Abstract
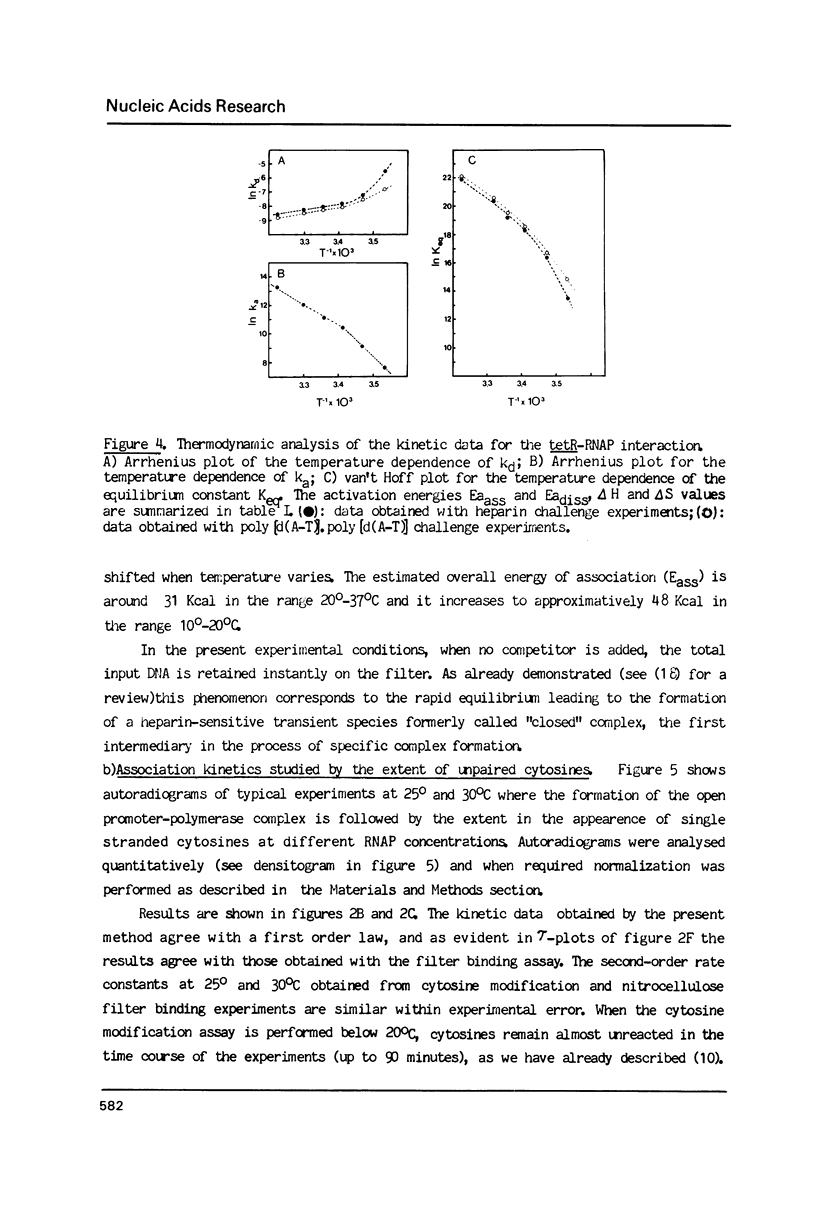
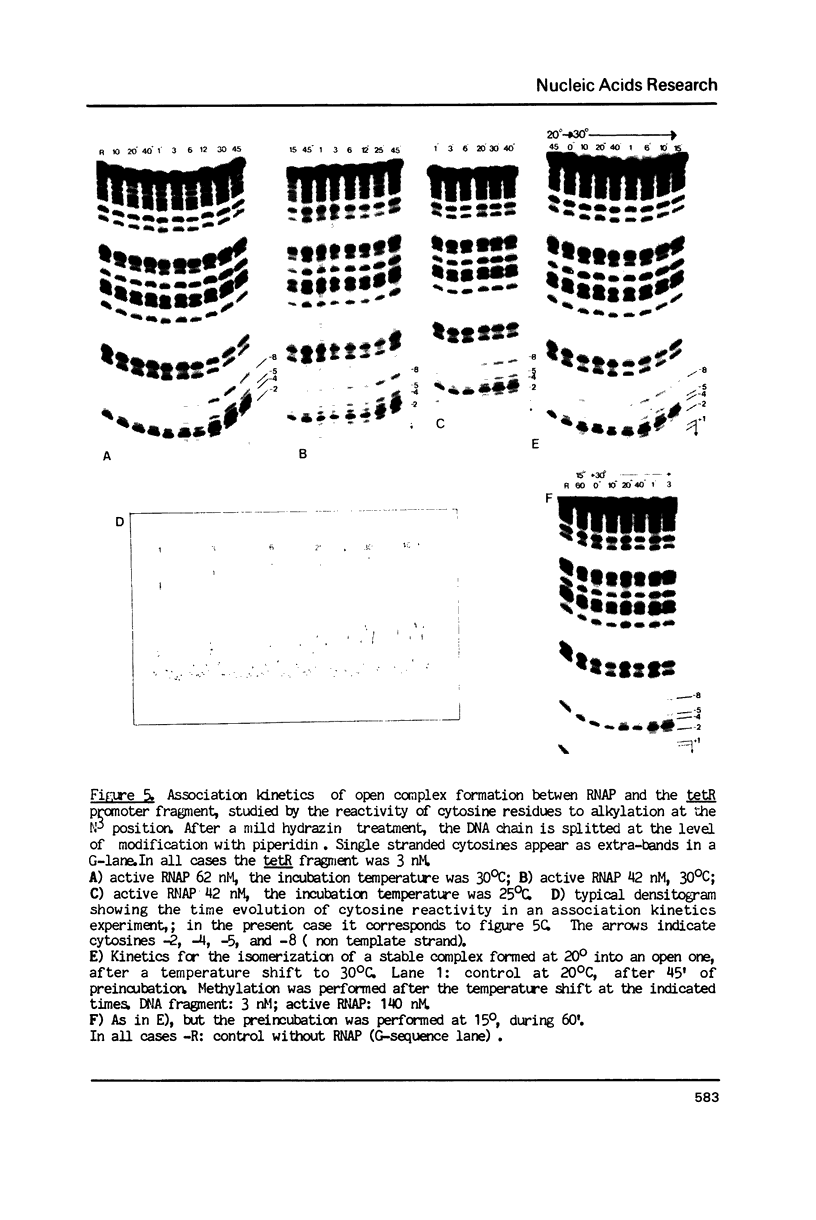
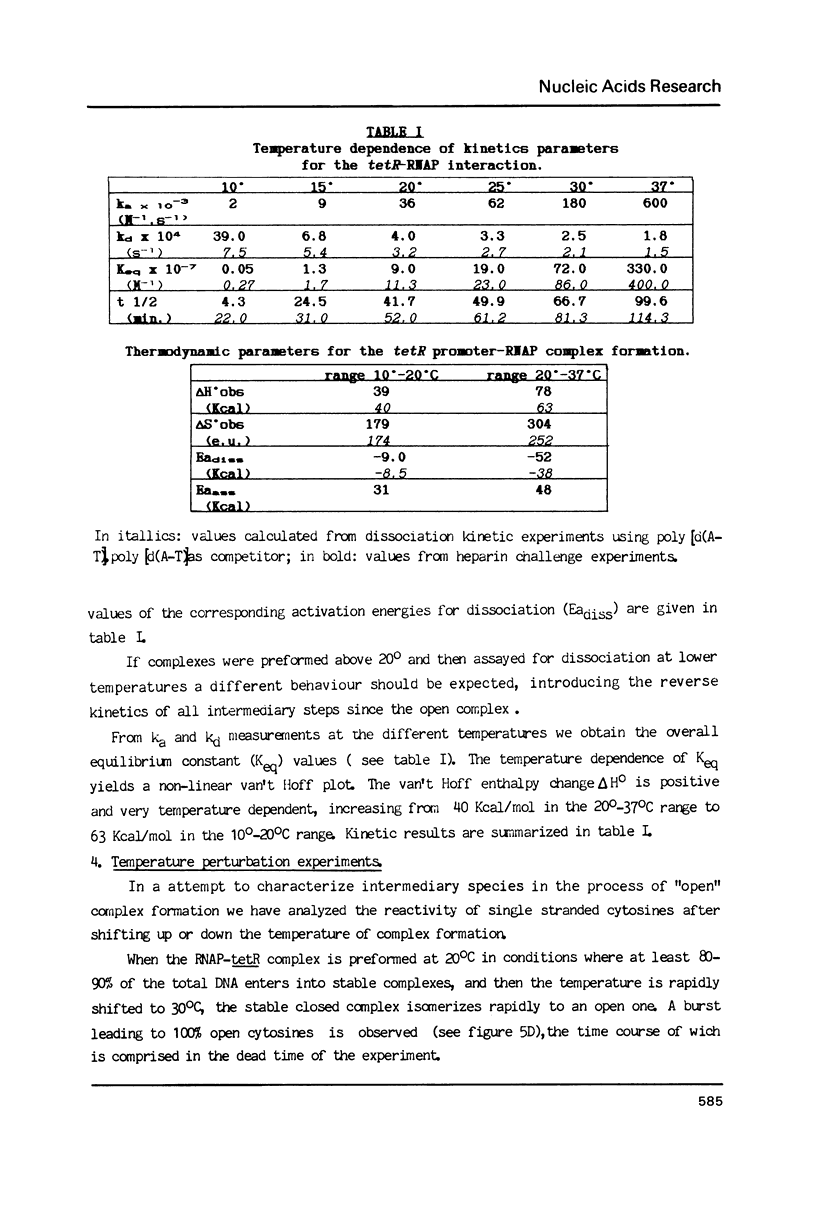
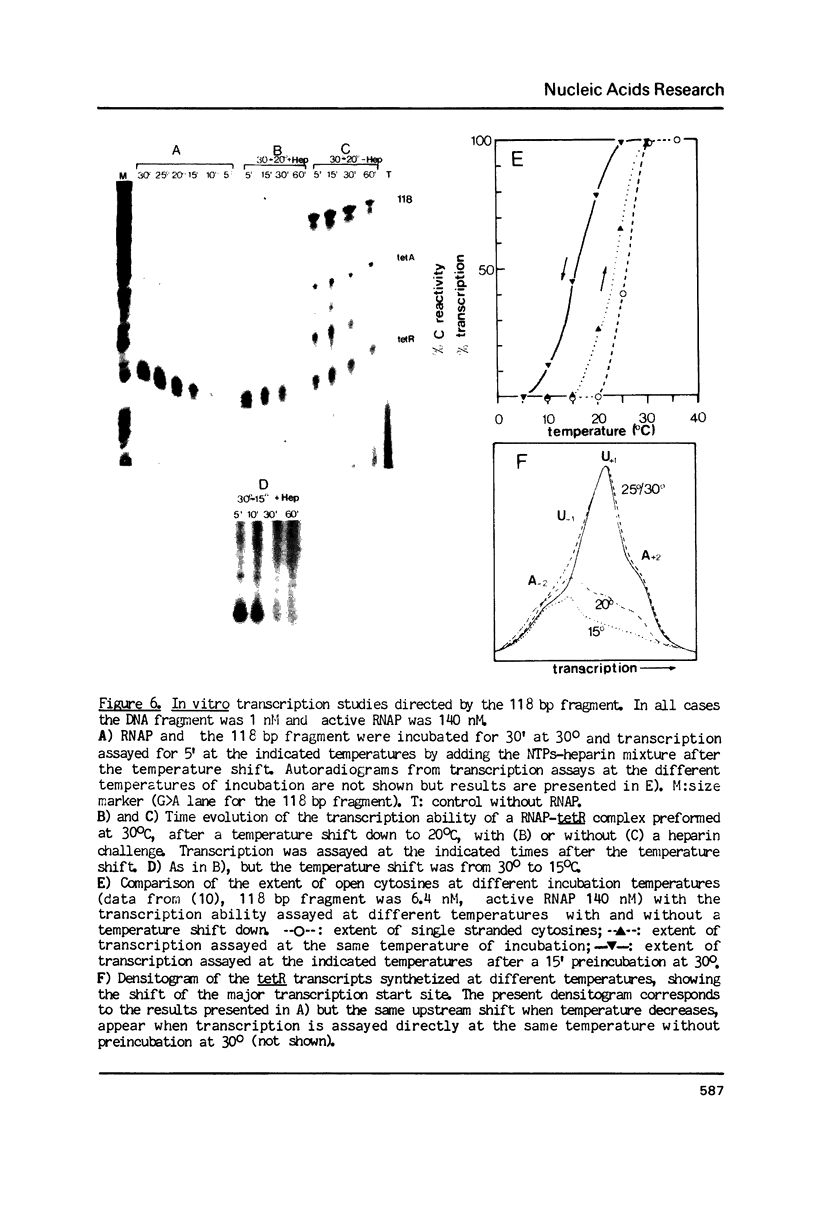
Kinetic, functional and structural studies of the recognition of the tetR promoter from pSC101 by E. coli RNA polymerase allowed the characterization of several steps in the specific complex formation and transcription initiation process. First, enzyme and DNA enter in a short life-time complex. An isomerization will convert this unstable complex into a closed stable one where RNA polymerase is tightly attached without establishing stable chemical contacts with the bases. In the next step, stable close contacts appear between both macromolecules involving mainly the downstream part of the promoter. A further isomerization will lead to an open complex where DNA is locally melted and the system is able to initiate transcription. This latter process is accompanied by changes in the upstream part of the promoter. Finally, in vitro transcription assays showed that the position of the major transcription start sites depends on temperature. From the reported results, it appears that the recognition event is a sequential process where different structural elements of the promoter, that can be located apart in the sequence, are involved in a concerted manner in each stage.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Buc H., McClure W. R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985 May 21;24(11):2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J., Nierman W. C., Wiggs J., Neff N. A quantitative assay for bacterial RNA polymerases. J Biol Chem. 1979 Oct 25;254(20):10061–10069. [PubMed] [Google Scholar]
- Duval-Valentin G., Ehrlich R. Interaction between E. coli RNA polymerase and the tetR promoter from pSC101: homologies and differences with other E. coli promoter systems from close contact point studies. Nucleic Acids Res. 1986 Mar 11;14(5):1967–1983. doi: 10.1093/nar/14.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich R., Larousse A., Jacquet M. A., Marin M., Reiss C. In vitro transcription initiation from three different Escherichia coli promoters. Effect of supercoiling. Eur J Biochem. 1985 Apr 15;148(2):293–298. doi: 10.1111/j.1432-1033.1985.tb08838.x. [DOI] [PubMed] [Google Scholar]
- Ehrlich R., Marin M., Larousse A., Gabarro-Arpa J., Schmitt B., Reiss C. Promoter recognition and transcription initiation in E. coli. Folia Biol (Praha) 1984;30(Spec No):105–118. [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. In vitro comparison of initiation properties of bacteriophage lambda wild-type PR and x3 mutant promoters. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6381–6385. doi: 10.1073/pnas.77.11.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melançon P., Burgess R. R., Record M. T., Jr Nitrocellulose filter binding studies of the interactions of Escherichia coli RNA polymerase holoenzyme with deoxyribonucleic acid restriction fragments: evidence for multiple classes of nonpromoter interactions, some of which display promoter-like properties. Biochemistry. 1982 Aug 31;21(18):4318–4331. doi: 10.1021/bi00261a022. [DOI] [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985 Aug 5;184(3):441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- Russell D. R., Miller P. D., Bennett G. N. In vitro characterization of hybrid promoters and altered tryptophan operon promoters. Biochemistry. 1985 Mar 12;24(6):1410–1417. doi: 10.1021/bi00327a019. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Strauss H. S., Boston R. S., Record M. T., Jr, Burgess R. R. Variables affecting the selectivity and efficiency of retention of DNA fragments by E. coli RNA polymerase in the nitrocellulose-filter-binding assay. Gene. 1981 Jan-Feb;13(1):75–87. doi: 10.1016/0378-1119(81)90045-7. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]