Abstract
Missing phenotype data can be a major hurdle to mapping quantitative trait loci (QTL). Though in many cases experiments may be designed to minimize the occurrence of missing data, it is often unavoidable in practice; thus, statistical methods to account for missing data are needed. In this paper we describe an approach for conjoining multiple imputation and QTL mapping. Methods are applied to map genes associated with increased breathing effort in mice after lung inflammation due to allergen challenge in developing lines of the Collaborative Cross, a new mouse genetics resource. Missing data poses a particular challenge in this study because the desired phenotype summary to be mapped is a function of incompletely observed dose-response curves. Comparison of the multiple imputation approach to two naive approaches for handling missing data suggest that these simpler methods may yield poor results: ignoring missing data through a complete case analysis may lead to incorrect conclusions, while using a last observation carried forward procedure, which does not account for uncertainty in the imputed values, may lead to anti-conservative inference. The proposed approach is widely applicable to other studies with missing phenotype data.
Keywords: multiple imputation, missing data, quantitative trait loci
1. Introduction
Interest in identifying the genetic basis of variation in quantitative traits continues to grow. In humans, family-based linkage studies were once the prominent study design to identify quantitative trait loci (QTLs), but the advent of genome-wide association studies (GWAS) using unrelated subjects has made QTL identification more feasible than ever, and numerous medically relevant traits have been mapped using this approach in the past few years. The use of model organisms to map QTLs is complementary to studies in humans and also offers some notable advantages. The vast genetic diversity of different natural populations can be exploited to uncover different regions of genome that harbor QTLs that would otherwise be obscure, and at the same time experimental populations can be designed to avoid potential biases due to population structure. Additionally, environmental variables can be tightly controlled to reduce undesired variance. Finally, model organisms provide the opportunity for experimentation that could not be performed in humans.
The mouse is often used for such purposes. Divergent parental strains are crossed to form F1 heterozygotes, and these F1 mice are then intercrossed (or backcrossed to a parental line) to form F2s with recombinant chromosomes. The trait, or phenotype, of interest is quantified, and statistical methods are employed to assess the association between the genotype and phenotype (Broman, 2001). Since the mice are studied in a common environment, differences in phenotype may be attributed to genetic discordance. This approach has been used for numerous traits of biomedical relevance, and through further experimentation, investigators can identify the specific genes underlying the phenotype of interest. Because a large effort is expended to breed mice for these types of studies, the mouse genetics community has developed resources to facilitate and expedite QTL mapping. In particular, recombinant inbred lines (RILs), generated by repeated (e.g. > 20 generations) brother-sister matings of mice produced from intercrosses, have been developed and are commercially available. These offer the advantage of being a renewable resource; that is, mice of the same genotype can be phenotyped for multiple traits across space and time. The Collaborative Cross (CC) is the newest and most powerful resource of this type. The CC was designed to overcome many limitations of previous QTL mapping approaches by capturing the maximal genetic diversity of inbred strains while creating a balanced population structure (Chesler et al., 2008; Churchill et al., 2004). Specifically, the CC is a panel of recombinant inbred lines derived from eight-way crosses of classical (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, and NZO/H1LtJ) and wild-derived inbred strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ). The strengths of the CC compared to other approaches (Roberts et al., 2007) and initial applications with the developing lines of the CC have recently been described (Aylor et al., 2011).
Here we use unrestrained whole body plethysmography (WBP) to phenotype a respiratory system trait in developing lines of the CC. WBP measures the amplitude and frequency of breathing and, from the features of the breathing waveform, an empirical parameter known as enhanced pause (Penh) is calculated. Measurements of Penh in the presence of aerosolized saline and then increasing concentrations of methacholine, a bronchoconstrictor, are used as measures of breathing effort, and this changes in the context of lung inflammation due to allergen challenge (Hamelmann et al., 1997). Penh is therefore often used in mouse studies of allergic airway disease, a model of human asthma. Results from previous QTL mapping experiments have been nicely summarized in two recent papers (Camateros et al., 2010; Leme et al., 2010). Zhang et al. (1999) used an F2 intercross, applied an ovalbumin-induced allergic model, and identified QTLs on chromosomes 9, 10, 11 and 17. Ackerman et al. (2005) identified interacting loci on chromosomes 2 and 6, and most recently Camateros et al. (2010) identified loci on several different chromosomes. We sought to exploit the diversity of the CC to identify new QTLs for Penh. This diversity was immediately apparent during the phenotyping process, as some mice had Penh values that were very high even at low doses of the bronchoconstrictor methacholine, and hence phenotyping was stopped in order to protect the well-being of the mouse. This led to a situation in which there was missing data for some mice, a result that has not been described in previous studies with the same goal.
Without missing data, the standard QTL mapping strategy would be to (i) fit a genetic model at each marker (for a large number of markers), (ii) obtain a test statistic that quantifies the discrepancy in the phenotype value across different genotype strains at each marker, and (iii) apply a permutation test to determine if the largest observed test statistic is statistically significant. If statistical significance is achieved, then this is considered evidence of a QTL at the marker having the maximal test statistic. In the situation where data are missing for some of the mice, this analysis must be adapted to appropriately account for the missing data.
If data necessary for quantifying the phenotype of interest is missing for a subset of mice, a significant loss of power and bias may be introduced unless the missing data are appropriately handled. Frequently the phenotype value is a numerical summary of several observed characteristics of the mice, e.g., area under the curve (AUC) values obtained from dose-response data in drug metabolism studies. In this case, when one or more of these observed characteristics are missing for a subset of mice, the desired phenotype measure (AUC) cannot be computed. An analysis that only uses data from those mice with complete measurements may yield biased results, depending on the missingness mechanism. Rather than perform the desired QTL mapping analysis on just the subset of mice for which the phenotype quantity is available, a model that describes the characteristics used to compute the phenotype summary may be developed and used to impute the values for these characteristics. Then, the complete dataset obtained from the imputation model may be used to impute the missing phenotype summary. The approach of multiple imputation (MI) is preferred to imputing a single dataset, because it provides a way to incorporate uncertainty in the imputed values into the analysis (Little and Rubin, 2002; Schafer, 1997).
While genotype imputation has become a widespread tool in genetic association studies (Li et al., 2009), imputation of missing phenotype data has been less commonly applied by the QTL mapping community. In particular, previous studies of Penh did not observe the same level of phenotypic diversity as seen in the CC mice, and so missing data did not pose a major challenge for these studies (Ackerman et al., 2005; Camateros et al., 2010; Leme et al., 2010; Zhang et al., 1999). Some prior work beyond the literature studying breathing effort in mice has explored methods for handling missing phenotype data in linkage analysis, family-based studies, and pedigree analysis (Ding and Laird, 2009; Fridley and Andrade, 2008; Fridley et al., 2003; Xing et al., 2003). In these studies the phenotype investigated was a univariate, continuous parameter rather than a function of incompletely observed dose-response curves, and so the development of a suitable phenotype imputation model was not a primary focus. Additionally, one feature not emphasized by these studies is how inferential methods that account for the imputation of missing phenotype data should control for multiple comparisons, which is an important goal in mapping studies seeking to identify significant QTLs among a large number of potential markers.
In this paper we describe the methodological challenges involved in incorporating MI in QTL mapping when there is substantial missingness of a complex phenotype. We focus on a particular experiment looking at the genetic factors that contribute to breathing effort, as quantified by Penh, in mice. Our work provides two main contributions. First, we develop a novel imputation model that captures the unique features of the data. Second, we develop a methodology for combining MI with a commonly used statistical analysis for QTL mapping. One of the features of this approach is that it is a generalization of the analysis that would have been conducted had there been no missing data. While our imputation model is specific to the experimental protocol for measuring Penh over increasing doses of the bronchoconstrictor, the conjoining of MI and QTL mapping may be generally applied to other experiments with missing phenotype data.
The outline of the paper is as follows. In section 2 we introduce the phenotype and genotype data and in section 3 we define the phenotype summary for QTL mapping. We describe the phenotype model used for MI in section 4 and the methodology for combining imputed datasets with the QTL analysis in section 5. The application of the methodology to QTL mapping of the Penh data is detailed and the results presented in section 6. We conclude with a discussion of the methods and results in section 7.
2. Data
One hundred and sixty-two male mice that were part of a collaboration, described in Aylor et al. (2011), were obtained and housed singly at the National Human Genome Research Institute. To distinguish between the fully inbred lines of the CC and the mice used for this study (which are not yet fully imbred), we refer to these mice as “preCC” mice. Given that the mice were not yet fully inbred, each mouse’s genome was unique, and biological replicates were precluded.
2.1. Genotype Data
Genotyping and haplotype assigment methods have been described by Aylor et al. (2011). Briefly, mice were genotyped using the Affymetrix Mouse Diversity Array (Yang et al., 2009a). SNP data were then used to infer haplotype probabilities using a hidden Markov model (Mott et al., 2000). That is, for each region of the genome, the probabilities that the region descended from each of the eight parental strains of the CC were estimated, and these founder haplotype probabilities were used (not genotypes) in our QTL model described in Section 5.1. For this study we consider just chromosome 8 in order to demonstrate the application of the proposed methodology. Because the marker density exceeded the total density of recombinations in the cross, it was possible to reduce the chromosome 8 genotype data to approximately 1,380 intervals within which estimated genotype probabilities were essentially constant, indicating a single haplotype for that region. Interval boundaries were defined at transitions in highest probability genotype, based on Baum-Welch output from a Hidden Markov model. In most intervals, haplotype probabilities were near 1 for the inferred states. The haplotype probabilities for each mouse sum to two (because each mouse inherited two CC alleles), and since the mice are the products of inbreeding but not yet fully inbred, most regions are homozygous for one CC strain.
2.2. Phenotype Data
Whole body plethysmography (WBP) phenotyping
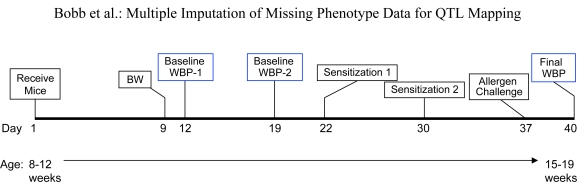
A longitudinal study design (Lofgren et al., 2006) was conducted in which each mouse was phenotyped for Penh using WBP at three time points, the first two being baseline measurements, and the third subsequent to an allergen challenge, as shown in Figure 1. The two baseline measurements were conducted within one week, and hence we assume that the baseline phenotypes are exchangeable. After acclimatization to the WBP chamber over 15–20 minutes, mice were phenotyped for Penh as follows. The mice were administered increasing doses (0, 3.1, 6.2, 12.5, and 25 mg/ml) of methacholine by nebulization over a 2.5 minute period. Following the nebulization period at each dose, mice were followed for five minutes, and average Penh over the five minute period was calculated.
Figure 1:
Timeline of study protocol on 162 preCC mice. Key phenotyping timepoints are outlined in blue. BW = body weight and WBP = whole body plethysmography.
Allergen challenge protocol
The protocol we employed is a slight modification of that described by Kelada et al. (2011). Low endotoxin Der p 1, from the Dermatophagoides pteronyssinus species of house dust mite, was purchased from Indoor Biotechnology. Mice were sensitized with 10 g Der p 1 by intra-peritoneal injection on days 22 and 30 of the study. On day 37, mice were challenged by oro-tracheal aspiration with 50 μg of Der p 1 diluted in 40 μl of saline. Peak inflammatory responses occurred 72 hours after airway challenge, and so mice were phenotyped at this time point.
The phenotyping protocol just described was designed to yield three methacholine-Penh dose-response curves for each mouse, including two replications at baseline (before the allergen challenge) and a single curve measured after the allergen challenge. The phenotype summaries, which are functions of the three dose-response curves, are described in section 3.
We next introduce some notation. Let Yijkl denote the Penh response for mouse i at dose dj of methacholine, for replication k, of condition l, where l ∈ {pre, post} is the pre- or post-allergen challenge condition. Denote the methacholine-Penh dose-response curve for mouse i of replication k and condition l by Yikl(d), where d is the dose, and note that Yijkl = Yikl(dj) for j = 1 to 5.
Table 1 summarizes the distribution of missingness of the dose-response curves. Given that a Penh value is missing for a particular dose, it is missing for all subsequent doses. Thus there are six possible missingness patterns for each methacholine-Penh curve: Penh missing at all 6 doses of methacholine; Penh observed at the first dose, but missing at doses 2 through 5; Penh observed at the first two doses, but missing at doses 3 through 5, and so on. Values at higher doses are missing in order to protect the well-being of the mouse. High concentrations of methacholine can lead to overstimulation of the airways, causing excessive narrowing and hence extreme difficulty breathing, and/or systemic cholinergic crisis. Additionally, we note that a subset of 40 mice were not phenotyped for the first repetition of the pre-condition (i.e. were only phenotyped at a single timepoint) due to a change in the WBP protocol.
Table 1:
Number of mice having each missingness pattern for phenotyping at the two repetitions of the pre-allergen challenge conditions (Pre1, Pre2) and at the post-allergen challenge condition. In each missingness pattern, a “1” in position j indicates that Penh was observed at dose j, while “0” denotes a missing value. For example, 15 mice had complete data at the second repetition of the pre-condition and were missing Penh at the last dose of the first repetition.
| Pre1 |
Pre2 |
Post |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 11111 | 11110 | 11100 | 11000 | 10000 | 00000 | Total | |||
| 11111 | 71 | 9 | 0 | 0 | 0 | 0 | 80 | 11111 | 81 |
| 11110 | 15 | 18 | 1 | 0 | 0 | 0 | 34 | 11110 | 44 |
| 11100 | 0 | 2 | 3 | 0 | 0 | 0 | 5 | 11100 | 25 |
| 11000 | 1 | 0 | 2 | 0 | 0 | 0 | 3 | 11000 | 6 |
| 10000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10000 | 0 |
| 00000 | 31 | 8 | 1 | 0 | 0 | 0 | 40 | 00000 | 6 |
| Total | 118 | 37 | 7 | 0 | 0 | 0 | 162 | Total | 162 |
To understand the potential impact this missing data may have on QTL mapping, we first provide a brief overview of commonly used terminology from the statistical literature on missing data. Historically, three classes of missing data mechanisms have been defined, which serve as a useful tool for selecting appropriate statistical analyses for handling missing data (Rubin, 1976; Little and Rubin, 2002). The first mechanism, missing completely at random (MCAR), occurs when the missing data come from the same underlying probability distribution as the observed data. The second mechanism, referred to as missing at random (MAR), arises when the probability that a data point is missing depends only on observed variables, which can include covariates (e.g. mouse body weight) and outcomes (e.g. observed Penh values Yijkl). Missing data are classified by the third mechanism, missing not at random (MNAR), when the probability that a data point is missing depends on unobserved variables. If the missingness mechanism is MCAR, then statistical analyses based only on individuals with complete data will yield unbiased results, though substantial loss of power may result under high levels of missingness. When the less stringent assumption of MAR holds, it is sufficient to posit a model for the outcome process and draw inference using multiple imputation or maximum likelihood methods. However if the mechanism is MNAR, then one must model the missingness mechanism as well.
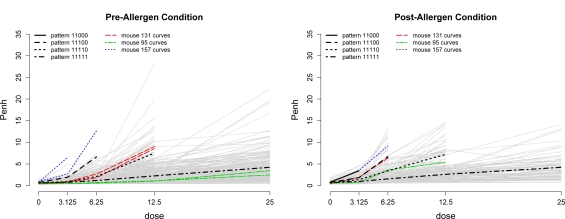
For the 40 mice that are completely missing dose-response curves from the first repetition of the pre-allergen challenge condition, it is plausible that these curves are MCAR, as the decision to change the WBP protocol was independent from the data-generating process. For these mice, the missing curves were completely imputed based on the imputation model described in section 4, which assumes the less restrictive MAR mechanism. We next investigate the potential missingness mechanism for the partially observed curves. Figure 2 displays the observed data for all mice in both the pre- and post-allergen challenge conditions. We observe that curves that have missing Penh values at the higher doses of methacholine tend to be increasing faster than those that are not missing Penh values. If the data were MCAR, then the rate of increase of curves with missing data would be the same as the rate of increase of the completely observed curves. It follows that the missingness mechanism is not MCAR for these partially observed dose-response curves, and so a complete case analysis would be inappropriate. However, the observed data cannot similarly be used to distinguish between MAR and MNAR. The approach for integrating multiple imputation with QTL mapping described in section 5 is valid under the MAR assumption.
Figure 2:
Observed mouse methacholine-Penh curves over five dosage levels in the pre-allergen challenge condition (left plot) and in the post-allergen challenge condition (right plot) are shown in gray. Trajectories from a sample of three mice are highlighted. Black lines denote the average of the population of curves with differing levels of missingness. Specifically, each black line corresponds to the average of the Penh values at each dose within each missingness pattern. Patterns are written such that a “1” in position j indicates that Penh was observed at dose j, while “0” denotes a missing value.
It is important to point out that there may be a philosophical objection to considering Penh values at the higher doses to be missing. In some cases, it is possible that the mouse may not have survived exposure to the bronchoconstrictor (methacholine) beyond a particular dose, but this dose of maximal tolerance is not observed by the study design. Thus the concept of “missing” may not be well-defined at the doses where Penh values have not been measured, if those values could never exist. Nonetheless, it may be reasonable to assume based on discussions with our collaborators that a Penh value at least one dose beyond the largest observed dose does exist. We will address this concern by considering alternative specifications of the phenotype summary (section 3) which do not depend on the highest doses, so as to mitigate reliance upon imputed Penh values at doses where their existence is questionable.
3. Phenotype Summaries for QTL Mapping
There are two primary phenotype summaries of interest for each mouse. The first phenotype , defined as the area under the curve for the pre-allergen challenge condition, quantifies breathing effort under normal physiologic circumstances. The second is the area under the curve for the pre-allergen condition subtracted from the area under the curve for the post-allergen condition. This latter phenotype quantifies the increased breathing effort when the lung is affected by airway inflammation. The two summaries and are correlated but are both unique biological entities of interest to the lung research community. Since mice have two replications of the pre-condition, we use the average area under the curve over the two replications. Specifically, we define the phenotype summaries of interest by
| (1) |
| (2) |
where is the average of the two pre-condition Penh measurements at dose d. For the purposes of calculating AUC for (1) and (2), we assume a linear interpolation of each dose-response curve between consecutive doses. The phenotypes of greatest scientific interest are and , with the integration through the fifth dose.
To demonstrate the potential gain achievable by an imputation approach versus a completers only analysis, we calculate the number of mice for whom and could be calculated using the observed data. Note that there are two ways to define a complete case for the phenotype summaries (1) and (2), since there are at most two repetitions of the pre-allergen challenge condition for each mouse. First, one could consider an exclusive criterion, where it is required that both precondition curves be observed at each dose up through d in order for (and hence since it is a function of ) to be computed. Alternatively, one could consider an inclusive criterion where is computed if either of the two pre-condition curves is observed through dose d. For this latter case, if there is only one measurement at a particular dose d, the value Ȳi(d) is set to be that measurement, and if there are are two measurements at that dose, the value Ȳi(d) is set to be the average of the two measurements. Using the inclusive criterion, out of the 162 mice, one could calculate for 127 mice and for 79 mice. Requiring the exclusive criterion, and could be calculated for only 71 mice and 53 mice, respectively.
If one were to conduct an analysis with only the complete cases, results may be biased since mice with complete data tend to have lower Penh values. To increase power and reduce the potential for bias in our analysis, we build a model for the methacholine-Penh curves that incorporates scientific knowledge of the shape of the relationship and use multiple imputation within the QTL mapping framework.
Alternative phenotype specifications
We considered sensitivity to the particular phenotype specifications and , which were calculated by integrating up to the fifth dose. There are a few reasons why alternative specifications to the original phenotypes might be desired. First, since the doses were not equally spaced, calculating area under the curve is more heavily influenced by observed Penh values at higher doses than at lower doses. Second, since there is more missing data at higher doses (Table 1), phenotype specifications based on fewer doses would be less sensitive to the imputation scheme and more subjects would be included in a complete case analysis leading to reduced bias. Finally, there is the potential philosophical issue regarding the imputation of Penh values that may not exist (described above). To address these concerns, we considered alternative phenotype specifications that are functions of Penh values at only the first three or four doses, namely and where we consider both j = 3 and j = 4.
4. Phenotype Model for Multiple Imputation
We developed a flexible parametric model to capture the nonlinear shape of the methacholine-Penh exposure-response relationship over the five dosage levels, before and after the allergen challenge. The model also incorporates mouse-specific covariate data, namely body weight, measured three days prior to the first Penh phenotyping (Figure 1), and the year phenotyping was conducted, as both were significantly associated with phenotype values. Since Penh must be strictly greater than zero, and since scientific knowledge of the range for Penh necessitates a finite upper bound, we modeled the transformed Penh as
| (3) |
where Zi1 is an indicator of year (2008 or 2009), Zi2 is body weight, and K is the upper bound. The maximum Penh value that we observed in the dataset was 28.2, and so we set the upper bound to be K = 30.
In the model (3), each mouse and each pre- or post-challenge condition was allowed to have its own knot til, which may be located at any of the last four doses. Note that a knot at the last dose corresponds to the case where a knot is not needed. The slope after the knot β4(til) was allowed to depend on the location of the knot. To account for the correlation of repeated Penh measurements from the same mouse, while at the same time accounting for the heterogeneity of the mouse-specific curves before versus after the allergen challenge, we assumed a random-effect distribution on the model coefficients. Specifically, we set β3il = β3l + bi and assumed (β0i, bi)′ ∼ N((β0, 0)′, Σ). We assumed the residuals to be independent with eijkl ∼ N(0, ) to capture the heterogeneity of the Penh response at different dose levels before and after the allergen challenge. Since some of the observed curves were non-monotone, the term eijkl can be thought of as measurement error from the true underlying curve, which is assumed to be monotonically increasing.
4.1. Prior Distributions
We assigned prior distributions for locations of the knots til; the model coefficients β1, β2, and β4(til); the parameters for the random-effect distribution β0, β3l, and Σ; and the heterogeneity parameters (j = 1, . . . , 5, l = pre, post). Specifically, we assumed that the knot locations come from a multinomial distribution, where the prior probability that the knot is at each of the four potential dose levels is 1/4. The parameters β0, β1, and β2 were each assigned independent N(0, 106) prior distributions. For the variance parameters, we assumed σjl ∼ unif(0, 100) and for Σ = var((β0i, bi)′), we assumed that σ(β0i) ∼ unif(0, 100), σ(bi) ∼ unif(0, 100), and cor(β0i, bi) ∼ unif(0, 1), where σ(·) denotes the standard deviation.
In order to incorporate the requirement that each mouse’s curve be monotone, we would require that β3il ≥ 0 and β3il + β4(til) ≥ 0. Rather than introducing this strong restriction on each curve, we elected to just have the population average curve be monotone by incorporating the restrictions β3l ≥ 0 and β3l + β4(t) ≥ 0, (j = 1, . . . , 5, and l = pre, post) on the prior distributions for these parameters. Thus we specified the prior distributions as
where I(·) is an indicator function.
4.2. Multiple Imputations
The model was used to impute M = 10 complete datasets from the posterior predictive distribution of Yijkl. Justification for the choice of M is provided in section 6.1. In particular, the mth complete dataset was obtained as follows. First randomly select a posterior sample s. Then, for each value of Yijkl that was missing in the original dataset, simulate a sample . Finally, set .
5. Incorporating MI in QTL Mapping
We first review a common approach for QTL mapping when there is complete data, and then we outline our methodology to integrate multiple sets of imputed data into the QTL mapping algorithm. We also consider some more routine methods for handling missing data that may be compared with the proposed multiple imputation approach.
5.1. QTL Mapping for Complete Data
Consider the case of no missing data. Let Yi be the phenotype summary for the ith mouse, i = 1, . . . , I. Let Zi be a vector of covariate data for mouse i and let Z be the matrix of covariate data having rows Z′i. Further, let Xp be the matrix of data for marker p (p = 1, . . . , P), where each row represents a mouse and each column (g = 1, . . . ,G) is the corresponding haplotype probability. In particular, a row Xip of the matrix Xp represents the vector of haplotype probabilities of mouse i having each of the eight CC parental haplotypes (see Section 2.1 and Aylor et al. (2011) for a description of haplotype inference methods). The goal of QTL mapping is to ascertain for which markers there is a difference in the phenotype value across the different genotypes. To do this, one conducts a hypothesis test for each marker and then computes a p-value that accounts for multiple testing of the P different markers.
To test the null hypothesis that there is no difference in the mean phenotype value across strains for marker p, we used an additive genetic model (described in Aylor et al. (2011)),
| (4) |
Note that G = 2 corresponds to the common scenario in which only two genotype classes are under consideration. The null hypothesis H0 : βgp = 0 for g = 1, . . . , G − 1 may be tested using an appropriate test statistic (e.g. likelihood ratio test, score test, or Wald test). For the remainder of this section, we will consider the multivariate Wald statistic, as this is the statistic that will be generalized in the analysis that takes into account multiple imputation. Denote the desired test statistic for the pth marker by . Thus, for the Wald test of the null hypothesis, we compute
| (5) |
where βp = (β1p, . . . , βG−1,p) and Σp is the estimated variance of the estimated parameter β̂p. A p-value may be computed from the appropriate asymptotic distribution (in this case ) or from a permutation distribution. Since a permutation test may be readily adapted to account for the testing of multiple markers, it is a practical choice.
A permutation test may be implemented as follows. For iterations r = 1, . . . , R, first permute the phenotype summaries for the I mice to obtain , keeping the design matrix Xp as well as the matrix of covariate data Z fixed. Then calculate the test statistic for the permuted summaries for each marker p. Identify the largest of the test statistics across markers at the rth iteration and denote this by . The p-value for the largest observed test statistic is then calculated as the proportion of iterations for which . P-values may similarly be obtained for the second largest observed test statistic, and so on.
5.2. QTL Mapping for Imputed Data
For each marker p and each imputed dataset m, the genetic model (4) is fit, yielding estimates of and the variance-covariance matrix . The generalization of the Wald test statistic for the pth marker that accounts for the variability in the imputed datasets is defined by
| (6) |
where
This test statistic is the multivariate analogue to the test statistic used for hypothesis tests of a one-dimensional parameter for multiply imputed datasets. However, unlike the univariate case, finding a suitable reference distribution for the multivariate analogue (6) is not straightforward. Approximate reference distributions have been proposed that are based on making additional assumptions about the between- and within-imputation covariance matrices Σ̄p and Bp, and which use an alternative estimate of the total variance Vp (for details, see Little and Rubin (2002) or Schafer (1997)). In addition to requiring additional assumptions, as with any asymptotic approximation it is also assumed that the sample size is large enough so that the approximation is reasonable.
An alternative to invoking large sample properties and applying the approximations necessary to derive an appropriate reference distribution is to use a permutation test. In addition to requiring fewer assumptions about the data, permutation testing may be conveniently employed to account for the testing of multiple genetic markers, as outlined above. The QTL mapping algorithm for complete data may be seamlessly integrated with multiple imputation by replacing the test statistic (5) used in the permutation testing procedure for complete data with from (6). In particular, we run the algorithm for R iterations as follows. For r = 1, . . . , R,
Randomly permute the labels of the I mice. Denote this permutation by j1, j2, . . . , jI.
- For each of the p markers,
- For each imputed dataset m, fit the additive genetic model (4) for the permuted outcomes . Note that the marker and covariate data (X1p, Z1); (X2p, Z2), . . . , (XIp, ZI) retain their original ordering. Obtain the estimates of and based on the permuted data.
- Calculate the permuted test statistic based on (6) using the estimates of , (m = 1, . . . , M) from the previous step.
Identify the largest permuted test statistic .
The p-value for the largest observed test statistic is then calculated as the proportion of iterations r for which , where is the maximal test statistic of the observed data. Note that the imputation model is fit just once prior to permutation testing, and that the same M imputed datasets are used throughout. In addition, as in the complete data setup, the order of the covariate data Z and marker data Xp remains unchanged throughout the procedure.
5.3. Other Approaches for Missing Data
Rather than developing an application-specific imputation model and incorporating the variability of multiple imputed datasets into QTL mapping, there are several alternative approaches to handle missing phenotype data.
We first consider a complete case analysis (CCA) where we perform the QTL mapping procedure separately for the subset of mice for which could be computed and for the subset for which could be calculated. With regards to the two repetitions of the pre-allergen challenge condition phenotyping, we used the inclusive criterion for defining a complete case (section 3). Of the 162 mice that were both genotyped and phenotyped, the statistic could be calculated for 127 mice and could be calculated for 79 mice. Additionally, we considered the last observation carried forward (LOCF) method (Heyting et al., 1992). While each of the 162 mice had at least one measurement in the pre-allergen condition, there were 6 mice without any post-allergen measurements. Thus, while the phenotype summary could be computed for each mouse under the LOCF imputation scheme, could be calculated for 156 of the 162 mice. For both CCA and LOCF, QTL mapping was performed using the permutation testing method for complete data described in section 5.1.
6. Application to Penh Data
6.1. Multiple Imputation
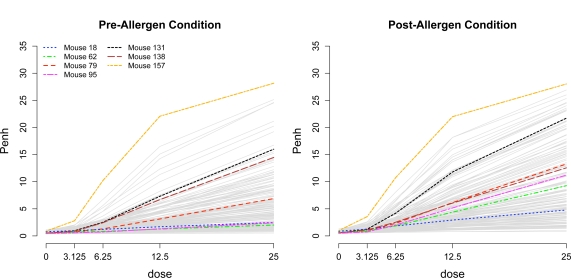
The phenotype model from section 4 was fit using WinBUGS version 1.4.3 (Lunn et al., 2000). We ran a single chain for 50000 iterations, with a burn-in of 10000, and we kept every fifth sample, for a total of S = 8000 posterior samples. The fitted methacholine-Penh curves are shown in Figure 3. Specifically, the fitted value of Yijkl (on the non-transformed scale) was estimated as K exp(μ^ijl)/{1 + exp(μ̂ijl)} where μ^ijl is the posterior mean of μijl = β0i + β1Zi1 + β2Zi2 + β3ildj + β4(til)(dj – til)+ from model (3). Note that since μijl is the median of the transformed Penh value log{Yijkl/(K – Yijkl)}, by taking a monotone transformation, it follows that K exp(μijl)={1 + exp(μijl)} is the median of Yijkl. We see that the Penh values in the post-allergen challenge condition are generally larger than in the pre-condition across the five doses of methacholine, as expected based on the change in lung function due to allergen challenge. However, this trend is not true for all mice; as shown in Figure 3, for example, values of the dose-response curve in the pre-condition exceed values in the post-condition for mouse 138. Additionally, these plots show that the variability in the methacholine-Penh curves increases at larger doses of methacholine.
Figure 3:
Fitted methacholine-Penh curves for the pre- and post-allergen challenge conditions. Curves from a sample of seven mice are highlighted.
To assess goodness-of-fit of the imputation model, we examined the posterior predictive intervals (PPI) of the methacholine-Penh dose-response curves. In particular, for each mouse i, each dose j, and each challenge condition l, we generated 8000 samples from the posterior predictive distribution of Yijkl using the retained MCMC samples, from which we calculated the 2.5th and 97.5th percentiles. We found that for the pre-condition, out of the 284 observed Penh values at the first dose, 97.2% were contained within the respective posterior-predictive interval; of the 284 observed Penh values at the second dose, 97.2% were within the PPI; at the third dose 97.9% of the 281 Penh values were within the PPI; of the 269 values at the fourth dose, 98.1% fell within the PPI; and of the 198 values at the fifth dose, 98.0% were contained in the PPI. For the post-condition, out of the 156 observed Penh values at the first dose, 98.7% were contained within the respective posterior-predictive interval; of the 156 observed Penh values at the second dose, 94.9% were within the PPI; 98.0% of the 150 Penh values at the third dose were within the PPI; of the 125 values at the fourth dose, all fell within the PPI; and of the 81 values at the last dose, 98.7% were within the PPI.
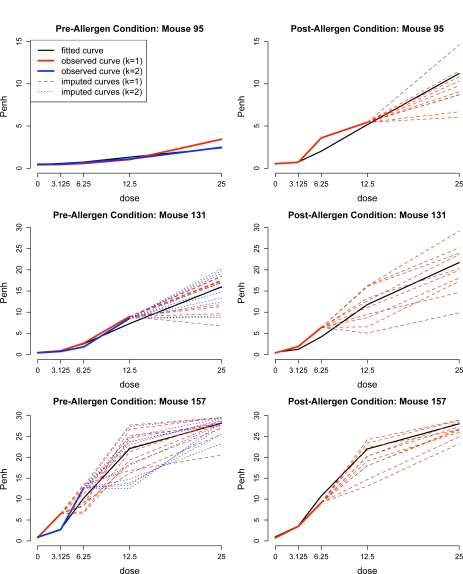
As described in section 4.2, M = 10 complete datasets were imputed. Figure 4 shows, for a sample of three mice, the fitted curves from the model and the corresponding imputed curves obtained from the model. We then applied the phenotype summary map from equations (1) and (2) to obtain for each mouse i and each complete dataset m (where Yi denotes either or ). As justification for the adequacy of M = 10 imputations, we applied the theory developed by Rubin (1987) to estimate the relative efficiency of an estimate based on M imputations to one based on an infinite number of imputations. We found that for the analysis with , the relative efficiency of multiple imputation with M = 10 datasets for estimating βp was ≥ 99% for each of the P markers; for the relative efficiency was ≥ 97% for each of the P markers.
Figure 4:
Mouse methacholine-Penh curves for three mice for the pre-allergen challenge condition (repetitions k = 1, 2) and post-condition. Black lines denote the fitted curves, i.e. the estimated median of Penh at each dose from fitting the phenotype model (3). Dashed lines denote the 10 imputed curves corresponding to each observed curve with missing values.
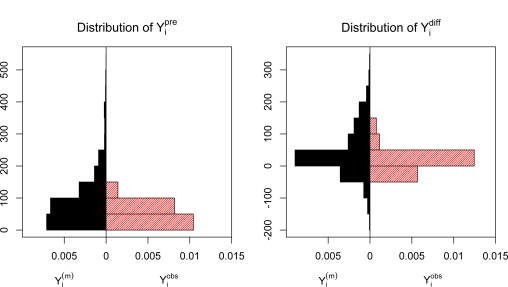
Comparing the distribution of the simulated from the posterior predictive distribution of Yi based on model (3) to the phenotype summaries calculated from mice with complete dose-response data shows that these distributions differ (Figure 5). In particular, the distribution of the imputed phenotype summaries has higher variability and wider tails than the observed phenotype summary values for both and . This can be explained by the fact that mice with missing Penh values tend to be those with larger Penh values at the higher doses, since phenotype summaries will only be large in absolute value if at least one of the pre- or post-allergen challenge curves has high Penh values.
Figure 5:
Distribution of phenotype summaries across mice for and . Solid histograms correspond to the distribution of the imputed across mice and imputations. Shaded histograms correspond to the distribution of the observed phenotype summaries calculated from the mice with complete data.
6.2. QTL Mapping
We implemented QTL mapping using the methods described in section 5 for the phenotype summaries and , j = 3, 4, 5. For the QTL mapping model (4), the covariates were Zi = (Zi1, Zi2) where Zi1, Zi2 were the covariates used in the imputation model (section 4).
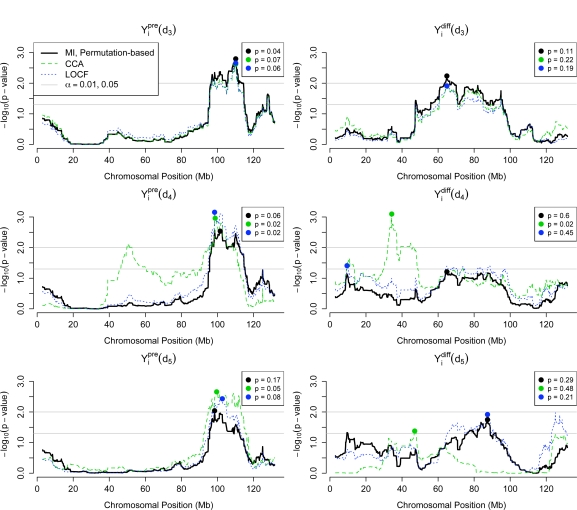
We first describe results from our primary analysis of incorporating multiple imputation within the QTL mapping framework through permutation testing (section 5.2) for the phenotype summaries and , and then we compare these results to results based on using CCA and LOCF as well as results based on the alternative phenotype specifications. Figure 6 shows the − log10(p-values) at each marker along the genome, for each of the different approaches and for each phenotype specification, that do not account for multiple testing. The p-values of the maximal observed test statistic for each approach, adjusted for multiple comparison, are included in the legend in the top right of each plot (Figure 6).
Figure 6:
Plot of – log10(p-values) across different approaches for QTL mapping of and , over specifications of the phenotype summaries based on integrating up to dose dj (j = 3, 4, 5). Plotted circles correspond to the location of the maximal test statistic across each approach, and legend in upper right lists p-values for , adjusted for multiple comparison. Horizontal lines denote significance corresponding to the 0.01 and 0.05 levels (on the p-value scale).
Incorporating multiple imputation
Using equation (6)x, we calculated the observed test statistics (p = 1, . . . , 1380) that account for the multiple imputation for each marker p. Permutation testing was based on R = 10000 permutations. We first examine the p-values that do not adjust for the fact that 1380 hypothesis tests (corresponding to the 1380 markers) were performed. For , there are two regions for which the p-value is less than 0.05, at chromosomal positions 95.7–109.0 megabases (Mb) and at 109.3–112.3 Mb, which consists of 124 and 24 consecutive markers, respectively; there is also a single marker having an unadjusted p-value below 0.01. The smallest p-value of 0.009 occurs for the marker with the maximal test statistic at 98.4 Mb. When we accounted for multiple comparison in the permutation test, the p-value for the largest observed test statistic was 0.17.
For , four regions of the genome were identified as having consecutive markers with unadjusted p-values less than 0.05, at 9.5–9.8 Mb, 76.2–79.8 Mb, 82.0–88.9 Mb, and 91.2–92.0 Mb, which consisted of 6, 36, 56, and 9 markers, respectively. The largest test statistic was (p unadjusted = 0.018) at 87.4 Mb. When we accounted for multiple comparison in the permutation test, the p-value for this largest observed test statistic was 0.29.
Alternative approaches
For the alternative approaches of complete case analysis (CCA), last observation carried forward (LOCF) imputation, and sensitivity analyses based on calculating the phenotypes using the first three or first four doses, we repeated the QTL mapping procedure.
We first observe that, consistent with the fact that there is much less missing data for calculating and than for calculating and , the discrepancy in results across the CCA, LOCF, and MI approaches is reduced for phenotype specifications based on fewer doses (Figure 6). The p-values adjusted for multiple comparison suggest that there may be a QTL for between chromosomal positions 95 Mb and 113 Mb. For a particular specification of (e.g. calculated by integrating up to the third, fourth, or fifth dose), the location of the QTL does not vary much across the approach used for handling missing data. However, using one of the more naive approaches (CCA or LOCF) may lead to overstating the statistical significance, since p-values are lower under both CCA and LOCF than under MI for and . For the phenotype , given the high adjusted p-values for the largest test statistic, and the inconsistency in the location of the largest effect across the three phenotype specifications, there is little evidence of a QTL. However, the CCA approach does identify a QTL at 34.3 Mb for , demonstrating that restricting to complete cases introduces bias that may yield false positives.
7. Discussion
In this paper we propose a framework for handling missing phenotype data in QTL mapping experiments through multiple imputation (MI), which we then apply to a study of genetic markers for allergic airway disease in mice. To conduct MI we develop a novel phenotype model describing the methacholine-Penh dose-response relation. We also compare the permutation method for QTL mapping with multiply-imputed datasets to two less sophisticated, but more commonly used, approaches for dealing with missing data. Moreover, we perform sensitivity analysis based on the phenotype summary specifications to assess the robustness of our conclusions to the degree of missingness present and to address a philosophical objection to the imputation of Penh values at doses where those values may not exist. This study focused on chromosome 8 as a “proof of concept,” in order to illustrate the proposed methodology for integrating MI with QTL mapping. In the future this approach will be applied to conduct QTL mapping for each of the remaining chromosomes.
The expectation-maximization algorithm (Dempster et al., 1977) and hidden Markov models have been used for dealing with missing genotype data in QTL mapping experiments (Broman and Sen, 2009). However, though some previous genetic studies have investigated approaches for handling missing phenotype data (Ding and Laird, 2009; Fridley and Andrade, 2008; Fridley et al., 2003; Xing et al., 2003), these types of approaches have not been widely adopted by the QTL mapping community. Generally, the experiment has either been conducted so that missing data are considered to be minimal and analyses ignore the problem, or ad hoc methods such as LOCF have been used without justification. However, these approaches may yield questionable results. Restricting to the subset of subjects (e.g. mice) for which there is complete data has the potential to lead to biased results if the missingness mechanism is not missing completely at random. On the other hand, though a single imputation approach precludes bias induced by a complete case analysis, it may introduce other biases if the imputation at the larger doses underestimates the values of the true curve. Further, by treating imputed values as observed data, single imputation may understate the variability underlying the imputed values which may lead to incorrect inference (Rubin, 1987).
In this study the range of Penh values observed (∼1–28) greatly exceeds those published in the literature (maximum values ∼10). This suggests that the genotypic diversity of the Collaborative Cross (CC) produces remarkable phenotypic diversity. In our case, the diversity of breathing effort phenotypes (Penh) was so high that the protocol had to be curtailed to prevent undue stress to the mice. This resulted in missing data, necessitating the phenotype imputation approach we have developed. Our results indicate that on chromosome 8 there is a potential QTL for baseline Penh when integrating up to 12 mg/ml methacholine (dose 4), between chromosomal positions 95 Mb and 113 Mb. The identified region contains 203 genes, including 195 that are protein-coding genes and several that are microRNA genes. This QTL is near a QTL previously reported by Camateros et al. (2010), lending support that it may be real. Validating this QTL and identifying the causal variants within this region will require additional experimentation as well as bioinformatic analyses incorporating complementary data (e.g. gene expression data, SNP data, as described in Aylor et al. (2011)).
It is worth noting that the experiment we conducted was limited by the availability of a relatively small set of CC lines (n = 162). When the CC lines are fully developed, more than 300 will be available, thereby providing a considerable increase in sample size as compared to the current study. At that time, biological replicates will also be available. Experimental noise or measurement variation can be reduced using multiple replicates, and therefore power will also increase.
The imputation model we developed flexibly captures the nonlinear shape of the methacholine-Penh dose-response curves, and achieves a good fit to the data. We additionally assessed sensitivity to the specification of the phenotype imputation model (3) and prior distributions, finding that the fitted curves were not highly affected and that the location of the potential QTL remained consistent across specifications. In comparing the proposed methods to two simpler approaches for handling missing phenotype data, our results demonstrate the potential pitfalls of using complete case analysis (CCA) in the presence of substantial missing data when the data are not missing completely at random. We found that while the CCA method implied the presence of a QTL for the phenotype , imputation approaches did not yield consistent results. Similarly, failing to account for the uncertainty in imputing a single dataset, as in LOCF, may lead to anti-conservative inference as suggested by the results for and .
The approach we described for conjoining MI with QTL mapping is easily generalizable to different study designs and QTL mapping analyses. For example, QTLs for diabetes-related traits are an area of considerable interest (Clee and Attie, 2007). Glucose tolerance tests are routinely performed in these studies, yet these measurements are inherently limited by an upper bound of the assay of ∼400–600 mg/dL (Jarvis et al., 2005; Pawlak et al., 2004). When such out of range values are observed, investigators typically invoke simple approaches to deal with the missing data, potentially leading to false positive or false negative results. In this situation, an imputation model suitable to the application may be developed, and then multiple imputation may be incorporated into QTL mapping using the methodology we have described. Additionally, while this paper focused a particular QTL mapping analysis, the approach for integrating multiple imputation with QTL mapping is not limited to the chosen analysis. For example, rather than testing the statistical significance of the largest observed test statistic max( ), one could easily adapt the permutation testing procedure to assess the significance of the largest series of consecutive markers having test statistic above a particular threshold.
We considered a two-stage analysis where, in the first stage we developed a model for the methacholine-Penh dose-response curves and performed multiple imputation, and in the second stage we performed QTL mapping to assess the association between a collection of genetic markers and the mouse-specific phenotype summaries. To incorporate multiple imputation within the QTL mapping analysis, we applied the combining rules developed by Rubin (1987) to obtain a test statistic that accounts for the variability of the imputed datasets. An alternative method is the “fuzzy p-values” approach of Thompson and Geyer (2007). Adapting this approach to our application would be an important direction of future work. A possible limitation of applying a two-stage analysis as we have done is that, given the hypothesis that genotype and phenotype are associated, it could be argued that the phenotype model for multiple imputation should include available genotype information. However, including the totality of genotype data would yield an ill-posed problem, with more predictors than responses. One solution would be to develop an overarching model that jointly describes both phenotype data (observed and unobserved) and genotype data, which would be used for QTL mapping.
Another direction of future work is to explore models for nonignorable missingness, as it is likely the assumption that the unobserved phenotype data are missing at random (MAR) is too simplistic. Since the decision to proceed to a higher dose was based on whether the mouse was observed to be struggling at the beginning of the phenotyping of that dose, the assumption that the data are MAR may not be plausible. This could be addressed using mixture models (Little, 1994) with models similar to (3) specified for each missing data pattern as given in Table 1. To implement this, one would need to assume that many of the parameters are the same across patterns. Sensitivity analysis could be conducted by varying the slope after the last observed dose (which is not identifiable from the observed data within a pattern).
There are several possible extensions of the methods developed here. Rather than consider a summary statistic (such as area under the curve) for the methacholine-Penh curves, one might conduct QTL mapping directly on the dose-response curves. This could be done either through a longitudinal analogue of (4) where the multiple Penh measurements Yij for mouse i at dose dj (j = 1, . . . , J) are modeled, or by adopting a functional data analysis framework (Ramsay and Silverman, 2005) to consider the entire mouse-specific curve Yi(d) as a functional outcome in the QTL mapping model. Functional mapping methods have been developed for other applications (Ma et al., 2002; Yang et al., 2009b), and approaches for incorporating multiple imputation within functional mapping could be examined in future work. To our knowledge the degree to which missing data differentially impacts QTL mapping when the target is a scalar summary (e.g. or ) versus repeated measurements (Yij) versus a continuous function (Yi(d)) has not been explored. Additionally, while our comparison of inference under the proposed multiple imputation approach to inference under CCA or LOCF is illustrative of the potential problems that may arise by using these more naive approaches, future work might conduct simulation studies to quantify the loss of power and bias that may result.
In previous QTL mapping experiments, we have seen little mention of approaches for dealing with missing phenotype data when the phenotype summary of interest is a complex biological parameter that is a function of missing data. While clearly it is preferable to design and conduct experiments in order to minimize the frequency of missing data, its occurrence is often unavoidable in practice. Consequently, developing statistical analyses that use as much information as possible, that limit potential biases, and that have high power to detect a QTL when one exists is an important task. The methods described in this paper, though developed to study airway disease in mice, are broadly applicable to QTL mapping experiments in the presence of missing phenotype data.
Contributor Information
Jennifer F. Bobb, Johns Hopkins Bloomberg School of Public Health
Daniel O. Scharfstein, Johns Hopkins Bloomberg School of Public Health
Michael J. Daniels, University of Florida
Francis S. Collins, National Human Genome Research Institute, National Institutes of Health
Samir Kelada, National Human Genome Research Institute, National Institutes of Health.
References
- Ackerman KG, Huang H, Grasemann H, Puma C, Singer JB, Hill AE, Lander E, Nadeau JH, Churchill GA, Drazen JM, Beier DR. “Interacting genetic loci cause airway hyperresponsiveness,”. Physiological Genomics. 2005;21:105–11. doi: 10.1152/physiolgenomics.00267.2004. [DOI] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, Mcmillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, Pardo-Manuel de Villena F, Churchill GA. “Genetic analysis of complex traits in the emerging Collaborative Cross,”. Genome Research. 2011:1–11. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K, Sen S. A Guide to QTL Mapping with R/qtl. Statistics for Biology and Health, Springer; 2009. chapter D. [DOI] [Google Scholar]
- Broman KW. “Review of statistical methods for QTL mapping in experimental crosses,”. Lab Animal. 2001;30:44–52. [PubMed] [Google Scholar]
- Camateros P, Marino R, Fortin A, Martin JG, Skamene E, Sladek R, Radzioch D. “Identification of novel chromosomal regions associated with airway hyperresponsiveness in recombinant congenic strains of mice,”. Mammalian Genome. 2010;21:28–38. doi: 10.1007/s00335-009-9236-z. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. “The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics,”. Mammalian Genome. 2008;19:382–9. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F. “The Collaborative Cross, a community resource for the genetic analysis of complex traits,”. Nature Genetics. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Clee SM, Attie AD. “The genetic landscape of type 2 diabetes in mice,”. Endocrine Reviews. 2007;28:48–83. doi: 10.1210/er.2006-0035. [DOI] [PubMed] [Google Scholar]
- Dempster A, Laird N, Rubin D, et al. “Maximum likelihood from incomplete data via the EM algorithm,”. Journal of the Royal Statistical Society. Series B (Methodological) 1977;39:1–38. [Google Scholar]
- Ding X, Laird N. “Family-based association tests with longitudinal measurements: handling missing data,”. Human Heredity. 2009;68:98–105. doi: 10.1159/000212502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley B, Rabe K, Andrade MD. “Imputation methods for missing data for polygenic models,”. BMC Genetics. 2003;4(Suppl 1):S42. doi: 10.1186/1471-2156-4-S1-S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Andrade MD. “Missing phenotype data imputation in pedigree data analysis,”. Genetic Epidemiology. 2008;32:52–60. doi: 10.1002/gepi.20261. [DOI] [PubMed] [Google Scholar]
- Hamelmann E, Schwarze J, Takeda K, Oshiba A, Larsen GL, Irvin CG, Gelfand EW. “Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography,”. American Journal of Respiratory and Critical Care Medicine. 1997;156:766–75. doi: 10.1164/ajrccm.156.3.9606031. [DOI] [PubMed] [Google Scholar]
- Heyting A, Tolboom JTBM, Essers JGA. “Statistical handling of drop-outs in longitudinal clinical trials,”. Statistics in Medicine. 1992;11:2043–2061. doi: 10.1002/sim.4780111603. [DOI] [PubMed] [Google Scholar]
- Jarvis J, Kenney-Hunt J, Ehrich T, Pletscher L, Semenkovich C, Cheverud J. “Maternal genotype affects adult offspring lipid, obesity, and diabetes phenotypes in LGXSM recombinant inbred strains,”. Journal of Lipid Research. 2005;46:1692. doi: 10.1194/jlr.M500073-JLR200. [DOI] [PubMed] [Google Scholar]
- Kelada SNP, Wilson MS, Tavarez U, Borate B, Kubalanza K, Carpenter DE, Whitehead G, Maruoka S, Brass DM, Wynn TA, Cook DA, Evans CM, Schwartz DA, Collins FS. “Strain-dependent genomic factors affect allergen-induced airway hyper-responsiveness in mice,”. American Journal of Respiratory Cell and Molecular Biology. 2011. [DOI] [PMC free article] [PubMed]
- Leme AS, Berndt A, Williams LK, Tsaih S-W, Szatkiewicz JP, Verdugo R, Paigen B, Shapiro SD. “A survey of airway responsiveness in 36 inbred mouse strains facilitates gene mapping studies and identification of quantitative trait loci,”. Molecular Genetics and Genomics. 2010;283:317–26. doi: 10.1007/s00438-010-0515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. “Genotype imputation,”. Annual Review of Genomics and Human Genetics. 2009;10:387. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R. “A class of pattern-mixture models for normal incomplete data,”. Biometrika. 1994;81:471. doi: 10.1093/biomet/81.3.471. [DOI] [Google Scholar]
- Little RJA, Rubin DB. Statistical Analysis with Missing Data, Second Edition. 2nd edition Wiley-Interscience; 2002. [Google Scholar]
- Lofgren JLS, Mazan MR, Ingenito EP, Lascola K, Seavey M, Walsh A, Hoffman AM. “Restrained whole body plethysmography for measure of strain-specific and allergen-induced airway responsiveness in conscious mice,”. Journal of Applied Physiology. 2006;101:1495–505. doi: 10.1152/japplphysiol.00464.2006. [DOI] [PubMed] [Google Scholar]
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. “WinBUGS–a Bayesian modelling framework: concepts, structure, and extensibility,”. Statistics and Computing. 2000;10:325–337. doi: 10.1023/A:1008929526011. [DOI] [Google Scholar]
- Ma C-X, Casella G, Wu R. “Functional mapping of quantitative trait loci underlying the character process: a theoretical framework,”. Genetics. 2002;161:1751–62. doi: 10.1093/genetics/161.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Talbot C, Turri M, Collins A, Flint J. “A method for fine mapping quantitative trait loci in outbred animal stocks,”. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12649. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak D, Kushner J, Ludwig D. “Effects of dietary glycaemic index on adiposity, glucose homoeostasis, and plasma lipids in animals,”. The Lancet. 2004;364:778–785. doi: 10.1016/S0140-6736(04)16937-7. [DOI] [PubMed] [Google Scholar]
- Ramsay J, Silverman B. Functional data analysis. Springer series in statistics, Springer; 2005. [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. “The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics,”. Mammalian Genome. 2007;18:473–81. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DB. “Inference and missing data,”. Biometrika. 1976;63:581–592. doi: 10.1093/biomet/63.3.581. [DOI] [Google Scholar]
- Rubin DB. Multiple imputation for nonresponse in surveys. Vol. 519. NY: Wiley; 1987. [DOI] [Google Scholar]
- Schafer JL. Analysis of incomplete multivariate data. Chapman & Hall/CRC; 1997. [DOI] [Google Scholar]
- Thompson EA, Geyer CJ. “Fuzzy p-values in latent variable problems,”. Biometrika. 2007;94:49–60. doi: 10.1093/biomet/asm001. [DOI] [Google Scholar]
- Xing C, Schumacher FR, Conti DV, Witte JS. “Comparison of missing data approaches in linkage analysis,”. BMC Genetics. 2003;4:S44. doi: 10.1186/1471-2156-4-S1-S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins L, Szatkiewicz J, Bell T, Paigen B, Graber J, de Villena F, Churchill G. “A customized and versatile high-density genotyping array for the mouse,”. Nature Methods. 2009a;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wu R, Casella G. “Nonparametric functional mapping of quantitative trait loci,”. Biometrics. 2009b;65:30–9. doi: 10.1111/j.1541-0420.2008.01063.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lefort J, Kearsey V, Lapa e Silva JR, Cookson WO, Vargaftig BB. “A genome-wide screen for asthma-associated quantitative trait loci in a mouse model of allergic asthma,”. Human Molecular Genetics. 1999;8:601–5. doi: 10.1093/hmg/8.4.601. [DOI] [PubMed] [Google Scholar]