Abstract
The inflammatory response to respiratory virus infection can be complex and refractory to standard therapy. Lactobacilli, when targeted to the respiratory epithelium, are highly effective at suppressing virus-induced inflammation and protecting against lethal disease. Specifically, wild-type mice primed via intranasal inoculation with live or heat-inactivated Lactobacillus plantarum or Lactobacillus reuteri were completely protected against lethal infection with the virulent rodent pathogen, pneumonia virus of mice (PVM); significant protection (60% survival) persisted for at least thirteen weeks. Protection was not unique to Lactobacillus species, and was also observed in response to priming with non-pathogenic gram-positive Listeria innocua. Priming with live lactobacilli resulted in diminished granulocyte recruitment, diminished expression of multiple proinflammatory cytokines (CXCL10, CXCL1, CCL2, and TNF) and reduced virus recovery, although we have demonstrated clearly that absolute virus titer does not predict clinical outcome. Lactobacillus priming also resulted in prolonged survival and protection against the lethal sequelae of PVM infection in MyD88 gene-deleted (MyD88−/−) mice, suggesting that the protective mechanisms may be Toll-like receptor-independent. Most intriguing, virus recovery and cytokine expression patterns in Lactobacillus-primed MyD88−/− mice were indistinguishable from those observed in control-primed MyD88−/− counterparts, In summary, we have identified and characterized an effective Lactobacillus-mediated innate immune shield, which may ultimately serve as critical and long-term protection against infection in the absence of specific antiviral vaccines.
Keywords: Inflammation, Cytokines, Leukocytes, Pneumovirus, Pharmabiotic
Introduction
Respiratory virus infections pose a major burden to society, both with regard to clinical illness and health care costs. Respiratory syncytial virus (RSV), a pneumovirus of the family Paramyxoviridae, is a chief cause of hospitalization for infants and young children and, likewise, results in significant morbidity and mortality in the elderly [1, 2]. In most infants and children, RSV bronchiolitis is self-limited, while in others, disease can progress to pneumonia and respiratory failure. Prematurity, multiple births, cardiovascular and pulmonary anomalies, and immunocompromised state are among the risk factors predisposing to severe RSV disease [3], although a recent prospective population-based surveillance study of acute RSV infection among children younger than 5 years of age indicated that most hospitalized children were previously healthy, with none of the documented risk factors [4]. The lack of a safe and effective vaccine for RSV and the expense and limited availability of prophylactic antibody therapy [5] represent serious hurdles in managing this significant public health problem.
In an effort to improve our understanding of the pathogenesis of severe pneumovirus infection in vivo, our laboratory has developed a model featuring the natural rodent pathogen, pneumonia virus of mice (PVM; [6, 7]). While no one animal model can replicate all the complexities of human disease, PVM has the advantage of undergoing robust replication and eliciting symptomatic disease in most inbred strains of mice [8]. With this model, we have documented molecular and cellular pathology, including granulocyte recruitment to the lung tissue and virus-induced production of proinflammatory cytokines; in doing so, we have highlighted parallels to the more severe forms of human RSV disease [9 – 11]. Using the PVM infection model, we have shown that morbidity and mortality depend largely on augmented inflammation, which can persist even when virus replication has been brought under control [12 – 14]. The importance of inflammation to the sequelae of severe respiratory virus infection has motivated our interest in immunomodulatory therapy [15] and, most recently, has led us to explore the immunotherapeutic potential of probiotic microbes and the expanding field of related microbial-derived biologics and biotherapies.
Probiotics are defined by the World Health Organization as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” [16, 17]; recently, the term “pharmabiotic” has been introduced so as to include inactivated microorganisms, biologicals that influence live organisms, and therapeutically-active metabolites [18]. Among the most familiar of this target group are microorganisms of the genus Lactobacillus, which includes over 100 species of gram-positive bacteria that form lactic acid as a product of carbohydrate metabolism. Lactobacilli are best known as minor components of the commensal microflora of the mammalian gastrointestinal (GI) tract, and they have been popularized by the food industry for the purposes of acidification, flavor enhancement, and nutrition. Multiple human studies have demonstrated the immunotherapeutic properties of probiotic lactobacilli as nutritional supplements targeted to the GI tract, including prevention of acute diarrhea, alleviation of the symptoms of allergy, and therapy for inflammatory bowel disease [19, 20]. In mouse model studies, colony forming units (cfu) of L. plantarum were administered orally prior to influenza virus challenge; however all mice eventually succumbed to the lethal sequelae of infection [21]. Likewise, a recent review of randomized controlled trials in which humans with respiratory tract infections were treated orally with lactobacilli indicates little to no effect on disease incidence, as well as conflicting data on the duration of symptoms [22].
Given that lactobacilli clearly modulate inflammation at the GI epithelium, it is conceivable that that the oral route of administration is not optimal for regulating local responses to infection with respiratory pathogens. As such, we considered the possibility that targeting live lactobacilli to the respiratory mucosa might result in a more effective immunomodulatory response, similar to the benefits observed when the intestinal mucosa is exposed directly to these organisms. To address this question, we primed the respiratory mucosae of mice with either Lactobacillus plantarum, an ecologically-flexible species first isolated from human saliva [23] or Lactobacillus reuteri, a common GI bacterium initially isolated from human feces [24], by direct intranasal inoculation with live microorganisms. We use the term “priming” to mean “providing a treatment that alters the capacity to respond to a second stimulus” (based on the definition in [25]), a phenomenon we observed upon administering lactobacilli directly to the respiratory epithelia of mice. Specifically, we found that mice primed with either of the two clinically-benign Lactobacillus species were completely protected from a subsequent lethal respiratory virus challenge. Here we explore the nature and characteristics of this response.
Materials and Methods
Mice
Wild-type BALB/c and C57BL/6 mice were purchased from Taconic Laboratories (Gaithersburg, MD). Homozygous MyD88−/− mice on a C57BL/6 genetic background were used with permission from Dr. Shizuo Akira [26]. All mouse experiments were performed in accordance with Animal Study Protocol LAD-8E, approved by the NIAID Animal Care and Use Committee.
Bacteria preparation and quantification
Lactobacillus plantarum NCIMB 8826 (ATCC BAA-793) and Lactobacillus reuteri F275 (ATCC 23272) were cultured overnight at 37°C in Difco Lactobacilli MRS Broth (BD Biosciences, Sparks, MD). Listeria innocua Seeliger (ATCC BAA-680) cultures were generated by overnight growth at 37°C in Sheep Brain Heart infusion medium (Thomas Scientific). Serial dilutions were plated and counted to correlate the number of colony-forming units per milliliter (cfu/mL) with the optical density at 600 nm (OD600). Bacterial cultures were harvested by centrifugation (5 min, 1500 rpm), washed with phosphate-buffered saline (PBS), and suspended in PBS with 1% bovine serum albumin (BSA) just prior to inoculation. Inactivated bacteria were prepared by washing in PBS and heating to 95°C for 30 min prior washing in PBS and resuspension in PBS with 1% BSA; inactivation was confirmed by overnight culture in appropriate culture broths.
Virus preparation and quantification
Virions of PVM strain J3666 passaged in vivo as described [27] were quantitated by dual standard curve qRT-PCR targeting the PVM SH gene [28] to determine both virion copies per unit volume for inoculation (PVMSH/μL) and virus recovery from infected mouse lung tissue (PVMSH/GAPDH). Specifically, RNA was prepared from mouse lung tissue that had been immersed and stored in RNAlater (Ambion, Austin, TX) and subsequently isolated with RNAzol Bee (Tel-Test, Friendswood, TX). Isolated RNA was treated with DNase I to remove genomic DNA contaminants. Reverse transcription was performed using a first-strand cDNA synthesis kit (Roche; catalog no. 1,483,188) with random primers and a no reverse transcriptase control. The quantitative PCR reactions were runin triplicate, with the ABI 2x TaqMan reagent, primer-probe mixes, and cDNA or plasmid standard in a 25-μl final volume(Applied Biosystems). Thermal cycling parameters for the ABI7500 absolute quantitation program (Applied Biosystems) include50°C for 2 min (UNG incubation), 95°C for 10 min (AmpliTaqGold activation), and 40 amplification cycles alternating 95°C for 15 s and 60°C for 1 min. Primer-probe mixes include (1) GAPDH-Vic (Applied Biosystems catalog no. 4308313) to target GAPDH cDNA and (2) PVMSH-Fam(custom design, primer 1, 5′-GCC GTC ATC AAC ACA GTG TGT-3′;primer 2, 5′-GCC TGA TGT GGC AGT GCT T-3′; probe 6FAM-CGC TGATAA TGG CCT GCA GCA-TAMRA) to target the PVM SH gene to target the virus genome. GAPDH standard curve includes serial dilutions (107, 106, 105, 104, 103 molecules/reaction) of mouse GAPDH coding sequence in pCMV Sport 6 (from ATCC cat no. 10539385). PVMSH standard curve includes serial dilutions(109, 108, 107, 106, 105 molecules per reaction) of the full-length PVM SH gene, GenBank (http://www.ncbi.nlm.nih.gov/nuccore) accession no. AY573815, in pBacPAK8. Experimental triplicate data pointsare interpolated to linear standard curves over the concentration ranges indicated. A sample calculation from data generated by this method is shown in Supplemental Figure 1.
Inoculations and sample collection
In our standard experimental protocol [Figure 1A], mice were anesthetized in 20% halothane in mineral oil and inoculated intranasally on days -14 and -7 with 109 cfu L. plantarum, L. reuteri, or control (PBS + 1% BSA) in a 50 μL volume. On day 0, Lactobacillus- or control-primed mice were inoculated with 50 μL PVM diluted 1:3000, to 2000 virion copies PVMSH per μL in Iscove’s Modified Dulbecco’s medium (IMDM). At selected time points, mice were sacrificed by cervical dislocation under anesthesia, and mice were subjected to trans-oral bronchoalveolar lavage (BAL) with PBS + 1% BSA, yielding 800 μL per mouse. Whole lung tissue was stored in RNAlater (Ambion, Austin, TX) for RNA isolation, detailed elsewhere [28] or blade-homogenized in ice-cold IMDM, followed by centrifugation at 4°C for preparation of clarified tissue homogenates. Clarified supernatants were stored in aliquots at −80°C until use.
Figure 1. Priming of the respiratory mucosa with live lactobacilli results in protection from an otherwise lethal virus infection.
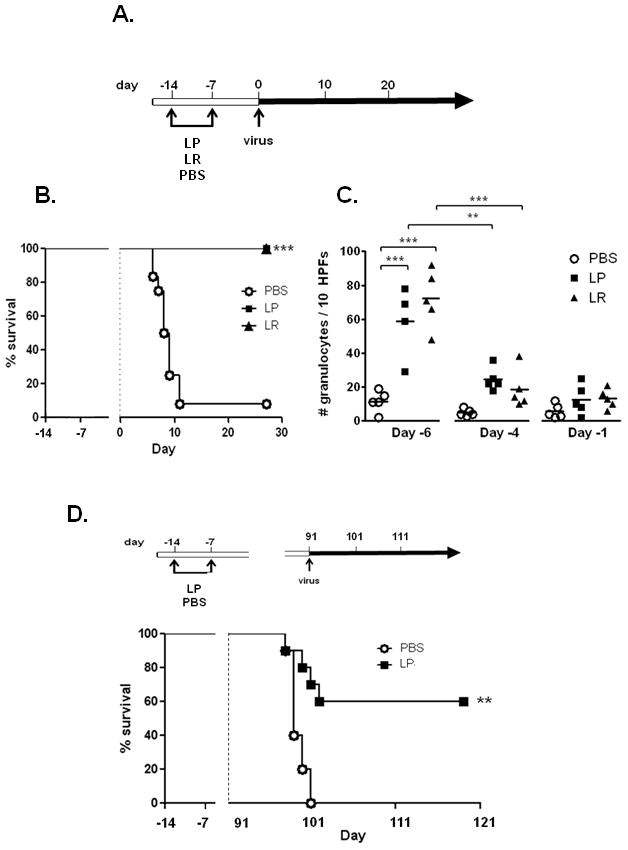
A. Standard experimental protocol. On days -14 and -7, BALB/c mice were inoculated intranasally with 109 cfu L. plantarum (LP), 109 cfu L. reuteri (LR), or PBS + 1% BSA vehicle control (PBS). On day 0, all mice were inoculated with pneumonia virus of mice (PVM; 2000 virion copies/μL). B. Survival of mice inoculated as indicated with LP, LR or PBS prior to virus infection, n = 10 mice per group, representative of at least two independent experiments; ***p < 0.001 C. Granulocytes detected in bronchoalveolar lavage (BAL) fluid at time points indicated (day -6, day -4, day -1 as per Fig. 1A; n = 5 – 7 mice per time point). Horizontal bars indicate mean values; statistical significance **p < 0.01; ***p < 0.001. D. Survival of mice primed with LP or PBS days -14 and -7 and challenged with PVM 13 weeks (91 days) later; n = 10 mice per group, **p < 0.01.
Histopathology
On day 7 of our treatment protocol, lungs of sacrificed mice were inflated trans-tracheally using 250 μL 10% phosphate-buffered formalin. The lungs and heart were removed and fixed overnight in 10% phosphate-buffered formalin at 4°C. Samples were paraffin-embedded, sectioned, and stained with hematoxylin and eosin (Histoserv, Inc., Germantown, MD).
Single cell suspensions
Single cell suspensions were generated from mouse lung tissue as described [29]. Lungs were perfused with 5 – 10 mL PBS + 10 mM EDTA and placed in digestion medium, consisting of Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen) supplemented with 5% fetal bovine serum (BioWhittaker, Walkersville, MD), 20 μg/mL grade II DNase I (Roche Diagnostics, Mannheim, Germany), and 40 μg/mL Collagenase D (Roche). The tissue was cut into ~3 mm3 pieces and incubated in digestion medium (37°C, 90 min, with rocking). Upon adding cold EDTA (10 mM) to the cell suspension, the cells were passed through a 40-μm Nylon cell strainer (BD Biosciences, Durham, NC) and washed with PBS supplemented with 0.3% BSA, 5 mM EDTA, and 20 μg/mL DNase I. Cells were centrifuged (5 min, 1500 rpm), red blood cells eliminated with 1 mL ACK Lysing Buffer (BioWhittaker) for 2 minutes as needed, and remaining cells washed again. All cell pellets from a given condition were pooled and resuspended in HBSS at a density of 106 cells/mL. Cells were treated as described below, fixed with PBS + 4% paraformaldehyde, washed, and stored at −80°C in PBS + 10% DMSO.
Flow cytometry
Prior to fixation, lung cells (~106 cells per condition) were blocked with anti-mouse CD16/CD32 mAb (BD Biosciences) and, as appropriate, stained with LIVE/DEAD violet-fluorescent reactive dye (Invitrogen, Eugene, OR) and/or DX-5 PE (BD Pharmingen; applied prior to fixation) in PBS + 1% BSA. Antibody-fluorochrome conjugates included B220 Alexa-Fluor 488, CD3 PE-Cy5, CD4 PerCP-Cy5.5, CD8 APC-Cy7, and GR1 APC (BD Pharmingen). Cells were stained for 30 minutes at 4°C, washed with PBS + 1% BSA, and analyzed using a LSR II flow cytometer in conjunction with FACSDiva software (BD Biosciences), with a minimum of 100,000 events collected per sample.
Cytokine expression
RNA from mouse lung tissue was isolated, pooled (12 μg total RNA per condition), and purified using the RT2 qPCR-grade RNA Isolation Kit (SA Biosciences, Frederick, MD). First-strand cDNA was generated using the RT2 First-strand Kit (SA Biosciences) and, in conjunction with the RT2 SYBR Green/ROX qPCR Master Mix (SA Biosciences), analyzed for differential gene expression using the RT2 Profiler Mouse Inflammatory Cytokines and Receptors PCR Array (SA Biosciences). ELISA analysis was performed to quantify immunoreactive proteins using Quantikine™ kits from R&D Systems (Minneapolis, MN) as per manufacturer’s instructions. Datapoints generated using lung tissue homogenates were normalized to total protein per sample, which was determined via BCA protein assay (Pierce, Rockford, IL) with BSA standards.
Microarray data
Our microarray dataset (M430-2 chip) documents the normalized expression profiles of 39,000 transcripts expressed in mouse lung response to PVM infection over time [30]. Profiles of specific chemokine transcripts were determined via algorithms within Genespring GX7.3 (Agilent Technologies).
Detection of granulocytes
Granulocytes were evaluated in BAL fluid at days indicated using modified Giemsa staining (Diff-Quik, Fisher Scientific, Pittsburgh, PA). To prepare cells for staining, BAL fluids were subjected to centrifugation, and the cell pellets were resuspended in PBS + 1% BSA. 105 cells were centrifuged onto slides using a Shandon Cytospin apparatus (Thermo Electron, Pittsburgh, PA). Following staining and mounting of cells, ten high-power fields were visually inspected by light microscopy.
Seroconversion
Mouse sera were isolated from tail bleeds via standard procedures and assessed for seroconversion (immunoreactivity to PVM antigens) using the SMART-M12 PVM Kit (Biotech Trading Partners, Encinitas, CA), per manufacturer’s guidelines.
Lactobacillus clearance
Mice were inoculated with 109 cfu L. plantarum or L. reuteri and sacrificed at 1, 2, 3 and 7 days thereafter; lungs were blade-homogenized in 1 mL sterile PBS + 1% BSA. Homogenates (100 μL) were plated at 1:1, 1:20, or 1:100 on MRS agar and incubated overnight at 37°C. Homogenates from PBS + BSA-treated mice were included as controls.
Statistical analysis
Data were evaluated using Welch’s t test, Mann-Whitney U test, ANOVA with Bonferroni’s multiple comparison test, or the log-rank test for survival curves, as appropriate. All statistical tests were included in the GraphPad Prism 5 software package (La Jolla, CA). Significant outliers were identified using the Grubbs test. All bar graphs indicate the mean ± the standard error of the mean (SEM).
Results
Priming of the respiratory mucosa with lactobacilli results in protection from lethal pneumovirus infection
The first question motivating our study was whether direct stimulation or priming of the respiratory mucosa with live lactobacilli would alter the outcome of a lethal pneumovirus infection. To explore this question, we devised a standard protocol whereby mice were primed with two intranasal inocula (on days -14 and -7, respectively), each containing 109 colony-forming units (cfu) of live L. plantarum or L. reuteri; these inocula were followed by intranasal challenge with pneumonia virus of mice (PVM) on day 0 [Fig. 1A]. We found that BALB/c mice primed in this fashion were fully protected from an otherwise lethal respiratory virus infection [Fig. 1B]. BALB/c mice responded to Lactobacillus priming with transient granulocyte recruitment [Fig. 1C]. Lactobacillus priming alone resulted in no acute mortality, and we observed complete clearance of live lactobacilli from the lung tissue within 7 days after inoculation (data not shown). Direct Lactobacillus priming of the respiratory mucosa resulted in protection from lethal sequelae of virus infection even when challenge was delayed from day 0 until day 7 or day 21 [Table 1] and we observed prolonged survival and significant long-term protection even when virus challenge was delayed until 91 days (13 weeks) after initial priming [Fig. 1D]. Priming with heat-inactivated L. plantarum (109 cfu equivalents) also generated protection against lethal virus challenge [Fig. 2A], as did priming with live or heat-inactivated gram-positive Listeria innocua [L. innocua; Fig. 2B], indicating that these findings are not unique to Lactobacillus species. Of note, neither live nor heat-inactivated L. plantarum were effective when administered therapeutically, three days after virus inoculation, rather than as a priming agent [Fig. 2C].
Table 1.
Survival of L. plantarum (LP)-primed BALB/c mice inoculated with PVM on day 0 as per standard protocol [Fig. 1A] or delayed seven, twenty-one or ninety-one days thereafter. Survival was determined at t = 30 days after each virus inoculation. All surviving mice (29 of a total 34) underwent seroconversion to PVM antigens (SMART-M12, see Methods).
| Virus inoculation on | day 0 | day 7 | day 21 | day 91 |
|---|---|---|---|---|
| Survival | 8/8 (100%) | 7/8a (88%) | 8/8 (100%) | 6/10 (60%) |
The single death among the mice inoculated on day 7 occurred 8 days after PVM inoculation. The full survival analysis of the mice inoculated 91 days after Lactobacillus priming is shown in Figure 1D.
Figure 2. Priming of the respiratory mucosa with heat-inactivated lactobacilli and other gram-positive bacteria.
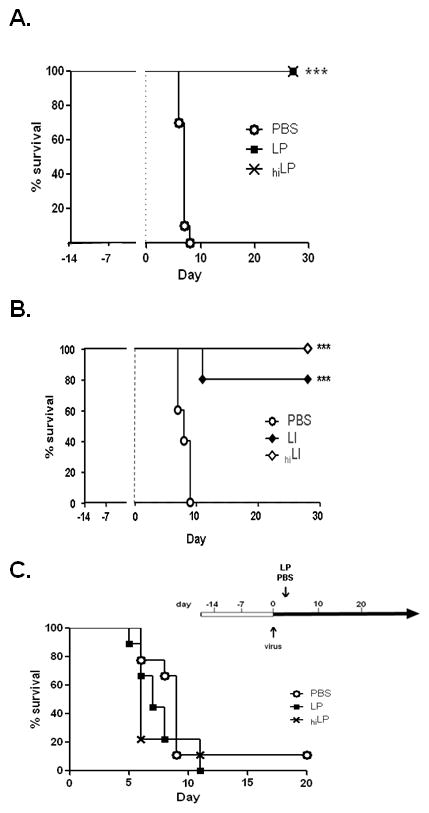
Standard experimental protocol is as per Fig. 1A unless otherwise noted. A. Survival of mice inoculated with LP, heat-inactivated (hi) LP (109 cfu-equivalents) or PBS prior to virus infection; B. Survival of mice inoculated with 109 cfu Listeria innocua (LI), 109 cfu-equivalents heat-inactivated L. innocua (LI), or PBS prior to virus infection; C. Survival of naïve BALB/c mice inoculated with PVM on day 0, then inoculated with lactobacilli (109 cfu LP, 109 cfu-equivalents heat-inactivated (hi) LP, or PBS + BSA) on day 3. Each group includes 10 or more mice per condition, representative of at least two independent experiments ***p < 0.001.
Virus recovery from lungs of Lactobacillus- and PBS-primed mice
Virus recovery from whole lung tissue was determined on days 3, 5 and 7 after PVM challenge [Fig. 3A]. Virus replication was detected in Lactobacillus-primed as well as PBS-primed mice, and seroconversion to PVM antigens was detected in all mice surviving infection (data not shown). Significantly fewer virion copies were detected in lungs of L. plantarum-primed compared to PBS-primed mice on all days evaluated (4 – 30-fold). Virus recovery from the lungs of L. reuteri-primed mice was also somewhat diminished compared to PBS-primed mice on day 3 (5-fold) and on day 7 (4-fold), but there was no significant difference between virus recoveries at peak. Despite this latter finding, all L. reuteri-primed mice survived virus infection (100%), whereas 95% of the PBS-primed mice did not [Fig. 1B]. These results suggest that Lactobacillus-mediated alteration of virus kinetics (replication and clearance) may not provide a sufficient explanation for differential survival. In order to evaluate the relationship between virus recovery and survival directly, ten L. plantarum (LP) and ten PBS (PBS + 1% BSA) primed mice were inoculated on day 0 with PVM (standard protocol, Fig. 1A), with the virus inoculum received by the control (PBS-primed) mice reduced to 600/μL, so that peak virus recoveries (day 5) would be comparable. Five mice were selected randomly from each group for determination of virus recovery. As anticipated, differences in peak virus recoveries were insignificant [Fig. 3B], yet none of the remaining PBS-primed, virus-infected mice survived beyond day 9, while all of the remaining LP-primed, virus-infected mice survived long-term [Fig. 3C, **p < 0.01]. From these findings we conclude that, while Lactobacillus priming modulates virus replication and clearance, virus recovery alone cannot predict disease outcome or long-term survival.
Figure 3. Virus recovery from lung tissue of Lactobacillus- and control-primed mice.
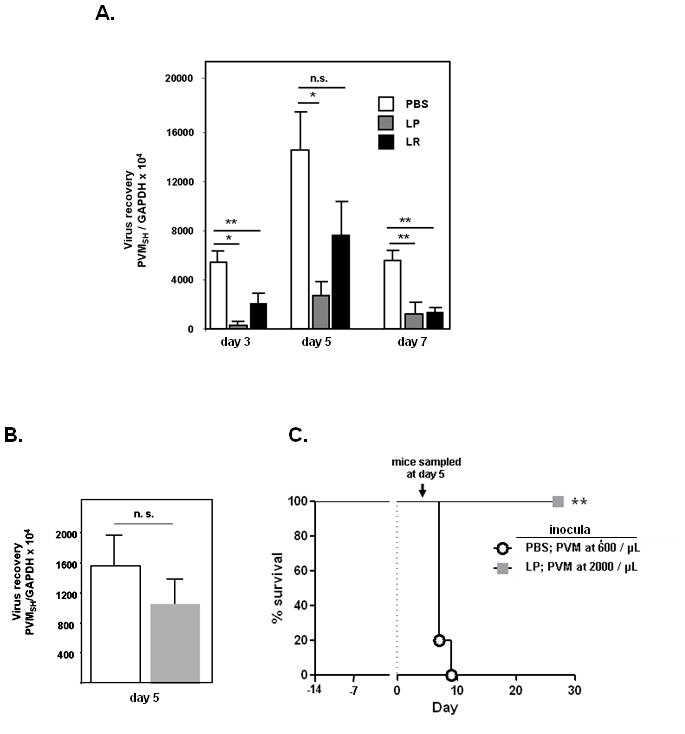
A. Virus recovery from lung tissue at days 3, 5 and 7 after inoculation with PVM from mice treated with either Lactobacillus spp. or PBS + BSA control, as determined by qRT-PCR with two standard curves [Suppl. Fig. 1]; n = 5 – 7 mice per timepoint per condition, *p < 0.05; **p < 0.01; n.s., not significant. B. and C. Survival of and virus recovery from Lactobacillus (LP) and PBS + BSA-primed mice. The virus (PVM) inoculum received by ten PBS + BSA -primed mice was reduced to 600 copies/μL so that peak recovery at day 5 would be comparable to that detected in the ten LP-primed mice. Virus recovery was evaluated in 5 mice selected randomly on day 5 from each group as shown, and % survival of 5 mice remaining is as shown; **p < 0.01.
Lung histopathology and differential leukocyte recruitment
In alignment with our previous studies [12, 13], lungs of control-primed, PVM-infected mice revealed a profound alveolitis, with widespread, diffuse granulocyte recruitment and early-onset edema [Figs. 4A–4C]. In contrast, the lungs of L. plantarum-primed, PVM-infected mice exhibited minimal inflammation peripherally and feature compact, peribronchiolar and perivascular cuff-like infiltrates, consistent with descriptions of induced bronchus-associated lymphoid tissue (iBALT; [Figs. 4D – 4F] [31, 32]).
Figure 4. Histopathologic analysis.
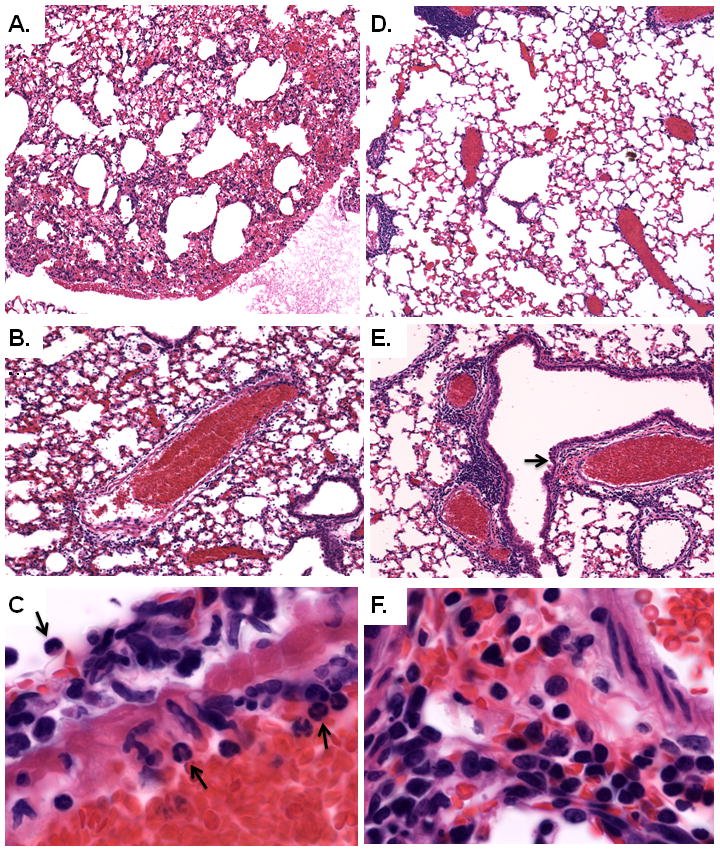
Hematoxylin and eosin-(H&E) stained lung tissue from mice primed with PBS + BSA (A., B., and C.) or L. plantarum in PBS + BSA (D., E., and F.) prior to virus infection; shown here at day 7 as per Fig. 1A. A. and B. Diffuse alveolar, bronchiolar, and perivascular inflammation is observed in lung tissue from PVM-infected, control-primed mice (original magnifications, 5X, and 10X, respectively). C. Arrows indicate infiltrating granulocytes (original magnification, 40X) seen exiting from the blood vessel in the center of the field in panel B. D. and E. Lung tissue from L. plantarum-primed, PVM-infected mice exhibits diminished alveolar inflammation and pronounced peribronchiolar and perivascular cuffing (original magnifications, 5X and 10X, respectively). F. Enlarged from E. (at arrow) dense lymphocyte-enriched inflammatory infiltrate, consistent with descriptions of induced bronchus-associated lymphoid tissue (iBALT; original magnification, 40X.)
Consistent with these histopathologic findings, priming with L. plantarum prior to PVM infection resulted in a 5-fold reduction in the fraction of GR1+ granulocytes [Fig. 5A and 5B] and a concomitant 2-fold increase in the fraction of lymphocytes in whole-lung single-cell suspensions compared to PBS-primed, PVM-infected controls [Fig. 5B]. Additional analysis of the lymphocyte populations revealed that Lactobacillus priming prior to PVM infection had no impact on the relative proportions of CD4+ T cells (CD3+CD4+CD8−), CD8+ T cells (CD3+CD4−CD8+), or B cells (B220+), while the fraction of NK cells (CD3−DX5+) was diminished [Fig. 5C].
Figure 5. Flow cytometric analysis of leukocyte subsets.
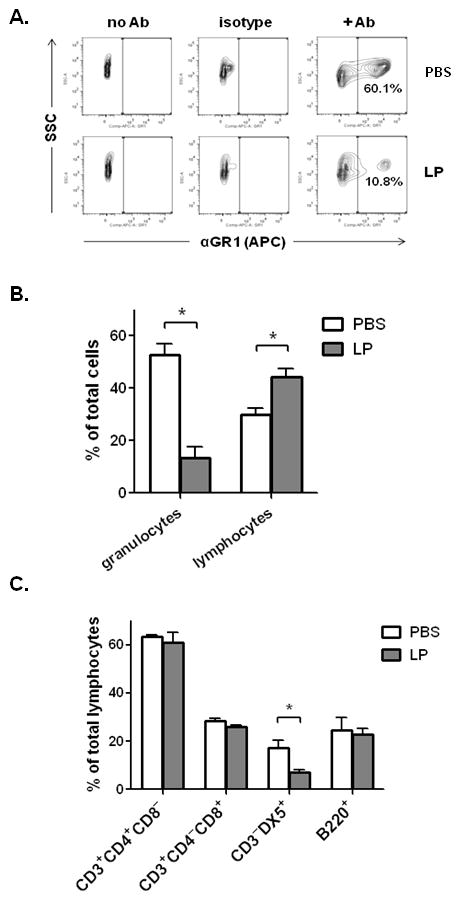
A. Flow cytometric analysis of whole-lung single-cell suspensions generated from control-(PBS + BSA) or L. plantarum (LP)-primed, PVM-infected mice (day 7, n = 5 – 6 mice per condition). Total lung cells were gated for side-scatter (SSC) and expression of the cell surface granulocyte marker GR1, shown with no-antibody and isotype-matched antibody controls. Data shown are representative of four independent experiments. B. Percentage of total viable lung cells identified as granulocytes (GR1+) or lymphocytes (identified by characteristic forward/side scatter) from PBS + BSA-primed, PVM-infected or L. plantarum-primed, PVM-infected mice. C. Percentage of total viable lymphocytes (in B.), with CD4+ T cell (CD3+CD4+CD8−), CD8+ T cell (CD3+CD4−CD8+) NK cell (CD3−DX5+) or B lymphocyte (B220+) immunophenotype; * = p < 0.05; ** = p < 0.01. Data shown are compiled from four independent experiments.
Suppression of virus-induced proinflammatory cytokines
Morbidity and mortality in response to severe respiratory virus infection can result in large part from uncontrolled amplification of proinflammatory signaling networks [33]. We observed that Lactobacillus priming of the respiratory mucosa resulted in suppression of multiple cytokines associated with the inflammatory pathology of respiratory virus infection. Specifically, priming with lactobacilli resulted in marked suppression of interferon-inducible protein (CXCL10/IP-10), monocyte-chemotactic protein 1 (CCL2/MCP-1), neutrophil-activating protein-3 (CXCL1/NAP-3), macrophage inflammatory protein-1γ (CCL9/MIP-1γ), tumor necrosis factor (TNF) and eotaxin-2 (CCL24) [Fig. 6A]. CCR1 is a prominent CC chemokine receptor detected on granulocytes [34]; differential detection of this transcript is consistent with the flow cytometric results shown in Figs. 4A and 4B. Of note, Lactobacillus-mediated suppression of CXCL10 and CCL2 was evident as early as day 4 after virus inoculation [Suppl. Fig. 2]. Cytokine mediators determined as not differentially expressed in response to Lactobacillus-priming included CCL3, CCL5, CXCL12, and TGF-β1. CXCL10, CCL2, and CXCL1 are expressed prominently in mouse lung tissue in response to PVM infection. These proinflammatory mediators are among those with transcriptional profiles that parallel respiratory dysfunction as detected in our PVM gene expression microarray database [30], which documents the full spectrum of transcriptional responses to the PVM pathogen in otherwise unmanipulated wild-type mice [Suppl. Fig. 3]. Immunoreactive CXCL10, CCL2, CXCL1, and TNF were detected in BAL fluid of PBS-primed, PVM-infected mice; expression was suppressed in mice primed with lactobacilli, most notably expression of CCL2 (~20-fold suppression) [Fig. 6B]. Interestingly, we detected expression of both interferon-β (IFN-β) and interleukin-10 (IL-10) in response to PVM infection, but neither of these cytokines, implicated in Lactobacillus-mediated immunomodulation in a variety of other settings [35 – 38], were detected as differentially expressed in response to Lactobacillus-priming in BALB/c mice [Fig. 6C].
Figure 6. Lactobacillus-mediated priming of the respiratory mucosa leads to suppression of multiple virus-induced proinflammatory cytokines.
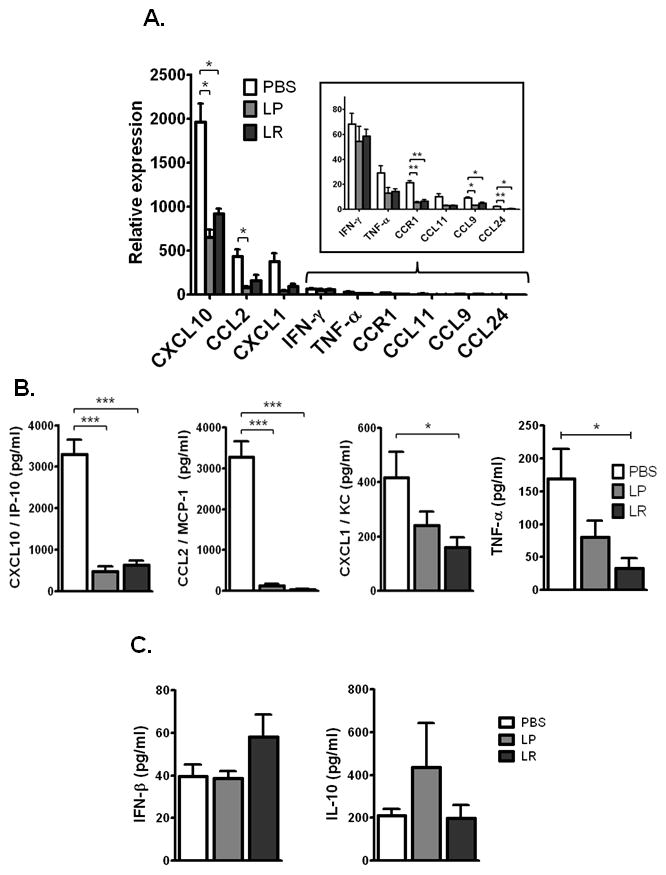
A. Expression of transcripts encoding proinflammatory mediators in lungs of Lactobacillus- or PBS-primed, PVM-infected mice, normalized to expression in Lactobacillus- or PBS-primed, uninfected mice, respectively (day 7 as per Fig. 1A, n = 4 mice per experimental group; data compiled from three independent experiments). B. Immunoreactive CXCL10, CCL2, CXCL1, and TNF detected in BAL fluid of Lactobacillus- or PBS-primed, PVM-infected mice; C. Immunoreactive IFN-β and IL-10 detected in BAL fluid of Lactobacillus- or PBS-primed, PVM-infected mice; n = 4 – 5 mice per experiment; data compiled from three independent experiments; * p < 0.05; ** p < 0.01; ***p < 0.001; LP, L. plantarum; LR, L. reuteri.
MyD88-dependent signaling, virus recovery and differential survival
Previous studies published by several groups have identified Lactobacillus-mediated immunomodulatory mechanisms that are dependent on Toll-like receptors (TLRs), primarily TLR2 and TLR4 [39]. As an initial exploration of the TLR dependence of Lactobacillus-mediated protection against virus infection, we carried out our inoculation protocol with mice devoid of MyD88 (MyD88−/−), the (near) universal TLR adapter protein. These mice were available to us on the C57BL/6 background. Mice of this background are less susceptible to the lethal sequelae of PVM infection than are BALB/c mice at the same virus inoculum [40]. Lactobacillus-mediated priming was effective at preventing lethal sequelae of PVM infection in the C57BL/6 background [Fig. 7A]. Although some mortality was observed among the L. plantarum-primed MyD88−/− mice at this virus inoculum, priming of the respiratory mucosa resulted in prolonged survival and significant long-term protection against the lethal sequelae of virus infection for a substantial fraction of mice [Fig. 7B]. Analysis of the differential survival [Suppl. Fig. 4] documents that TLR-MyD88 signaling did not contribute substantially to Lactobacillus-mediated protection. Similar to what was observed in experiments performed with control- and L. plantarum-primed wild-type BALB/c mice [Fig. 3A], Lactobacillus priming of C57BL/6 mice resulted in diminished virus recovery from lung tissue [~5-fold; Fig. 7C]. Virus recovery from lungs of control-primed MyD88−/− mice was not significantly different from that measured in the control-primed wild-type C57BL/6 mice. Likewise, virus recovery from lungs of L. plantarum-primed MyD88−/− mice was indistinguishable from that of the control-primed MyD88−/− mice, despite clear evidence of differential survival within this genotype [Fig. 7B]. These results are consistent with those shown in Fig. 3, and stand in support of our earlier conclusion, that although Lactobacillus-priming has a significant impact on virus recovery, virus titer in lung tissue does not serve as a predictor of disease outcome or long-term survival.
Figure 7. Lactobacillus-primed MyD88 gene-deleted (MyD88−/−) mice are also protected against the lethal sequelae of PVM infection.
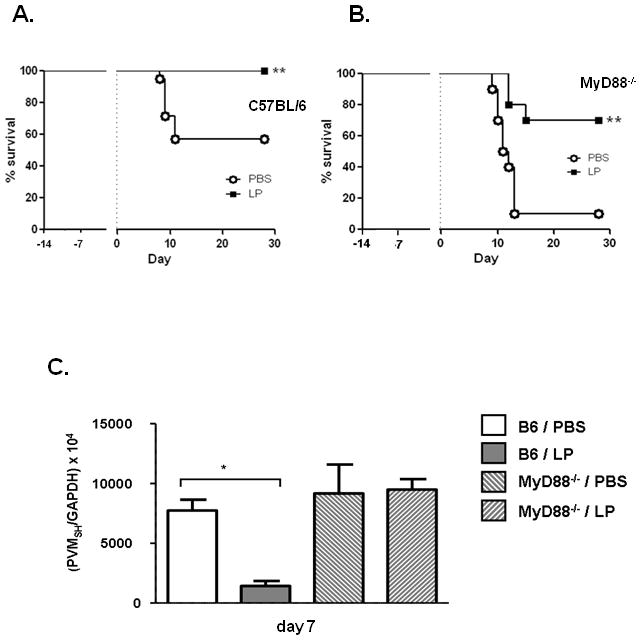
A. Survival of C57BL/6 mice primed with 109 cfu L. plantarum or PBS + BSA prior to PVM infection; n = 20 mice per group. B. Survival of MyD88−/− mice primed with 109 cfu L. plantarum or PBS + 1% BSA control prior to PVM infection; n = 8 – 10 mice per group. Data shown are representative of three independent experiments, ** = p < 0.01; LP = L. plantarum. C. Virus recovery from lung tissue determined by qRT-PCR of L. plantarum-primed and PBS-primed C57BL/6 and MyD88−/− mice on day 7 after PVM infection (as per Fig. 1A); n = 5 – 8 mice per timepoint per condition, *p < 0.01; data compiled from two independent experiments.
MyD88-dependent signaling, virus infection and proinflammatory cytokines
Initial identification of specific proinflammatory mediators that were differentially expressed in Lactobacillus-primed, PVM-infected vs. control-primed, PVM-infected lung tissue from BALB/c mice was determined by PCR array of 84 potential transcripts and confirmed by ELISA [Fig 6, see Methods]. As shown in Fig. 8, differential cytokine expression in L. plantarum-primed vs. control-primed, PVM-infected wild-type C57BL/6 mice is comparable to what was determined in wild-type BALB/c mice. Specifically, we observed profound suppression of CCL2 and CXCL10 [Fig. 8A and 8B], and moderate suppression of CXCL1 and TNF [Fig. 8C and 8D]. Of note, significant suppression of interleukin-10 was observed in L. plantarum-primed, PVM-infected C57BL/6 mice [Fig. 8E]; this was not observed in BALB/c mice. All of these cytokines were detected in lung tissue of PVM-infected MyD88−/− mice, albeit at significantly reduced concentrations as compared to PVM-infected C57BL/6 wild-type mice (save for TNF, which is detected at levels that are not statistically different from control-primed, PVM-infected wild-type C57BL/6 mice). Interestingly, Lactobacillus-mediated cytokine suppression was not detected in MyD88−/− mice. Thus, differential cytokine expression is not a crucial feature of Lactobacillus-mediated protection in the absence of TLR-MyD88 receptor signaling.
Figure 8. Lactobacillus-priming of MyD88−/− mice has no impact on virus-mediated production of inflammatory cytokines.
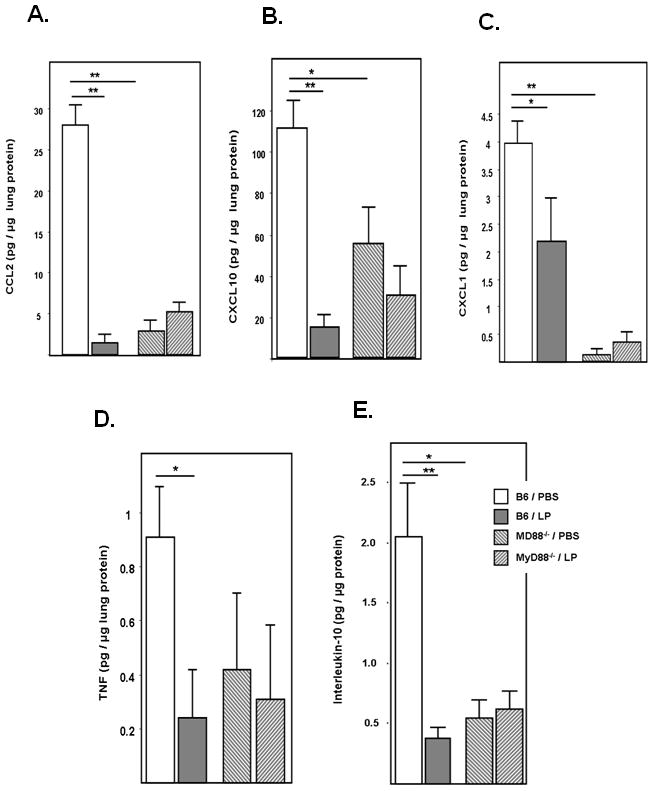
Immunoreactive A. CCL2, B. CXCL1, C. CXCL10, D. IL-10, and E. TNF detected in lung tissue homogenates from C57BL/6 and MyD88−/− mice at day 7 after virus infection; n = 5 – 8 mice per time point per condition, *p < 0.05; **p < 0.01; data compiled from two independent experiments.
Discussion
In this study we demonstrate that priming of the respiratory mucosa with non-pathogenic gram-positive microorganisms results in full protection from the otherwise lethal sequelae of severe pneumovirus infection. Protection is observed in response to both live and heat-killed Lactobacillus strains (L. plantarum and L. reuteri) and live and heat-killed gram-positive Listeria innocua, which, although not pathogenic, had not been considered previously as a potential probiotic or pharmabiotic microorganism. Administration of live lactobacilli directly to the respiratory mucosa resulted in diminished virus recovery at multiple time points and prominent suppression of virus-induced proinflammatory mediators.
Although virus recovery is diminished in response to Lactobacillus priming, similar to results obtained by several other groups in experiments with influenza virus [41 – 43], here we build on these findings, as we examine the unique responses to priming and the specific relationship to virus recovery and cytokine production. First, we have determined clearly that virus recovery does not correlate with disease outcome. Specifically, virus inocula were altered so that peak virus recoveries in Lactobacillus-primed and control-primed wild-type mice were comparable to one another. Nevertheless, Lactobacillus-primed mice proceeded to survive long-term, and the control-primed mice succumbed to the lethal sequelae of virus infection. These findings are consistent with our earlier studies on the pathogenesis of PVM infection in wild-type mice, in which we determined that virus recovery alone cannot predict outcome of disease, and that the virus-induced inflammatory response can lead to lethal sequelae even after initial replication is blunted with effective antiviral therapy [12, 13]. Given our current understanding of the immunomodulatory capacity of lactobacilli at the GI mucosa, including modulation of cytokine production by target epithelial cells [16 – 17, 19, 44], our primary hypothesis was that the major protective role of lactobacilli at the respiratory mucosa was directly related to their ability to suppress local virus-induced inflammatory responses.
Upon further analysis, we found the relationship between virus-induced inflammation and Lactobacillus-mediated protection to be more complex than previously anticipated. We observed that mice devoid of the gene encoding the MyD88-TLR signaling adaptor were also protected from the lethal sequelae of PVM infection after priming with L. plantarum. Clearly, this does not rule out potential roles played by TLR-mediated signaling pathways that circumvent MyD88 or other pattern-recognition receptors that may play a role in sensing Lactobacillus species [45, 46]; TLR-desensitization has been observed as a mechanism that explains reduced inflammatory responses observed after recuperation from influenza virus [47]. The data on TLR signaling in response to lactobacilli is complex and, to date, no consensus findings or aligning principles have emerged. For example, Hisbergues and colleagues [48] reported that mouse bone marrow-derived dendritic cells produce both IL-10 and IL-12 in response to L. plantarum via TLR-2/MyD88-dependent and TLR4/MyD88-independent pathways. At the same time, Ichikawa and colleagues [49] reported that cells from mouse spleen and mesenteric lymph node produce IL-12 in response to L. paracasei via pathways that are MyD88-dependent, but completely independent of TLR2, TLR4 or TLR9. In contrast, Chung and colleagues [50] found that L. casei-mediated protection in the mouse model of dextran-sulfate sodium-induced colitis was primarily dependent on signaling via TLR4. At this point in time, it is not clear whether responses are defined by an individual Lactobacillus species or strain, unique to a given organ or tissue, or whether they are specified by mouse genotype or functional overlap of specific signaling pathways.
Of note, this is the also the first documented evaluation of PVM infection in vivo in MyD88 gene-deleted mice. Virus recovery from MyD88−/− mouse lung tissue was similar to that reported by Bhoj and colleagues [51] in their study of RSV clearance. However, in contrast to these findings, we observed accelerated replication of PVM in MyD88−/− eosinophilic granulocytes from bone marrow cultures [52]. The role of individual TLRs in mediating responses to pneumovirus infection remains incompletely explored [reviewed in 53]; TLR 2/6, TLR4, and TLR7 have all been implicated in augmenting immune responses in the RSV challenge mouse model, while closely-related mouse pathogens Sendai virus (parainfluenza virus 1) and PVM do not engage TLR4 [54, 55].
Similar to findings described for wild-type mice, Lactobacillus-priming promoted survival in PVM-infected MyD88−/− mice, but in contrast, promoted no differential expression of proinflammatory mediators, CCL2, CXCL1, CXCL10, TNF. These cytokines are all mediators that are prominently suppressed in association with Lactobacillus-mediated protection in wild-type mice. Although there are no unique roles attributed to any of these mediators in the pathogenesis of pneumovirus disease, collectively they have been implicated in promoting disease severity in a variety of settings. For example, the chemokine CCL2 has been detected in BAL fluid from RSV-infected infants [9, 56], in RSV-challenged mice [57], and levels of this mediator correlate with respiratory dysfunction in the PVM infection model [30]. Similarly, although CXCL10 has not received as much attention vis-à-vis the pathogenesis of pneumovirus-induced bronchiolitis [58], McNamara and colleagues [9] examined a series of BAL samples from infants intubated secondary to RSV bronchiolitis and detected CXCL10 as one of the two predominant chemokines. TNF has likewise been associated with severe RSV disease in several human studies [59, 60]. Overall, our findings introduce an interesting and important conundrum, as it is clear that Lactobacillus-mediated protection is associated with diminished levels of multiple proinflammatory cytokines, but, given the fact that protection can be elicited in MyD88−/− mice without evidence for this suppression, it is clear that this may be only one aspect of the overall protective mechanism. Among the possibilities, Lactobacillus priming may elicit multiple parallel, potentially degenerate responses that may compensate for one another (e.g. MyD88-independent pathways eliciting protection in equal force, duplicating the efforts of the derailed MyD88-dependent pathways). Given the variety and complexity of potential immunostimulants presented by unfractionated gram-positive organisms (i.e. peptidoglycan, lipoteichoic acid, membrane proteins), this is a reasonable possibility that will require careful mechanistic dissection.
In summary, we have observed that priming of the respiratory mucosa with clinically-benign gram-positive Lactobacillus species results in markedly diminished inflammatory responses upon challenge with a pneumovirus pathogen, and Lactobacillus-primed hosts are fully protected from the otherwise lethal sequelae of a severe respiratory virus infection. We demonstrate that virus recovery from lung tissue is not a predictor of disease outcome. Our goal is to characterize the protective responses at the molecular level and at the same time to determine a means by which lactobacilli might be used as a broad-spectrum respiratory mucosal immunomodulatory agent. These lactobacilli, or components thereof, may serve as an effective innate immune shield to provide critical protection against pneumovirus, and perhaps other severe respiratory virus infections, prior to the development of safe, specific, and reliable vaccines.
Supplementary Material
Acknowledgments
The authors thank Dr. S. Akira (Osaka University, Japan) for his gracious gift of MyD88 gene-deleted mice for use in these experiments, Dr. Catherine Ptaschinski (University of Newcastle, Australia) for her protocol for generating single-cell suspensions, Dr. Alfonso Gozalo and the staff of NIAID 14BS for care of the mice used in these studies, Dr. Kirk Druey for careful review of the manuscript, and Mr. Ricardo Dreyfuss of Medical Arts Branch, National Institutes of Health, for preparation of the photomicroscopic images.
Footnotes
Work presented in this manuscript is supported by funds from the NIAID Division of Intramural Research Project Z01-AI000943 (HFR) and Children’s Miracle Network of Central New York (JBD).
References
- 1.Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falsey AR, Walsh EE. Respiratory syncytial virus infection in elderly adults. Drugs Aging. 2005;22:577–587. doi: 10.2165/00002512-200522070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welliver RC. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. J Pediatr. 2003;143(5 Suppl):S112–S117. doi: 10.1067/s0022-3476(03)00508-0. [DOI] [PubMed] [Google Scholar]
- 4.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009;360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olszewska W, Openshaw P. Emerging drugs for respiratory syncytial virus infection. Expert Opin Emerg Drugs. 2009;14:207–217. doi: 10.1517/14728210902946399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg HF, Domachowske JB. Pneumonia virus of mice: severe respiratory infection in a natural host. Immunol Lett. 2008;118:6–12. doi: 10.1016/j.imlet.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easton AJ, Domachowske JB, Rosenberg HF. Pneumonia virus of mice. In: Cane P, editor. Perspectives in Medical Virology. Vol. 12. 2006. pp. 299–319. [Google Scholar]
- 8.Anh DB, Faisca P, Desmecht DJ. Differential resistance/susceptibility patterns to pneumovirus infection among inbred mouse strains. Am J Physiol Lung Cell Mol Physiol. 2006;291:L426–L435. doi: 10.1152/ajplung.00483.2005. [DOI] [PubMed] [Google Scholar]
- 9.McNamara PS, Flanagan BF, Hart CA, Smyth RL. Production of chemokines in the lungs of infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis. 2005;191:1225–1232. doi: 10.1086/428855. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod Pathol. 2007;20:108–119. doi: 10.1038/modpathol.3800725. [DOI] [PubMed] [Google Scholar]
- 11.Bataki EL, Evans GS, Everard ML. Respiratory syncytial virus and neutrophil activation. Clin Exp Immunol. 2005;140:470–477. doi: 10.1111/j.1365-2249.2005.02780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonville CA, Easton AJ, Rosenberg HF, Domachowske JB. Altered pathogenesis of severe pneumovirus infection in response to combined antiviral and specific immunomodulatory agents. J Virol. 2003;77:1237–1244. doi: 10.1128/JVI.77.2.1237-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonville CA, Lau VK, DeLeon JM, Gao JL, Easton AJ, Rosenberg HF, Domachowske JB. Functional antagonism of chemokine receptor CCR1 reduces mortality in acute pneumovirus infection in vivo. J Virol. 2004;78:7984–7989. doi: 10.1128/JVI.78.15.7984-7989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventre K, Randolph A. Ribavirin for respiratory syncytial virus infection of the lower respiratory tract in infants and young children. Cochrane Database Syst Rev. 2004:CD000181. doi: 10.1002/14651858.CD000181.pub2. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg HF, Bonville CA, Easton AJ, Domachowske JB. The pneumonia virus of mice infection model for severe respiratory syncytial virus infection: identifying novel targets for therapeutic intervention. Pharmacol Ther. 2005;105:1–6. doi: 10.1016/j.pharmthera.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Borchers AT, Selmi C, Meyers FJ, Keen CL, Gershwin ME. Probiotics and immunity. J Gastroenterol. 2009;44:26–46. doi: 10.1007/s00535-008-2296-0. [DOI] [PubMed] [Google Scholar]
- 17.Vaarala O. Immunological effects of probiotics with special reference to lactobacilli. Clin Exp Allergy. 2003;33:1634–1640. doi: 10.1111/j.1365-2222.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- 18.Shanahan F. Physiological basis for novel drug therapies Used to Treat the Inflammatory Bowel Diseases I. Pathophysiological basis and prospects for probiotic therapy in inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2005;288:G417–G421. doi: 10.1152/ajpgi.00421.2004. [DOI] [PubMed] [Google Scholar]
- 19.Boirivant M, Strober W. The mechanism of action of probiotics. Curr Opin Gastroenterol. 2007;23:679–692. doi: 10.1097/MOG.0b013e3282f0cffc. [DOI] [PubMed] [Google Scholar]
- 20.Lomax AR, Calder PC. Probiotics, immune function, infection and inflammation: a review of the evidence from studies conducted in humans. Curr Pharm Des. 2009;15:1428–1518. doi: 10.2174/138161209788168155. [DOI] [PubMed] [Google Scholar]
- 21.Maeda N, Nakamura R, Hirose Y, Murosaki S, Yamamoto Y, Kase T, Yoshikai Y. Oral administration of heat-kills Lactobacillus plantarum L-137 enhances protection against influenza virus infection by stimulation of type I interferon production in mice. Int Immunopharmacol. 2009;9:1122–1125. doi: 10.1016/j.intimp.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Vouloumanou EK, Makris GC, Karageorgopoulos DE, Falagas ME. Probiotics for the prevention of respiratory tract infections: a systematic review. Int J Antimicrob Agents. 2009;34:197. e1–197.e10. doi: 10.1016/j.ijantimicag.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Hols P, Slos P, Dutot P, Reymund J, Chabot P, Delplace B, Delcour J, Mercenier A. Efficient secretion of the model antigen M6-gp41E in Lactobacillus plantarum NCIMB 8826. Microbiology. 1997;143:2733–2741. doi: 10.1099/00221287-143-8-2733. [DOI] [PubMed] [Google Scholar]
- 24.Kandler O, Stetter K, Köhl R. Lactobacillus reuteri sp. nov., a new species of heterofermentative lactobacilli. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt. 1980;1:264–269. [Google Scholar]
- 25.Mondofacto Medical Dictionary Search Online. at http://www.mondofacto.com/dictionary.
- 26.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 27.Domachowske JB, Bonville CA, Gao JL, Murphy PM, Easton AJ, Rosenberg HF. The chemokine macrophage-inflammatory protein-1 alpha and its receptor CCR1 control pulmonary inflammation and antiviral host defense in paramyxovirus infection. J Immunol. 2000;165:2677–2682. doi: 10.4049/jimmunol.165.5.2677. [DOI] [PubMed] [Google Scholar]
- 28.Percopo CM, Qiu Z, Phipps S, Foster PS, Domachowske JB, Rosenberg HF. Pulmonary eosinophils and their role in immunopathologic responses to formalin-inactivated pneumonia virus of mice. J Immunol. 2009;183:604–612. doi: 10.4049/jimmunol.0802270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garvy BA, Qureshi MH. Delayed inflammatory response to Pneumocystis carinii infection in neonatal mice is due to an inadequate lung environment. J Immunol. 2000;165:6480–6486. doi: 10.4049/jimmunol.165.11.6480. [DOI] [PubMed] [Google Scholar]
- 30.Bonville CA, Bennett NJ, Koehnlein M, Haines DM, Ellis JA, DelVecchio AM, Rosenberg HF, Domachowske JB. Respiratory dysfunction and proinflammatory chemokines in the pneumonia virus of mice (PVM) model of viral bronchiolitis. Virology. 2006;349:87–95. doi: 10.1016/j.virol.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Bienenstock J, McDermott MR. Bronchus- and nasal-associated lymphoid tissues. Immunol Revs. 2005;206:22–31. doi: 10.1111/j.0105-2896.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 32.Foo SY, Phipps S. Regulation of inducible BALT formation and contribution to immunity and pathology. Mucosal Immunol. 2010 doi: 10.1038/mi.2010.52. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 33.Welliver RC., Sr The immune response the respiratory syncytial virus: friend or foe? Clin Rev Allergy Immunol. 2008;34:163–173. doi: 10.1007/s12016-007-8033-2. [DOI] [PubMed] [Google Scholar]
- 34.Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 35.Mengheri E. Health, probiotics, and inflammation. J Clin Gastroenterol. 2008;42(Suppl 3 part 2):S177–S178. doi: 10.1097/MCG.0b013e31817eedc4. [DOI] [PubMed] [Google Scholar]
- 36.Torii A, Torii S, Fujiwara S, Tanaka H, Inagaki N, Nagai H. Lactobacillus acidophilus strain L-92 regulates the production of Th1 cytokine and well as Th2 cytokines. Allergol Int. 2007;56:293– 301. doi: 10.2332/allergolint.O-06-459. [DOI] [PubMed] [Google Scholar]
- 37.Kitazawa H, Matsumura K, Itoh T, Yamaguchi T. Interferon induction in murine peritoneal macrophage by stimulation with Lactobacillus acidophilus. Microbiol Immunol. 1992;36:311– 315. doi: 10.1111/j.1348-0421.1992.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 38.Kopp MV, Goldstein M, Dietschek A, Sofke J, Heinzmann A, Urbanek R. Lactobacillus GG has in vitro effects on enhanced interleukin-10 and interferon-gamma release of mononuclear cells but no in vivo effects in supplemenented mothers and their neonates. Clin Exp Allergy. 2008;38:602–610. doi: 10.1111/j.1365-2222.2007.02911.x. [DOI] [PubMed] [Google Scholar]
- 39.Lakatos PL, Fischer S, Lakatos L, Gal I, Papp J. Current concept on the pathogenesis of inflammatory bowel disease-crosstalk between genetic and microbial factors: pathogenic bacteria and altered bacterial sensing or changes in mucosal integrity take “toll”? World J Gastroenterol. 2006;28:1829–1841. doi: 10.3748/wjg.v12.i12.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anh DB, Faisca P, Desmecht DJ. Differential resistance/susceptibility patterns to pneumovirus infection among inbred strains of mice. Am J Physiol Lung Cell Mol Physiol. 2006;291(3):L426–435. doi: 10.1152/ajplung.00483.2005. [DOI] [PubMed] [Google Scholar]
- 41.Hori T, Kyoshima J, Shida K, Yasui H. Effect of intranasal administration of Lactobacillus casei Shirota on influenza infection of upper respiratory tract in mice. Clin Diagn Lab Immunol. 2001;8:593–597. doi: 10.1128/CDLI.8.3.593-597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harata G, He F, Hiruta N, Kawase M, Kubota A, Hiramatsu M, Yausi H. Intranasal administration of Lactobacillus rhamnosus GG protects mice from H1N1 influenza virus infection by regulating respiratory immune responses. Lett Appl Microbiol. 2010;50:5:97–602. doi: 10.1111/j.1472-765X.2010.02844.x. [DOI] [PubMed] [Google Scholar]
- 43.Izumo T, Maekawa T, Ida M, Noguchi A, Kitagawa Y, Shibata H, Yasui H, Kiso Y. Effect of intranasal administration of Lactobacillus pentosus S-PT84 on influenza virus infection in mice. Int Immunopharmacol. 2010;10:1:101–1106. doi: 10.1016/j.intimp.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Rautava S, Kalliomaki M, Isolauri E. New therapeutic strategy for combating the increasing burden of allergic disease: probiotics – a nutrition, allergy, mucosal immunology and intestinal microbiota (NAMI) research group report. J Allergy Clin Immunol. 2005;116:31–37. doi: 10.1016/j.jaci.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Zeuthen LH, Fink LN, Frøkliaer H. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology. 2008;124:489–502. doi: 10.1111/j.1365-2567.2007.02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohamadzadeh M, Klaenhammer TR. Specific Lactobacillus species differentially activate Toll-like receptors and downstream signals in dendritic cells. Expert Rev Vaccines. 2008;7:1155–1164. doi: 10.1586/14760584.7.8.1155. [DOI] [PubMed] [Google Scholar]
- 47.Diderlaurent A, Goulding J, Patel S, Snelgrove R, Low L, Bebien M, Lawrence T, van Rijt LS, Lambrecht BN, Sirard JC, Hussell T. Sustained desensitization to bacterial Toll-like receptor ligands after resolution of respiratory influenza infection. J Exp Med. 2008;205:323–329. doi: 10.1084/jem.20070891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hisbergues M, Magi M, Rigaux P, Steuve J, Garcia L, Goudercourt D, Pot B, Pestel J, Jacquet A. In vivo and in vitro immunomodulation of Der p 1 allergen-specific response by Lactobacillus plantarum bacteria. Clin Exp Allergy. 2007;37:1286–1295. doi: 10.1111/j.1365-2222.2007.02792.x. [DOI] [PubMed] [Google Scholar]
- 49.Ichikawa S, Fujii R, Fujiwara D, Komiyama Y, Kaisho T, Sakaguchi M, Konishi Y. MyD88 but not TLR2, 4 or 9 is essential for IL-12 induction by lactic acid bacteria. Biosci Biotechnol Biochem. 2007;71:3026–3032. doi: 10.1271/bbb.70414. [DOI] [PubMed] [Google Scholar]
- 50.Chung YW, Choi JH, Oh TY, Eun CS, Han DS. Lactobacillus casei prevents the development of dextran sulphate sodium-induced colitis in Toll-like receptor 4 mutant mice. Clin Exp Immunol. 2008;151:182–189. doi: 10.1111/j.1365-2249.2007.03549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2008;105:14046–14051. doi: 10.1073/pnas.0804717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyer KD, Percopo CM, Fischer ER, Gabryszewski SJ, Rosenberg HF. Pneumoviruses infect eosinophils and elicit MyD88-dependent release of chemoattractant cytokines and interleukin-6. Blood. 2009;114:2649–2656. doi: 10.1182/blood-2009-01-199497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klouwenberg PK, Tan L, Werkman W, van Bleek GM, Coenjaerts FEJ. The role of Toll-like receptors in regulating the immune response against respiratory syncytial virus. Crit Revs Immunol. 2009;29:531–550. doi: 10.1615/critrevimmunol.v29.i6.40. [DOI] [PubMed] [Google Scholar]
- 54.Van der Sluijs KF, van Delden L, Nijhuis M, Schuurman R, Florquin S, Jansen HM, Lutter R, van der Poll T. Toll-like receptor 4 is not involved in host defense against respiratory tract infection with Sendai virus. Immunol Lett. 2003;89:201–206. doi: 10.1016/S0165-2478(03)00138-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faisca P, Tran Anh DB, Thomas A, Desmecht D. Suppression of pattern-recognition receptor TLR4 sensing does not alter lung responses to pneumovirus infection. Microbes Infect. 2006;8:621–627. doi: 10.1016/j.micinf.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Garofalo RP, Patti J, HIntz KA, Hill V, Ogra PL, Welliver RC. Macrophage inflammatory protein-1alpha (not T helper type 2 cytokines) is associated with severe forms of respiratory syncytial virus bronchiolitis. J Infect Dis. 2001;184:393–399. doi: 10.1086/322788. [DOI] [PubMed] [Google Scholar]
- 57.Culley FJ, Pennycook AM, Tregoning JS, Hussell T, Openshaw PJ. Differential chemokine expression following respiratory virus infection reflects Th1- or Th2-biased immunopathology. J Virol. 2006;80:4521–4527. doi: 10.1128/JVI.80.9.4521-4527.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindell DM, Lane TE, Lukacs NW. CXCL10/CXCR3-mediated responses promote immunity to respiratory syncytial virus infection by augmenting dendritic cell and CD8(+) T cell efficacy. Eur J Immunol. 2008;38:2168–2179. doi: 10.1002/eji.200838155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puthothu B, Bierbaum S, Kopp MV, Forster J, Heinze J, Weckmann M, Krueger M, Heinzmann A. Association of TNF-alpha with severe respiratory syncytial virus infection and bronchial asthma. Pediatr Allergy Immunol. 2009;20:157– 163. doi: 10.1111/j.1399-3038.2008.00751.x. [DOI] [PubMed] [Google Scholar]
- 60.McNamara PS, Flanagan BF, Selby AM, Hart CA, Smyth RL. Pro-and anti-inflammatory responses in respiratory syncytial virus bronchiolitis. Eur Respir J. 2004;23:106–112. doi: 10.1183/09031936.03.00048103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


