Abstract
Protein conformational switches alter their shape upon receiving an input signal, such as ligand binding, chemical modification, or change in environment. The apparent simplicity of this transformation—which can be carried out by a molecule as small as a thousand atoms or so—belies its critical importance to the life of the cell as well as its capacity for engineering by humans. In the realm of molecular switches, proteins are unique because they are capable of performing a variety of biological functions. Switchable proteins are therefore of high interest to the fields of biology, bio-technology, and medicine. These molecules are beginning to be exploited as the core machinery behind a new generation of biosensors, functionally regulated enzymes, and “smart” biomaterials that react to their surroundings. As inspirations for these designs, researchers continue to analyze existing examples of allosteric proteins. Recent years have also witnessed the development of new methodologies for introducing conformational change into proteins that previously had none. Herein we review examples of both natural and engineered protein switches in the context of four basic modes of conformational change: rigid-body domain movement, limited structural rearrangement, global fold switching, and folding–unfolding. Our purpose is to highlight examples that can potentially serve as platforms for the design of custom switches. Accordingly, we focus on inducible conformational changes that are substantial enough to produce a functional response (e.g., in a second protein to which it is fused), yet are relatively simple, structurally well-characterized, and amenable to protein engineering efforts.
Keywords: allostery, biosensors, protein design, protein engineering, protein folding
Introduction
Protein conformational switches are polypeptides that undergo a significant change in structure upon receiving an input signal. Input stimuli can consist of covalent modifications, such as phosphorylation or cleavage of the polypeptide backbone, or more commonly, molecular recognition events. Examples of the last type include absorbing a photon, binding a drug, or engaging an entire cell via a surface receptor. The resulting conformational change establishes the output response. For instance, the structural change may modulate enzymatic activity or expose new surface for the protein to interact with other molecules. Many proteins bind other molecules and/or become covalently modified; however, it is the property of stimulus-induced structural change that defines the more select cohort of conformational switches.
Switchable proteins are of special interest for two reasons. First, they are of fundamental importance to biology. They can regulate cellular signaling pathways, including those involved in growth control, development, sensory signal mediation, and pathogen infection. For example, the β2 adrenergic receptor (β2AR) is a member of the large family of G protein coupled receptors (GPCRs) which respond to a variety of signals, such as light, odorants, hormones, and neurotransmitters. When an agonist binds to the extracellular face of β2AR, two of the seven membrane-spanning α-helices are displaced toward the cytoplasm by as much as 14Å.[1] This movement generates a binding site at the cytoplasmic face of β2AR for the G protein heterotrimer (Gsαβγ). Interaction with β2AR causes Gsαβγ to exchange its bound GDP for GTP then dissociate into Gα-GTP and Gβγ subunits. Gα-GTP and Gβγ then go on to bind and activate their downstream targets.
Another class of functionally important switches is the viral protein “harpoons” that mediate fusion of the viral membrane with that of the host. Influenza haemagglutinin (HA) is the protein responsible for anchoring the virus to the cellular receptor. Once the docked virus is endocytosed into the cell, the pH inside the endosome decreases and a proton binding event triggers a dramatic conformational change in HA.[2] The three lobes of HA splay apart to reveal a buried triple-stranded coiled-coil stalk. A loop-to-helix folding transition causes the coiled-coil to extend and flip outwards, thrusting the hydrophobic fusion peptide (which resides at the tip of the newly lengthened stalk) into the vacuolar membrane. In this conformation HAs are proposed to distort and ultimately bridge the viral and vacuolar membranes, although details remain unclear. Similar conformational changes are thought to occur in gp41 (HIV),[3] spike protein (SARS),[4] GP2 (Ebola virus),[5] and functionally homologous proteins from other viruses. The triggering conditions differ (decrease in pH, binding to receptor, proteolytic cleavage, redox change), but the basic harpooning action is likely to be conserved.
The second reason why switchable proteins are of interest is that their switching mechanisms, once understood, can be modified by the tools of protein engineering to create molecules with new functionalities. Research in this area is largely motivated by two related applications: functional regulation and biosensing. The goal of functional regulation is to be able to turn on, turn off, or alter the biological function of a target protein by application of a desired stimulus. By imbuing such regulatory capability into, for example, transcription factors or signaling proteins, it might be possible to program developmental pathways or rewire cellular signaling circuitry. Biosensors operate on a similar principle, but the emphasis is placed on detecting the stimulus. Accordingly, the conformational change itself is of primary interest. It is designed to transduce the input event, which typically takes place on a recognition domain, to a reporter element for detection. The reporter can be an electronic device or a separate biological element, such as a fluorescent protein or enzyme. Use of protein switches allows one to achieve this communication in a number of creative ways, including by integrating recognition and reporting functions in a single molecule.
As in many areas of engineering, our attempts to build switches often draw heavily from nature’s designs. Several studies of structures in the Protein Data Bank (PDB) have surveyed the types of structural changes that occur in response to external perturbation.[6] These changes can be grouped into the following broad and sometimes overlapping categories: 1) rigid-body domain movement (most commonly hinge or shear-type motions), 2) limited structural rearrangement (in which a segment of the structure moves or becomes ordered/disordered), and 3) large-scale structural rearrangement in which the protein can switch global folds. A fourth category, binding-induced folding of intrinsically disordered proteins (IDPs), is now recognized as a switching mechanism used by many regulatory and signaling proteins.[7] IDPs illustrate that changes in structure and dynamics often go hand-in-hand, but this is not always the case. NMR studies of retroviral matrix proteins,[8] PDZ domain,[9] eglin c,[10] and others[11] have shown that ligand binding can decrease or even increase dynamic motions at the active site as well as at distant locations, in the absence of significant structural changes. The same phenomenon has been observed as a result of post-translational modifications, such as phosphorylation.[12] These proteins represent a class of “entropic switches” the potential of which for design remains untested.
Herein we review current efforts to develop protein conformational switches. The number of existing switches and design strategies is too large to discuss in a single treatise; the reader is referred to other articles for additional perspectives.[13] Moreover, the size and complexity of natural switches such as β2AR and HA (notwithstanding the fact that they are membrane bound) restricts their practical use as templates for design. At the opposite end of the spectrum are proteins that undergo functional switching with only minor changes in conformation. The engineering potential of these molecules is limited as well. For instance, EF-hand domains mediate calcium binding in a large family of signaling and transport proteins. The simplest family members, exemplified by calbindin D9k, contain two EF-hands that bind Ca2+ cooperatively. The binding mechanism involves allosteric coupling of conformational substates,[14] but the difference between the bound and free structures of calbindin D9k is too small to be useful. The conformational change must first be amplified before the EF-hand can serve as a calcium-triggered functional switch. Natural as well as artificial means for doing so are discussed in the sections below. As a rule of thumb, for a conformational change to be considered exploitable it should involve unfolding, rigid-body reorientation, or otherwise substantial movement of at least one secondary structural element. We therefore focus on switching mechanisms that are simple enough to be amenable to engineering efforts, yet are of sufficient magnitude to warrant such endeavors. We concentrate on some well-characterized cases from each of the four categories of conformational change.
Rigid-Body Domain Movement
Open-to-closed motions are exemplified by rigid-body displacement of two or more domains with respect to each other, often by a hinge-type action. Secondary structure is typically not affected. In a comprehensive study of 839 non-redundant pairs of ligand-free and ligand-bound proteins in the PDB, Amemiya et al.[6e] found that 325 exhibited structural changes that were linked to ligand binding. Of these 325 proteins, half underwent domain and/or open-to-closed movements while the rest displayed local changes (e.g., rearrangement of a surface loop).
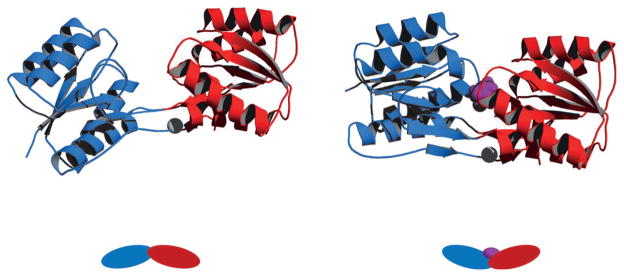
Domain movement is particularly agreeable to biosensor development. The general idea is to attach reporter groups, for example, environment or distance-sensitive fluorophores, at positions that detect the movement. Periplasmic binding proteins (PBPs) embody this design. PBPs are a superfamily of nonenzymatic receptors the members of which are found in all kingdoms of life. They are versatile scaffolds for bio-sensor development, because they bind a variety of small molecules with high specificity and dissociation constants (Kd) in the nM to μM range.[15] Substrates for PBPs include ribose,[16] maltose,[17] glucose/galactose,[18] allose,[19] lactate,[20] cellulose,[21] amino acids,[22] vitamin B12,[23] iron siderophores,[24] nickel metallophores,[25] copper,[26] phosphonate,[27] and glutathione.[28] Figure 1 illustrates the “venus flytrap” action of E. coli ribose binding protein (RBP). The structure consists of N- and C-terminal lobes that are predominantly in an open conformation in the ligand-free state (Figure 1). The substrate binding pocket is located in the hinge region between the domains. Ribose binding causes the two domains to close by about 30°, clamping down on the substrate and almost completely occluding it from the solvent.
Figure 1.
Rigid-body domain movement exemplified by E. coli RBP in its open (ribose-free; left) and closed (ribose-bound; right) states.[122] Amino and carboxy domains are colored blue and red, respectively, and ribose is shown in purple. Residue 265, located in the hinge region and a site of environmentally sensitive fluorophore attachment, is depicted as a grey sphere. PDB codes are 1URP (left) and 2DRI (right).
Establishing the conformational change goes only half way toward constructing a functional switch. In biosensor applications it is necessary to couple the change to a detection event. Fluorescence is often the method of choice as it offers sensitive and potentially ratiometric output. Intrinsic protein fluorescence is seldom useful, because tryptophan and tyrosine absorb and emit in the crowded ultraviolet range, have relatively low quantum yields, photobleach easily, and may not report on the binding event in their native locations. Thus, two general approaches have emerged for engendering a conformation-dependent fluorescence response. The first is to introduce a chemical fluorophore at a site at which the environment is perturbed by the domain movement. Although choosing dyes and locations can be guided by computational approaches (and structural intuition), it is still an empirical exercise. To illustrate, De Lorimer et al. placed eight different dyes at a total of 67 positions among ten PBPs.[29] Of these, 24 of the locations resulted in a robust change in intensity and/or a ratiometric response, and only one or a few of the dyes could produce that result at a given site. A more recent study of phosphonate binding protein found that the IANBD fluorophore was able to report on domain closure when placed at two of 31 positions. In RBP, one of the best combinations was found to be 4-fluoro-7-aminosulfonylbenzofurazan attached to Cys265,[16d] which is located in the hinge region (Figure 1).
The second strategy for visualizing conformational change is to employ donor and acceptor groups that give rise to distance-dependent fluorescence phenomena such as quenching, excimer formation, and Förster resonance energy transfer (FRET). The first two processes require that donor and acceptor come into contact, whereas FRET response is sigmoidal over a relatively large distance centered at the Förster radius (R0). The R0 values of most donor/acceptor pairs (30–80 Å) are comparable to the diameters of small to medium-sized proteins, making FRET suitable for detecting rigid-body domain movements. Additional advantages of FRET are that the output is often ratiometric, and FRET pairs can be genetically encoded in the form of fluorescent proteins (FPs). The disadvantages are that it can be technically difficult to introduce donor and acceptor dyes into the same protein, and the change in donor–acceptor distance must be substantial to be detected by FRET. In the case of PBPs, the location of the N- and C-termini near the tips of the respective domains allows FP donors and acceptors to be appended to ends of the protein in order to detect domain closure, as was demonstrated for RBP,[16a] maltose binding protein (MBP),[30] glucose/galactose binding protein,[31] and phosphonate binding protein.[27] For glucose binding protein, two halves of the protein aequorin can replace the FPs to provide bioluminescent output.[32] Glucose binding brings the two aequorin fragments together which allows them to bind and reconstitute the functional molecule.
The bacterial actin analogue ParM is another example of a protein that undergoes a rigid-body, open-to-closed movement on interacting with its cognate ligands (ADP or GDP).[33] The nucleotide binding site is again located at the interface between two domains, which rotate by about 25° to close down on the substrate. Cysteine was introduced at six locations, and subsequent labeling with a single diethylaminocoumarin dye revealed that the fluorescence of one construct increased by three fold in the presence of ADP.[34] The same researchers subsequently improved their design by introducing two tetramethylrhodamine groups so that they stack and quench each other’s fluorescence when ParM is in the open state.[33b] Domain closure separates the dyes, causing fluorescence to increase by 15-fold.
The Fc region of immunoglobulin E (IgEFc) engages a high-affinity receptor (FcεRI) on the surface of mast cells and basophils. IgEFc is a homodimer of three subdomains: Cε2, Cε3, and Cε4. X-ray structural analysis revealed that IgEFc adopts a crooked conformation in its free state, with (Cε2)2 bending back against (Cε3)2 and (Cε4)2, and that bending increases upon binding to FcεRI.[35] To visualize this change Hunt et al. fused FP donor and acceptor groups to the N- (Cε2) and C-terminus (Cε4) of IgEFc.[36] FRET efficiency was substantial in the absence of receptor and increased significantly in its presence. The IgEFc biosensor is useful for investigating the molecular bases of antigen recognition and allergic response. The complexity of the conformational change, however, suggests that exploiting it as a general sensing mechanism would be challenging.
The cis/trans isomerization about X-Pro peptide bonds can effect conformational switching by altering the direction of the polypeptide chain. The Crk family of adaptor proteins employs Pro isomerism to toggle between open and closed conformational states. Crks mediate formation of a number of protein complexes by binding to specific phospho-Tyr-containing motifs (via SH2 domains) and Pro-rich peptides (via SH3 domains) on target proteins. Cellular Crk consists of an SH2 domain followed by two SH3 domains (SH3N and SH3C). The SH3 domains are connected by a 50 amino acid linker, in which the highly conserved Pro238 abuts SH3C. cis-Pro238 orients the n-src loop so that Pro238, Phe239, and Ile270 interact with SH3N.[37] The resulting intramolecular interaction between SH3N and SH3C inhibits Crk from binding its ligands. When Pro238 isomerizes to trans, the n-src loop rearranges, Phe239 and Ile270 swing toward SH3C, and Crk is free to engage its targets. Similarly, a Pro switch in the SH2 domain of interleukin-2 tyrosine kinase (Itk) remodels the SH2 binding surface, such that it has a greater affinity for phospho-Tyr peptides or the Itk SH3 domain in the trans and cis states, respectively.[38] As a final example, cis/trans isomerization of a single Pro acts as a molecular hinge for gating the 5-hydroxytryptamine type 3 (5-HT3) receptor[39] and probably other members of the Cys-loop family of ion channels.[40] Pro8* lies in the all-helical transmembrane domain of 5-HT3 at the interface with the extracellular domain. It is at the tip of a loop that connects the channel-lining M2 helix to another transmembrane helix. The trans-to-cis isomerization of Pro8* is thought to shift the M2 helix and open the pore. Pro isomerization constitutes a modest conformational rearrangement; it affects the geometry of just one bond. Nevertheless, because Pro8* is situated at the end of an α-helix, and because α-helices are relatively rigid, isomerization establishes a pivoting action that results in substantial movement of the other end of the M2 helix.
With respect to the engineering potential of Pro switches, it is clear that peptide bond isomerization can provide leverage for transducing an input signal to a structural change. Peptidylprolyl cis/trans isomerases (PPIases) including cyclophilins, FK506 binding proteins, and parvulins add further interest to such designs. PPIases can exert control at the level of switching rate and phosphorylation status of the amino acid preceding Pro (the Pin1 PPIase only catalyzes isomerization of phospho-Ser and phospho-Thr prolyl bonds).[41] It is less clear how one can link Pro isomerization to a specific stimulus. For example, the neurotransmitter binding site in 5-HT3 is distant from Pro8* and the mechanism by which binding leads to isomerization is not known. One promising approach is to modify the polypeptide backbone with a photoresponsive chemical group that undergoes cis/trans isomerization upon absorption of light, as we will discuss in the section on limited structural rearrangement below.
Domain motions of a receptor protein are not only useful for biosensing, but also for modulating the function of another protein. To do so requires a physical interaction between the two proteins. Indeed, the principle challenge in such efforts is to determine how to best connect the molecules to maximize interplay of their conformational responses. End-to-end fusion with a long peptide linker is the method of choice when the goal is to completely decouple their functions. Increasing the points of contact, for example, by inserting one protein into the middle of a second protein using short linkers, increases the likelihood that structural or dynamic changes in one domain will be propagated to the other (Figure 2). With that objective in mind the following considerations can be helpful.
Figure 2.
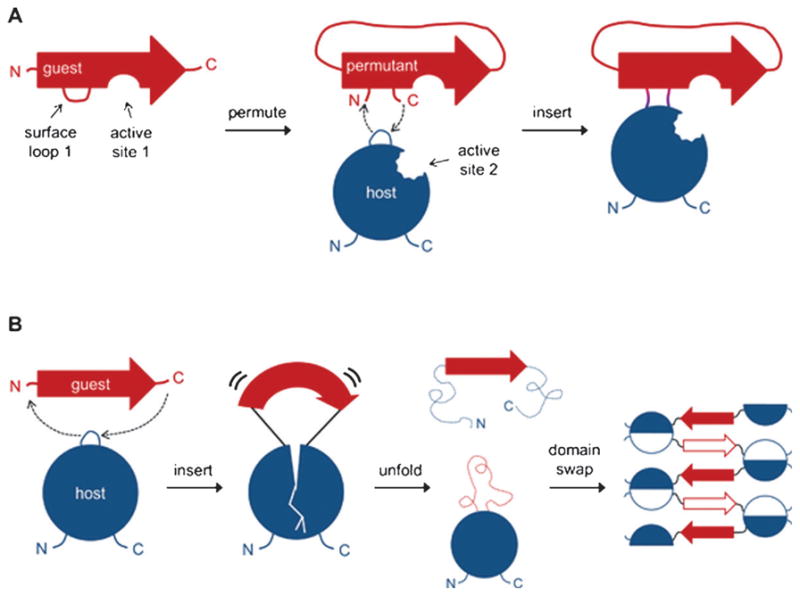
Domain-in-domain insertion schemes for effecting conformational coupling. A) Domain insertion with circular permutation. The guest is first circularly permuted so that its new N- and C-termini are near its binding/active site. The permutant is then inserted into a surface loop near the binding/active site of the host. This placement maximizes the chance that binding-induced conformational changes of one domain will be transduced to functional changes in the other domain. B) Mutually exclusive folding. A guest with a long N-to-C terminal distance is inserted into a surface loop of a host (left-most structure). If the linkers used to join the two proteins are sufficiently short (second structure from left), the guest stretches and unfolds the host, or the host compresses and unfolds the guest (second structure from right). The host domains of some MEF constructs are able to refold by domain swapping with identically unfolded hosts to generate dimers and higher order oligomers (right-most structure).
Interdomain communication can be maximized by inserting the receptor protein into the reporter near the re-porter’s active site, or inserting the reporter into the receptor at a location near the receptor’s binding site.
To avoid perturbing structure and stability it is generally advisable that the insertions take place in surface loops rather than in interior locations or secondary structure elements.
Circularly permuting the guest protein (see Figure 3) prior to inserting it into the host provides a more gentle and versatile means for effecting coupling (Figure 2A). Circular permutation exchanges the position of the original N- and C-termini with that of a site elsewhere in the protein, usually chosen to be a surface loop. This rearrangement serves two purposes. First, it guarantees that the new N- and C-termini of the guest will be close in space, thereby increasing the number of host sites into which it can be inserted (including secondary structure elements) without disrupting the structure of the host. Second, being able to choose the permutation site introduces a valuable combinatorial aspect to the process.
The length of the peptides used to fuse the two proteins is a critical variable. Very long linkers decouple any potential interaction, whereas excessively short linkers may lead to the “mutually exclusive folding” (MEF) effect in which the guest protein stretches the host protein at the point of insertion, the host simultaneously compresses the guest, and one unfolds the other (Figure 2B).[42] MEF represents a special case of interdomain fusion and it is discussed in a separate section below. One can therefore vary the junction points on both receptor and reporter proteins, along with the lengths and sequences of linker peptides, to optimize interdomain coupling.
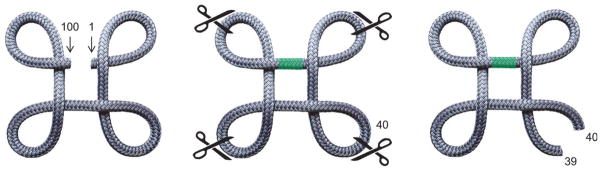
Figure 3.
Circular permutation changes the topology of a protein but frequently not its structure or function. The left structure represents the WT fold of a 100-amino acid protein. Circular permutation can be conceptualized by first joining residue 100 to residue 1 with a flexible peptide linker (green), generating a hypothetical fully circularized protein (center structure). Permutation is completed by clipping the protein at a location of choice, typically chosen to be a surface loop. Cleaving the protein at position 40 (right structure) shifts the N-and C-termini of the protein to positions 40 and 39, respectively, and effectively exchanges the positions of the termini with that of a surface loop without significantly perturbing the overall fold.
As an illustration of this procedure, Ostermeier and coworkers took a library-based approach by inserting random circular permutants of TEM1 β-lactamase (cpBLA) into random positions in MBP.[43] Their goal was to create a hybrid protein in which catalytic activity of the cpBLA domain is regulated by sugar binding to the MBP domain. They screened a library of approximately 106 variants by plating transformed E. coli on media containing maltose and different concentrations of ampicillin. Several switches were found in which maltose acted as either a positive or negative effector.[43a] (They also discovered one enzyme that was non-competitively inhibited by Zn2+, despite the fact that neither parent protein possesses zinc-binding activity.[44] This serendipitous result demonstrates that unanticipated structural changes can give rise to novel switching properties, particularly when fusing proteins in a combinatorial fashion.) The catalytic efficiency of one positive effector variant, RG13, was similar to that of wild-type (WT) BLA in the presence of saturating maltose and decreased by 25-fold in the absence of sugar.[43b]
A recent NMR study of RG13 provides clues to the structural basis for MBP-cpBLA coupling.[45] Chemical shift analysis suggests that the MBP domain is unperturbed by the cpBLA domain except for the α-helix into which cpBLA was inserted. Resonances in the cpBLA active site were largely unchanged in the maltose-bound and maltose-free proteins except for Glu166. The authors speculated that Glu166 (believed to be the general base for β-lactam acylation)[46] is properly positioned in the presence of maltose and displaced in its absence. However, the pattern of maltose-dependent chemical shift changes in the cpBLA domain does not immediately suggest a mechanism by which movement of Glu166 is coupled to domain closure of the MBP domain.
With respect to functional switches, one of the more versatile stimuli is light.[47] Phytochromes are photoswitches that regulate a variety of signal transduction pathways in plants, fungi, and bacteria.[48] Phytochrome switches are reversible: transformation from the inactive (Pr) to the active (Pfr) structure is triggered by absorption of red light by Pr, and reversed by absorption of far-red light by Pfr. The photosensory core consists of three domains [Per-ARNT-Sim (PAS), GAF, and PHY]. The bilin chromophore is bound to the central GAF domain. Absorption of a red photon induces isomerization about a double bond in bilin. Recent structures suggest that chromophore isomerization alters the relative orientations of GAF and PHY, a motion possibly mediated by rearrangement of the helices that connect them.[49] The photosensory core of phytochrome has not yet been fused to another protein to achieve functional regulation in a manner akin to MBP and BLA. However, several designs have linked phytochrome-interacting factors (proteins that bind specifically to Pfr) to other molecules to generate photoswitchable protein-protein interactions[50] that modulate gene expression,[51] actin polymerization,[52] protein splicing,[53] and protein localization.[54]
Limited Structural Rearrangement
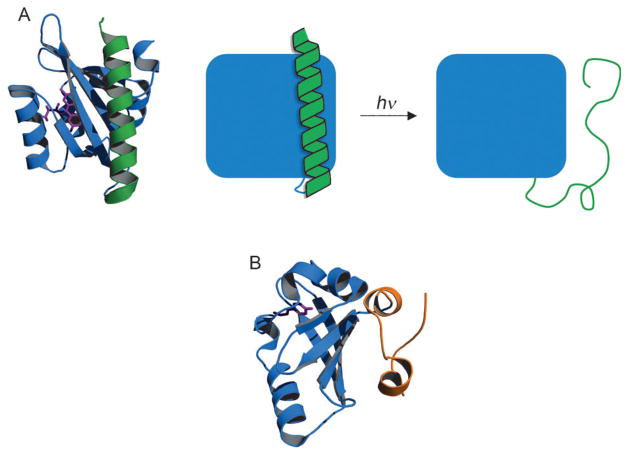
Continuing with the theme of light-activated proteins, several photoswitches exhibit localized conformational changes upon photon absorption. Photosensory domains of the light-oxygen-voltage (LOV) class transduce photon absorption to phototropic and other blue light responses in plants and algae. The structure of LOV2 from oat phototropin 1 consists of a PAS core domain to which an approximate 20-residue C-terminal helix (Jα) is loosely associated (Figure 4A).[55] When LOV2 absorbs blue light, the noncovalently bound flavin mononucleotide (FMN) chromophore forms a covalent adduct with a Cys residue. Structural changes originating from FMN are transmitted to Jα causing it to unfold (Figure 4A). A recent paper reported that Jα unfolding is mediated by unfolding of the N-terminal helix, but the latter event has yet to be capitalized upon.[56] The PAS core of LOV2 remains largely unperturbed.
Figure 4.
Photoswitching by limited structural rearrangement. Dark states of A) LOV2[123] and B) PYP[124] are shown. Photon absorption causes the C-terminal Jα helix of LOV2 (green) and the N-terminal 25 residues of PYP (orange) to lose structure and detach from the core domains (blue). Chromophores are depicted in purple. PDB codes of LOV2 and PYP are 2V0U and 1NWZ, respectively.
The location of Jα at the very end of LOV2 proved to be providential, because Jα can be used to transduce conformational changes between LOV2 and an effector protein fused to its C-terminus. Sosnick, Moffat, and co-workers were the first to employ this design.[57] Their approach was to attach an effector protein that contains an N-terminal α-helix (the DNA-binding protein Trp repressor; TrpR) to the C-terminus of LOV2, such that the two proteins share Jα and compete for its binding. The system is thermodynamically tuned such that LOV2 has a greater affinity for Jα in the dark state. The effector protein is rendered inactive by the absence of its N-terminal helix. Photon absorption then shifts the relative binding affinity, allowing the target protein to steal Jα from LOV2. Some structural (and amino acid sequence) overlap between LOV2 and TrpR is needed to establish this competition. To that end they progressively truncated the N-terminal helix of TrpR. The construct that showed the best photoactivation joined Jα of the LOV2 domain to the middle of the N-terminal α-helix of the TrpR domain. This variant, named LovTAP, exhibited a 5.6-fold increase in DNA binding affinity after illumination.
In a later study, the same researchers attempted to enhance coupling between photon absorption and DNA binding by rational design.[58] They considered mutations that were predicted to preferentially stabilize either Jα-LOV2 docking in the dark (off) state or Jα undocking in the lit (on) state. Two mutations in the Jα helix, G528A and N538E, stabilized docking in the dark state and increased the dynamic range of photoswitching from 5.6 to about 70.
Photoactive yellow protein (PYP) is a photoreceptor used by Halorhodospira halophilia to detect and avoid blue light. In the dark state, PYP is fully folded and the covalently bound p-coumaric acid chromophore is in the trans configuration. Photon absorption triggers trans-to-cis isomerization followed by transfer of a proton from Glu46 to the chromophore. The presence of strain in the chromophore and the negative charge on the buried Glu46 causes the first 25 residues of PYP to unfold, exposing Glu46 and p-coumaric acid to the solvent and relieving the unfavorable interactions (Figure 4B).[59] Woolley and colleagues created a light-activated DNA-binding switch conceptually analogous to LovTAP by fusing the GCN4 DNA binding domain to the N-terminus of PYP.[60] As with LovTAP, they were able to improve photoactivation by rational stabilization of the dark state.[61]
LovTAP and the GCN4-PYP chimera require some degree of homology between the ends of the input and output domains. Localized structural unfolding can, however, achieve coupling in other ways as well. Crosson et al. replaced the oxygen-sensing PAS B domain of the histidine kinase FixL with the LOV domain from the YtvA photoreceptor to generate YF1, a kinase that is regulated by light instead of oxygen.[62] Further analysis suggested that switching occurs by means of a rotary mechanism in which Jα mediates a 40–60° twist of a coiled-coil that links the input and output domains.[63] The authors proposed that Jα and the N-terminal helix of the kinase unite to form a continuous signaling helix. Because PAS domains are widespread and sense a wide range of stimuli, this mix-and-match strategy can potentially be used to introduce photoswitching into other PAS-containing proteins or to integrate multiple signal responses into a single protein.[64]
Rac1 is a member of the Rho family of signaling GTP-ases. Wu et al. created photoactivatable Rac1 (PA-Rac1) by fusing LOV2 to the N-terminus of Rac.[65] In the dark state, the close proximity of the two domains appears to sterically inhibit Rac1 from interacting with its effector (p21-activated kinase). Jα unfolding releases this occlusion. Functional photoswitches such as PA-Rac are a boon for the in vivo manipulation of biochemical pathways (“optogenetics”), because light can be applied to specific locations in the cell with almost no side effects (excluding radiation damage at sub-visible wavelengths).[66]
LOV and PYP depend on bound cofactors to trigger photoswitching. Chemically modifying a protein’s backbone with a photoresponsive element is an alternative approach that can in principle be applied to proteins that do not contain natural chromophores. Several recent designs draw parallels to the cis/trans Pro switches discussed in the previous section. Originally generated by cleaving ribonuclease A into two fragments using subtilisin, ribonuclease S (RNase S) is a longstanding model protein for folding studies. RNase S is the native complex formed by binding and folding of the S-peptide (residues 1–20) and the S-protein (residues 21–124). Fischer and co-workers synthesized an S-peptide variant that contained a thioxylated peptide bond (C=S in place of C=O) between Ala4 and Ala5.[67] The thioxylated S-peptide was >99% trans in reconstituted RNase S, and, being nearly isosteric with the peptide bond, did not significantly diminish activity of the enzyme. Illumination with 254 nm light increased the cis-thioxopeptide bond content to about 30% without causing the S-peptide to dissociate. Initial rates of substrate hydrolysis decreased by a similar amount, suggesting that cis-thioxo-RNase S is catalytically inert and that photocontrol of catalytic activity had been partially achieved. A similar result was reported for the functional interaction between thioxylated kinin and its receptor.[68] Thioxo and selenoxo[69] modifications establish valuable triggers for the proven cis/trans switching mechanism. The principle limitation is that the isomerizing groups must be introduced by synthetic methods and this restricts their placement to peptides or very small proteins. In addition, they can be destabilizing (particularly to α-helices)[70] and they may only partially isomerize upon irradiation.
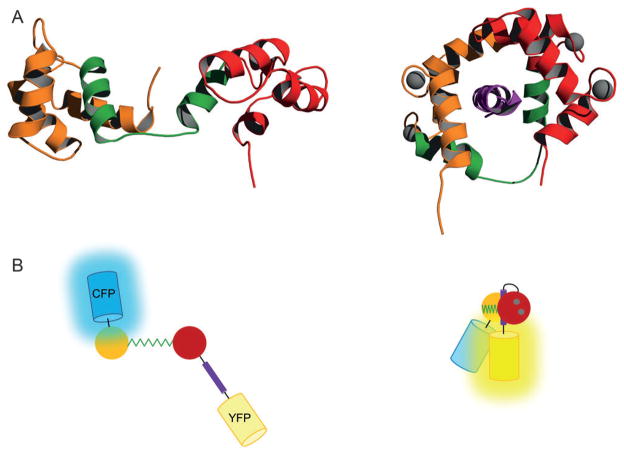
Perhaps the most successful examples of switches that undergo limited conformational rearrangements are the designs based on the calmodulin (CaM) scaffold. CaM is an intermediary messenger protein that links the functions of numerous target enzymes, ion channels, and other proteins to intracellular calcium concentration. CaM is composed of two helical domains connected by a stretch of about 27 amino acids (Figure 5A). Each domain contains two Ca2+ -binding EF-hands. In the absence of metal, the domains and linker lend CaM a roughly dumb-bell shaped appearance. Calcium binding triggers a unique structural transformation that extends the linker into a flexible α-helix and creates a binding surface from portions of the linker and EF-hand domains. Chiefly hydrophobic in character, this pocket is capable of recognizing up to several hundred regulatory peptide and protein sequences.[71] The two EF-hand domains wrap tightly around the target peptide, resulting in a compact, globular complex (Figure 5A). Approaches that capitalized on CaM’s distinctive conformational change to generate a family of fluorescent calcium sensors, including cameleons (Figure 5B),[72] pericams,[73] and camgaroos,[74] were reviewed elsewhere.[75]
Figure 5.
Calcium-triggered structural rearrangement of A) CaM and B) the cameleon biosensor. Structures of the calcium-free[125] and Ca2+ /peptide-bound[126] proteins are depicted at left and right, respectively. The N-terminal EF-hand is in orange, the C-terminal EF-hand is in red, the linker region is in green, and calcium ions are grey spheres. The CaM-binding M13 peptide is in purple. Cameleon was created by fusing the M13 peptide to the C-terminus of CaM, then attaching cyan FP (CFP) and yellow FP (YFP) to the amino and carboxy termini of the fusion protein. CFP and YFP are far apart in calcium-free cameleon; consequently, exciting CFP produces mostly cyan photon emission. When the CaM domain of cameleon binds calcium it wraps around the attached M13 peptide and brings CFP and YFP into close proximity. Exciting CFP now produces fewer cyan photons and more yellow photons due to efficient FRET between CFP and YFP. PDB codes are 1DMO (left) and 2BBM (right).
What if it were possible to alter the binding specificity of proteins that possess natural switching mechanisms, or graft completely new binding sites onto them? If so, then one could make new biosensors by modifying CaM or PBPs to recognize new ligands and simply recycling the detection methodologies outlined above. Similarly, if catalytic function can be engineered into an existing switch then one can hope to create allosterically regulated enzymes. By combining basic physical principles with modern computational tools, DeGrado and colleagues did just that.[76] Their goal was to convert CaM into an enzyme that catalyzes Kemp elimination in response to calcium binding. Carbon acid deprotonation is facilitated by a carboxylate base in a non-aqueous environment. To that end they introduced a Glu residue into the mostly hydrophobic pocket formed by the EF-hand domain and the linker in the Ca2+-bound state. The resulting enzyme, named AlleyCat, exhibited a kcat/KM value of 5.6M−1s−1 in 10 mM Ca2+, and a 25-fold lower catalytic efficiency in the absence of metal. Recent successes in computational design of enzyme active sites[77] indicate that it may be feasible to expand the repertoire of existing allosteric enzymes and binding proteins to include new functional switches.
Global Fold Switching
Most amino acid sequences fold to a single structure that corresponds to the global minimum of free energy. Members of the newly recognized class of “metamorphic” proteins,[78] however, have the singular ability to interconvert between two alternate native conformations, the energy minima of which are relatively closely spaced. The distinction between the conformational changes exhibited by metamorphic proteins and those of PBP, LOV, PYP, CaM, and so forth, is that the latter changes can be regarded as perturbations of a single native structure, whereas the former involve conversion between two unrelated folds. Lymphotactin (Ltn) is the archetypical example of a metamorphic protein. At low temperatures and in the presence of NaCl, Ltn adopts the canonical α/β chemokine fold.[79] In this form its function is to bind its target GPCR. At high temperature and no salt, Ltn takes on a dimeric all-β structure, the purpose of which is to bind glycosaminoglycan groups found in the extracellular matrix. Virtually all of the tertiary contacts are different in the two conformations,[79] and the entire molecule needs to unfold in order to transform from one state to the other.[78a] The finding that the two forms are equally populated at physiological temperature and in the presence of physiological NaCl concentration suggests that Ltn is poised to carry out either function in the cell. Other examples of metamorphic proteins include the spindle checkpoint protein Mad2,[80] Cro transcription factors,[81] and the sugar-binding protein tachylectin-2.[82]
Only a handful of proteins are known to possess natural fold-switching behavior. Several studies, however, suggest that it may be feasible to engineer this capability into other proteins. Proteins can be induced to switch conformations by changing a relatively small number of amino acids, as shown for Cro proteins,[81,83] Arc and Cro repressors,[84] metal-dependent switches,[85] and Streptococcus protein G.[86] Recent work of Bryan and Orban with protein G stands out as an example of this phenomenon. Protein G contains two types of domains: GA and GB (Figure 6). GA (45 structured amino acids) adopts a three-helical fold and binds human serum albumin, while GB (56 structured amino acids) takes on a mixed α/β fold and binds the constant region of IgG. No significant sequence homology exists between GA and GB. Alexander et al. set out to determine the minimum number of mutations that are sufficient to convert one fold to the other. In a series of studies they progressively increased sequence identity until they found that the GA and GB folds remained autonomous and stable (ΔGfold ≥−3 kcal mol−1) when all but three (positions 20, 30, and 45) of the 56 structured amino acids were made to be the same.[87] In other words, two polypeptides that are 95% identical can fold into two completely different and stable structures! Commonizing the residues at positions 20 and 30 produced a 98% identical pair in which GA98 and GB98 still adopted the respective all-α and α/β folds (Figure 6), but each was destabilized to the point where 10% of the molecules were unfolded. This instability allowed the single remaining “mutation” at position 45 (Leu in GA98 and Tyr in GB98) to dictate the switch between folds—a remarkable result.
Figure 6.
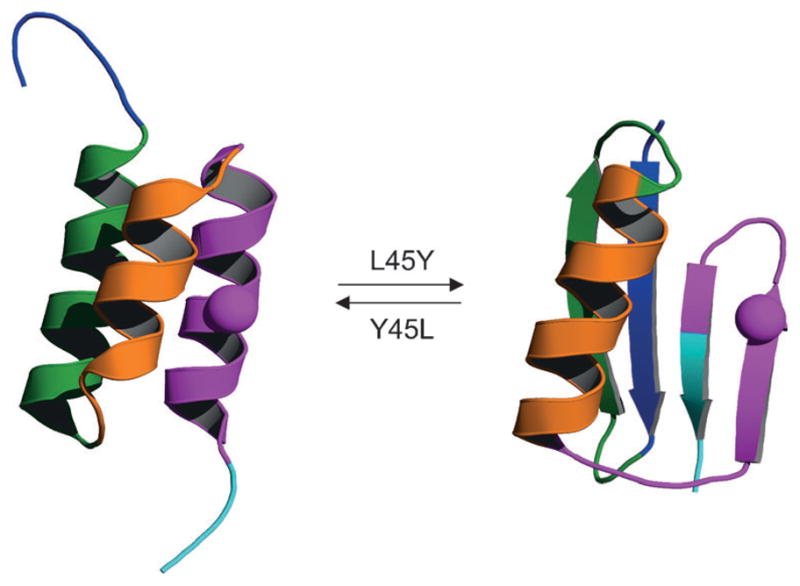
Fold-switching of protein G. GA98 (left) and GB98 (right) differ only by a single amino acid at position 45 (sphere), but they adopt two completely different folds. Amino acid sequences are colored identically in the two structures. PDB codes are 2LHC (left) and 2LHD (right).
Changing the identity of an amino acid is not a practical trigger for a conformational switch. Nonetheless, the above study creates an impact, because it demonstrates that a protein can be tweaked so that it is poised between two folds.[88] One can imagine that, instead of mutation, binding energy or a change in solution conditions (c.f. Ltn) could tip the balance to the other fold.
Folding–Unfolding Switches
The most dramatic conformational change that a protein can undergo is also one that all proteins experience: folding and unfolding. Folding makes for a particularly effective switching mechanism for several reasons. First, because a protein’s native function is typically abolished upon denaturation, the unfolded form constitutes a default off-state in functional switches. Second, so many of a protein’s properties change as a result of folding–unfolding (structure, dynamics, charge distribution, etc.) that it is likely that a means to detect binding can be devised when designing biosensors. Third, a protein constantly cycles through all its possible conformations—from native to globally unfolded to all states in between—according to their Boltzmann distributions. Thus, even a protein, the structure of which appears immutable, is already a reversible switch, though it spends most of its time in one state.
It is now recognized that a significant fraction of proteins in the cell, perhaps as high as one-half,[89] contain regions of intrinsic disorder. An IDP lacks well-defined structure, either in part or in whole, until it engages its cognate ligand(s). The advantages that “unstructure” confers to molecular recognition are not clear; some possibilities include increasing binding promiscuity, increasing the rate of macro-molecular association, and offering multiple sites for post-translational modification (e.g. phosphorylation).[7a,b,d] Breast cancer type 1 susceptibility protein (BRCA1) is an 1863-amino acid protein that functions in DNA repair, cell cycle checkpoint control, and other processes that help maintain genome integrity.[90] The N-terminal RING domain (residues 1–103) and the C-terminal BRCT domain (residues 1646–1863) are structured, but the central region of about 1500 amino acids is intrinsically disordered.[91] The central region binds a number of proteins involved in the DNA damage response including p53.[92] Cissell et al. utilized the 219–498 fragment of BRCA1 as an intrinsically disordered binding element to detect p53.[93] Binding of p53 induced the BRCA1 fragment to fold, which was then detected by strategic placement of environmentally sensitive tetramethylrhodamine or dansyl dyes.
When attempting to design a conformational switch, one may draw on the axiom that it is easier to induce a protein to unfold than to change native conformations. There are two main methods by which one can tip the thermodynamic balance of stability towards the unfolded state: substitution of one or a few amino acids (frequently in the tightly packed core of the protein) and truncation of amino acids from either terminus. Truncation can be beneficial because it does not require prior knowledge of 3D structure, and the degree of destabilization can be tuned to an extent by deleting amino acids one at a time.[94] Kohn and Plaxco demonstrated these principles by progressively truncating amino acids from the C-terminus of the SH3 domain (57 amino acids) from Fyn tyrosine kinase.[95] It unfolded after the fourth deletion (CΔ4). Binding the cognate peptide induced CΔ4 to fold, which was monitored by fluorescence of the intrinsic Trp residue or that of a BODIPY group previously attached to a Cys side chain.
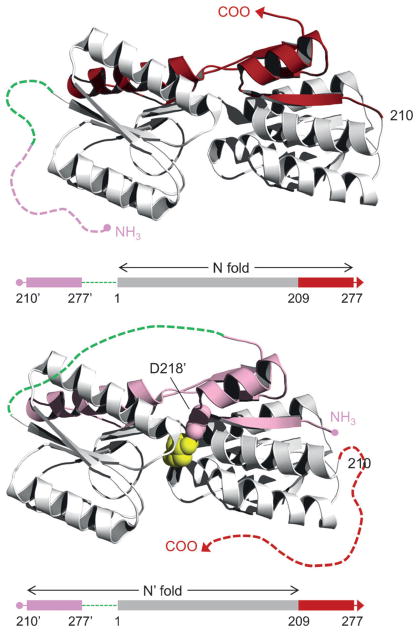
“Alternate frame folding” (AFF) incorporates elements of binding-induced folding and fold switching.[96] It is a technique for modifying an existing binding protein so that it undergoes a binding-induced (or cleavage-induced) conformational change. We have applied AFF to convert barnase into an artificial zymogen[97] and calbindin D9k and RBP into fluorescent sensors for calcium[96,98] and ribose (unpublished), respectively. The main steps in this process are illustrated for RBP in Figure 7. First, a segment from the carboxy terminus is duplicated and appended to the amino terminus using a peptide linker long enough to span the ends of the WT protein (alternately, a segment from the amino terminus can be copied and joined to the carboxy terminus). The length of this segment is determined by two factors: 1) it must contain at least one amino acid critical for ligand binding, and 2) it should end at a surface loop. RBP contains many surface loops, but it is generally recommended to choose the one closest to the key binding residue (Asp218) in order to minimize length. The twin segment establishes a second “frame” of protein folding. RBP can fold to the WT native structure (N-fold) using amino acids 1–277. It can also fold using amino acids (210′–277′)-linker-(1–209) to yield the circularly permuted structure (N′-fold). The duplicate peptides are expected to extend from the amino or car-boxy termini (in the N- and N′-fold, respectively) as disordered tails. The protein cannot adopt N and N′ conformations simultaneously, because they compete for residues 1–209. It consequently interconverts between the two forms with the equilibrium distribution determined by their relative stabilities. Asp218 is mutated to Ala in the more stable of the two forms (N-fold in this case) so that ligand binding shifts the population to the N′-fold. Finally, the conformational change is detected by placing fluorophores at the N-terminus and in the loop where the permutation took place. These sites are naturally suited to FRET because they are spatially close in N′ and are predicted to be more distant in N.
Figure 7.
Conformational switching by alternate frame folding. Schematics of the N-fold and N′-fold of RBP-AFF are depicted at top and bottom, respectively. Amino acid sequences are shown below each structure. The critical binding residue (D218) and permutation site (position 210) are indicated. Residues 210–277 (red) are duplicated and appended to the amino terminus using a flexible peptide linker (green). The duplicated amino acids (pink) are indicated by prime superscripts but are otherwise numbered identically to those of the parent sequence. The binding-knockout mutation D218A is introduced into the red sequence. In the absence of ligand, the N-fold (residues 1–277) predominates over the N′-fold (residues 210′–277′, the linker, and residues 1–209). Addition of ribose (yellow), and the binding energy provided by D218′ (pink spheres) in the remodeled binding pocket, drives the shift from the N-fold to the N′-fold. The protein cannot simultaneously adopt N- and N′-folds because they compete for a shared sequence (grey). The dashed lines denote that the pink segment is unstructured in the N-fold and the red segment is unstructured in the N′-fold. Placement of distance-sensitive fluorophores at the amino terminus and at position 210 links the conformational change to an optical output.
Calbindin D9k (75 amino acids) corresponds to just one of the EF-hand domains in CaM. It does not appreciably change structure on calcium binding.[99] Nevertheless, Stratton et al. converted calbindin to a fluorescent calcium sensor (calbindin-AFF) by the AFF method.[96] Subsequent NMR structural studies verified the fold shift and demonstrated that the duplicate peptides in calbindin-AFF do indeed extend from N and N′ as disordered tails.[98b] This result is significant, because it suggests that similar binding-induced fluorescence changes can be elicited from other AFF-based sensors. AFF, however, is not limited to biosensor design. Mitrea et al. used it to convert barnase into a functional switch (barnase-AFF) that is activated by proteolytic cleavage, that is, an artificial zymogen.[97] Barnase was modified as in Figure 7, except a key catalytic residue was mutated instead of a ligand-binding residue, and an HIV-1 protease recognition sequence was inserted into the loop that bore the permutation site. Protease cleavage triggered the shift from the catalytically inert N conformation to the active N′ form, increasing kcat/KM by about 250-fold.
Folding–unfolding switches appear to be major players in cellular mechano-transduction pathways. Mechano-sensing proteins are stimulated by mechanical forces, for example, those that are generated by fluid flow, membrane deformation, and stresses at cell-cell and cell-extracellular matrix junctions. The giant protein titin is one of the most well studied examples of a mechano-sensor.[100] By integrating actin and myosin filaments at sarcomeres, titin is responsible for passive elasticity of the sarcomere as well as transducing muscle stress into a host of signaling responses. Its springlike properties are established by the linear array of repeating Ig and fibronectin-like domains that unfold and refold with muscle load. It also contains a single titin kinase domain (TK), the catalytic activity of which is controlled by a unique force-triggered mechanism. The C-terminal tail of TK consists of two α-helices and a β-strand. In the unstressed state, the tail inhibits TK by wrapping around the catalytic domain and occluding the nucleotide binding site.[101] The structurally related myosin light-chain kinases are activated when CaM binds to their tails and exposes the ATP pocket. TK and twitchin kinase domains are not activated by CaM binding, but rather by mechanical unfolding of the inhibitory tail.[102] The approximate 30 pN gating force required to unfold the tail without unfolding the catalytic domain is consistent with the pulling force generated by the six myosin heads in the crown of a myosin filament. Cadherins,[103] integrins,[104] filamin A,[105] PKD domains of polycistin-1,[106] and the negative regulatory region of Notch receptor[107] are other examples of mechano-sensors that appear to operate by force-induced unfolding, whereas proteins such as the MscL ion channel[108] and the AT1 and other GPCRs[109] likely respond to mechanical stimuli with conformational changes that do not involve unfolding.
As with all of the switching mechanisms discussed here, a basic question is how to incorporate a natural phenomenon—force-induced unfolding in this case—into new switch designs. Mutually exclusive folding is one such approach.[42,110] MEF is a special class of protein-in-protein fusion. Rather than attempting to make the insertion structurally compatible to both host and guest (e.g. by circularly permuting the guest), a guest is chosen with an N-to-C distance at least twice as long as the end-to-end distance of the loop in the host (Figure 2B, left-most structure). The guest stretches the host at the point of insertion, while the host simultaneously compresses the ends of the guest (Figure 2B, second structure from left). The more thermodynamically stable of the two domains unfolds the less stable domain and renders it inactive (Figure 2B, second structure from right). One can take advantage of the intrinsic coupling between folding and binding to generate a protein that switches between active and inactive conformations depending on ligand. Using the GCN4 DNA binding domain (75 Å N-to-C distance) as the guest and the ribonuclease barnase (10 Å end-to-end loop distance) as the host, Ha et al. created a chimera in which the RNase activity of the barnase domain was regulated by DNA binding to the GCN4 domain.[111]
More recently we showed that MEF can also be a switch for triggering self-assembly.[112] When ubiquitin was inserted into each of six different surface loops of barnase, the fusion proteins formed dimers, trimers, tetramers, and higher-order oligomers. The X-ray structure of one of the insertion variants revealed a long, linear polymer in which the binding interface between protomers consisted of domain-swapped barnase subunits (Figure 2B, right-most structure). The molecular stress imposed on the monomeric barnase domain by ubiquitin was relieved by unfolding of the barnase domain followed by intermolecular refolding with identically disrupted barnase subunits. The structures of both ubiquitin and barnase domains in the swapped oligomer were virtually identical to their WT counterparts, suggesting that it may be possible to create self-assembling oligomers and polymers that retain and integrate the functions of the parent molecules.
One distinction of the MEF design is that it may be modular. Any stable guest protein with a moderately long N-to-C distance should be able to perform the same stretching function as ubiquitin and GCN4. As to the host, there is no reason to expect that barnase is unique in its ability to domain swap upon forced unfolding. On the other hand, Peng and Li created an MEF chimera in which the 27th Ig domain of titin was inserted into a surface loop of the GB5 protein.[113] They were able to directly witness unfolding of GB5 by Ig in real time, but subsequent refolding/domain swapping of GB5 was not observed. Therefore, the principles that determine whether MEF generates a self-assembly switch (Figure 2B, right-most structure) or a functional switch (Figure 2B, second structure from right) are largely unknown.
The main concerns with folding–unfolding-based switches are that unfolded proteins tend to misfold and/or aggregate, particularly in complex environments such as the inside of a cell, and degradation can be a concern. Natural IDPs have evolved the ability to remain soluble when unfolded, likely due (at least in part) to their atypically high ratio of charged to hydrophobic amino acids.[89] Destabilization of native proteins, on the other hand, is implicated as one of the root causes of amyloidoses and other aggregation-related diseases.[114] SH3 CΔ4 remained monomeric and soluble at >100 μM concentration and it was not degraded when expressed in bacterial cells. Calbindin-AFF and the MEF constructs were similarly well behaved, but the extent to which unfolded forms of large proteins retain these favorable characteristics remains to be determined.
Adjusting the Properties of Conformational Switches
The defining characteristics of any switch include reversibility, response time, sensitivity (how much stimulus is required for a response), dynamic range (over what range of stimulus concentration the switch responds), and gain (signal change between on and off states). These properties can be understood—and perhaps manipulated—by recognizing that conformational switching originates from the transitions between structural substates. According to this view, the on-and off-states are part of the native ensemble of structures; they are local minima in a free-energy surface that are separated by a distinct energy barrier. Protein switches thus cycle between on and off states as one of their natural dynamic modes. This notion leads to the following conclusions.
Reversibility and response time
Conformational switches are usually reversible, though the forward and reverse rates are highly variable. For example, apo-MBP interconverts between the open state (95% populated at equilibrium) and the closed state (5% populated) on a timescale of ns–μs.[115] PBPs therefore bind their ligands with rates approaching the diffusion limit.[116] The overall response time (i.e., how fast a PBP sensor can respond to a change in ligand concentration) is limited by the off-rate, which can be slow for high-affinity PBPs. The ParM open-to-closed change, on the other hand, appears to be slower and ADP binds with an on-rate of 105M−1s−1.[117] Folding-based switches are likewise inherently reversible as long as the unfolded proteins do not aggregate or become degraded. Unfolded SH3 CΔ4 refolds with a rate constant of 102 s−1, which is comparable to the rate at which it responds to its ligand.[95] In other instances the recognition event may be rapid, but the overall rate of switching is limited by a slower process that was introduced by fusion with a reporter domain or other modification. The first 25 amino acids of PYP unfold within a few milliseconds after photoexcitation,[118] but the corresponding sequence from the PYP-GCN4 chimera does not fully unfold and detach from the PYP domain in the absence of the DNA ligand.[61] Though calbindin binds calcium with a diffusion-limited rate of 109M−1s−1,[119] calbindin-AFF switches conformations on the timescale of seconds.[96] The difference is apparently due to an unfolding step being at least partially rate-limiting.
In some cases it has been possible to adjust response times using rational or random mutagenesis. For switches in which the rates are limited by local or global unfolding, for example, calbindin-AFF and Ltn,[78a] introducing a destabilizing mutation generally increases unfolding rates and this strategy hastened the response time of calbindin-AFF.[98a] Structural characterization of intermediates in the LOV phototransduction pathway allowed Moffat and colleagues to make mutants in which the LOV2 photocyle was accelerated or decelerated by up to 80-fold.[47] The comparatively slow rate of Pro cis/trans isomerization (typically 100–10−3 s−1) has been suggested to control the timing of various cellular processes. PPIases accelerate this rate by up to 106-fold. Pro isomerization is among the only example of a switching mechanism for which the response time can be sped up by an enzyme.
Gain and sensitivity
According to the conformational substate view, gain and sensitivity are determined by the free-energy difference between on- and off-states. High gain requires that the latter be of much lower energy than the former in the absence of stimulus so that nearly all molecules are initially in the off position. The trade-off is that sensitivity may suffer: the amount of effector required to invert the populations (i.e. turn on the switch) is proportional to their energetic difference. One can turn this relationship into an advantage by rationally adjusting energy levels (thermodynamic tuning). For example, many biosensor applications demand that Kd match the concentration range of analyte to be detected. A PBP may bind glucose with micromolar Kd, but it would fail as a blood sugar monitor for diabetes, since it would always be saturated by the millimolar glucose content present in serum. One can attempt to raise Kd by mutating residues in the binding pocket, but these substitutions often alter specificity. An alternate approach is to employ binding-induced folding. Because binding energy is used to invert the unfolded (off) and folded (on) populations, the apparent Kd is weaker than the intrinsic Kd of the native protein by a factor of (1+Kfold)/Kfold, in which Kfold = exp(−ΔGfold/RT). To illustrate, Kd of SH3 CΔ4 was 100-fold higher than that of WT SH3,[95] and the calcium affinity of calbindin-AFF was tuned in both directions by introducing a destabilizing point mutation into either the N or N′ frame.[98a] The same strategy was used to suppress cytotoxic RNase activity of barnase-AFF in its uncleaved form to nearly undetectable levels.[97] For the micromolar Kd glucose sensor mentioned above, one could achieve the requisite 103-fold reduction in glucose affinity by destabilizing the WT PBP by about 4 kcalmol−1. The principle of thermodynamic tuning applies to the other examples presented here as well, although in practice it is challenging for switches for which mechanisms are not well characterized (e.g. LOV2, PYP) or for which populations are not easily perturbed (Pro switches).
Finally, dynamic range remains a limitation of current switch designs. For biosensors, simple one-site binding dictates that the minimal useful signal change (which typically means inverting the populations from >10:1 off:on to >10:1 on:off) occurs over less than a 102-fold change in effector concentration. Many assays specify that the switch respond over broader or narrower concentration regimes. Vallée-Bélisle et al. recently showed that dynamic range can be extended many orders of magnitude by combining variants of the same switch that possess different binding affinities.[120] Although they demonstrated this principle by altering stability (and therefore Kd) of a DNA aptamer by means of base-pair substitution, the same thermodynamic tuning strategy can in principle be applied to protein switches. For applications that demand a steep response curve over a narrow concentration range, they introduced a receptor that binds the target with high affinity but does not undergo conformational switching. This “depletant” acts as a sink to prevent free ligand from accumulating until the total target concentration exceeds that of the depletant.
Summary and Outlook
Proteins are unique in nature in that they incorporate a large array of conformational change modes into an equally diverse repertoire of biological functions. Cells and viruses employ protein conformational switches for molecular recognition, environmental sensing, and regulating signaling pathways involved in development, homeostasis, immunity, infection, and other cellular processes. Structural and biochemical elucidation of some of these switching mechanisms has enabled researchers to begin adapting them for use in biosensing and functional regulation. As more structures become available, the main challenges shift to altering specificity of existing binding domains, and learning how to transduce natural conformational changes within a binding domain to a functional or optical output module. The number of ways in which two (or more) proteins can be joined is combinatorial. Existing studies, few as they are, suggest that subtle structural changes in one domain can alter function of an attached domain in a manner that is not anticipated or even understood. Thus, future efforts will likely benefit from the collaboration of library-based screening methods and computational simulations.
A parallel tack is to exploit the conformational change common to all proteins—the folding–unfolding reaction—to convert ordinary proteins into switches. Natural IDPs that fold upon binding provide a source of prospective biosensors as well as an inspiration for like designs. An altogether new class of switch can be envisioned by once again fusing two (or more) proteins, then using the physics that link binding to folding (and perhaps folding to unfolding) to establish communication between domains within the fusion protein, as well as between the fusion protein and its environment. Hilser and colleagues recently presented a model of allostery that recapitulates the intricate switching behavior shown by some natural systems, whereupon a given ligand can act as both an antagonist and an agonist.[121] The physical manifestation of their formalism is a multidomain protein in which the domains interact favorably or unfavorably when in their high-affinity states. The allosteric response is greatest when one or more of the domains folds upon binding; no additional conformational change is needed. The possibility of creating complex switches by incorporating this concept with some of the designs discussed herein is especially captivating.
Acknowledgments
This work was supported by NIH. grant R01GM069755 to S.N.L.
Biographies

Jeung-Hoi Ha was born in Seoul (South Korea), and received her B.Sc. in Biochemistry at Yonsei University. She obtained her Ph.D. in Biochemistry at the University of Wisconsin-Madison under the supervision of Professor M. Thomas Record, Jr. Her Ph.D. work focused on the hydrophobic effect and thermodynamics of the lac repressor–operator interaction. She received postdoctoral training first from Professor Ronald Raines at the University of Wisconsin-Madison, then from Professor David McKay in the Department of Structural Biology at Stanford University. At Stanford, she investigated the structure and mechanism of the HSC70 molecular chaperone. She moved to the State University of New York Upstate Medical University in 1996. She is a Senior Research Scientist in the Department of Biochemistry & Molecular Biology. She and Dr. Loh jointly manage Dr. Loh’s research group and are known as the left and right brains of the laboratory.

Stewart N. Loh received his B.Sc. in Chemistry from the University of Utah and his Ph.D. in Biochemistry from the University of Wisconsin-Madison, under the supervision of Professor John Markley. His Ph.D. training involved applying nuclear magnetic resonance techniques, including amide hydrogen exchange and hydrogen/deuterium isotope fractionation measurements, to investigate folding of staphylococcal nuclease. He was a Damon Runyon-Walter Winchell Postdoctoral Fellow in the laboratory of Professor Robert L. Baldwin in the Biochemistry Department at Stanford University, where he studied the kinetic folding mechanisms of apomyoglobin and ribonuclease A. He joined the State University of New York Upstate Medical University as an Assistant Professor in 1996. He and Dr. Ha have continued to study the thermodynamics and kinetics of protein folding since then, and have specifically concentrated on folding-based switching mechanisms for the past ten years. He became an Associate Professor in 2002 and a Full Professor of Biochemistry and Molecular Biology in 2008. Dr. Loh’s current research focus is to apply principles of protein folding and unfolding to convert ordinary proteins into molecular switches for use in biology, biotechnology, and medicine.
References
- 1.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison SC. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 4.Beniac DR, deVarennes SL, Andonov A, He R, Booth TF. PLoS One. 2007;2:e1082. doi: 10.1371/journal.pone.0001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 6.a) Gerstein M, Krebs W. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Flores S, Echols N, Milburn D, Hespenheide B, Keating K, Lu J, Wells S, Yu EZ, Thorpe M, Gerstein M. Nucleic Acids Res. 2006;34:D296–301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Qi G, Lee R, Hayward S. Bioinformatics. 2005;21:2832–2838. doi: 10.1093/bioinformatics/bti420. [DOI] [PubMed] [Google Scholar]; d) Seeliger F, Leeb T, Peters M, Brugmann M, Fehr M, Hewicker-Trautwein M. Vet Pathol. 2003;40:530–539. doi: 10.1354/vp.40-5-530. [DOI] [PubMed] [Google Scholar]; e) Amemiya T, Koike R, Fuchigami S, Ikeguchi M, Kidera A. J Mol Biol. 2011;408:568–584. doi: 10.1016/j.jmb.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 7.a) Wright PE, Dyson HJ. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Babu MM, van der Lee R, de Groot NS, Gsponer J. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]; c) Fisher CK, Stultz CM. Curr Opin Struct Biol. 2011;21:426–431. doi: 10.1016/j.sbi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tompa P. Curr Opin Struct Biol. 2011;21:419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 8.a) Chen K, Bachtiar I, Piszczek G, Bouamr F, Carter C, Tjandra N. Biochemistry. 2008;47:1928–1937. doi: 10.1021/bi701984h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Proc Natl Acad Sci USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Proc Natl Acad Sci USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzeng SR, Kalodimos CG. Curr Opin Struct Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Volkman BF, Lipson D, Wemmer DE, Kern D. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 13.a) Ambroggio XI, Kuhlman B. Curr Opin Struct Biol. 2006;16:525–530. doi: 10.1016/j.sbi.2006.05.014. [DOI] [PubMed] [Google Scholar]; b) Ostermeier M. Curr Opin Struct Biol. 2009;19:442–448. doi: 10.1016/j.sbi.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ostermeier M. Protein Eng Des Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]; d) Wright CM, Heins RA, Ostermeier M. Curr Opin Chem Biol. 2007;11:342–346. doi: 10.1016/j.cbpa.2007.04.011. [DOI] [PubMed] [Google Scholar]; e) Vallée-Bélisle A, Plaxco KW. Curr Opin Struct Biol. 2010;20:518–526. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Koide S. Curr Opin Biotechnol. 2009;20:398–404. doi: 10.1016/j.copbio.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Golynskiy MV, Koay MS, Vinkenborg JL, Merkx M. ChemBioChem. 2011;12:353–361. doi: 10.1002/cbic.201000642. [DOI] [PubMed] [Google Scholar]; h) Stratton MM, Loh SN. Protein Sci. 2011;20:19–29. doi: 10.1002/pro.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mäler L, Blankenship J, Rance M, Chazin WJ. Nat Struct Biol. 2000;7:245–250. doi: 10.1038/73369. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer MA, Hellinga HW. Curr Opin Struct Biol. 2004;14:495–504. doi: 10.1016/j.sbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 16.a) Lager I, Fehr M, Frommer WB, Lalonde S. FEBS Lett. 2003;553:85–89. doi: 10.1016/s0014-5793(03)00976-1. [DOI] [PubMed] [Google Scholar]; b) Björkman AJ, Mowbray S. J Mol Biol. 1998;279:651–664. doi: 10.1006/jmbi.1998.1785. [DOI] [PubMed] [Google Scholar]; c) Ravindranathan KP, Gallicchio E, Levy RM. J Mol Biol. 2005;353:196–210. doi: 10.1016/j.jmb.2005.08.009. [DOI] [PubMed] [Google Scholar]; d) Vercillo NC, Herald KJ, Fox JM, Der BS, Dattelbaum JD. Protein Sci. 2007;16:362–368. doi: 10.1110/ps.062595707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Medintz IL, Deschamps JR. Curr Opin Biotechnol. 2006;17:17–27. doi: 10.1016/j.copbio.2006.01.002. [DOI] [PubMed] [Google Scholar]; b) Dattelbaum JD, Looger LL, Benson DE, Sali KM, Thompson RB, Hellinga HW. Protein Sci. 2005;14:284–291. doi: 10.1110/ps.041146005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borrok MJ, Zhu Y, Forest KT, Kiessling LL. ACS Chem Biol. 2009;4:447–456. doi: 10.1021/cb900021q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnusson U, Chaudhuri BN, Ko J, Park C, Jones TA, Mowbray SL. J Biol Chem. 2002;277:14077–14084. doi: 10.1074/jbc.M200514200. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama N, Takeda K, Miki K. J Mol Biol. 2009;392:559–565. doi: 10.1016/j.jmb.2009.07.043. [DOI] [PubMed] [Google Scholar]
- 21.Cuneo MJ, Beese LS, Hellinga HW. J Biol Chem. 2009;284:33217–33223. doi: 10.1074/jbc.M109.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Trakhanov S, Vyas NK, Luecke H, Kristensen DM, Ma J, Quiocho FA. Biochemistry. 2005;44:6597–6608. doi: 10.1021/bi047302o. [DOI] [PubMed] [Google Scholar]; b) Sack JS, Trakhanov SD, Tsigannik IH, Quiocho FA. J Mol Biol. 1989;206:193–207. doi: 10.1016/0022-2836(89)90532-9. [DOI] [PubMed] [Google Scholar]; c) Bermejo GA, Strub MP, Ho C, Tjandra N. Biochemistry. 2010;49:1893–1902. doi: 10.1021/bi902045p. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bermejo GA, Strub MP, Ho C, Tjandra N. J Am Chem Soc. 2009;131:9532–9537. doi: 10.1021/ja902436g. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Scirè A, Marabotti A, Staiano M, Iozzino L, Luchansky MS, Der BS, Dattelbaum JD, Tanfani F, D’Auria S. Mol BioSyst. 2010;6:687–698. doi: 10.1039/b922092e. [DOI] [PubMed] [Google Scholar]
- 23.a) Karpowich NK, Huang HH, Smith PC, Hunt JF. J Biol Chem. 2003;278:8429–8434. doi: 10.1074/jbc.M212239200. [DOI] [PubMed] [Google Scholar]; b) Locher KP, Borths E. FEBS Lett. 2004;564:264–268. doi: 10.1016/S0014-5793(04)00289-3. [DOI] [PubMed] [Google Scholar]
- 24.Chu BC, Vogel HJ. Biol Chem. 2011;392:39–52. doi: 10.1515/BC.2011.012. [DOI] [PubMed] [Google Scholar]
- 25.Cavazza C, Martin L, Laffly E, Lebrette H, Cherrier MV, Zeppieri L, Richaud P, Carriere M, Fontecilla-Camps JC. FEBS Lett. 2011;585:711–715. doi: 10.1016/j.febslet.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 26.a) Zhang L, Koay M, Maher MJ, Xiao Z, Wedd AG. J Am Chem Soc. 2006;128:5834–5850. doi: 10.1021/ja058528x. [DOI] [PubMed] [Google Scholar]; b) Arnesano F, Banci L, Bertini I, Thompsett AR. Structure. 2002;10:1337–1347. doi: 10.1016/s0969-2126(02)00858-4. [DOI] [PubMed] [Google Scholar]; c) Hussain F, Sedlak E, Wittung-Stafshede P. Arch Biochem Biophys. 2007;467:58–66. doi: 10.1016/j.abb.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Alicea I, Marvin JS, Miklos AE, Ellington AD, Looger LL, Schreiter ER. J Mol Biol. 2011;414:356–369. doi: 10.1016/j.jmb.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergauwen B, Elegheert J, Dansercoer A, Devreese B, Savvides SN. Proc Natl Acad Sci USA. 2010;107:13270–13275. doi: 10.1073/pnas.1005198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Lorimier RM, Smith JJ, Dwyer MA, Looger LL, Sali KM, Paavola CD, Rizk SS, Sadigov S, Conrad DW, Loew L, Hellinga HW. Protein Sci. 2002;11:2655–2675. doi: 10.1110/ps.021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehr M, Frommer WB, Lalonde S. Proc Natl Acad Sci USA. 2002;99:9846–9851. doi: 10.1073/pnas.142089199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fehr M, Lalonde S, Lager I, Wolff MW, Frommer WB. J Biol Chem. 2003;278:19127–19133. doi: 10.1074/jbc.M301333200. [DOI] [PubMed] [Google Scholar]
- 32.Teasley Hamorsky K, Ensor CM, Wei Y, Daunert S. Angew Chem. 2008;120:3778–3781. doi: 10.1002/anie.200704440. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:3718–3721. doi: 10.1002/anie.200704440. [DOI] [PubMed] [Google Scholar]
- 33.a) van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kunzelmann S, Webb MR. Biochem J. 2011;440:43–49. doi: 10.1042/BJ20110349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunzelmann S, Webb MR. J Biol Chem. 2009;284:33130–33138. doi: 10.1074/jbc.M109.047118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.a) Wan T, Beavil RL, Fabiane SM, Beavil AJ, Sohi MK, Keown M, Young RJ, Henry AJ, Owens RJ, Gould HJ, Sutton BJ. Nat Immunol. 2002;3:681–686. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]; b) Holdom MD, Davies AM, Nettleship JE, Bagby SC, Dhaliwal B, Girardi E, Hunt J, Gould HJ, Beavil AJ, McDonnell JM, Owens RJ, Sutton BJ. Nat Struct Mol Biol. 2011;18:571–576. doi: 10.1038/nsmb.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunt J, Keeble AH, Dale RE, Corbett MK, Beavil RL, Levitt J, Swann MJ, Suhling K, Ameer-Beg S, Sutton BJ, Beavil AJ. J Biol Chem. 2012 doi: 10.1074/jbc.M111.331967. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Nat Chem Biol. 2011;7:51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Severin A, Joseph RE, Boyken S, Fulton DB, Andreotti AH. J Mol Biol. 2009;387:726–743. doi: 10.1016/j.jmb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 40.Lee WY, Sine SM. Nature. 2005;438:243–247. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- 41.(a) Schiene-Fischer C, Aumüller T, Fischer G. Top Curr Chem. 2012 doi: 10.1007/128_2011_151. in press. [DOI] [PubMed] [Google Scholar]; b) Lu KP, Finn G, Lee TH, Nicholson LK. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]; c) Liou YC, Zhou XZ, Lu KP. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radley TL, Markowska AI, Bettinger BT, Ha J-H, Loh SN. J Mol Biol. 2003;332:529–536. doi: 10.1016/s0022-2836(03)00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.a) Guntas G, Mansell TJ, Kim JR, Ostermeier M. Proc Natl Acad Sci USA. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guntas G, Mitchell SF, Ostermeier M. Chem Biol. 2004;11:1483–1487. doi: 10.1016/j.chembiol.2004.08.020. [DOI] [PubMed] [Google Scholar]; c) Guntas G, Ostermeier M. J Mol Biol. 2004;336:263–273. doi: 10.1016/j.jmb.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Liang J, Kim JR, Boock JT, Mansell TJ, Ostermeier M. Protein Sci. 2007;16:929–937. doi: 10.1110/ps.062706007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright CM, Majumdar A, Tolman JR, Ostermeier M. Proteins Struct Funct Bioinf. 2010;78:1423–1430. doi: 10.1002/prot.22657. [DOI] [PubMed] [Google Scholar]
- 46.Shoichet BK. Nat Biotechnol. 2007;25:1109–1110. doi: 10.1038/nbt1007-1109. [DOI] [PubMed] [Google Scholar]
- 47.Möglich A, Moffat K. Photochem Photobiol Sci. 2010;9:1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- 48.a) Rockwell NC, Su YS, Lagarias JC. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nagatani A. Curr Opin Plant Biol. 2010;13:565–570. doi: 10.1016/j.pbi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 49.a) Ulijasz AT, Cornilescu G, Cornilescu CC, Zhang J, Rivera M, Markley JL, Vierstra RD. Nature. 2010;463:250–254. doi: 10.1038/nature08671. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang X, Kuk J, Moffat K. Proc Natl Acad Sci USA. 2009;106:15639–15644. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toettcher JE, Gong D, Lim WA, Weiner OD. Methods Enzymol. 2011;497:409–423. doi: 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 52.Leung DW, Otomo C, Chory J, Rosen MK. Proc Natl Acad Sci USA. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyszkiewicz AB, Muir TW. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 54.Toettcher JE, Gong D, Lim WA, Weiner OD. Nat Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harper SM, Neil LC, Gardner KH. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 56.Zayner JP, Antoniou C, Sosnick TR. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.02.037. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strickland D, Moffat K, Sosnick TR. Proc Natl Acad Sci USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.a) Vreede J, Juraszek J, Bolhuis PG. Proc Natl Acad Sci USA. 2010;107:2397–2402. doi: 10.1073/pnas.0908754107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xie A, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. Biochemistry. 2001;40:1510–1517. doi: 10.1021/bi002449a. [DOI] [PubMed] [Google Scholar]; c) Bernard C, Houben K, Derix NM, Marks D, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NA. Structure. 2005;13:953–962. doi: 10.1016/j.str.2005.04.017. [DOI] [PubMed] [Google Scholar]; d) Cheng G, Cusanovich MA, Wysocki VH. Biochemistry. 2006;45:11744–11751. doi: 10.1021/bi0608663. [DOI] [PubMed] [Google Scholar]
- 60.a) Morgan SA, Al-Abdul-Wahid S, Woolley GA. J Mol Biol. 2010;399:94–112. doi: 10.1016/j.jmb.2010.03.053. [DOI] [PubMed] [Google Scholar]; b) Morgan SA, Woolley GA. Photochem Photobiol Sci. 2010;9:1320–1326. doi: 10.1039/c0pp00214c. [DOI] [PubMed] [Google Scholar]
- 61.Fan HY, Morgan SA, Brechun KE, Chen YY, Jaikaran AS, Woolley GA. Biochemistry. 2011;50:1226–1237. doi: 10.1021/bi101432p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crosson S, Rajagopal S, Moffat K. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 63.Möglich A, Ayers RA, Moffat K. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Möglich A, Ayers RA, Moffat K. J Mol Biol. 2010;400:477–486. doi: 10.1016/j.jmb.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 65.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.a) Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang X, He L, Wu YI, Hahn KM, Montell DJ. Nat Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wildemann D, Schiene-Fischer C, Aumuller T, Bachmann A, Kiefhaber T, Lucke C, Fischer G. J Am Chem Soc. 2007;129:4910–4918. doi: 10.1021/ja069048o. [DOI] [PubMed] [Google Scholar]
- 68.Huang Y, Cong Z, Yang L, Dong S. J Pept Sci. 2008;14:1062–1068. doi: 10.1002/psc.1042. [DOI] [PubMed] [Google Scholar]
- 69.Huang Y, Jahreis G, Lucke C, Wildemann D, Fischer G. J Am Chem Soc. 2010;132:7578–7579. doi: 10.1021/ja1019386. [DOI] [PubMed] [Google Scholar]
- 70.Reiner A, Wildemann D, Fischer G, Kiefhaber T. J Am Chem Soc. 2008;130:8079–8084. doi: 10.1021/ja8015044. [DOI] [PubMed] [Google Scholar]
- 71.Shen X, Valencia CA, Szostak JW, Dong B, Liu R. Proc Natl Acad Sci USA. 2005;102:5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 73.Nagai T, Sawano A, Park ES, Miyawaki A. Proc Natl Acad Sci USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.a) Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]; b) Demaurex N, Frieden M. Cell Calcium. 2003;34:109–119. doi: 10.1016/s0143-4160(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 76.Korendovych IV, Kulp DW, Wu Y, Cheng H, Roder H, DeGrado WF. Proc Natl Acad Sci USA. 2011;108:6823–6827. doi: 10.1073/pnas.1018191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.a) Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]; b) Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.a) Tyler RC, Murray NJ, Peterson FC, Volkman BF. Biochemistry. 2011;50:7077–7079. doi: 10.1021/bi200750k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Murzin AG. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 79.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Proc Natl Acad Sci USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.a) Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]; b) Luo X, Yu H. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. Proc Natl Acad Sci USA. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Proc Natl Acad Sci USA. 2010;107:7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anderson WJ, Van Dorn LO, Ingram WM, Cordes MH. Protein Eng Des Sel. 2011;24:765–771. doi: 10.1093/protein/gzr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.a) Anderson TA, Cordes MH, Sauer RT. Proc Natl Acad Sci USA. 2005;102:18344–18349. doi: 10.1073/pnas.0509349102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mossing MC, Sauer RT. Science. 1990;250:1712–1715. doi: 10.1126/science.2148648. [DOI] [PubMed] [Google Scholar]
- 85.a) Ambroggio XI, Kuhlman B. J Am Chem Soc. 2006;128:1154–1161. doi: 10.1021/ja054718w. [DOI] [PubMed] [Google Scholar]; b) Hori Y, Sugiura Y. J Am Chem Soc. 2002;124:9362–9363. doi: 10.1021/ja026577t. [DOI] [PubMed] [Google Scholar]; c) Cerasoli E, Sharpe BK, Woolfson DN. J Am Chem Soc. 2005;127:15008–15009. doi: 10.1021/ja0543604. [DOI] [PubMed] [Google Scholar]
- 86.Dalal S, Regan L. Protein Sci. 2000;9:1651–1659. doi: 10.1110/ps.9.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.a) Alexander PA, He Y, Chen Y, Orban J, Bryan PN. Proc Natl Acad Sci USA. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Alexander PA, He Y, Chen Y, Orban J, Bryan PN. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Alexander PA, Rozak DA, Orban J, Bryan PN. Biochemistry. 2005;44:14045–14054. doi: 10.1021/bi051231r. [DOI] [PubMed] [Google Scholar]; d) He Y, Chen Y, Alexander P, Bryan PN, Orban J. Proc Natl Acad Sci USA. 2008;105:14412–14417. doi: 10.1073/pnas.0805857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He Y, Chen Y, Alexander PA, Bryan PN, Orban J. Structure. 2012;20:283–291. doi: 10.1016/j.str.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uversky VN, Dunker AK. Biochim Biophys Acta Proteins Proteomics. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.a) O’Donovan PJ, Livingston DM. Carcinogenesis. 2010;31:961–967. doi: 10.1093/carcin/bgq069. [DOI] [PubMed] [Google Scholar]; b) Huen MS, Sy SM, Chen J. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A, Arrowsmith CH. J Mol Biol. 2005;345:275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 92.Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber BL, El-Deiry WS. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 93.Cissell KA, Shrestha S, Purdie J, Kroodsma D, Deo SK. Anal Bioanal Chem. 2008;391:1721–1729. doi: 10.1007/s00216-007-1819-5. [DOI] [PubMed] [Google Scholar]
- 94.Flanagan JM, Kataoka M, Shortle D, Engelman DM. Proc Natl Acad Sci USA. 1992;89:748–752. doi: 10.1073/pnas.89.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kohn JE, Plaxco KW. Proc Natl Acad Sci USA. 2005;102:10841–10845. doi: 10.1073/pnas.0503055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stratton MM, Mitrea DM, Loh SN. ACS Chem Biol. 2008;3:723–732. doi: 10.1021/cb800177f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mitrea DM, Parsons L, Loh SN. Proc Natl Acad Sci USA. 2010;107:2824–2829. doi: 10.1073/pnas.0907668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.a) Stratton MM, Loh SN. Proteins Struct Funct Bioinf. 2010;78:3260–3269. doi: 10.1002/prot.22833. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stratton MM, McClendon S, Eliezer D, Loh SN. Biochemistry. 2011;50:5583–5589. doi: 10.1021/bi102040g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Skelton NJ, Kordel J, Chazin WJ. J Mol Biol. 1995;249:441–462. doi: 10.1006/jmbi.1995.0308. [DOI] [PubMed] [Google Scholar]
- 100.Gautel M. Pfluegers Arch. 2011;462:119–134. doi: 10.1007/s00424-011-0946-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mayans O, van der Ven PF, Wilm M, Mues A, Young P, Furst DO, Wilmanns M, Gautel M. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 102.a) Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schafer LV, Brandmeier B, Grater F, Grubmuller H, Gaub HE, Gautel M. Proc Natl Acad Sci USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gräter F, Shen J, Jiang H, Gautel M, Grubmuller H. Biophys J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.a) Leckband DE, le Duc Q, Wang N, de Rooij J. Curr Opin Cell Biol. 2011;23:523–530. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]; b) Gomez GA, McLachlan RW, Yap AS. Trends Cell Biol. 2011;21:499–505. doi: 10.1016/j.tcb.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 104.Schwartz MA, DeSimone DW. Curr Opin Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kesner BA, Ding F, Temple BR, Dokholyan NV. Proteins. 2010;78:12–24. doi: 10.1002/prot.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.a) Forman JR, Yew ZT, Qamar S, Sandford RN, Paci E, Clarke J. Structure. 2009;17:1582–1590. doi: 10.1016/j.str.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dalagiorgou G, Basdra EK, Papavassiliou AG. Int J Biochem Cell Biol. 2010;42:1610–1613. doi: 10.1016/j.biocel.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 107.Chen J, Zolkiewska A. PLoS One. 2011;6:e22837. doi: 10.1371/journal.pone.0022837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kumamoto CA. Nat Rev Microbiol. 2008;6:667–673. doi: 10.1038/nrmicro1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mederos y Schnitzler M, Storch U, Gudermann T. Curr Opin Pharmacol. 2011;11:112–116. doi: 10.1016/j.coph.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 110.a) Cutler T, Loh SN. J Mol Biol. 2007;371:308–316. doi: 10.1016/j.jmb.2007.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cutler T, Mills BM, Lubin DJ, Chong LT, Loh SN. J Mol Biol. 2009;386:854–868. doi: 10.1016/j.jmb.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ha J-H, Butler JS, Mitrea DM, Loh SN. J Mol Biol. 2006;357:1058–1062. doi: 10.1016/j.jmb.2006.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ha J-H, Karchin JM, Walker-Kopp N, Huang L-S, Berry EA, Loh SN. J Mol Biol. 2012;416:495–502. doi: 10.1016/j.jmb.2011.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peng Q, Li H. J Am Chem Soc. 2009;131:13347–13354. doi: 10.1021/ja903480j. [DOI] [PubMed] [Google Scholar]
- 114.Luheshi LM, Dobson CM. FEBS Lett. 2009;583:2581–2586. doi: 10.1016/j.febslet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 115.Tang C, Schwieters CD, Clore GM. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 116.Miller DM, III, Olson JS, Pflugrath JW, Quiocho FA. J Biol Chem. 1983;258:13665–13672. [PubMed] [Google Scholar]
- 117.Kunzelmann S, Webb MR. ACS Chem Biol. 2010;5:415–425. doi: 10.1021/cb9003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pan D, Philip A, Hoff WD, Mathies RA. Biophys J. 2004;86:2374–2382. doi: 10.1016/S0006-3495(04)74294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martin SR, Linse S, Johansson C, Bayley PM, Forsen S. Biochemistry. 1990;29:4188–4193. doi: 10.1021/bi00469a023. [DOI] [PubMed] [Google Scholar]
- 120.Vallée-Bélisle A, Ricci F, Plaxco KW. J Am Chem Soc. 2012;134:2876–2879. doi: 10.1021/ja209850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Motlach HN, Hilser VJ. Proc Natl Acad Sci USA. 2012 in press. [Google Scholar]
- 122.Björkman AJ, Binnie RA, Zhang H, Cole LB, Hermodson MA, Mowbray SL. J Biol Chem. 1994;269:30206–30211. [PubMed] [Google Scholar]
- 123.Halavaty AS, Moffat K. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 124.Getzoff ED, Gutwin KN, Genick UK. Nat Struct Biol. 2003;10:663–668. doi: 10.1038/nsb958. [DOI] [PubMed] [Google Scholar]
- 125.Zhang M, Tanaka T, Ikura M. Nat Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 126.Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]