Abstract
The copper-catalyzed azide-alkyne cycloaddition reaction is widely used for the connection of molecular entities of all sizes. A protocol is provided here for the process with biomolecules. Ascorbate is used as reducing agent to maintain the required cuprous oxidation state. Since these convenient conditions produce reactive oxygen species, five equivalents of a copper-binding ligand is used with respect to metal. The ligand both accelerates the reaction and serves as a sacrificial reductant, protecting the biomolecules from oxidation. A procedure is also described for testing the efficiency of the reaction under desired conditions for purposes of optimization, before expensive biological reagents are used.
Keywords: click chemistry, azides, alkynes, bioconjugation, proteins, nucleic acids, copper
Introduction
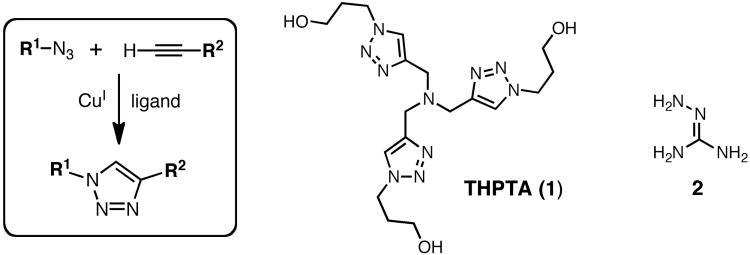
A reliable set of procedures for the copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction (Figure 1) is described here, based on investigations of the reaction mechanism and optimization using accelerating ligands for bioconjugative applications.(Chan et al., 2004; Lewis et al., 2004; Himo et al., 2005; Rodionov et al., 2005; Sen Gupta et al., 2005; Chan and Fokin, 2007; Rodionov et al., 2007a; Rodionov et al., 2007b; Hong et al., 2008; Hong et al., 2009; Buckley et al., 2010; Presolski et al., 2010) In practical terms, “bioconjugation” is usually used to signify making covalent bonds to biological molecules (principally oligopeptides, proteins, oligonucleotides, nucleic acids, and fatty acids), assemblies of biological molecules (such as lipid bilayers and vesicles, virus particles, and protein aggregates of many kinds), and living systems (usually cells in culture). However, these procedures are also applicable to polymers, having been particularly important for the attachment of poly(ethylene glycol) to many compounds, or to any molecules that are handled in aqueous solvents at low concentrations. High concentrations and nonaqueous reaction conditions demand a different catalyst formulation for demanding situations, as as been discussed elsewhere.(Presolski et al., 2010)
Figure 1.
The general CuAAC reaction, and structures of accelerating ligand THPTA (1) and aminoguanidine additive 2.
The basic CuAAC process requires only copper ions in the +1 oxidation state. These may be supplied by a discrete CuI complex,(Rostovtsev et al., 2002; Tornøe et al., 2002) by metallic copper(Rostovtsev et al., 2002) or copper-impregnated materials(Lipshutz et al., 2006) that expose cuprous ions to the reaction solution, or, most conveniently, by a mixture of a CuII salt and a reducing agent, sodium ascorbate being by far the most popular.(Rostovtsev et al., 2002) The development of accelerating ligands for the reaction is primarily driven by the need to maintain a sufficient concentration of CuI in solution, since copper ions can undergo fast and debilitating redox and disproportionation reactions if not properly bound by chelating ligands.
Copper-Catalyzed Azide–Alkyne Cycloaddition for Coupling of Cargo-Azide to Biomolecule-Alkyne
The basic procedure below describes the ligation of a functional (“cargo”) azide to a biomolecule alkyne. It can equally well be used in the reversed sense (biomolecule-azide + cargo-alkyne), and the cargo can also be a biomolecule, of course. Aminoguanidine is recommended when side reactions between dehydroascorbate and protein side chains (principally arginine) are to be suppressed, but is not otherwise helpful and should be omitted if possible. Tris(3-hydroxypropyltriazolyl-methyl)amine] (THPTA, structure 1, Figure 1), serves the dual purpose of protecting biomolecules from hydrolysis by Cu(II) byproducts, and sacrificially intercepting the radicals and/or peroxides derived from O2/Cu/ascorbate that oxidize histidine and other residues. An excess of this ligand does not dramatically slow the reaction, so more than five equivalents can be used if necessary. Note that other water-soluble versions of the tris(triazolylmethyl)amine motif have been reported(Hong et al., 2008; Soriano del Amo et al., 2010) and are probably suitable.
Materials and stock solutions
All of the materials are commercially available from standard suppliers, except for the ligand (THPTA). THPTA can be made by the published procedure,(Hong et al., 2009) or is available from the authors in small quantities until it becomes commercially available.
CuSO4: 20 mM (in water)
Ligand THPTA (1): 50 mM (in water)
Sodium ascorbate: 100 mM (make fresh before using by adding 1 mL of water to 20 mg).
Aminoguanidine hydrochloride (2): 100 mM (add 1 mL of water to 11 mg)
Cargo-azide: 5 mM
Biomolecule-alkyne: as desired
Buffer: 100 mM potassium phosphate, pH 7 (see note below regarding buffers)
Final concentrations
CuSO4: 0.10 mM (Note: can be adjusted as desired between 50 and 250 μM)
Ligand 1: 0.50 mM (ligand to copper ratio is 5:1)
Sodium ascorbate: 5 mM
Aminoguanidine: 5 mM
Biomolecule-alkyne: successfully done with 2 μM and higher; this example is for 50 μM alkyne.
Cargo-azide: approx. 2-fold excess with respect to alkyne groups on the biomolecule, down to 20 μM (in other words, if the alkyne concentration is very low, more than two equivalents of azide are needed for fast reaction).
Performing the CuAAC reaction
In a 2 mL Eppendorf tube, combine the reagents in the following order:
Biomolecule-alkyne + buffer to make 432.5 μL of solution that is 57.8 μM in alkyne.
10 μL of Cargo-azide
A premixed solution of CuSO4 (2.5 μL) and 1 (5.0 μL).
25 μL of aminoguanidine.
25 μL of sodium ascorbate.
Close the tube (to prevent more oxygen from diffusing in), mix by inverting the tube several times, or attach to a slow rotisserie (approx. 30 rotations per minute). Allow the reaction to proceed for an hour.
Workup depends on your application. Copper ions can be removed by washing or performing dialysis with solutions of buffered ethylenediamine tetraacetic acid (EDTA). The addition of an excess of EDTA relative to Cu also serves to stop the reaction when it is not desirable to simply expose the mixture to air and allow any remaining reducing agent to be used up (thus generating reactive oxygen species in the process, as described above). Copper-adsorbing resins such as Cuprisorb® are useful in cases of preparative organic synthesis, but tend to bind biomolecules and thus be of lesser value for bioconjugation. Thus, in such cases the conjugates are purified directly after the reaction in such a way as to leave small molecules behind.
Support protocol: determining the efficiency of bioconjugation CuAAC with a fluorogenic probe
Introduction
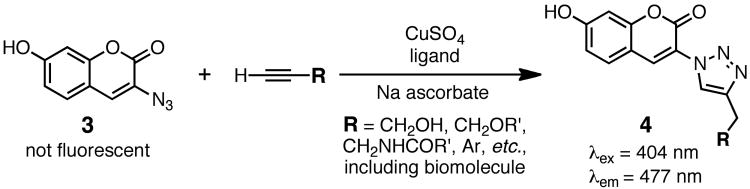
It is often helpful to test the reactivity of your desired biomolecule-alkyne in a way that allows an easy readout. For this purpose the fluorogenic coumarin azide 3 of Wang and coworkers is usually employed (Figure 2).(Sivakumar et al., 2004) A convenient assessment can be made of CuAAC efficiency under a particular set of conditions by first reacting 3 with an excess of a small-molecule “model” alkyne such as propargyl alcohol or phenylacetylene to ensure completion of the click reaction. The resulting solution is then diluted to the same concentration at which the biomolecule alkyne will be used. (For a better control, also include the biomolecule without its alkyne appendage to provide a good mimic of the bioconjugation environment.) The fluorescence of that solution can be used to define 100% reaction. The CuAAC reaction on the desired biomolecule-alkyne can then be performed and the fluorescence intensity used directly to estimate the progress of the reaction. The assumption that the fluorescence wavelength and intensity of triazole 4 will not be much changed when attached to the biomolecule is usually a reasonable one, as has been demonstrated previously.(Hong et al., 2009)
Figure 2.
CuAAC reaction with Wang's fluorogenic azide 3, used to investigate reaction conditions before using the expensive biomolecule or cargo reagent(s).
The example here describes a representative procedure for optimizing CuAAC conditions for a protein (“X”) that has been decorated with an aliphatic terminal alkyne group, designated “X-alkyne,” to be used at a concentration of 25 μM (1 mg/mL of a 40-kDa protein bearing one alkyne). Compound 3 serves as a surrogate for the cargo-azide, to be used at a concentration of 50 μM (two equivalents with respect to alkyne). The buffer used here should be the same buffer that will be used in the desired conjugation process.
Step 1: Establishing an approximate fluorescent quantitation standard
Materials and stock solutions
The materials used here are identical to those above, except for the “cargo-azide,” which is compound 3. This molecule is available from Glen Research (www.glenresearch.com, catalog number 50-2004-92), or it can also be made by the published procedure.(Sivakumar et al., 2004) The solid or DMSO stock solution of this compound should be stored in a refrigerator and protected from light.
Final concentrations
CuSO4: 0.25 mM
Ligand 1: 1.25 mM (ligand to copper ratio is 5:1)
Sodium ascorbate: 5 mM
Aminoguanidine: 5 mM
Propargyl alcohol (as the model alkyne): 500 μM
Azide 3: 100 μM
Performing the CuAAC reaction
In a 2 mL Eppendorf tube, combine the reagents in the following order:
Propargyl alcohol + buffer to make 446.2 μL of solution that is 560 μM in alkyne.
10 μL of azide 3 stock solution (5 mM)
A premixed solution of CuSO4 (6.3 μL of 20 mM stock) and 1 (12.5 μL of 50 mM stock).
25 μL of sodium ascorbate (100 mM stock).
Close the tube (to prevent more oxygen from diffusing in), mix by inverting the tube several times, or attach to a slow rotisserie (approx. 30 rotations per minute). Allow the reaction to proceed for an hour.
Dilute the reaction mixture by a factor of 4 with buffer to obtain a solution approximately 25 μM in triazole 4. In this dilution step, include protein X if possible (or a surrogate protein such as bovine serum albumin) so that the final concentration of protein in the diluted solution is also 25 μM. Read the fluorescence intensity at 477 nM (excitation wavelength 404 nm), which should be substantially greater than that of a 25 μM solution of 3.
Note: Routine users of the CuAAC bioconjugation reaction may wish to simply make a supply of triazoles 4 using propargyl alcohol and phenylacetylene, to act as standards for reactions involving aliphatic and aromatic biomolecule-alkynes, respectively. These pure compounds can then be diluted to the appropriate concentration in the buffer/protein mixtures of interest to establish an approximate expected fluorescence intensity readout of a complete click reaction under desired conditions.
Step 2: Determining CuAAC efficiency
Materials
Identical to those above.
Final concentrations
CuSO4: 0.10 mM (Note: can be adjusted as desired between 50 and 250 μM)
Ligand 1: 0.50 mM (ligand to copper ratio is 5:1)
Sodium ascorbate: 5 mM
Aminoguanidine: 5 mM
Biomolecule-alkyne: 25 μM
Azide 3: 50 μM
Performing the CuAAC reaction
Biomolecule-alkyne + buffer to make 437.5 μL of solution that is 28.6 μM in alkyne.
5 μL of azide 3
A premixed solution of CuSO4 (2.5 μL) and 1 (5.0 μL).
25 μL of aminoguanidine.
25 μL of sodium ascorbate.
Close the tube (to prevent more oxygen from diffusing in), mix by inverting the tube several times, or attach to a slow rotisserie (approx. 30 rotations per minute). Allow the reaction to proceed for an hour.
Read the fluorescence intensity of the mixture at 477 nM (excitation wavelength 404 nm) and compare to the intensity observed for the model reaction as described above. The observation of substantially lower fluorescence suggests the need for adjustment of the reaction conditions.
Commentary
Background Information
The marriage of azides and alkynes in biological molecules has emerged from an appreciation of the insights made possible by the application of earlier bioorthogonal reactions in chemical biology.(Lemieux and Bertozzi, 1998; Prescher and Bertozzi, 2005) A bioorthogonal reaction is performed in the presence of, or with the participation of, biological molecules, in which the molecular components of the native structures do not participate. For example, Bertozzi's use of the Staudinger ligation to attach carbohydrate molecules to proteins helped popularize the use of azides in this approach, the application being notable for the fact that efficient connections were achieved without engaging the many hydroxyl, carboxylic acid, amine, indole, imidazole, thioether, and even thiol groups present on one or both of the participating partners.(Hang and Bertozzi, 2001)
Azides and alkynes are particularly useful because they are small and unobtrusive. Lacking the ability to engage in strong hydrogen bonding, acid-base, hydrophobic, coulombic, dipolar, or π-stacking interactions, they are unlikely to perturb the biological molecules to which they are attached, as long as their density of presentation on such scaffolds is not overwhelmingly great. An outstanding example of their invisible nature is provided by the growing number of examples in which azide- or alkyne-derivatized nutrients or cofactors are taken up and incorporated into biological molecules by living cells.(Kiick et al., 2002; Prescher et al., 2004; Laughlin et al., 2006; Baskin and Bertozzi, 2007; Gierlich et al., 2007; Hsu et al., 2007; Stabler et al., 2007; Wirges et al., 2007; Ning et al., 2008; Chang et al., 2009; Banerjee et al., 2010b; Breidenbach et al., 2010; Rangan et al., 2010) While exquisitely selective, the uncatalyzed 1,3-dipolar cycloaddition reaction of standard azides and alkynes is quite slow unless the alkyne is rendered sufficiently electron-deficient to open up conjugate addition pathways and thus compromise its bioorthogonality.(Huisgen, 1962; Huisgen, 1989) Copper catalysis of the reaction between azides and terminal alkynes represents one solution to this problem, first described independently in 2002 by the groups of Meldal in Denmark (in the context of solid-phase peptide modification)(Tornøe et al., 2002) and Sharpless/Fokin in the U.S. (in the context of solution-phase reactivity).(Rostovtsev et al., 2002) This reaction has come to represent “click chemistry” for many, although other reactions are also included in this concept.(Kolb et al., 2001) Another general solution to the azide-alkyne rate problem is to make the alkyne highly strained in a ring structure, and is sometimes referred to as “copper-free click chemistry.” This ligation method is not discussed here, but the reader should be aware that recent developments from the Bertozzi laboratory(Jewett et al., 2010) have made this process fast enough to be applicable to most bioconjugation conditions. In such situations, the major practical differences between the two triazole-forming reactions are the size of the participating alkyne group, favoring the CuAAC process which uses only a small and easily-installed terminal alkyne group, vs. the need for a catalyst, which of course only the CuAAC reaction requires.
Critical Parameters
We have found in reviewing the literature and discussing click chemistry with others that several practices of questionable value are sometimes employed in the execution of the CuAAC reaction; several are listed below in approximate order of frequency. Note that these practices are not necessarily poisonous: the reaction usually works anyway, just not at optimal rates.
The use of strong base in nonaqueous conditions. Hünig's base (iPr2NEt) is often used and yet is unnecessary and likely to diminish cycloaddition rates. The CuAAC reaction involves both the deprotonation of alkyne and reprotonation of Cu-triazolyl intermediate. While alkynes are not very acidic, there is almost never a need to accelerate Cu-acetylide formation with base, since this is already a very fast process.
Failure to use an appropriate accelerating ligand. One of the reasons for the popularity of the CuAAC reaction is its permissive nature: a wide variety of functional groups in substrates and solvates are tolerated, and ligands are often not required. However, when higher temperatures cannot be used, or rate accelerations beyond those that can be achieved by heating are required, accelerating ligands can solve may problems, since the CuAAC reaction is strongly aided by the correct coordination environment.
Failure to use rudimentary methods to protect reactions from oxygen. Even when excess sodium ascorbate is present to maintain a sufficient concentration of cuprous ions, it is useful to at least cap reactions to minimize oxygen exposure. Otherwise, copper will catalyze the oxidation of ascorbate, eventually depleting the reducing agent, killing the CuAAC catalyst, promoting CuII-mediated alkyne-alkyne (Glaser) coupling, and generating larger amounts of reactive oxygen spcies than is necessary. While many CuAAC reactions are fast enough to withstand these challenges, why take the chance? Safety note: This suggestion refers to reactions involving small amounts or low concentrations of azide and alkyne reagents, as are present in most bioconjugation situations. Capping CuAAC reactions involving high concentrations of reagents can be dangerous, as the reaction releases approximately 50 kcal/mol of energy.
Unrealistic expectations. Even a reaction as fast and reliable as CuAAC cannot connect two reactants to each other in a few hours if both are present in low nanomolar concentrations. In such situations, either the concentration of one of the reaction partners must be increased, or the two partners must be engineered to interact with each other in a preorganized fashion, such as the binding of two complementary oligonucleotide chains. Note also in this protocol, and in most CuAAC bioconjugation cases, the copper complex is not used a catalyst, but rather is present in stoichiometric or excess amounts. This is because rate is dependent on copper concentration in a non-obvious way: for most catalysts of the type described here, a threshold behavior has been observed, such that little reactivity occurs below 50 μM in Cu, and maximal activity is reached at approximately 250 μM Cu.(Rodionov et al., 2005; Presolski et al., 2010)
The use of cuprous iodide as a copper source. Iodide ions are good ligands for CuI, and can either interfere with Cu-acetylide formation or divert the complexes into unproductive aggregates. This is not to say that CuI cannot be used, but it is not recommended when maximal rates are desired.
The use of TCEP as reducing agent. While we reported this procedure early on,(Wang et al., 2003) we later found that the Cu-binding and azide-reducing properties of phosphines can interfere with the CuAAC reaction. When ascorbate cannot be used, hydroxylamine can function as a reducing agent for CuII, or Cu wire can be used to protect a pre-formed CuI complex if the reaction mixture is protected from excessive exposure to oxygen.
-
Use of non-optimal buffers or solvent mixtures. With the catalyst described here, the reaction has been shown to proceed well over a broad pH range;(Presolski et al., 2010) pH 7 or thereabouts is recommended for most cases. Buffers that contain very high concentrations (greater than approximately 0.2 M) of chloride ion are to be avoided because chloride can compete for Cu at these concentrations. For the same reason of Cu binding, Tris buffers can slow CuAAC reactions. Cu-phosphate complexes are often insoluble, but if the Cu source is pre-mixed with the ligand, such complexes do not form (or do not precipitate) even in phosphate-based buffers, and reaction rates are high. Phosphate, acetate, HEPES, or MOPS buffers are commonly employed, but others are almost certainly suitable as long as they do not contain Cu-binding species.
Similarly, the role of DMSO and other coordinating co-solvents is an important one to appreciate. See reference (Presolski et al., 2010) for a complete account, the overall lesson of which is that when high concentrations of such co-solvents are required (greater than approximately 30-50% of the solvent volume), a different ligand is suggested.
Failure to turn up the heat. The CuAAC process benefits greatly from higher temperatures; even modest increases that are tolerated by some biological molecules can bring about good results. We speculate that many cases in which CuAAC bioconjugation does not work well suffer from the sequestering of the metal by competing coordinating species in solution, such as donor solvent molecules or donor groups in protein or other species present.(Presolski et al., 2010) In such situations, the great kinetic lability of CuI centers may be compromised, and a little heat may be all that is needed to shake the Cu ions loose to do their job.
Safety notes. While happily very rare, especially in bioconjugation situations for which the amounts of azides used tend to be small and the molecules to which they are attached are large, hazardous practices with azides should be mentioned here. Of greatest concern is the potential for the generation of hydroazidoic acid (HN3) during the synthesis of organic azides, generally from an electrophile and sodium azide. When working up such reactions that contain excess inorganic azide, exposure to acid will generate HN3, which is volatile, highly toxic, and explosive. In general, therefore, such workups should avoid acid; in large amounts azide ion should be quenched by nitration.(Clusius and Effenberger, 1955; Stedman, 1960) In a fume hood, treat a stirred aqueous solution containing no more than 5% azide ion with sodium nitrite (approximately 7 mL of 20% aqueous NaNO2 per gram NaN3, representing a 40% excess). Aqueous sulfuric acid (20% solution) is then added slowly with stirring until the reaction mixture is acidic. Note that it is important for sodium nitrite to be introduced first. When the evolution of nitrogen oxides (toxic – keep this reaction in the hood) ceases, the acidic solution is tested with starch iodide paper. If it turns blue, excess nitrite is present, indicating complete azide decomposition.
Although not generally used with biomolecules, chlorinated solvents (particularly CH2Cl2 and CHCl3) should be avoided with azide ion, since it is possible to generate CH2(N3)2 and CH(N3)3, which can be highly explosive when concentrated, such as in the trap of a vacuum line. Similarly, small molecule-azides should never be isolated away from solvent in significant quantities, such as by distillation, precipitation, or recrystallization.
Troubleshooting
Troubleshooting suggestions are presented in Table 1, in the form of problems presented to us by users of the CuAAC reaction over the past several years.
Table 1.
Troubleshooting guide for bioconjugation by the CuAAC click reaction.
| Problem | Cause | Solution |
|---|---|---|
| A dextran-alkyne at 25 mM would not click in water, even when accelerating ligands were used. | Collapse of hydrophobic regions, burying the alkynes and making them inaccessible; similar challenges can occur with proteins and oligonucleotides | (a) Perform the reaction in denaturing or solvating conditions such as the use of lots of DMSO, along with appropriate ligand (ligand 7 in (Presolski et al., 2010)). |
| Low yields in complex biological systems, such as with the product resulting from enzymatic incorporation of an alkyne into RNA | Potential failure of one or more steps preceding the CuAAC step. | Perform a test reaction with coumarin azide 3. If little or no reaction is observed, add propargyl alcohol. Continued failure of the reaction suggests that the biological substrate is sequestering the copper catalyst, in which case excess Cu can be added. It is also possible to release active Cu by the addition of Zn2+. |
| Substrates are incompatible with ascorbate (such as showing excessive sensitivity to reactive oxygen species or dehydroascorbate byproduct formation, not solved by the recommended use of excess ligand and aminoguanidine) | Oxidation, binding, etc.? | (a) Use hydroxylamine as reducing agent for CuII (typically 10 mM). (b) Use a CuI complex such as CuBr, CuOAc, or CuOTf, and protect the reaction from air. (c) Generate CuI electrochemically.(Hong et al., 2008) |
| Invitrogen Click-It kit doesn't work | Various | Use the procedure outlined in this protocol |
| Failure of this protocol involving protein, DNA, RNA, gold nanoparticles, or other species that can bind Cu ions | Sequestration of Cu away from azide and alkyne reactants | (a) Use excess Cu and ligand. (b) Add ZnII as a sacrificial metal to whatever is removing Cu from the reaction. (c) Use alternative ligand as previously described,(Presolski et al., 2010) in the proper ligand:Cu ratio of 1:1 or 2:1. |
| CuAAC doesn't work in fresh cell lysate or under other conditions that may contain free thiols | Strong Cu-thiolate binding ties up the metal. (CuAAC with THPTA usually tolerates glutathione up to 1 mM, but not more than that.) | Using an accelerating ligand will help, and excess Cu, ZnII or NiII can often occupy the thiols and leave some CuI free to mediate the CuAAC reaction. |
| Failure of His6-tagged proteins to engage in CuAAC ligation | the His-tag binds copper | Use excess copper or sacrificial metals, such as ZnII or NiII; may also change to a FLAG or other peptide tag. |
| DNA damage observed, sometimes more seriously when the reaction mix is vortexed. | Oxidation from excess reactive oxygen species (and, in one case, the use of cupric nitrate) | Use CuSO4 + ascorbate, but minimize agitation of the solution and keep it capped as much as possible. In extremely sensitive cases, CuOAc with Cu wire provides the mildest conditions |
| Reactions are slow with CuBr, CuI, TCEP | Low solubility and bad reducing agent | Use CuSO4, sodium ascorbate, and DMSO, DMF, or NMP as co-solvent (up to 10%). These solvents appear to be biocompatible and help to dissolve most small molecules of interest. |
Anticipated Results
We have used this CuAAC protocol for the attachment of a wide variety of ligands (small molecules,(Astronomo et al., 2010; Hong et al., 2010; Pokorski et al., 2011) proteins,(Banerjee et al., 2010a) DNA,(Cigler et al., 2010) and organic polymers(Manzenrieder et al., 2011)) to a wide variety of biological molecules, such as the surfaces of virus-like particles. In many cases, the attachments result in polyvalent displays of triazoles on the biomolecular scaffold, and so yield can be difficult to quantify. However, when yields can be measured, CuAAC reactions usually give quantitative or near-quantitative yields, with excellent recovery of the desired conjugates, showing that the reaction conditions do not induce substantial cleavage of biological molecules.
Time Considerations
In general, the CuAAC reaction is easy to set up and perform. When the reactants are present in sufficient concentration (greater than 10 μM each), the reaction is performed properly, and no unexpected sequestration of copper ions takes place, the CuAAC reaction can be expected to provide quantitative yields of triazoles within an hour or two at room temperature. The time required for workup and purification varies from minutes for simple precipitation or molecular weight cutoff filtration to separate biomolecules from the catalyst and small-molecule reagents, to hours for chromatographic techniques.
Acknowledgments
This work was supported by The Skaggs Institute for Chemical Biology and the NIH (RR021886). We are especially grateful to the discoverer of the solution-phase CuAAC reaction, Prof. Valery V. Fokin, for his insights and contributions, and we thank the many coworkers and colleagues who have contributed to our understanding of the reaction. These include Prof. K. Barry Sharpless, Dr. Valentin Rodionov, Dr. Reshma Jagasia, Dr. Warren Lewis, Dr. Andrew Udit, Dr. Yeon-Hee Lim, Dr. So-Hye Cho, Dr. David Díaz, and Dr. Sayam Sen Gupta.
Literature Cited
- Astronomo RD, Kaltgrad E, Udit AK, Wang SK, Doores KJ, Huang CY, Pantophlet R, Paulson JC, Wong CH, Finn MG, Burton DR. Defining Criteria for Oligomannose Immunogens for HIV Using Icosahedral Virus Capsid Scaffolds. Chem Biol. 2010;17:357–370. doi: 10.1016/j.chembiol.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Liu A, Voss N, Schmid S, Finn MG. Multivalent Display and Receptor-Mediated Endocytosis of Transferrin on Virus-Like Particles. Chembiochem. 2010a;11:1273–1279. doi: 10.1002/cbic.201000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PS, Ostapchuk P, Hearing P, Carrico I. Chemoselective Attachment of Small Molecule Effector Functionality to Human Adenoviruses Facilitates Gene Delivery to Cancer Cells. J Am Chem Soc. 2010b;132:13615–13617. doi: 10.1021/ja104547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Bertozzi CR. Bioorthogonal Click Chemistry: Covalent Labeling in Living Systems. QSAR Comb Sci. 2007;26:1211–1219. [Google Scholar]
- Breidenbach MA, Gallagher JEG, King DS, Smart BP, Wu P, Bertozzi CR. Targeted metabolic labeling of yeast N-glycans with unnatural sugars. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0911247107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BR, Dann SE, Heaney H. Experimental Evidence for the Involvement of Dinuclear Alkynylcopper(I) Complexes in Alkyne-Azide Chemistry. Chem Eur J. 2010;16:6278–6284. doi: 10.1002/chem.201000447. [DOI] [PubMed] [Google Scholar]
- Chan TR, Fokin VV. Polymer-Supported Copper(I) Catalysts for the Experimentally Simplified Azide-Alkyne Cycloaddition. QSAR Comb Sci. 2007;26:1274–1279. [Google Scholar]
- Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as Copper(I)-Stabilizing Ligands in Catalysis. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- Chang P, Chen X, Smyrniotis C, Hu T, Bertozzi CR, Wu P. Metabolic Labeling of Sialic Acids in Living Animals with Alkynyl Sugars. Angew Chem Int Ed. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigler P, Lytton-Jean AKR, Anderson DG, Finn MG, Park SY. DNA-controlled assembly of a NaTl lattice structure from gold nanoparticles and protein nanoparticles. Nat Mater. 2010;9:918–922. doi: 10.1038/nmat2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clusius K, Effenberger E. Reaktionen mit N-15. 21. Die Einwirkung von Nitrit auf Stickstoffwasserstoffsaure. Helv Chim Acta. 1955;38:1843–1847. [Google Scholar]
- Gierlich J, Gutsmiedl K, Gramlich PME, Schmidt A, Burley GA, Carell T. Synthesis of Highly Modified DNA by a Combination of PCR with Alkyne-Bearing Triphosphates and Click Chemistry. Chem Eur J. 2007;13:9486–9494. doi: 10.1002/chem.200700502. [DOI] [PubMed] [Google Scholar]
- Hang HC, Bertozzi CR. Chemoselective Approaches to Glycoprotein Assembly. Accounts Chem Res. 2001;34:727–736. doi: 10.1021/ar9901570. [DOI] [PubMed] [Google Scholar]
- Himo F, Lovell T, Hilgraf R, Rostovtsev VV, Fokin VV, Noodleman L, Sharpless KB. Copper(I)-Catalyzed Synthesis of Azoles. DFT Predicts Unprecedented Reactivity and Intermediates. J Am Chem Soc. 2005;127:210–216. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- Hong V, Presolski SI, Ma C, Finn MG. Analysis and Optimization of Copper-Catalyzed Azide-Alkyne Cycloaddition for Bioconjugation. Angew Chem, Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong V, Steinmetz NF, Manchester M, Finn MG. Labeling Live Cells by Copper-Catalyzed Alkyne-Azide Click Chemistry. Bioconjugate Chem. 2010;21:1912–1916. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong V, Udit AK, Evans RA, Finn MG. Electrochemically Protected Copper(I)-Catalyzed Azide-Alkyne Cycloaddition. Chem Bio Chem. 2008;9:1481–1486. doi: 10.1002/cbic.200700768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci USA. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisgen R. 1,3-Dipolar Cycloadditions Past and Future. Angew Chem, Int Ed Engl. 1962;2:565–632. [Google Scholar]
- Huisgen R. Kinetics and Reaction Mechanisms: Selected Examples from the Experience of Forty Years. Pure Appl Chem. 1989;61:613–628. [Google Scholar]
- Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J Am Chem Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of Azides into Recombinant Proteins for Chemoselective Modification by the Staudinger Ligation. Proc Natl Acad Sci USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Agard NJ, Baskin JM, Carrico IS, Chang PV, Ganguli AS, Hangauer MJ, Lo A, Prescher JA, Bertozzi CR. Metabolic labeling of glycans with azido sugars for visualization and glycoproteomics. Glycobiology. 2006;415:230–250. doi: 10.1016/S0076-6879(06)15015-6. [DOI] [PubMed] [Google Scholar]
- Lemieux GA, Bertozzi CR. Chemoselective Ligation Reactions with Proteins, Oligosaccharides and Cells. Trends Biotechnol. 1998;16:506–513. doi: 10.1016/s0167-7799(98)01230-x. [DOI] [PubMed] [Google Scholar]
- Lewis WG, Magallon FG, Fokin VV, Finn MG. Discovery and Characterization of Catalysts for Azide-Alkyne Cycloaddition by Fluorescence Quenching. J Am Chem Soc. 2004;126:9152–9153. doi: 10.1021/ja048425z. [DOI] [PubMed] [Google Scholar]
- Lipshutz BH, Frieman BA, Tomaso AE., J Copper-in-Charcoal (Cu/C): Heterogeneous, Copper-Catalyzed Asymmetric Hydrosilylations. Angew Chem, Int Ed. 2006;45:1259–1264. doi: 10.1002/anie.200503149. [DOI] [PubMed] [Google Scholar]
- Manzenrieder F, Luxenhofer R, Retzlaff M, Jordan R, Finn MG. Stabilization of Virus-Like Particles with Poly(2-oxazoline)s. Angew Chem Int Ed. 2011;50:2601–2605. doi: 10.1002/anie.201006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing Metabolically Labeled Glycoconjugates of Living Cells by Copper-Free and Fast Huisgen Cycloadditions. Angew Chem, Int Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorski JK, Breitenkamp K, Liepold LO, Qazi S, Finn MG. Functional Virus-Based Polymer-Protein Nanoparticles by Atom Transfer Radical Polymerization. J Am Chem Soc. 2011;133:9242–9245. doi: 10.1021/ja203286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Bertozzi CR. Chemistry in Living Systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- Presolski SI, Hong V, Cho SH, Finn MG. Tailored Ligand Acceleration of the Cu-Catalyzed Azide-Alkyne Cycloaddition Reaction: Practical and Mechanistic Implications. J Am Chem Soc. 2010;132:14570–14576. doi: 10.1021/ja105743g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan KJ, Yang YY, Charron G, Hang HC. Rapid visualization and large-scale profiling of bacterial lipoproteins with chemical reporters. J Am Chem Soc. 2010;132:10628–10629. doi: 10.1021/ja101387b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VO, Fokin VV, Finn MG. Mechanism of the Ligand-Free CuI-Catalyzed Azide-Alkyne Cycloaddition Reaction. Angew Chem, Int Ed. 2005;44:2210–2215. doi: 10.1002/anie.200461496. [DOI] [PubMed] [Google Scholar]
- Rodionov VO, Presolski S, Díaz DD, Fokin VV, Finn MG. Ligand-Accelerated Cu-Catalyzed Azide-Alkyne Cycloaddition: a Mechanistic Report. J Am Chem Soc. 2007a;129:12705–12712. doi: 10.1021/ja072679d. [DOI] [PubMed] [Google Scholar]
- Rodionov VO, Presolski S, Gardinier S, Lim YH, Finn MG. Benzimidazole and Related Ligands for Cu-Catalyzed Azide-Alkyne Cycloaddition. J Am Chem Soc. 2007b;129:12696–12704. doi: 10.1021/ja072678l. [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sen Gupta S, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Accelerated Bioorthogonal Conjugation: A Practical Method for the Ligation of Diverse Functional Molecules to a Polyvalent Virus Scaffold. Bioconjugate Chem. 2005;16:1572–1579. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. A Fluorogenic 1,3-Dipolar Cycloaddition Reaction of 3-Azidocoumarins and Acetylenes. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan A, Levy M, Liu Y, Marlow FL, Wu P. Biocompatible copper(I) catalysts for in vivo imaging of glycans. J Am Chem Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler CL, Sun XL, Cui X, Wilson JT, Haller CA, Chaikof EL. Chemo- and Bioorthogonal Surface Re-Engineering of Pancreatic Islets. J Am Chem Soc. 2007 doi: 10.1021/bc7002814. submitted. [DOI] [PubMed] [Google Scholar]
- Stedman G. Mechanism of the Azide-Nitrite Reaction. Part IV. J Chem Soc. 1960:1702–1709. and references therein. [Google Scholar]
- Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. joc. 2002;67:3057–3062. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by Copper(I)-Catalyzed Azide-Alkyne [3+2] Cycloaddition. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- Wirges CT, Gramlich PME, Gutsmiedl K, Gierlich J, Burley GA, Carell T. Pronounced Effect of DNA Hybridization on Click Reaction Efficiency. QSAR Comb Sci. 2007;26:1159–1164. [Google Scholar]