPs associated with those genes. Then they load the weighted genes onto a comprehensive human PPI network to construct a node-weighted PPI. Subsequently, they use a dense module-searching algorithm to identify sub-networks that maximize the proportion of low P-value nodes. The novelty of this method is to use all the SNP P-values in the calculation; this ‘moderate-significance’ SNPs can positively contribute to the networks if they interact with highly significant SNPs, while highly significant SNPs that only ‘talk’ to low significance SNPs are downgraded. For example, in the analysis of breast cancer GWAS, interesting genes like SMAD3, whose individual association is not significant (p ≥ 0.10), are identified as it is recruited into a dense sub-network that overall is significant.
Liu et al. [52] take a related approach for analyzing GWAS data from an obstructive sleep apnea (OSA) GWAS study. They first generate a tissue-specific protein–protein interactome from adipose tissue, as inflammation of adipose tissue and its relationship to obesity are key variables of the disease. They then search this interactome d for OSA related sub-networks by mapping P-values from the GWAS study to proteins of the interactome, similar to the approach above. In this case, the Cytoscape plug-in jactivemodule, originally developed to analyze microarray data [17], is re-purposed to detect a large significant sub-network within that interactome. The jactivemodule combines the network structure and associated P-value of each protein to extract potentially meaningful sub-networks. A highly significant subnetwork with 203 proteins and 324 interactions is identified by this method (Figure 4). Note that many of the nodes have modest P-values (low z-scores), and would not be seen as significant in a conventional GWAS. Another Cytoscape plugin MCODE is applied to explore the protein complexes or other modules present in the sub-network identified by jactivemodule. GO functional categories are analyzed for enrichment in the sub-network, the detected functions included insulin receptor signaling pathway, and negative regulation of tyrosine phosphorylation of STAT3 proteins. This is of interest as STAT3 tyrosine phosphorylation is noted to be critical for interleukin protein production in the inflammatory response and STAT family members are implicated in several processes relevant to tumor growth, providing a novel link between OSA and cancer.
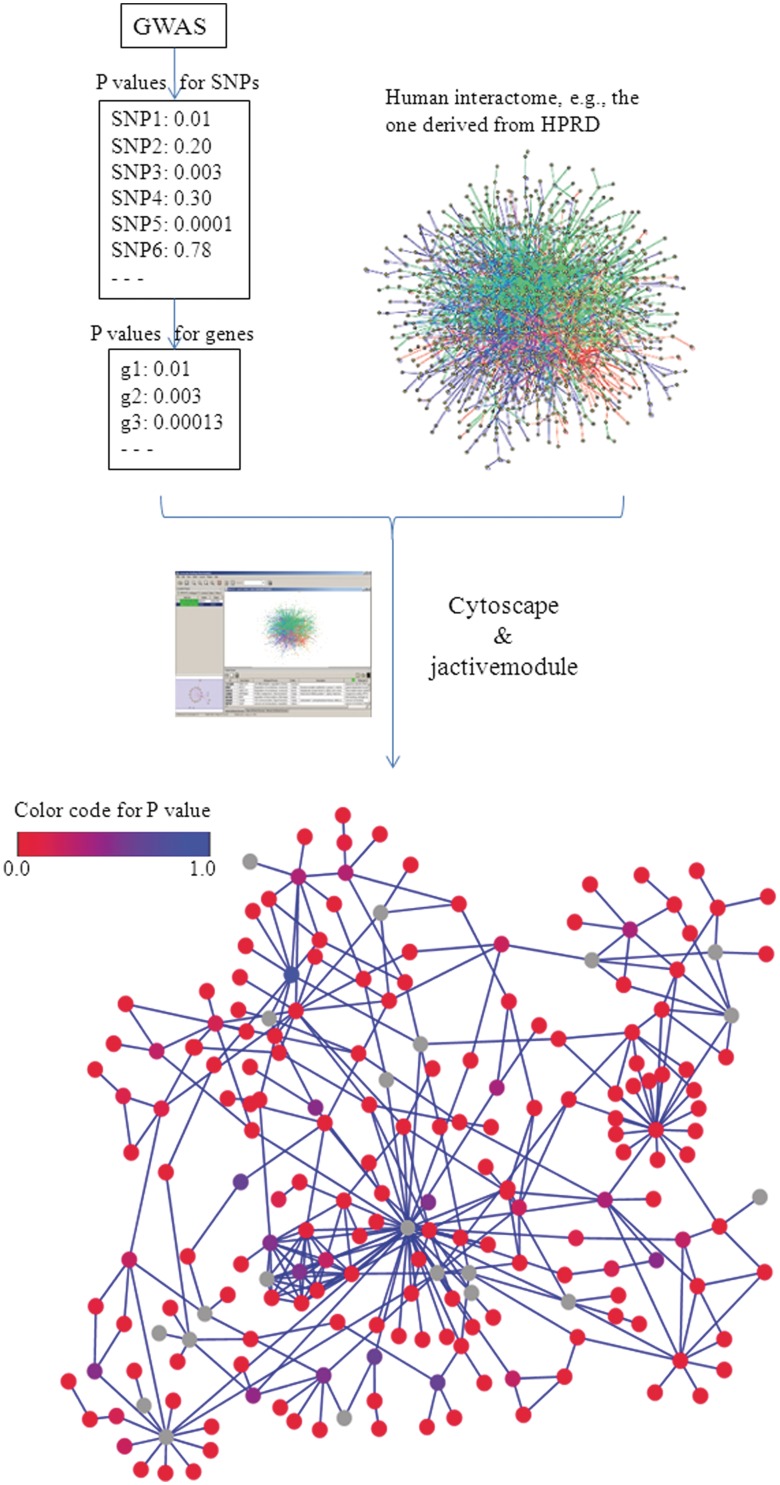
Figure 4:
Workflow for network detection. Networks are identified by jactivemodule using P-values from GWAS study (see text). Briefly, based on the P-values for each SNPs from GWAS, each gene has been assigned a P-value, then, they are superposed on the human PPI interactome derived from HPRD, finally, Cytoscape and jactivemodule are used to identify the network that is enriched with significant P-values. The color represents the P-values and nodes with gray color indicate that the P-values are missing from GWAS.
Overall, many recent studies apply GWAS data in an unbiased manner and use most or all of the data to probe for interactions of interest. This is an important next step in GWAS analysis, as it identifies additional genes that likely contribute to phenotype. However, the newly identified targets, missed in conventional GWAS, are not yet proven to recover missing phenotype. Construction of classifiers with these new targets, and testing in various cohorts is required to assure that significant progress is being made with these integrative approaches. Use of PPI networks is also likely to be promising in identifying epistatic interactions among two or more functionally related genes.
LOOKING AHEAD: MECHANISTIC IN SILICO MODELS TO GUIDE PERSONALIZED MEDICINE
An emerging frontier in systems and network approaches to medicine comes from recent advances in the ability to model large-scale biochemical reaction networks in human systems. Thus far, we have discussed multiple network approaches to aid the development of systems medicine; these approaches have primarily used networks generated via high-throughput interaction data and statistical network inference from high-throughput measurements (e.g. transcriptomes and GWAS data). However, biochemical reaction networks differ fundamentally from these networks in that they are based explicitly on our detailed understanding of the underlying chemical mechanisms in the cell [53], while the networks generated using high-throughput data are generally far less detailed or complete. These types of biochemical networks, particularly for metabolism, have been highly successful in modeling microbial systems [54, 55], as well as in smaller-scale human systems [56, 57]. These types of models offer strong potential for linking genotype with observed phenotypes quantitatively, and methods for their reconstruction are now quite advanced [58]. Most significantly, as biochemical reaction networks are based explicitly on the underlying chemical mechanisms of the system, the rules of physics and chemistry such as mass-energy balances and thermodynamics can be applied directly. Their basis in mechanism also means that they are generally built from the bottom up and involve ‘forward-calculations’ [59] based on linking well-characterized components together and computing the consequences of observed experimental data on the rest of the system, rather than on statistical learning or data-fitting. Thus, they can be used as a strong basis to interpret high-throughput data and offer the long-term potential to be a powerful means to address fundamental challenges of personalized medicine where particular disease-relevant perturbations in patients can be unique (and thus not easy to address from purely statistical approaches) (Figure 5).
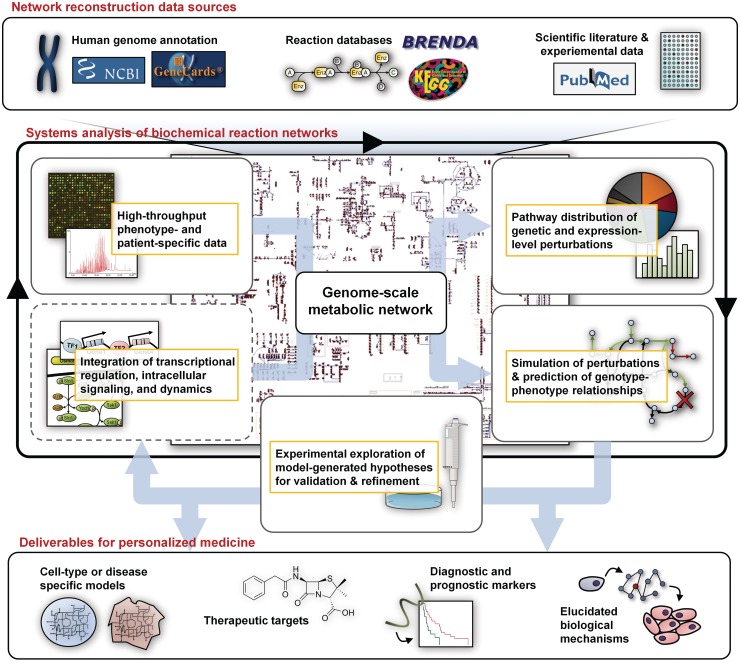
Figure 5:
Biochemical networks for personalized medicine. Biochemical reaction networks are rooted in the mechanistic interactions that comprise biological pathways; as such, these networks are carefully constructed from a wealth of genomic and metabolic databases, as well as from detailed experimental and literature data. Once networks have been constructed and curated—to ensure mass and charge balance, and to minimize gaps in connectivity—they serve as a powerful platform for interpreting high-throughput data. Not only can the network provide functional pathway context for genetic, transcriptomic or other perturbations, but through constraint-based modeling, these perturbations can be directly related to emergent phenotypes. Incorporating information from transcriptional regulation and intracellular signaling can lead to improved ability of the model to replicate in vitro and in vivo conditions. The iterative process of generating and experimentally testing simulated predictions leads to a refined and accurate model that holds great promise for facilitating personalized medicine.
IN SILICO METABOLIC MODELS AT THE GENOME-SCALE
Metabolic networks are the most comprehensive and well-understood class of biochemical reaction networks today. While large-scale maps of known metabolism in humans have existed for quite some time, it is only in the past few years that computational models of metabolism at the genome-scale in humans have been made [60, 61]. These initial models represent globally all the known metabolic potential encoded in the human genome. This global map is now serving as a starting point for generating specific metabolic models of each of the cell types of the human body, with the first genome-scale reconstructions having been completed for hepatocytes [62, 63]. Additionally, core models have been made of interactions between three distinct types of neurons and astrocytes [64]. These models of interacting cells can demonstrate differential effects of genetic perturbations on neuron subtypes and different regions of the brain—providing the ability to evaluate modifications in neurodegenerative diseases such as Alzheimer's [64] or Leigh's syndrome [65]. Even more recently, methods have been developed to integrate genome-scale metabolic networks with genome-scale transcriptional regulatory networks in a semi-automated framework [66], and efforts are underway to also elucidate genome-scale signaling networks in a similar manner [67].
APPLICATIONS OF GENOME-SCALE METABOLIC MODELS TO DISEASE
Genome-scale in silico models of human metabolism are already being applied to important medical questions, and their continued development holds great potential for long-term significant impact for personalized medicine. One of the best-known hallmarks of cancer and one of its most pervasive features is the Warburg effect, where cancer cells shift their metabolism to less energetically efficient glycolysis based energy production, which is potentially adaptive to a hypoxic tumor micro-environment, instead of the greater adenosine triphosphate producing oxidative phosphorylation generally used in normal cells. Using the first genome-scale metabolic model for cancer, evidence showed that this adaptation could be computed directly from cancer cells adapting to having higher growth rates [68]. Importantly, the model also captured three distinct metabolic phases commonly seen in tumor progression, as well as other key metabolic changes such as a preference for glutamine uptake over other amino acids.
Biochemical reaction network models also show promise in providing predictions of biomarkers grounded in disease mechanisms. One intriguing early study focused on predicting metabolites whose concentration in the blood was predicted to change based on inborn genetic errors in metabolic enzymes [69]. Genetic mutations that cause enzyme defects can be simulated and the highest confidence metabolic changes predicted, which give rise to highly enriched hypotheses for metabolite-based biomarkers. One important aspect of this type of approach is that the mechanism related to the biomarker change is explicitly described via the model, and thus represents more than only observed statistical association. This study predicted 233 high confidence biomarker changes reflecting 176 possible enzyme defects from mutations.
Another major area of use is for predicting drug targets. For example, the first generic genome-scale network model for cancer metabolism was used to predict 52 cytostatic drug targets [70]. Around 40% of these were already targeted by known, approved or experimental anti-cancer therapeutics, leaving over half that were predicted to be candidates for novel interventions. More importantly, the model also predicted synthetic lethal combinations of drugs, which are difficult to screen comprehensively in experiments.
Another highly useful aspect of such models is their ability to predict drug off-target effects when combined with structural protein analysis. This approach is two-pronged, with structural bioinformatics enabling the prediction of protein-drug off targets based on ligand binding sites, while the metabolic model enables the system-level prediction of what effects these putative interactions would have on the system and which would be predicted to affect e.g. cell viability. A kidney metabolic model predicted causal drug off-targets that were experimentally shown to impact renal function in patients with gene deficiencies that may cause observed side effects from clinical trials. The model also predicted genetic risk factors for drug treatment that corresponded to both known and unknown renal metabolic disorders.
CONCLUSION
This review highlights recent trends in network biology analysis of -omics data for to understand the mechanistic basis of disease. The power of functional network frameworks for analyzing disparate sets of -omics data is increasingly clear in the articles reviewed here. We expect network biology to play an increasingly important role in informing research studies, including clinical trails and drug development. A challenge for practitioners in the field is to provide robust tools that can be used by the non-specialist permitting rapid growth of the approaches. In addition, network-based models and related network biomarkers must be tested outside their initial discovery cohorts to provide robust clinical predictions.
Key Points.
Network biology approaches are rapidly overtaking gene, gene set, and even pathway analyses as the provide a functional framework for analyzing multiple types of -omics data.
Network approaches in the analysis of genome-wide association data can ‘rescue’ potentially interesting associations that appear insignificant due to multiple hypothesis testing corrections.
Modeling of biochemical networks is rapidly improving, providing potential connections between explicit cellular models and genome-wide data.
FUNDING
The National Center for Research Resources (Clinical and Translational Science Awards UL1-RR024989); National Institute for Drug Abuse (P20-DA26113); National Cancer Institute (P30-CA-043703); National Science Foundation (CCF-0953195 and IIS-0916102); an NCI Howard Temin Pathway to Independence Award in Cancer Research (R00-CA126184) and the Grand Duchy of Luxembourg-Institute for Systems Biology Consortium.
Acknowledgements
The authors would like to acknowledge James Eddy for making Figure 5.
Biographies
Gurkan Bebek is an Instructor in the Center for Proteomics and Bioinformatics at Case Western Reserve University and a Visiting Scientist at the Cleveland Clinic Genomics Medicine Institute. His research focuses on integrating diverse biomedical data.
Mehmet Koyutürk is Assistant Professor of Electrical Engineering and Computer Science and Proteomics and Bioinformatics at Case Western Reserve University. His research focuses on algorithm development for systems and network biology.
Nathan D. Price is an Associate Professor at the Institute for Systems Biology in Seattle, WA and is an Affiliate Associate Professor in the Departments of Bioengineering and Computer Science & Engineering at the University of Washington.
Mark R. Chance is Vice Dean for Research and Professor of Genetics and General Medical Sciences at Case Western Reserve University. He established the Center for Proteomics and Bioinformatics at the University in 2005.
References
- 1.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Gavin AC, Bosche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, Chiba T, Ozawa R, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98(8):4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartman JLt, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291(5506):1001–4. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 5.Ho Y, Gruhler A, Heilbut A, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 6.Huang LS, Sternberg PW. Genetic dissection of developmental pathways. WormBook. 2006:1–19. doi: 10.1895/wormbook.1.88.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong AH, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 8.Bebek G, Orloff M, Eng C. Microenvironmental genomic alterations reveal signaling networks for head and neck squamous cell carcinoma. J Clin Bioinforma. 2011;1(1):21. doi: 10.1186/2043-9113-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32(Suppl):502–8. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- 10.Liang P, Pardee AB. Analysing differential gene expression in cancer. Nat Rev Cancer. 2003;3(11):869–76. doi: 10.1038/nrc1214. [DOI] [PubMed] [Google Scholar]
- 11.Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 12.Iorio F, Bosotti R, Scacheri E, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci USA. 2010;107(33):14621–6. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1(1):107–29. [Google Scholar]
- 16.Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ideker T, Ozier O, Schwikowski B, et al. Discovering regulatory and signaling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl. 1):S233–40. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 18.Suthram S, Dudley JT, Chiang AP, et al. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol. 2010;6(2):e1000662. doi: 10.1371/journal.pcbi.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang HY, Lee E, Liu YT, et al. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nibbe RK, Koyutürk M, Chance MR. An integrative -omics approach to identify functional subnetworks in human colorectal cancer. PLoS Comput Biol. 2010;6(1):e1000639. doi: 10.1371/journal.pcbi.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury SA, Nibbe RK, Chance MR, et al. Subnetwork state functions define dysregulated subnetworks in cancer. J Comput Bio. 2011;18(3):263–81. doi: 10.1089/cmb.2010.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Mol Syst Biol. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkinson J, Wang X, Zheng T, et al. Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC Syst Biol. 2008;2:10. doi: 10.1186/1752-0509-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mani KM, Lefebvre C, Wang K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dao P, Colak R, Salari R, et al. Inferring cancer subnetwork markers using density-constrained biclustering. Bioinformatics. 2010;26(18):i625–31. doi: 10.1093/bioinformatics/btq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury SA, Koyuturk M. Identification of dysregulated subnetworks in complex phenotypes. Pac Symp Biocomput. 2010:133–44. doi: 10.1142/9789814295291_0016. [DOI] [PubMed] [Google Scholar]
- 27.Dao P, Wang K, Collins C, et al. Optimally discriminative subnetwork markers predict response to chemotherapy. Bioinformatics. 2011;27(13):1205–13. doi: 10.1093/bioinformatics/btr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutkowski J, Ideker T. Protein networks as logic functions in development and cancer. PLoS Comput Biol. 2011;7(9):e1002180. doi: 10.1371/journal.pcbi.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 30.De Bakker PI, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):127–3. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 31.Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26(4):445–55. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manilio TA. Genome-wide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 33.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187(2):367–83. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 36.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37(4):413–7. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 37.Ritchie MD. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann Hum Genet. 2011;75(1):172–82. doi: 10.1111/j.1469-1809.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun YV, Kardia SL. Identification of epistatic effects using a protein-protein interaction database. Hum Mol Genet. 2010;19(22):4345–52. doi: 10.1093/hmg/ddq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herold C, Steffens M, Brockschmidt FF, et al. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics. 2009;25(24):3275–81. doi: 10.1093/bioinformatics/btp596. [DOI] [PubMed] [Google Scholar]
- 40.Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23(5):561–6. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Bardes EE, Aronow BJ, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;(Web Server issue):W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Dushlaine C, Kenny E, Heron EA, et al. The SNP ratio test: pathway analysis of genome-wide association datasets. Bioinformatics. 2009;25(20):2762–3. doi: 10.1093/bioinformatics/btp448. [DOI] [PubMed] [Google Scholar]
- 43.Braun R, Buetow K. Pathways of distinction analysis: a new technique for multi-SNP analysis of GWAS data. PLoS Genet. 2011;7(6):e1002101. doi: 10.1371/journal.pgen.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26(8):1057–63. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goh KI, Cusick ME, Valle D, et al. The human disease network. PNAS. 2007;104(21):8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lage K, Karlberg OE, Størling ZM, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25(3):309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 47.Köhler S, Bauer S, Horn D, et al. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82(4):949–58. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanunu O, Magger O, Ruppin E, et al. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6(1):e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erten S, Bebek G, Ewing R, et al. DADA: degree-aware algorithms for network-based disease gene prioritization. BMC BioData Mining. 2011;4:19. doi: 10.1186/1756-0381-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erten S, Bebek G, Koyutürk M. Disease gene prioritization based on topological similarity in protein-protein interaction networks. Proceedings in 15th International Conference, Research in Computational Bolecular Biology (RECOMB'11) 2011 doi: 10.1089/cmb.2011.0154. LNCS 6577:54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia P, Zhen S, Long J, et al. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27(1):95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Patel S, Nibbe R, et al. Systems biology analyses of gene expression and genome wide association study data in obstructive sleep apnea. Pac Symp Biocomput. 2011:14–25. doi: 10.1142/9789814335058_0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price ND, Shmulevich I. Biochemical and statistical network models for systems biology. Curr Opin Biotechnol. 2007;18(4):365–70. doi: 10.1016/j.copbio.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feist AM, Herrgard MJ, Thiele I, et al. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7(2):129–43. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2(11):886–97. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 56.Thiele I, Price ND, Vo TD, et al. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. J Biol Chem. 2005;280(12):11683–95. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 57.Price ND, Schellenberger J, Palsson BO. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys J. 2004;87(4):2172–86. doi: 10.1529/biophysj.104.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brenner S. Sequences and consequences. Philos Trans R Soc Lond B Biol Sci. 2010;365(1537):207–12. doi: 10.1098/rstb.2009.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duarte NC, Becker SA, Jamshidi N, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104(6):1777–82. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma H, Sorokin A, Mazein A, et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol. 2010;6:401. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gille C, Bolling C, Hoppe A, et al. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol. 2010;6:411. doi: 10.1038/msb.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewis NE, Schramm G, Bordbar A, et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat Biotechnol. 2010;28(12):1279–85. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vo TD, Paul Lee WN, Palsson BO. Systems analysis of energy metabolism elucidates the affected respiratory chain complex in Leigh's syndrome. Mol Genet Metab. 2007;91(1):15–22. doi: 10.1016/j.ymgme.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Chandrasekaran S, Price ND. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2010;107(41):17845–50. doi: 10.1073/pnas.1005139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hyduke DR, Palsson BO. Towards genome-scale signalling-network reconstructions. Nat Rev Genet. 2010;11(4):297–307. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- 68.Shlomi T, Benyamini T, Gottlieb E, et al. Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the Warburg effect. PLoS Comput Biol. 2011;7(3):e1002018. doi: 10.1371/journal.pcbi.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shlomi T, Cabili MN, Ruppin E. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol Syst Biol. 2009;5:263. doi: 10.1038/msb.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Folger O, Jerby L, Frezza C, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:527. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]