Summary
The centromere is a specialized chromosomal structure that regulates chromosome segregation. Centromeres are marked by a histone H3 variant. In budding yeast, the histone H3 variant Cse4 is present in a single centromeric nucleosome. Experimental evidence supports several different models for the structure of centromeric nucleosomes. To investigate Cse4 copy number in live yeast we developed a new method coupling fluorescence correlation spectroscopy and calibrated imaging. We find that centromeric nucleosomes have one copy of Cse4 during most of the cell cycle, whereas two copies are detected at anaphase. The proposal of an anaphase coupled structural change is supported by Cse4-Cse4 interactions, incorporation of Cse4, and the absence of Scm3 in anaphase. Nucleosome reconstitution and ChIP suggests both Cse4 structures contain H2A/H2B. The increase in Cse4 intensity and deposition at anaphase are also observed in Candida albicans. Our experimental evidence supports a cell cycle coupled oscillation of centromeric nucleosome structure in yeast.
Keywords: Cse4/CENP-A, centromere, yeast, Scm3, nucleosome
Introduction
The centromere in all eukaryotic organisms plays a critical role in chromosome segregation in mitosis and meiosis. Centromeres are the site where the kinetochore is built. The kinetochore mediates the attachment of chromosomes to spindle microtubules. The centromere is defined by specific DNA sequences as well as by a specialized chromatin structure. Although centromere proteins are evolutionarily conserved among all organisms, the DNA sequence at the centromere is not conserved. Centromeres range in size from the ~125bp found in budding yeast to kb in S. pombe to several Mb in humans. Centromeres in the budding yeast Saccharomyces cerevisiae are short, simple, and consist of common sequence elements (CDE I, CDE II and CDE III) (Fitzgerald-Hayes et al., 1982). This sequence is the DNA component of a single Cse4-containing nucleosome at the centromere of each chromosome (Camahort et al., 2009; Cole et al., 2011; Furuyama and Biggins, 2007; Meluh et al., 1998). In contrast to the variability between centromeric DNA sequences, all eukaryotic centromeres are universally marked by a centromere specific histone variant (CenH3). This variant is called CENP-A in humans, CID in flies and Cse4 in budding yeast. This variant is essential for kinetochore formation and proper chromosome segregation (Henikoff and Dalal, 2005; Meluh and Koshland, 1997). Cse4 can functionally substitute for CENP-A (Wieland et al., 2004), suggesting that the structure of CenH3 nucleosomes is evolutionarily conserved.
Although it is clear that a histone variant replaces H3 at centromeres and these nucleosomes are very important for proper chromosome segregation, their structure is unclear. Since these nucleosomes specify the centromere, they are likely to have unique characteristics. Several models have been proposed for the structure of these nucleosomes, including octasomes, hemisomes/heterotypic tetrasomes and hexasomes (Black and Cleveland, 2011). The most conventional model is an octameric configuration, having two copies of H4, H2B, H2A, and Cse4 (Camahort et al., 2009; Conde e Silva et al., 2007; Foltz et al., 2006; Kingston et al., 2011; Palmer et al., 1987; Palmer and Margolis, 1985; Shelby et al., 1997; Zhou et al., 2011) and DNA wrapping with a conventional left-handed wrap, (Sekulic et al., 2010). The hemisome/heterotypic tetrasome model is a highly unique model based initially on experimental evidence from Drosophila S2 cells (Dalal et al., 2007), and further supported by additional evidence in yeast (Furuyama and Henikoff, 2009) and human cells (Dimitriadis et al., 2010). This model proposes that a single copy of each histone is present in the nucleosome and DNA is wrapped in a right-handed configuration. A third proposed model is the hexasome, in which a tetramer of Cse4 and H4 is joined by 2 copies of the non-histone protein Scm3 (Mizuguchi et al., 2007; Xiao et al., 2011). Additional models (tetrasome, trisome and reversome) have also been proposed but they lack substantial experimental evidence (Black and Cleveland, 2011).
The timing of deposition of the centromeric H3 variant with respect to the cell cycle varies in different species. Photobleaching of Cse4-GFP in budding yeast in anaphase showed that the GFP signal did not recover until the following S phase, (Pearson et al., 2004), suggesting Cse4 is deposited in S phase. In humans, CENP-A is expressed during G2, after S-phase is completed, but deposition occurs in late telophase to early G1 phase (Jansen et al., 2007). In Drosophila embryos, CID deposition takes place at anaphase (Schuh et al., 2007) but during metaphase in S2 cells (Mellone et al., 2011). In Schizosaccharomyces pombe, CENP-A (Cnp1) appears to be able to load both in a replication dependent and independent manner (Takahashi et al., 2000; Takahashi et al., 2005; Takayama et al., 2008). In Arabidopsis, it is reported that loading of CENP-A occurs mainly in G2 phase (Lermontova et al., 2006). Together these reports suggest that the timing of CENP-A deposition varies in different organisms.
We present evidence for a structural oscillation of centromeric nucleosomes in budding yeast. We developed a unique method to quantify the number of Cse4-EGFP molecules per centromeric nucleosome cluster in vivo using fluorescence correlation spectroscopy (FCS) coupled with calibrated avalanche photodiode (APD) -confocal imaging. Interestingly, when we quantified the number of Cse4-EGFP molecules per centromere cluster we find ~16 Cse4-EGFP/ cluster at G1/S/M/telophase and ~32 at anaphase. Since budding yeast have 16 chromosomes and each centromere contains one nucleosome (Camahort et al., 2009; Furuyama and Biggins, 2007; Henikoff and Henikoff, 2012), our results suggest that one copy of Cse4 is present per nucleosome at G1/S/M/telophase and two copies of Cse4 are present per nucleosome at anaphase. Measuring the distance between the spindle pole bodies (SPBs) reveals that the apparent structural change occurs during early anaphase and is complete by anaphase B. Furthermore, fluorescence resonance energy transfer (FRET) measurements and sequential co-immunoprecipitation are both consistent with two copies of Cse4 per centromeric nucleosome at anaphase. Both Cse4 structures contain H2A but the anaphase structure lacking the histone chaperone Scm3. Taken together, our experimental evidence supports a cell cycle coupled oscillation between an octasome and a hemisome at the centromere.
Results
Cse4 –EGFP intensity doubles during anaphase B
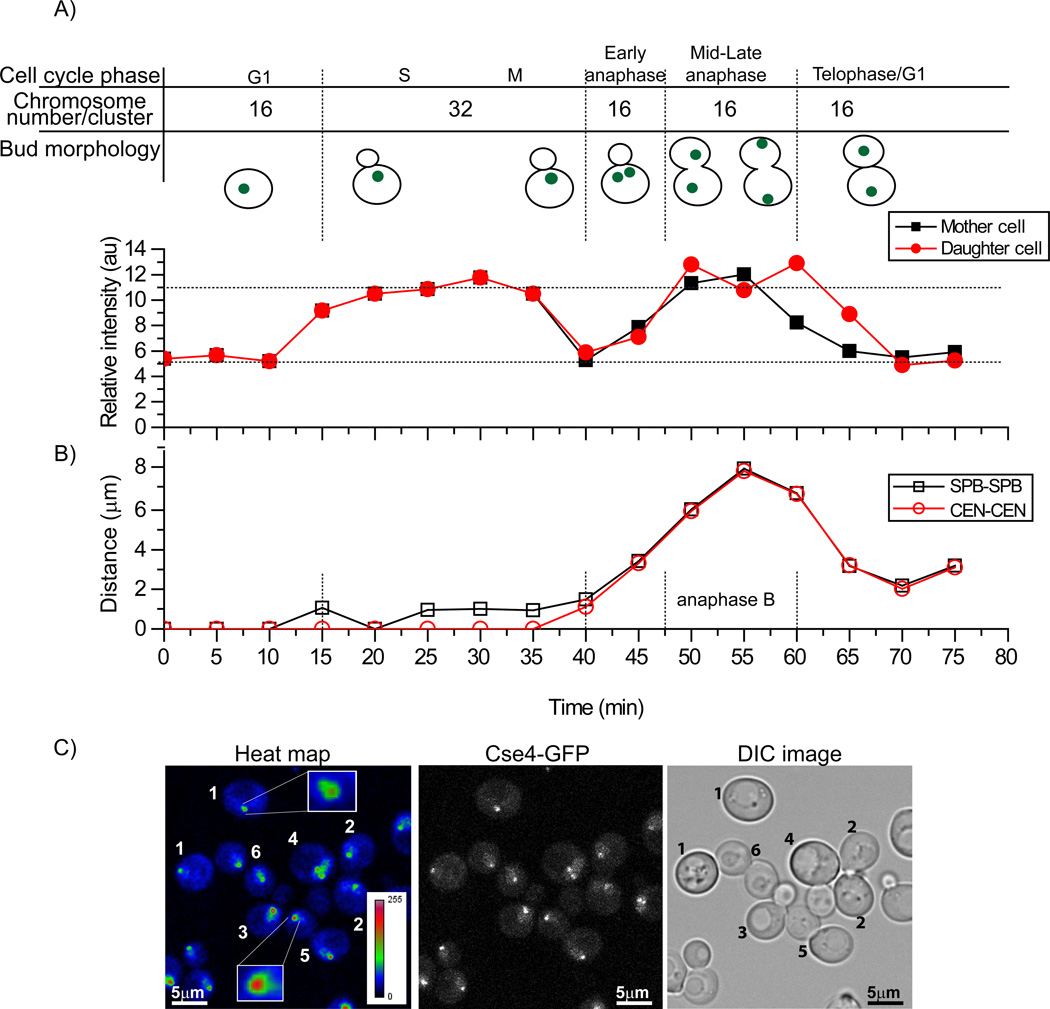
In budding yeast, centromeres are clustered throughout the mitotic cell cycle (Jin et al., 1998; Jin et al., 2000). Thus, centromeric Cse4-EGFP is visible as a single focus in the nucleus of living yeast cells. We measured the centromeric fluorescence intensity of Cse4-EGFP per nucleus throughout the cell cycle. Since all sister centromeres are grouped within one cluster until metaphase, and then separate into two centromere clusters in anaphase, we expected a 50% drop of the fluorescence intensity per cluster in anaphase compared to the value observed in late S /M. Subsequently, the fluorescence intensity is expected to increase only in the next S phase (Pearson et al., 2004). When clusters initially separated, we observed a drop in fluorescent intensity. However, as anaphase progressed we observed a 2-fold increase in the same cell cycle (Figure 1A, Movie S1 & Supplemental figure 1) compared to G1/telophase. Cell cycle staging was based on the bud morphology and centromere cluster position. To specifically assign the cell cycle phase when this intensity increase occurred, we used Spc42-mCherry as a marker for the spindle pole body (SPB) to demarcate the cell cycle stage. The distance between the SPBs was measured in 3 dimensions. Since the distance between SPBs increases slowly from 1–2 µm during G2/M and then rapidly to 4–10 µm during anaphase B (Yeh et al., 1995), we compared the distance between the SPB and Cse4-EGFP intensity over the cell cycle. As shown in Figure 1, intensity doubled at anaphase B.
Figure 1. Cse4-EGFP intensity doubles at anaphase B.
A) Time-lapse series of a centromere cluster followed from G1 to telophase indicates that Cse4 intensity doubles in anaphase. Quantification of centromere localized Cse4-EGFP-fluorescence shows that at anaphase the intensity doubled compared to G1 and telophase (n=14). B) The spindle pole body (SPB) was used as a cell cycle stage marker and the distances were measured between SPBs and centromere clusters in 3D using Image J. C) Anaphase centromeric clusters are brighter than clusters in G1/telophase cells. The left panel shows a heat map for centromeric Cse4-EGFP brightness in different stages of cells. The numbers in the image indicate cell cycle phase based on bud morphology: 1) G1, 2) telophase/G1, 3) M, 4) early S phase, 5) M and 6) late anaphase. Centromeric clusters in M and late anaphase are brighter than those in G1 or telophase. See Supplemental Figure 1 and Movie S1.
Next we specifically examined the brightness differences between G1 and anaphase B cells in the same imaging focus. We collected a z-stack image of an asynchronous cell culture where we visualized cells in many stages of the cell cycle, generated maximum intensity projection of the entire z stacks, and then developed a heat map (Figure 1C). In Figure 1C there is a clear difference in brightness between the centromeric spots of anaphase (cell 6) versus G1 cells (cell 1). Single metaphase clusters were also brighter than the separated or G1/telophase clusters because at this stage 2 copies of each chromosome are present. This measurement suggested that the intensity of the centromere cluster doubled during anaphase.
Cse4-Cse4 interaction is restricted to anaphase at centromeres
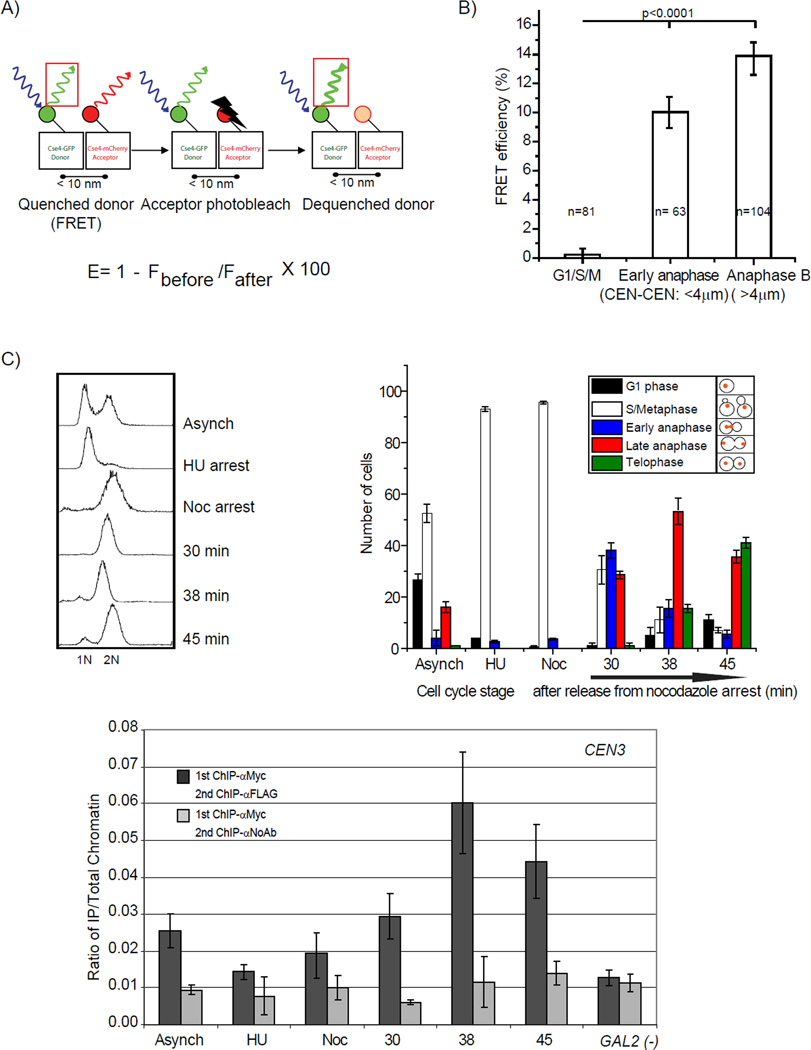
Since the brightness measurements suggested the doubling of intensity in anaphase B, we examined this result further by additional methods. We previously reported that Cse4-Cse4 interaction is critical for Cse4 function (Camahort et al., 2009). We used FRET to ask whether Cse4-Cse4 interaction could be detected in the centromere cluster and if so, when it occurred. We made a diploid strain which has one copy of Cse4 tagged with EGFP and a second copy tagged with mCherry. The fluorescence intensity of the Cse4-EGFP was measured at centromeres from the most intense focal slice of a z-stack. Immediately following the collection of the initial z-stack, the Cse4-mCherry in the entire cell was irreversibly photobleached using 561 nm excitation. The intensity of Cse4-EGFP in the centromere was re-measured after acceptor photobleaching. In the case where the donor (Cse4-EGFP) is undergoing FRET (Figure 2A), irreversible photobleaching of the acceptor probe will result in increased fluorescence of the donor (Cse4-EGFP) (see Experimental procedures for more details).
Figure 2. Cse4-Cse4 interactions are restricted to anaphase.
A) Simplified schematic of acceptor photobleaching. FRET can occur when a donor (e.g. Cse4-EGFP) and acceptor (e.g. Cse4-mCherry) are very close to each other (<10µm). If the acceptor is photobleached, emission from the donor will increase. B) FRET between Cse4-EGFP and Cse4-mCherry occurs only at anaphase. The FRET efficiency was measured in cycling cells (n=257). The cell cycle stage was defined by measuring the distance between centromere clusters (see Figure 1). FRET was not detected for G1, S or M phase cells. Following centromere cluster separation, FRET is observed. Error bars represent ± the standard deviation. C. Cse4-Cse4 interaction occurs only during anaphase. Sequential ChIP was performed on MNase-treated chromatin from strain MM118 using primers that amplify 125-bp of CEN3 sequence (Krassovsky et al., 2012). The signal from each XChIP has been divided by the signal obtained with total chromatin. The GAL2 gene serves as a negative control for Cse4 localization. Error bars represent ± the average deviation.
We measured the FRET efficiency in cycling cells (n=257), and noticed that cells in anaphase B had high levels of FRET while G1/S/M phase cells (n=146) did not show FRET (Figure 2B). In early anaphase cells we also observed FRET (Figure 2B). The simplest interpretation is that the FRET signal is due to Cse4-Cse4 interaction in a single nucleosome during anaphase. However, we cannot rule out the possibility of interactions between nucleosomes. In other phases of the cell cycle, where no FRET is observed, the simplest interpretation is that either there is one copy of Cse4 per centromeric nucleosome or Cse4 is not in a configuration to exhibit FRET with other Cse4 molecules. We note that similarly sized clusters were used for FRET analysis over the cell cycle.
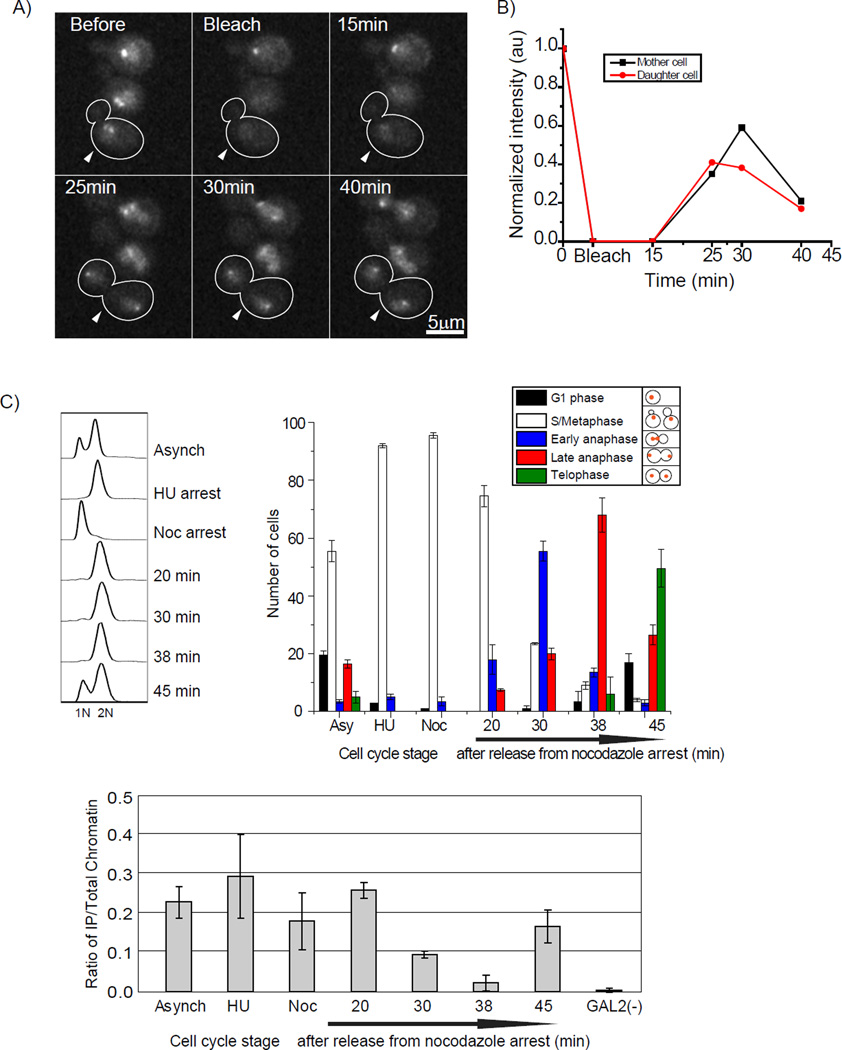
To further test Cse4-Cse4 interactions at the centromere during anaphase, we used a sequential ChIP procedure (Camahort et al., 2009). Yeast cells expressing Cse4-Myc and Cse4-FLAG were staged in the cell cycle. Using MNase digested chromatin, the Cse4-Myc protein was immunoprecipitated, and once eluted from the beads, we performed a second immunoprecipitation with anti-Flag antibody. If the two proteins are present in the same nucleosome at the centromere, then this procedure should immunoprecipitate centromeric DNA. We tested whether CEN DNA was enriched relative to a control in which the anti-Flag antibody was omitted from the second step. We found that CEN DNA was enriched in the anaphase samples, but not in an S phase or G2/M sample (Figure 2C), consistent with the interaction between Cse4 molecules at centromeres being confined to anaphase.
There are 2 copies of Cse4 per centromere at anaphase and 1 copy at G1/S/M/telophase
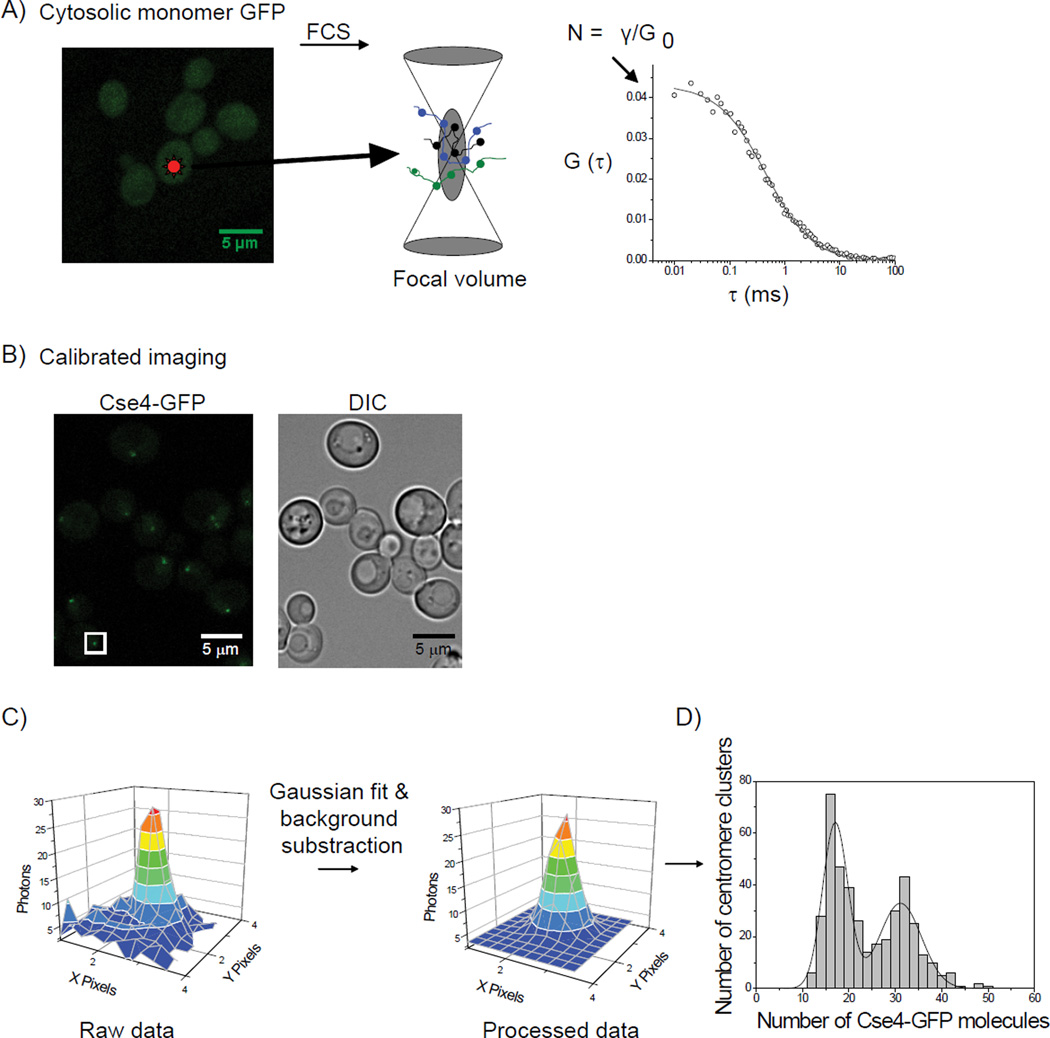
Since the brightness analysis and FRET measurements suggested a change in copy number and structure of Cse4 nucleosomes at anaphase, we wanted to further determine the exact Cse4 copy number at centromere clusters throughout the cell cycle. To quantify the number of Cse4-EGFP molecules in the yeast centromere, we developed a new and unique method to quantify the protein numbers in live cells using FCS and calibrated imaging (Figure 3) and used it to quantify the number of Cse4-EGFP molecules per centromere cluster for cycling cells (n=420). We observed two distribution peaks, one at ~16 Cse4-EGFP molecules per cluster and another at ~32 (Figure 4A).
Figure 3. Experimental design to determine the number of Cse4-EGFP per centromere.
A) A yeast strain expressing cytosolic EGFP was analyzed by fluorescence correlation spectroscopy (FCS) to determine the average number of EGFP molecules in a focal volume. The number of molecules in the focal volume in FCS is determined by γ/G0, where G0 is the amplitude of the correlation curve propagated to τ= 0. γ was determined to be 0.27, consistent with values published for FCS with 1-photon excitation. B) Calibrated images were acquired for cytosolic EGFP (A) and a yeast strain expressing Cse4-EGFP. C) To determine number of Cse4-EGFP per centromere, it was necessary to distinguish the emission emanating from the point centromere cluster from the background nuclear Cse4 signal. Therefore, we selected the z-slice where each centromere cluster was best in focus and fit the profile to a 2D Gaussian distribution, and selected the value of the peak minus the background as the intensity. D) γ from a point source is equal to 1; therefore we directly compared the intensity of the centromere cluster to the intensity we obtained for cytosolic EGFP using identical imaging parameters. With the known number of molecules of cytosolic EGFP from the FCS measurement, comparison allows for calculation of the number of Cse4-EGFP per centromere cluster. See Supplemental Figures 2 and 3.
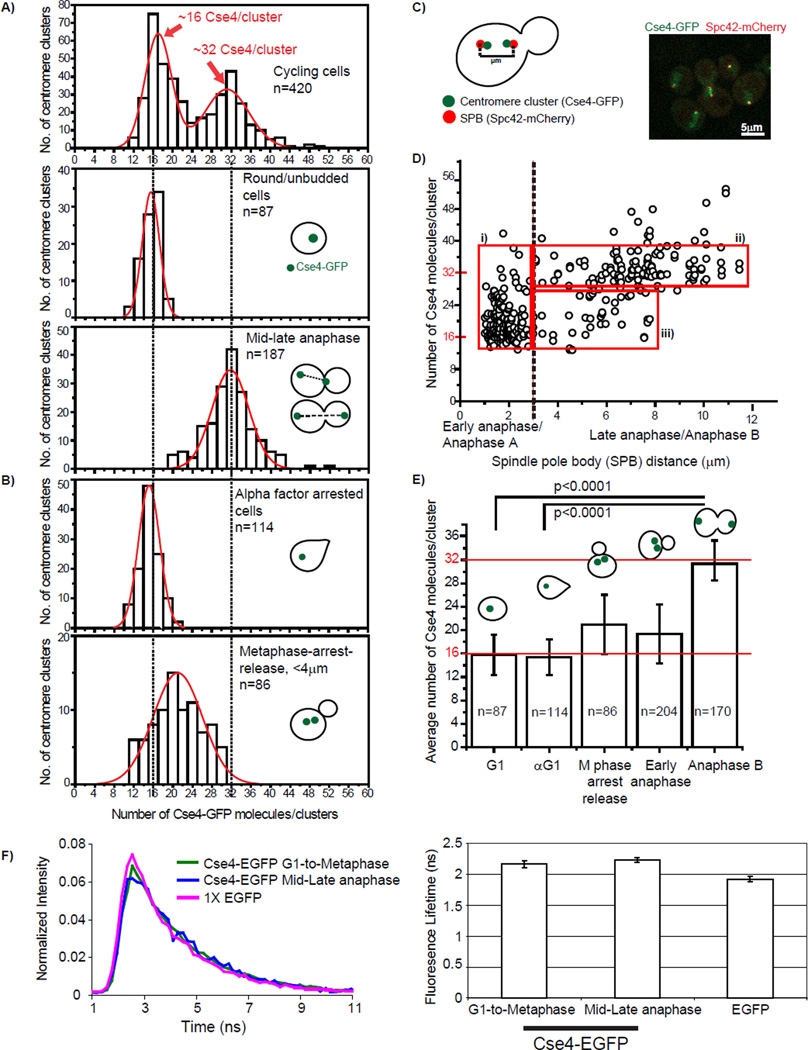
Figure 4. Two copies of Cse4 per centromere at anaphase and one copy of Cse4 for G1/telophase.
The stoichiometry of Cse4 per centromere cluster was measured in vivo, using FCS combined with calibrated imaging. A) A histogram is shown for Cse4-GFP/clusters in cycling cells (top panel) (n=420). Based on bud morphology and centromere cluster position, these cells were sorted into the G1 population (middle panel, n=87) and the mid to late anaphase population (bottom panel, n=187). B) Cells were arrested with either α factor in the top panel (G1), n=114, or nocodazole and released immediately prior to imaging in the bottom panel (early anaphase), n=80. Histograms are plotted for Cse4-EGFP/cluster in arrested cells. C) A schematic representation of how we measured distance between the SPBs to define the cell cycle stage is shown. The right side panel contains an image of live cells with centromere clusters (Cse4-EGFP) and SPBs (Spc42-mCherry). D) The number of Cse4 copies per cluster is plotted as a function of SPB distance. The three red boxes indicate populations of cells with the given SPB distances and the range of Cse4 copies per centromere clusters; i) 15–36, ii) 28–36 and iii) 15–26. E) The average number of Cse4 copies per cluster for A, B, and D. Error bars represent ± the standard deviation. P values are calculated by standard student’s t-test. F) The fluorescent lifetime measurements show similar lifetimes for EGFP and Cse4-EGFP and Cse4-EGFP in different stages of the cell cycle. The left panel shows the normalized average fluorescence decay for Cse4-EGFP centromere clusters in G1 to M (blue), mid-late anaphase (red), and unmodified EGFP from nuclear regions. The right panel shows the bar plot for fluorescent lifetimes from single exponential fits of Cse4-EGFP centromere clusters in G1 to M, mid to late anaphase, and regions in yeast nuclei expressing EGFP alone. Error bars represent ± the standard error of the mean. See Supplemental Figures 4, 5, and 6.
To determine the correspondence between the number of Cse4s/centromeric nucleosome and cell cycle stage, we sorted the cells based on the bud morphology and centromere cluster position. In the case of G1 cells, the distribution centered at ~16 Cse4-EGFP per cluster (Figure 4A middle panel), suggesting only one copy of Cse4 per centromeric nucleosome. In mid and late anaphase cells the distribution centered at ~32 Cse4-EGFP per cluster (Figure 4A bottom panel). In order to rule out the possibility that changes in EGFP fluorescence over the cell cycle caused two distributions, we examined the copy number of another inner kinetochore protein, Mif2/CENP-C. The average Mif2 copy number was similar in G1 and late anaphase (Supplemental figure 2), suggesting there is one copy per centromere throughout the cell cycle. This data helps rule out that the two distributions for Cse4 are due to a fluctuation in Cse4-EGFP signal over the cell cycle. In order to minimize the error from the cluster size variation we evaluated similarly sized clusters (Supplemental Figure 3).
To further confirm the Cse4 copy number at G1, we arrested the cells with α factor and counted the Cse4-EGFP molecules. The arrested cells had ~16 copies per centromere cluster (Figure 4B top panel), consistent with the round/unbudded cells from an asynchronous culture. Since we observed Cse4-EGFP brightness increasing during early anaphase, we next examined the Cse4 copy number during the transition period. We synchronized the cells with hydroxyurea, then with nocodazole, then released them immediately prior to imaging. The distribution centered at ~21 Cse4-EGFP molecules per cluster (Figure 4B bottom panel), consistent with these clusters being in a transition period.
We confirmed the karyotype of our Cse4-EGFP haploid strain and a diploid strain using qPCR (Supplemental Figure 4A). For the diploid strain we observed ~32 copies of Cse4/centromere cluster for cells in G1 phase and ~64 copies of Cse4/centromere cluster for cells in late anaphase (Supplemental Figure 4B). We note that the Cse4-EGFP strain from the UCSF collection is aneuploid; the distribution is shifted as expected (Supplemental Figure 4).
To define the precise cell cycle stage where the transition from one to two copies of Cse4 occurs, we monitored the position of SPBs (Figure 4C) and plotted the number of Cse4-EGFP per cluster as a function of SPB distance (Figure 4D). Two major distributions were evident and we classified them into two groups based on the SPB distance: 1) < 3 µm (early anaphase) and 2) >3 µm (anaphase B). At a SPB distance of >3 µm, the fluorescence measurements suggest that there are two Cse4-EGFP copies per centromere. In the <3 µm group, the brightness for the centromeric clusters ranged from 15 to 36 Cse4-EGFP copies per cluster (Figure 4Di). In the >3 µm group, the majority of cells ranged from 28–36 Cse4-EGFP copies per cluster (Figure 4D ii). Interestingly, a few centromeric clusters were in the brightness range from 15 to 26 at higher SPB distances (4–8 µm) (Figure 4Diii). One possible explanation for this variability could be a structural change at telophase when the centromeric clusters retract from the longest spindle position. Our results suggest that the transition from one to two copies of Cse4/nucleosome occurs in the <3µm group, which is during early anaphase.
To ensure that differences in fluorescence intensity of Cse4-EGFP and cytosolic free EGFP were due to differences in protein concentration and not quenching, fluorescent lifetime images were obtained using pulsed two-photon excitation at 920 nm and the same detection setup as used for confocal imaging. We did not detect differences in fluorescent lifetime between EGFP and Cse4-EGFP or between Cse4-EGFP in different phases of the cell cycle (Figure 4F).
Recently two groups have reported that budding yeast have 4–8 copies of Cse4 per centromere (Coffman et al., 2011; Lawrimore et al., 2011). One challenge is to try to reconcile this data with our own. One group measured Cse4 copy number in both G1 and anaphase cells, finding that the copy number doubled in anaphase (Coffman et al., 2011), in nice agreement with our results. To explain the differences observed in Lawrimore et al., and Coffman et al., versus our studies, we explored three possibilities: 1) aneuploidy, 2) differences in centromeric chromatin in different strain backgrounds and 3) differences in the molecular ruler used. We used qPCR and found no aneuploidy (Supplemental Figure 5A). Lawrimore et al. and Coffman et al shared strains, so the analysis is applicable to both studies. We used strains A and B from Lawrimore et al., to examine Cse4-GFP over the cell cycle. Both strains behaved nearly identically to our primary strain in terms of both Cse4-GFP copy number and the oscillation in copy number over the cell cycle (Supplemental Figure 5) indicating there are not major differences in centromeric chromatin behavior or the number of Cse4 molecules in the centromere cluster in different strain backgrounds. Recent ChIP seq data confirms that there is a single Cse4 nucleosome per centromere (Henikoff and Henikoff, 2012). The last possibility we explored was how the molecular ruler used by Lawrimore et al, composed of GFP on a glass slide (1 GFP), motB-GFP in E. coli (~22 GFP) and viral coat protein VLP2/6 (~120 GFP), compared to our measurements. While the fluorescence of GFP on a glass slide was technically challenging to measure, its fluorescence lifetime was similar to EGFP expressed in yeast, and these measurements were consistent with our Cse4-EGFP measurements (data not shown). In contrast, E. coli motB-GFP had a shorter lifetime than Cse4-EGFP in yeast (Supplemental Figure 6), suggesting mot-B should not be used to quantify Cse4-GFP. We did not analyzeVLP2/6-GFP. The FCS/calibrated imaging method we have developed in conjunction with its verification using known standards in the nuclear pore complex (see Figure 6), all conducted in live yeast, constitute an accurate measuring technique for determining the number of Cse4 molecules in the centromere cluster.
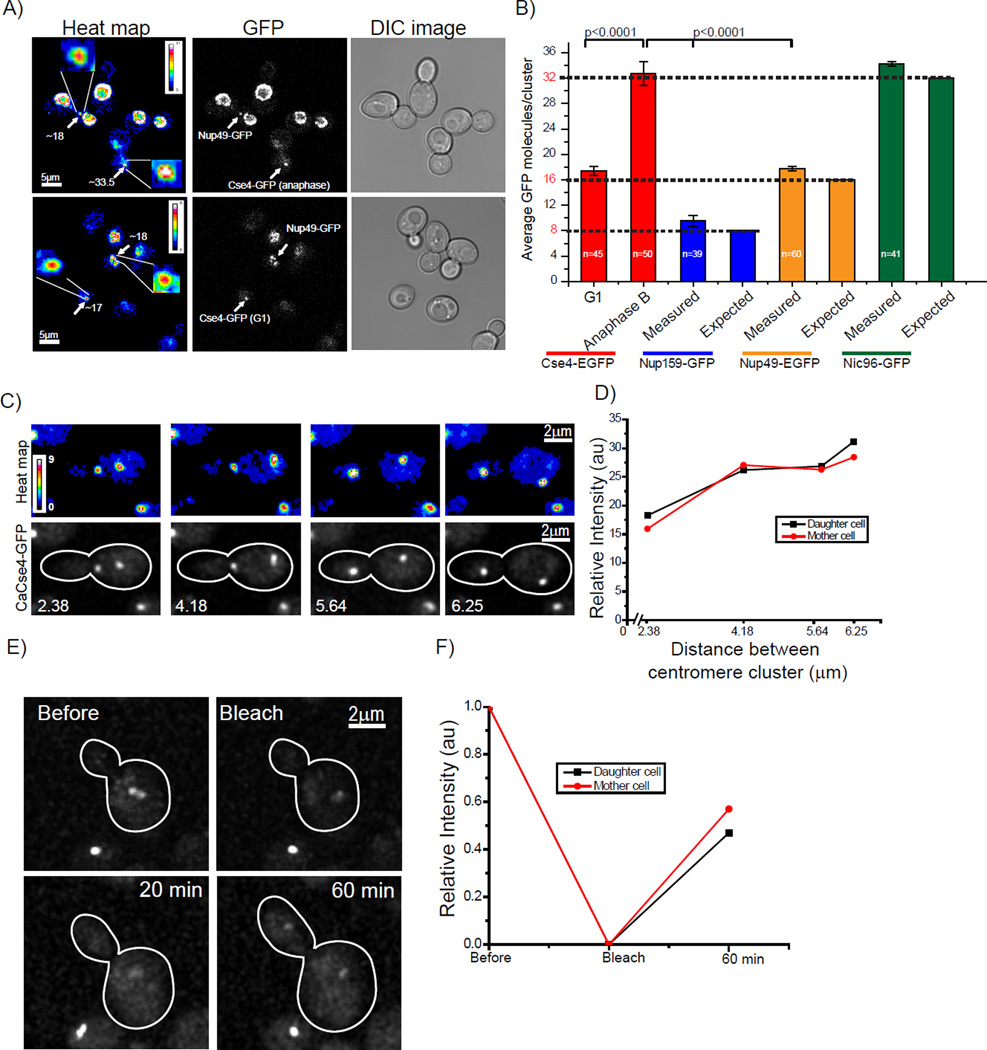
Figure 6. Fluorescence measurements of Cse4 relative to NPC components and Cse4 in C. albicans.
A) The intensity of a Cse4 cluster and Nup49 in a single NPC were compared in a single image. A heat map was developed by maximum intensity projection of all z stacks. Cell cycle stages for Cse4-EGFP expressing cells were determined by bud morphology. Clusters of Cse4-EGFP had a comparable intensity to Nup49-EGFP in G1 (top panel). However, Cse4-EGFP clusters at anaphase showed twice the intensity of Nup49-EGFP (bottom panel). B) We used FCS in conjunction with calibrated imaging (Figure 3) to calculate the number of GFP copies per focus. The number of Nup159, Nup49, and Nic96 molecules in a single NPC is predicted to be 8, 16, and 32 respectively (Alber et al., 2007a; Alber et al., 2007b; Cronshaw et al., 2002; Rout et al., 2000; Wente and Rout, 2010). The average number of Nup159-EGFP, Nup49-EGFP, and Nic96-EGFP molecules in a single NPC was ~8 (n=17), ~16 (n=21), and ~32 (n=29) compared to ~16 copies/cluster of Cse4-EGFP at G1 (n=45) and ~32 copies/cluster in anaphase B (n=50). Error bars represent ± the standard deviation. P values are calculated by standard student’s t-test. C) The doubling of Cse4 intensity in anaphase is observed in Candida albicans. CaCse4-GFP intensity was analyzed specifically from early anaphase to late anaphase (anaphase B). A heat map (top panel) was developed by maximum intensity projection of all z slices of a z-stack (bottom panel). D) Quantification of CaCse4-GFP intensity. The intensity of CaCse4-GFP doubled at anaphase B. This is anexample (n=5) of fluorescent intensity from early anaphase to anaphase B. E) Cells expressing CaCse4-GFP were photobleached (bleach) immediately following centromere cluster separation. The recovery of fluorescence was monitored at 20 and 60 mins. F) The starting fluorescence (pre-bleach) intensity was normalized to 1 and the recovery is shown as a function of time. The average recovery from 4 sets was 42% +/− 3%. See Movie S3 and S4 and Supplemental Figure 7.
Cse4 can be deposited during anaphase
The transition during early anaphase was very surprising to us. It seemed paradoxical to consider a structural change at anaphase, when previous data indicated deposition of Cse4 during S phase (Pearson et al., 2004). We decided to use FRAP to examine whether Cse4-EGFP recovers in the same cell cycle after photobleaching. We followed live cells where two centromeric clusters had just separated. We photobleached the centromere clusters. After 25 minutes we observe recovery of Cse4 foci, when cells were in anaphase B. The average recovery of each cluster was ~38% (n=6), although there was some variability (Figure 5 and Movie S2). We also confirmed this recovery by bleaching whole cells (data not shown). In summary, all movies show recovery in anaphase when photobleaching is performed upon clusters that have just separated. If we bleach later in anaphase we do not observe recovery until the next S phase (data not shown), consistent with the previous report (Pearson et al., 2004).
Figure 5. Timing of Cse4 deposition at early anaphase corresponds with absence of Scm3.
A) Cells expressing Cse4-EGFP were photobleached (0 min) immediately following centromere cluster separation. The recovery of fluorescence was monitored at 15, 25, 30, and 40 mins. The white arrow indicates the cell being followed. B) The starting fluorescence (pre-bleach) intensity was normalized to 1 and the recovery is shown as a function of time. The average recovery from 14 sets was 37 +/− 4 %. See movie S2. C) Scm3 disappears from centromere during early to late anaphase. Quantitative PCR (qPCR) results of the Scm3-3HA XChIP for the 125-bp region of CEN3 (Krassovsky et al., 2012). Cells were double synchronized, first with hydroxyurea and second with nocodazole and after release time points were taken and cell cycle stage was scored by DAPI staining and cell morphology. Error bars represent ± the average deviation. GAL2 is a negative control for Scm3 localization. Without antibody, the XChIP/qPCR signal was <10% of the total signal; this has been subtracted from the values presented.
The deposition of Cse4 observed during S phase is likely to represent the assembly of new Cse4 nucleosomes with DNA replication. In contrast, the incorporation of new Cse4 during anaphase indicates instability associated with Cse4 nucleosomes that likely corresponds with the transition from one copy to two copies, consistent with the ability of centromeric Cse4 nucleosomes to undergo a transition during this window of the cell cycle. Because Cse4 does not recover in telophase or G1 following bleaching in mid-anaphase (Pearson et al., 2004), the transition from two copies of Cse4 back to one may not require new deposition.
Scm3 is a Cse4 nucleosome assembly factor that is present at centromeres. To explore the how Scm3 might contribute to the transition, we conducted ChIP in staged cells. We found a very short window in anaphase during which the signal for Scm3 decreases (Figure 5C) (Mishra et al., 2011). The timing suggests that the absence of Scm3 coincides with the transition to the structure containing two copies of Cse4. The disappearance of Scm3 at centromeres during mitosis has also been reported in S. pombe (Pidoux et al., 2009; Williams et al., 2009), suggesting this behavior is evolutionarily conserved. Furthermore, structural studies show that Cse4/CENP-A uses the same interface to interact with Scm3/HJURP and a second molecule of Cse4/CENP-A (Cho and Harrison, 2011; Hu et al., 2011), consistent with the absence of Scm3/HJURP during the period of time that the structure contains two copies of Cse4/CENP-A.
Brightness comparison between Cse4 and Nup49, a component of the NPC
Extensive work in the yeast Saccharomyces cerevisiae and in vertebrates has elucidated the molecular architecture of the nuclear pore complex (NPC). The subunits of the NPC, known as nucleoporins, are present at 8, 16, or 32 copies (Alber et al., 2007a; Alber et al., 2007b; Cronshaw et al., 2002; Rout et al., 2000; Wente and Rout, 2010). We compared the Cse4-EGFP centromere cluster brightness to a single NPC, marked with Nup49-EGFP, which is predicted to be present at 16 copies per NPC. Single NPCs are most easily visualized during anaphase (Antonin et al., 2008; Shcheprova et al., 2008; Winey et al., 1997). We mixed cells expressing Cse4-EGFP and Nup49-EGFP on a slide and collected z-stack images, generated maximum intensity projections of the entire z stacks, and then developed a heat map. We found that during anaphase, centromeric clusters were brighter than a single Nup49-EGFP containing NPC (Figure 6A top panel), whereas during G1, centromeric clusters were similar in brightness to the single NPC (Figure 6A bottom panel). This comparison strongly suggests that in G1 the number of Cse4-EGFP molecules in a centromere cluster is comparable to the number of Nup49-EGFP molecules in a single NPC, and that in anaphase the number of Cse4-EGFP molecules in a centromere cluster is larger than the number of Nup49-EGFP molecules.
Next we used our unique FCS calibration method to count Nup159 (8 copies), Nup49, and Nic96 (32 copies) in a single NPC. We collected data for the Nups along with Cse4 in a similar experimental set up. The distribution for Nup159-EGFP, Nup49-EGFP, and Nic96-EGFP per NPC centered around 9.5, 17.7, and 34.2 while the distribution for Cse4-EGFP centered around 17.3 at G1 and around 32.7 at anaphase B (Figure 6B). To further confirm that we were quantifying the signal from single NPCs, we used a strain expressing Nup49-EGFP and collected data from several NPC “spots” during anaphase, including only diffraction limited signals but also including signals in more crowded regions on the anaphase bridge where it was possible that multiple NPCs were present. We could detect spots that had signal in multiples of 16 (Supplemental Figure 7), demonstrating our ability to identify single NPCs. This data establishes that the FCS method can count EGFP tagged proteins with reasonable accuracy and provides further support for the claim that the copy number of Cse4-EGFP oscillates between 16 and 32 molecules per cluster.
Many studies have used Cse4-EGFP copy number as a metric for the copy number of a protein of interest. We suggest that the Nups may provide a better measuring stick since 1) the copy number does not change over the cell cycle, and 2) the copy number should not vary with ploidy.
The anaphase behavior of Cse4 is evolutionarily conserved in C. albicans
To ask if our observations in budding yeast were evolutionarily conserved in another yeast species, we used Candida albicans, which has a simple regional centromere. Each centromere, which contains no defined sequence, is composed of a mean of four CENP-A containing nucleosomes and one kinetochore with one microtubule attachment site (Allshire and Karpen, 2008; Joglekar et al., 2008; Mishra et al., 2007). The intensity of Cse4-GFP centromere clusters doubled at late anaphase (Figure 6D, Movie S3), similar to our observations in budding yeast. Photobleaching performed upon centromere clusters separated in early anaphase showed recovery of CaCse4 foci in late anaphase (Figure 6E, Movie S4). The average recovery was ~42% (n=3) (Figure 6F). Thus, the anaphase behavior of Cse4 is conserved in a regional centromere across 180 million years of evolution.
The biochemical properties of Scm3-assembled Cse4 nucleosomes are distinct from Nap1-assembled Cse4 nucleosomes
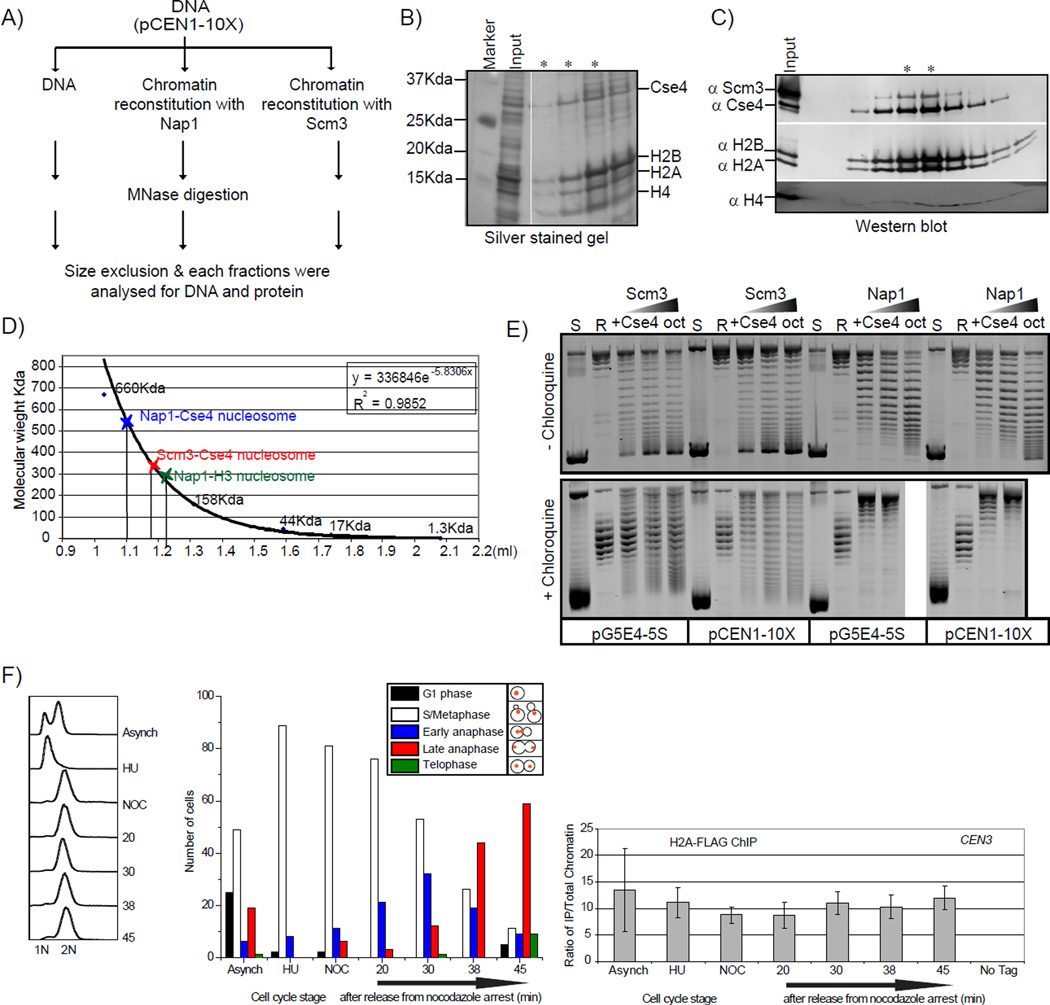
Cse4/CENP-A containing nucleosomes can be assembled in vitro (Camahort et al., 2009; Kingston et al., 2011; Shelby et al., 1997; Shivaraju et al., 2011; Yoda et al., 2000). Scm3 is a Cse4-specific nucleosome assembly factor (Shivaraju et al., 2011). Similarly, the mammalian ortholog of Scm3, HJURP, facilitates deposition of CENP-A/H4 tetramers on DNA (Shuaib et al., 2010). We compared Cse4 nucleosomes assembled by Scm3 and a canonical histone chaperone, Nap1. Chromatin was reconstituted using Scm3 or Nap1, digested with MNase, and subjected to gel filtration chromatography to isolate mononucleosomes (Figure 7A). We examined the proteins in the fractions where the DNA size was ~100–150 bp. In a control reaction lacking histones only small fragments of DNA are observed (<100 bp). Fractions from the both the Nap1 and Scm3 reconstitutions had all four histones (Figure 7B). In addition, the presence of Scm3 suggests Scm3 remains associated with the Cse4 mononucleosomes (Figure 7C). The mononucleosomes assembled with Scm3 displayed a different elution profile than those assembled with Nap1, suggesting the two species may be different. Nap1 assembled Cse4 mononucleosomes behaved as larger species (530+/−10 kDa) than Scm3 assembled mononucleosomes (330 +/− 10 kDa) (Figure 7D). The elution of Nap1 assembled H3 mononucleosomes is shown for comparison. H3 is 15 kDa while Cse4 is 27 kDa which in part explains the difference in Nap1 assembled Cse4 and H3 nucleosomes. In addition, the discrepancy between the elution profile of Cse4 mononucleosomes and their expected size could be attributed to the shape of the complexes (Dechassa et al., 2011). Two possible explanations could account for these observations: 1) the Scm3 nucleosomes could be a compacted octasome species or 2) the complex could be a hemisome.
Figure 7. Biochemical properties of Scm3 assembled and Nap1 assembled Cse4 nucleosomes.
Centromeric nucleosomes were assembled on a plasmid containing 10 copies of CEN1 or 5S positioning sequence (Shivaraju et al., 2011). A) Schematic representation of the mononucleosome purification is shown. Chromatin was reconstituted on the plasmid containing 10 copies of CEN1, MNase treated for 5 minutes, and the mixture was fractionated on a superdex-200 column using a Smart system. The mononucleosome fraction was recovered for further analysis. Protein content was analyzed by SDS-PAGE (B) for Nap1 Cse4 assembled mononucleosomes or western blotting (C) for Scm3 assembled Cse4 mononucleosomes. D) A molecular size standard was run on the same column used for fractionation, and an apparent MW standard curve was created. Error is estimated to be +/− 10 kDa. The MW of an assembled nucleosome is calculated to be ~530 kD for Nap1 assembled and ~320 kDa for Scm3 assembled nucleosomes. The calculated weight for an octamer and DNA is ~229 kDa. E) Chromatin assembly reactions were performed by incubating the relaxed CEN1 or 5S plasmid separately with the indicated proteins. Topoisomers are separated on an agarose gel without chloroquine (top panel) or with chloroquine (bottom panel). The Scm3 assembled nucleosomes do not exhibit the same shift in topoisomers observed in the Nap1 assembled nucleosomes. F) H2A-FLAG ChIP was conducted as in Figure 5C except that crosslinking was omitted and the chromatin was treated with MNase prior to immunoprecipitation. The results from a single timecourse are shown; the experiment was repeated twice with similar results. As a control we performed the ChIP on a strain without a FLAG tag on H2A (no tag).
One of the distinguishing features of the octasome and hemisome is the direction of DNA wrapping on these nucleosomes (Furuyama and Henikoff, 2009; Zhou et al., 2011). We evaluated the supercoiling of DNA on Cse4 nucleosomes assembled by Nap1 or by Scm3 usinga standard plasmid supercoiling assay (Shivaraju et al., 2011). Once chromatin is assembled on a plasmid, the proteins can be removed but the topology will be maintained. The supercoils will reflect the direction the DNA was wrapped. On a normal agarose gel we can measure the extent of nucleosome assembly by the observation of topoisomers (Furuyama and Henikoff, 2009; Prunell, 1998). If the topoisomers are electrophoresed in the presence of chloroqine, which reduces the twist of DNA (Furuyama and Henikoff, 2009; Prunell, 1998), we can determine if the supercoiling is positive or negative. Since the linking number (Lk) is fixed in a covalently closed plasmid, the reduction in twist (Tw) must be compensated for by an increase in writhe (Wr), (ΔLk =ΔTw + ΔWr) (Prunell, 1998). In a chloroquine containing gel, plasmids containing negative supercoils run slower because of the addition of positive writhe compared to relaxed plasmid.
Chloroquine intercalation caused topoisomers induced by Nap1 with Cse4 octamers to move slower than initially relaxed (R) circular plasmid. Therefore, these topoisomers were negatively supercoiled, as expected for an octasome species. Strikingly, the topoisomers induced by Scm3 did not show a simple pattern that would indicate 100% negative or positive supercoiling (Figure 7E). However, the supercoils that formed with Scm3 appeared to be more positive compared to those assembled with Nap1. If our assembly reactions contained some nucleosomes that have DNA wrapped in a right handed configuration, or even some nucleosomes that simply have less left handed wrapping (e.g. less DNA wrapped), this could account for the observed results. We speculate that DNA could be wrapped differently around a Cse4 histone complex by using different chaperones.
The Cse4 nucleosomes we have reconstituted in vitro possess H2A and H2B. A distinguishing feature of the hexasome model is the absence of H2A and H2B. To address whether H2A is absent at centromeres at any stage over the cell cycle, we carried out a ChIP experiment with staged cells. We found that H2A was present at centromeres throughout the cell cycle, including anaphase cells (Figure 7F). This result suggests that both of the Cse4 containing structures in vivo contain H2A. This result is inconsistent with a hexasome.
Discussion
To fully appreciate the behavior of centromeric nucleosomes in yeast, we analyzed their composition in live cells, using several fluorescent microscopy methods and chromatin biochemistry. Our data suggest that there are 32 Cse4 molecules per centromere cluster at anaphase B and 16 Cse4s per centromere cluster during the rest of the cell cycle. By using the SPB as a cell cycle phase marker, we defined the transition point as early anaphase. Although unexpected, these results are further supported by independent brightness measurements in budding yeast and C. albicans, comparisons to Nups, FRET and sequential ChIP analysis showing Cse4-Cse4 interaction in anaphase, FRAP showing Cse4 deposition in anaphase. We show that both structures have H2A but the anaphase structure lacks Scm3. Taken together our data is consistent with a cell cycle coupled oscillation between a hemisome structure from G1 to metaphase and an octasome structure during anaphase. A cell cycle-coupled oscillation between hemisomes and octasomes has also been demonstrated in human cells (Y. Dalal, cosubmitted). There has been a heated debate over the structure of centromeric nucleosomes with strong experimental support for several models. Our studies help to reconcile part of the debate, since the structure appears to be dynamic.
Our FCS/calibrated imaging and FRET data showed a transition from the one Cse4/nucleosome structure to the two molecule structure during early anaphase. In this stage we detected some clusters with intermediate values between 16 and 32 Cse4 copies. This suggests that the structural transition of all the centromeric nucleosomes in all the chromosomes is not completely concurrent. Moreover, the doubling of the Cse4-EGFP intensity measurement over the cell cycle took several minutes. Sister chromosomes start to move toward the spindle poles in anaphase A, and rapid elongation of the spindle separates the centromere clusters/spindle poles at anaphase B (He et al., 2000; Straight et al., 1997; Yeh et al., 1995). Kinetochore–microtubule attachments impose sufficient tension and force on sister chromosomes to transiently separate centromeric chromatin during prometaphase (He et al., 2000). We speculate that the loss of Scm3 in combination with tension and force on centromeric nucleosomes could influence the structural transition (Bancaud et al., 2007; Lavelle et al., 2009). The transition back to the one copy structure appears to occur in telophase, and we speculate that this transition may normally rely on Scm3 but not new Cse4 deposition. In centromeres with tens to hundreds of CENP-A nucleosomes, the force exerted on CENP-A nucleosomes may be more variable such that the existence of the hemisome is mainly controlled by chaperone activity.
A common denominator in the structural changes observed in yeast and human cells is the presence of an octasome when Scm3/HJURP is absent. We speculate that the hemisome may be specified by Scm3, and additional chaperones may be involved in octasome formation. Structural studies are consistent with this proposal since the absence of Scm3 would expose the domain of Cse4 needed for dimerization and octasome formation (Cho and Harrison, 2011; Hu et al., 2011). Also consistent with this proposal, additional chaperones have been reported to be associated with the centromeric histone H3 variant (Furuyama et al., 2006). While Scm3 is required for Cse4 nucleosome assembly (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007), depletion of Scm3 following Cse4 nucleosome assembly still activates the spindle assembly checkpoint, indicating that the presence of Scm3 at centromeres is required to pass the checkpoint and suggesting that the structure of this nucleosome may be carefully monitored (Shivaraju et al., 2011). Furthermore, when Scm3 is overexpressed, its presence at centromeres does not oscillate, and chromosome loss occurs (Mishra et al., 2011), suggesting the oscillation is important for euploidy. Further studies will be required to understand how chaperone dynamics regulate the structure of centromeric nucleosomes over the cell cycle, and how this contributes to chromosome segregation.
Experimental procedures
Yeast strains
The S. cerevisiae and C. albicans strains used in this study are listed in Supplemental Table 1. Karyotyping by qPCR as previously described (Pavelka et al., 2010) was done to examine the ploidy level of strains used in microscopy studies (Supplemental Figure 4 and 5).
Microscopic techniques
All microscope images were acquired using a Carl Zeiss LSM-510 Confocal microscope (Jena, Germany), outfitted with a ConfoCor 3 module and two single-photon counting avalanche photodiodes (APD’s). A C-Apochromat 40× 1. 2 NA water objective was used. A HFT 488/561 main dichroic allowed excitation of GFP (488 nm laser line) and mCherry (561 nm laser). A secondary NFT 565 beam splitter was used as an emission dichroic. After passage through a 505–550 nm BP or LP 580 filter for GFP and mCherry, respectively, photon counts were collected on APDs in single photon counting mode. For cell cycle series/movies the cells were maintained on 2% agar pads at room temperature. For more details of FCS, FRAP and FRET measurements see supplemental experimental procedures.
Cell staging, crossliked and MNase ChIP
For more details of cell synchronization-release experiments, FACS and nuclear morphology analyses, formaldehyde crosslinked and MNase ChIP/qPCR see supplemental experimental procedures
Expression and purification of recombinant protein
Histone chaperones Scm3 and Nap1 and yeast recombinant histones (H3, H4, H2A, H2B, and Cse4) were individually expressed in E. coli and purified as previously described (Luger et al., 1997; Shivaraju et al., 2011).
Reconstitution of protein complexes and in vitro chromatin assembly
Cse4-containing histone octamers were reconstituted using established protocols (Luger et al., 1999). The assembly of chromatin was performed as previously described (Camahort et al., 2009; Ito et al., 1997; Shivaraju et al., 2011). For more information see supplemental experimental procedures.
Supplementary Material
Highlights.
A new method enables copy measurements of fluorescent proteins in live cells
Cse4-Cse4 interactions are restricted to anaphase at centromeres
Cse4 can be deposited at centromeres during anaphase
Scm3 is absent from Cse4 nucleosomes at anaphase
Acknowledgements
We thank Jerry Workman, Christophe Lavelle and Sue Jaspersen for valuable discussions. We thank Swaminathan Venkatesh for topoI. We thank Dan Bradford for technical assistance and all of the members of the Gerton lab for their help and support. We thank Yamini Dalal and Kerry Bloom for sharing unpublished data and valuable discussions, and a special thanks to Minh Bui for composing the graphical abstract. This research was supported by NIH R01GM080477 and SIMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Letters. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Bancaud A, Wagner G, Conde e Silva N, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome Chiral Transition under Positive Torsional Stress in Single Chromatin Fibers. Molecular Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic Centromere Propagation and the Nature of CENP-A Nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 Is Essential to Recruit the Histone H3 Variant Cse4 to Centromeres and to Maintain a Functional Kinetochore. Molecular Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 Is Part of an Octameric Nucleosome in Budding Yeast. Molecular Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. The Journal of Cell Biology. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proceedings of the National Academy of Sciences. 2011;108:12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing Nucleosomes: Easier Disassembly versus Exclusive Centromeric Localization. Journal of Molecular Biology. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. The Journal of Cell Biology. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric Structure of Centromeric Nucleosomes in Interphase Drosophila Cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proceedings of the National Academy of Sciences. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proceedings of the National Academy of Sciences. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Centromeric Nucleosomes Induce Positive DNA Supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Asthana S, Sorger PK. Transient Sister Chromatid Separation and Elastic Deformation of Chromosomes during Mitosis in Budding Yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Current Opinion in Genetics & Development. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. 'Point' Centromeres of Saccharomyces Harbor Single CenH3 Nucleosomes. Genetics. 2012 doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A–histone H4 heterodimer in complex with chaperone HJURP. Genes & Development. 2011 doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-Containing and ATP-Utilizing Chromatin Assembly and Remodeling Factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. The Journal of Cell Biology. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q-w, Trelles-Sticken E, Scherthan H, Loidl J. Yeast Nuclei Display Prominent Centromere Clustering That Is Reduced in Nondividing Cells and in Meiotic Prophase. The Journal of Cell Biology. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci. 2000;113:1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. The Journal of Cell Biology. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JSY, Singleton MR. Biophysical Characterization of the Centromere-specific Nucleosome from Budding Yeast. Journal of Biological Chemistry. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proceedings of the National Academy of Sciences. 2012;109:243–248. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle C, Recouvreux P, Wong H, Bancaud A, Viovy J-L, Prunell A, Victor J-M. Right-Handed Nucleosome: Myth or Reality? Cell. 2009;139:1216–1217. doi: 10.1016/j.cell.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. The Journal of Cell Biology. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. Loading of Arabidopsis Centromeric Histone CENH3 Occurs Mainly during G2 and Requires the Presence of the Histone Fold Domain. The Plant Cell Online. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MMY, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of Molecular Biology. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila Centromeric Chromatin Proteins during Mitosis. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002068. e1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Development. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p Is a Component of the Core Centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Mishra P, Baum M, Carbon J. Centromere size and position in <i>Candida albicans</i> are evolutionarily conserved independent of DNA sequence heterogeneity. Molecular Genetics and Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002303. e1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and Histones CenH3-H4 Assemble the Core of Centromere-Specific Nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Palmer D, O'Day K, Wener M, Andrews B, Margolis R. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. The Journal of Cell Biology. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, Margolis RL. Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol Cell Biol. 1985;5:173–186. doi: 10.1128/mcb.5.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable Kinetochore-Microtubule Attachment Constrains Centromere Positioning in Metaphase. Current Biology. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JKR, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission Yeast Scm3: A CENP-A Receptor Required for Integrity of Subkinetochore Chromatin. Molecular Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A. A Topological Approach to Nucleosome Structure and Dynamics: The Linking Number Paradox and Other Issues. Biophysical Journal. 1998;74:2531–2544. doi: 10.1016/S0006-3495(98)77961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The Yeast Nuclear Pore Complex. The Journal of Cell Biology. 2000;148:635–652. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into Centromeres during Early Embryonic Anaphase. Current Biology. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into Centromeric Chromatin Requires a Cooperative Array of Nucleosomal DNA Contact Sites. The Journal of Cell Biology. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Camahort R, Mattingly M, Gerton JL. Scm3 is a centromeric nucleosome assembly factor. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proceedings of the National Academy of Sciences. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in Living Budding Yeast: Anaphase A But No Metaphase Plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 Centromere Connector for Localizing a CENP-A-Like Protein in Fission Yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360:595–607. doi: 10.1098/rstb.2004.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell. 2008;19:682–690. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harbor Perspectives in Biology. 2010;2 doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional Complementation of Human Centromere Protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission Yeast Scm3 Mediates Stable Assembly of Cnp1/CENP-A into Centromeric Chromatin. Molecular Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, Mastronarde DN. Nuclear Pore Complex Number and Distribution throughout the Saccharomyces cerevisiae Cell Cycle by Three-Dimensional Reconstruction from Electron Micrographs of Nuclear Envelopes. Molecular Biology of the Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 Binds to AT-Rich DNA to Organize Atypical Centromeric Nucleosome of Budding Yeast. Molecular Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. The Journal of Cell Biology. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou B-R, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.