Abstract
Aerobic methanotrophic bacteria can use methane as their sole energy source. The discovery of “Ca. Methylacidiphilum fumariolicum” strain SolV and other verrucomicrobial methanotrophs has revealed that the ability of bacteria to oxidize CH4 is much more diverse than has previously been assumed in terms of ecology, phylogeny, and physiology. A remarkable characteristic of the methane-oxidizing Verrucomicrobia is their extremely acidophilic phenotype, growing even below pH 1. In this study we used RNA-Seq to analyze the metabolic regulation of “Ca. M. fumariolicum” SolV cells growing at μmax in batch culture or under nitrogen fixing or oxygen limited conditions in chemostats, all at pH 2. The analysis showed that two of the three pmoCAB operons each encoding particulate methane monoxygenases were differentially expressed, probably regulated by the available oxygen. The hydrogen produced during N2 fixation is apparently recycled as demonstrated by the upregulation of the genes encoding a Ni/Fe-dependent hydrogenase. These hydrogenase genes were also upregulated under low oxygen conditions. Handling of nitrosative stress was shown by the expression of the nitric oxide reductase encoding genes norB and norC under all conditions tested, the upregulation of nitrite reductase nirK under oxygen limitation and of hydroxylamine oxidoreductase hao in the presence of ammonium. Unraveling the gene regulation of carbon and nitrogen metabolism helps to understand the underlying physiological adaptations of strain SolV in view of the harsh conditions of its natural ecosystem.
Keywords: Methylacidiphilum, methane, RNA-Seq, Verrucomicrobia, metabolic regulation, nitrogen, pMMO
Introduction
Methanotrophs are an unique group of microorganisms that can use methane (CH4) as sole carbon and energy source (Hanson and Hanson, 1996). Methanotrophs are found both in aerobic and anaerobic natural environments (Hanson and Hanson, 1996; Boetius et al., 2000; Raghoebarsing et al., 2006; Conrad, 2009). Aerobic methane-oxidizing bacteria are represented by members of the Alphaproteobacteria, the Gammaproteobacteria, the Verrucomicrobia, and the NC10 phylum (Hanson and Hanson, 1996; Op den Camp et al., 2009; Ettwig et al., 2010). “Candidatus Methylomirabilis oxyfera,” a representative of the latter phylum and growing anaerobically in the absence of oxygen, has the unique ability to produce intracellular oxygen through an alternative denitrification pathway (Ettwig et al., 2010).
During aerobic CH4 oxidation, energy is conserved during the oxidation of methanol, formaldehyde, and formate (Hanson and Hanson, 1996; Chistoserdova et al., 2009). In the oxidation of methanol, electrons are transferred to a membrane bound electron transport chain via a pyrroloquinoline quinone cofactor to cytochrome c and the bc1 complex by the enzyme methanol dehydrogenase. During formaldehyde and formate oxidation, NAD is reduced to NADH and transferred to NADH-oxidoreductase complex I (nuo genes). Electrons flow via the membrane protein complexes, Nuo, bc1, to the cytochrome c oxidases and produce a proton motive force that is converted to the cellular energy carrier ATP by the ATPase enzyme complex.
Verrucomicrobial methanotrophs were isolated from volcanic areas in Italy, New Zealand, and Russia (Dunfield et al., 2007; Pol et al., 2007; Islam et al., 2008) and, the genus name “Methylacidiphilum” was proposed since 16S rRNA gene sequences of the three independent isolates had 98–99% sequence identity (Op den Camp et al., 2009). Although environmental clone libraries from many ecosystems show a large abundance and biodiversity of Verrucomicrobia (Wagner and Horn, 2006), little is known about their in situ physiology. There are now several verrucomicrobial genome assemblies available (van Passel et al., 2011) including two of the verrucomicrobial methanotrophs (Hou et al., 2008; Khadem et al., 2012). The genome data of strains V4 and SolV showed some similarities but also major differences in the C1-utilization pathways compared to proteobacterial and NC10 methanotrophs. The functional significance of these differences can only be validated by a combination of physiological and expression studies.
Physiological studies of “Ca. M. fumariolicum” strain SolV have demonstrated that this microorganism was able to grow with ammonium, nitrate, or dinitrogen gas as nitrogen source (Pol et al., 2007; Khadem et al., 2010). 13C-labeling studies showed that strain SolV growing on CH4, fixed CO2 into biomass exclusively via the Calvin Benson Bassham (CBB) cycle (Khadem et al., 2011). Based on these results we expect that genes involved in nitrogen fixation are only expressed in the absence of ammonium/nitrate and genes involved in the CBB cycle are constitutively expressed. To evaluate this in more detail, analysis of the complete set of transcripts (the transcriptome) and their quantity present in cells grown under different condition is needed.
With the development of microarrays (Malone and Oliver, 2011) high-throughput quantification of the transcriptome became possible, improving the low throughput mRNA data from Northern blots or reverse-transcription PCR (RT-PCR) analysis. More recently, next generation sequencing has been shown to be a very powerful method to analyze the transcriptome of cells by what is known as RNA-Seq (Wang et al., 2009). Furthermore, this technique can detect transcripts without corresponding genomic sequences and can detect very low abundance transcripts (Croucher and Thomson, 2010; Malone and Oliver, 2011).
In this study we used RNA-Seq to analyze the genome wide transcriptome of “Ca. M. fumariolicum” SolV cells grown under different conditions at pH 2. Expression profiles of exponentially growing SolV batch cultures (at μmax) were compared to nitrogen fixing or oxygen limited chemostat cultures and used to unravel the gene and genome regulation of carbon and nitrogen metabolism which may reflect the underlying physiological adaptations of SolV.
Materials and Methods
Organism and medium composition for growth
“Ca. Methylacidiphilum fumariolicum” strain SolV used in this study was originally isolated from the Solfatara volcano, Campi Flegrei, near Naples, Italy (Pol et al., 2007).
Preparation and composition of the growth medium (pH 2) was described previously (Khadem et al., 2010). Mineral salts composition and concentration were changed for oxygen limited SolV chemostat cultures: 0.041 g l−1 MgCl2·6H2O was added (instead of 0.08 g l−1) and CaHPO4·2H2O was replaced by 0.138 g l−1 NaH2PO4.H2O to limit precipitation.
Chemostat cultivation
Chemostat cultivation of strain SolV under nitrogen fixing condition at pH 2 was performed as described previously (Khadem et al., 2010). Growth yield and stoichiometry of CH4 conversion to CO2 of strain SolV were also determined for oxygen limited SolV chemostat cultures. The chemostat liquid volume was 300 ml and the system was operated at 55°C with stirring at 900 rpm with a stirrer bar. The chemostat was supplied with medium at a flow rate of 5.1 ml h−1, using a peristaltic pump. Culture liquid level was controlled by a peristaltic pump actuated by a level sensor. A gas mixture containing (v/v) 5.8% CH4, 2.3% O2, 0.4% N2, and 91.1% CO2 was supplied to the chemostat by mass flow controllers through a sterile filter and sparged into the medium just above the stirrer bar. Oxygen concentrations in the liquid were measured with a Clarke-type electrode.
After steady state was reached, CH4 and O2 consumption and CO2 production were determined by measuring the ingoing and outgoing gas flows and the gas concentrations. The outgoing gas passed through a sterile filter at a flow rate of 11.9 ml h−1, and contained (v/v) a mixture of approximately 4.8% CH4, 0.72% O2, 0.7% N2, and 92.7% CO2. The dissolved oxygen concentration (dO2) was below 0.03% oxygen saturation.
To determine biomass dry weight concentration, triplicate 5 ml samples from the culture suspension were filtered through pre-weighed 0.45 μm filters and dried to constant weight in a vacuum oven at 70°C. After steady state, both chemostats were sampled for mRNA isolation and Illumina sequencing.
Batch cultivation
Cells of SolV grown at maximal growth rate (μmax), without any nitrogen, O2, and CH4 limitation were obtained in 1 liter serum bottles, containing 50 ml medium (with 4 mM ammonium, 2% fangaia soil extract, and at pH 2, Khadem et al., 2010) and sealed with red butyl rubber stoppers. Incubations were performed in duplicate and contained in (v/v) 10% CH4, 5% CO2, and 18% O2 at 55°C with shaking at 180 rpm. Exponentially growing cells were collected for mRNA isolation and Illumina sequencing.
Gas and ammonium analyses
Gas samples (100 μl) were analyzed for methane (CH4), carbon dioxide (CO2), and oxygen (O2) on an Agilent series 6890 gas chromatograph (GC) equipped with Porapak Q and Molecular Sieve columns and a thermal conductivity detector as described before (Ettwig et al., 2008).
Ammonium concentrations were measured using the orthophthaldialdehyde (OPA) method (Taylor et al., 1974).
Transcriptome analysis
The draft genome sequence of strain SolV (Khadem et al., 2012) was used as the template for the transcriptome analysis. Cells were harvested by centrifugation and 3.1 mg dry weight cells were used for isolation of mRNA, and subsequent synthesis of cDNA (328 ng) was done as described before (Ettwig et al., 2010). The cDNA was used for Illumina sequencing (RNA-Seq) as described before (Ettwig et al., 2010; Kartal et al., 2011). Expression analysis was performed with the RNA-Seq Analysis tool from the CLC Genomic Workbench software (version 4.0, CLC-Bio, Aarhus, Danmark) and values are expressed as RPKM (Reads Per Kilobase of exon model per Million mapped reads; Mortazavi et al., 2008).
Results and Discussion
Physiology of “Ca. M. fumariolicum” SolV growing with and without nitrogen source and under oxygen limitation
Prior to the expression studies the physiological properties of strain solV were examined in batch and chemostat continuous culture. These studies showed that strain SolV in batch culture had a maximum growth rate of 0.07 and 0.04 h−1, with ammonium or nitrate as nitrogen source, respectively (Table 1). In the absence of ammonium and nitrate “Ca. M. fumariolicum” SolV cells were able to fix atmospheric N2 only at headspace oxygen concentration below 1% (Khadem et al., 2010). The additional reduction steps of nitrate to ammonium could explain the observed increase in doubling time with nitrate compared to ammonium. The slower growth rate with N2 as nitrogen source was expected, since N2 fixation is an endergonic process, which needs about 16 mol ATP per mol N2 fixed (Dixon and Kahn, 2004). Based on the μmax data obtained, strain SolV seems to prefer ammonium, which is also the most likely nitrogen source in its natural environment.
Table 1.
Description of batch and chemostat cultures of “Ca. Methylacidiphilum fumariolicum” SolV.
| Culture | Nitrogen source | Growth rate (h−1) | Doubling time (h) | Yield (g DW/mol CH4) | Limitation | O2 concentrationb (%) |
|---|---|---|---|---|---|---|
| Batch | Ammoniuma | μmax = 0.07 | 10 | 6.5 | No | 18 |
| Nitrate | μmax = 0.04 | 17 | n.d. | No | 18 | |
| N2 | μmax = 0.025 | 27 | n.d. | Nitrogen | <1 | |
| Chemostat | N2a | μ = 0.017 | 40 | 3.5 | Methane | 0.5 |
| Ammoniuma | μ = 0.017 | 40 | 4.9c | Oxygen | <0.03 |
aCells used for transcriptome analysis. In all other Tables referred to as “Cells at μmax,” “N2 fixation,” and “O2 limitation.”
bFor batch cultures initial headspace oxygen concentrations are given, while for the chemostat cultures measured dissolved oxygen concentration (dO2) is expressed as % oxygen saturation (100% equals 800 μmol/l at 55°C).
cCalculated from OD600 comparisons at steady state.
Continuous cultivation of strain SolV cells in a chemostat at pH 2 under nitrogen fixing conditions, was performed at dissolved oxygen concentrations (dO2) equal to 0.5% oxygen saturation and without ammonium or nitrate (Table 1). Growth was limited by CH4 liquid-gas transfer in this chemostat culture (Khadem et al., 2010). The growth rate (0.017 h−1) is 68% of the μmax (0.025 h−1) obtained in N2 fixing batch cultures.
For continuous cultivation of strain SolV under oxygen limitation and in the presence of excess methane and ammonium, the chemostat was supplied with medium at a dilution rate of 0.017 h−1 (Table 1). This resulted in a dO2 equal to 0.03% oxygen saturation (<0.24 μmol/l).
After a steady state was obtained in the chemostat, the stoichiometry of CH4 oxidation, and cell yield under N2 fixing and O2 limiting conditions were determined. Under O2 limitation the stoichiometry of CH4 oxidation was the same as reported, for excess ammonium and O2 (Pol et al., 2007). However, under N2 fixing conditions, a slightly higher consumption of O2 and production of CO2 was found (Khadem et al., 2010). This coincides with the lower cell yield of the nitrogen fixing chemostat culture of strain SolV (Table 1).
Three of the above described physiological conditions were selected for a genome wide transcriptome analysis (Table 1), e.g., exponentially growing cells (batch culture at μmax) and cells from nitrogen or oxygen limited chemostat cultures.
Whole genome transcriptome analysis of “Ca. M. fumariolicum” SolV
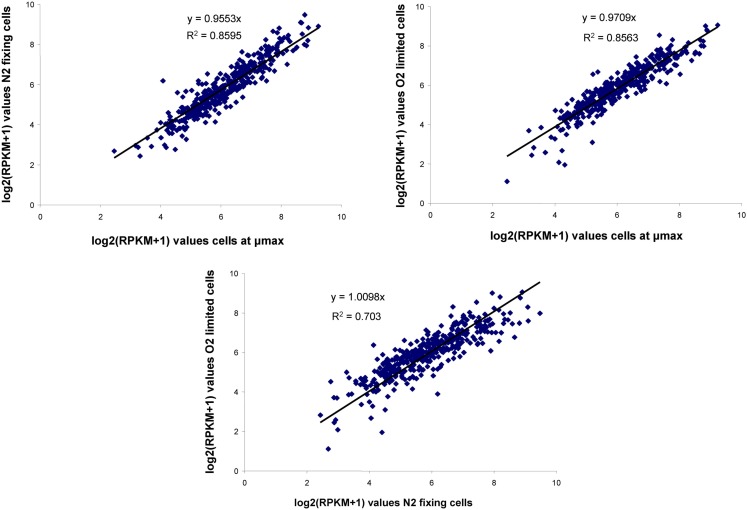
The SolV transcriptome was characterized using RNA-Seq. RNA was prepared from the three different cell cultures (see above), converted to cDNA and sequenced. The Illumina Genome Analyzer reads (75 bp) were first mapped to the ribosomal RNA operon and mapped reads were discarded. The unmapped reads (3.5 × 106, 3.2 × 106, and 2.0 × 106 reads for μmax, N2 fixing, and O2 limited cells, respectively) were mapped to the CDS, tRNA, and ncRNA sequences extracted from the genome sequence of strain SolV (Khadem et al., 2012). The total number of reads obtained and mapped for each sampled culture together with the calculated expression levels (RPKM) are provided File S1 in Supplementary Material (RNA-Seq_SolV.xls). We selected a set of 394 housekeeping genes (in total 443 kbp) involved in energy generation, in ribosome assembly, carbon fixation (CBB cycle), C1 metabolism (except for pmo), amino acid synthesis, cell wall synthesis, translation, transcription, DNA replication, and tRNA synthesis for the three conditions, to compare baseline expression levels (File S2 in Supplementary Material; Housekeeping genes.xls). For this gene set all ratios of expression between conditions were >0.5 and <2. The robustness of the transcriptome data were checked by the method of Chaudhuri et al. (2011) in which the expression levels (log2(RPKM + 1)) of the 394 gene set for the three conditions were plotted against each other. This resulted in correlation coefficients of 0.70, 0.86, and 0.86 (Figure A1 in Appendix), which are only slightly lower than those of technical replicates as reported by Chaudhuri et al. (2011).
In the following paragraphs, the differences in expression pattern under the various cultivation conditions with respect to energy, carbon, nitrogen, and hydrogen metabolism of strain SolV will be presented and discussed.
Energy metabolism
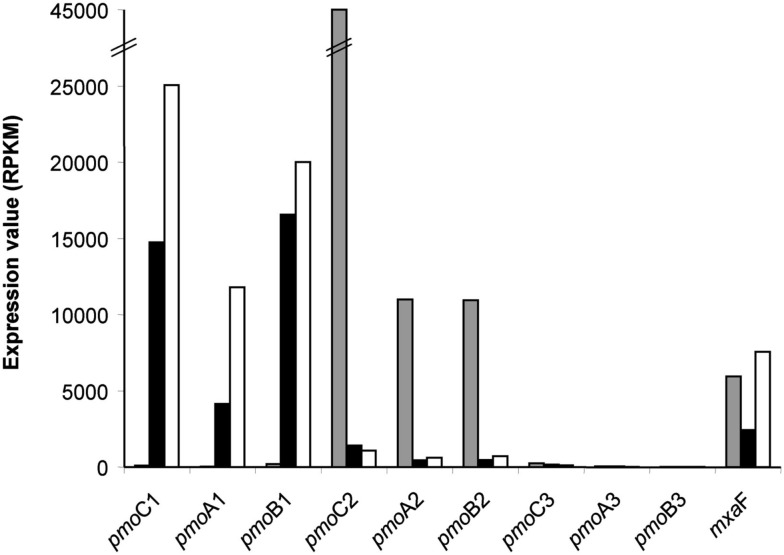
Genes involved in CH4 oxidation pathway (Hanson and Hanson, 1996; Chistoserdova et al., 2009) and their RPKM values are presented in (Table 2). In the genome data of the verrucomicrobial methanotrophs no genes encoding for the soluble cytoplasmic form of the methane monooxygenase (sMMO) were found (Hou et al., 2008; Khadem et al., 2012). However, three pmoCAB operons, encoding for the three subunits of particulate membrane-associated form (pMMO) were predicted. Transcriptome analysis of “Ca. M. fumariolicum” SolV showed differential expression of two of the three different operons. One of the pmoCAB operons (pmoCAB2) was highly expressed (RPKM values 10.9 × 103 to 45 × 103, Figure 1) in cells growing at μmax with excess ammonium and oxygen (initial headspace concentration of 18%). The other two pmoCAB operons were hardly expressed under these culture conditions (RPKM 21–253). The cells from CH4 limited N2 fixing chemostat culture and the O2 limited chemostat culture with dO2 of 0.5 and 0.03% oxygen saturation, respectively, showed a remarkable different expression pattern of the pmoCAB operons. Under these conditions the pmoCAB1 operon was highly expressed (RPKM values 4.1 × 103 to 25 × 103) while expression of the pmoCAB2 operon was down regulated 40 times compared to the batch culture. The pmoCAB3 operon was hardly expressed in cells from the two chemostat cultures, expression values being identical to that of the cells at μmax. Although other factors like growth rate, cell density, etc., could have an effect, the results point to a regulation of the pmoCAB1/pmoCAB2 genes by the oxygen concentration. Since the pmoCAB3 operon was not expressed under the conditions tested, other growth conditions have to be tested to elucidate the regulation and function of this pMMO. In a recent study, qPCR was used to investigate expression of the four pmoA genes of “Ca. M. kamchatkense” Kam1 (Erikstad et al., 2012). The pmoA2 gene was 35-fold stronger expressed than the other copies. Suboptimal temperature and pH conditions did not change this pattern. Other limitations were not tested. Grow on methanol resulted in a 10-fold decreased expression of pmoA2.
Table 2.
Transcription of genes involved in oxidation of CH4 in “Ca. M. fumariolicum” strain SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM) |
||
|---|---|---|---|---|---|
| Cells at μmax | N2 fixing cells | O2 limited cells | |||
| Methane monooxygenase_1 | pmoC1 | Mfum_790003 | 90 | 14764 | 25054 |
| pmoA1 | Mfum_790002 | 37 | 4148 | 11804 | |
| pmoB1 | Mfum_790001 | 181 | 16550 | 20004 | |
| Methane monooxygenase_2 | pmoC2 | Mfum_780001 | 45059 | 1405 | 1087 |
| pmoA2 | Mfum_770004 | 10994 | 454 | 598 | |
| pmoB2 | Mfum_770003 | 10930 | 467 | 712 | |
| Methane monooxygenase_3 | pmoC3 | Mfum_480006 | 253 | 148 | 101 |
| pmoA3 | Mfum_480005 | 45 | 44 | 24 | |
| pmoB3 | Mfum_480007 | 21 | 17 | 15 | |
| Methanol dehydrogenase | mxaF/xoxF | Mfum_190005 | 5945 | 2434 | 7554 |
| Periplasmic binding protein | mxaJ | Mfum_190004 | 941 | 1548 | 629 |
| Cytochrome c family protein | mxaG | Mfum_190003 | 760 | 513 | 522 |
| Coenzyme PQQ synthesis proteins | pqqB | Mfum_80011 | 483 | 288 | 677 |
| pqqC | Mfum_80010 | 589 | 645 | 1243 | |
| pqqD | Mfum_710019 | 32 | 26 | 55 | |
| pqqD | Mfum_80009 | 108 | 118 | 195 | |
| pqqE | Mfum_80008 | 525 | 1078 | 766 | |
| pqqF | Mfum_690050 | 527 | 339 | 737 | |
| NADPH:quinone reductase | qor1 | Mfum_270035 | 330 | 236 | 1092 |
| qor2 | Mfum_300032 | 608 | 690 | 509 | |
| qor3 | Mfum_820025 | 117 | 77 | 128 | |
| Zn-dependent alcohol dehydrogenase | adhP1 | Mfum_310051 | 216 | 397 | 218 |
| adhP2 | Mfum_680019 | 165 | 96 | 196 | |
| Aldehyde dehydrogenase | dhaS1 | Mfum_940074 | 58 | 68 | 73 |
| dhaS2 | Mfum_840001 | 1345 | 1008 | 1337 | |
| 7,8-Dihydropteroate synthase | folP1 | Mfum_690066 | 199 | 162 | 236 |
| folP2 | Mfum_940066 | 163 | 236 | 133 | |
| Formate-tetrahydrofolate ligase | fhs | Mfum_300027 | 286 | 279 | 371 |
| Bifunctional protein | folD | Mfumv1_210029 | 162 | 89 | 112 |
| GTP cyclohydrolase | folE | Mfumv1_990006 | 993 | 501 | 1135 |
| Formate dehydrogenase | fdsA | Mfumv1_80015 | 987 | 645 | 763 |
| fdsB | Mfumv1_80014 | 675 | 905 | 520 | |
| fdsC | Mfumv1_80013 | 298 | 75 | 182 | |
| fdsD | Mfumv1_80016 | 313 | 260 | 62 | |
| Formate dehydrogenase | fdh | Mfumv1_50001 | 233 | 186 | 257 |
| Methylamine dehydrogenase | mauA | Mfumv1_700106 | 45 | 24 | 101 |
| mauB | Mfumv1_700109 | 220 | 199 | 168 | |
The mRNA expression is expressed as RPKM according to Mortazavi et al. (2008). Changes in expression in the chemostat cultures (N2 fixing cells or O2 limited cells) compared to batch culture cells growing at μmax are indicated by shading: up regulation >2 times (dark gray), down regulation <0.5 (light gray).
Figure 1.
Expression of the three pmoCAB operons, encoding the three subunits of the particulate methane monooxygenase (pMMO). Values from batch culture cells growing at μmax (  ), N2 fixing cells (■), and O2 limited cells (□) are expressed as RPKM (Mortazavi et al., 2008). Expression of mxaF (xoxF) encoding methanol dehydrogenase is shown for comparison.
), N2 fixing cells (■), and O2 limited cells (□) are expressed as RPKM (Mortazavi et al., 2008). Expression of mxaF (xoxF) encoding methanol dehydrogenase is shown for comparison.
Also some proteobacterial methanotrophs are known to contain multiple copies of pmo operons (Semrau et al., 1995; Murrell et al., 2000). Within sequenced genomes of gammaproteobacterial methanotrophs two nearly sequence-identical copies of pmoCAB1 were found. It is thought that sequence-identical copies have arisen through gene duplications and insertion. Mutation studies in Methylococcus capsulatus Bath have demonstrated that both pMMO’s were required for growth (Stolyar et al., 1999). More sequence-divergent copies (pmoCAB2) were shown to be widely distributed in alphaproteobacterial methanotrophs (Yimga et al., 2003). Recently it was found that some genera of gammaproteobacterial methanotrophs also posses a sequence-divergent particulate methane monooxygenase, depicted pXMO (Tavormina et al., 2011). Unlike the CAB gene order of the pmo operon the pxm operon shows an ABC gene order. The presence of sequence-divergent copies suggests alternative physiological function under different environmental conditions. Methylocystis sp. strain SC2 was shown to possess two pMMO isozymes, encoded by pmoCAB1 and pmoCAB2 operons. The pmoCAB1 operon was expressed by strain SC2 at mixing ratios >600 ppmv CH4, while growth and concomitant oxidation of methane at concentrations <600–700 ppmv was due to the expression of pmoCAB2 (Baani and Liesack, 2008). In this case the methane concentration seems to control the up- and down regulation of the different pMMOs.
The second step in CH4 oxidation pathway is the conversion of methanol to formaldehyde by methanol dehydrogenase. Methanol dehydrogenase activity in strain SolV could be demonstrated but the gene cluster encoding this activity seems to be rather different compared to proteobacterial methanotrophs. The mxaFJGIRSACKLDEH cluster encoding the methanol dehydrogenase (mxaFI), a cytochrome (mxaG), a solute binding protein (mxaJ), and accessory proteins (Chistoserdova et al., 2003; Ward et al., 2004; Chen et al., 2010) was absent in the verrucomicrobial methanotrophs (Hou et al., 2008; Khadem et al., 2012) and found to be replaced by a mxaFJG operon. In addition the gene cluster pqqABCDEF encoding proteins involved in biosynthesis of the methanol dehydrogenase cofactor pyrroloquinoline quinone was present. The expression of these genes did not vary much under the conditions tested (Table 2; Figure 1).
Formaldehyde, the product from the methanol dehydrogenase, is a key intermediate in methanotrophs. It may be oxidized for energy and detoxification, or fixed into cell carbon via the ribulose monophosphate pathway (RuMP) or serine cycle (see below, Hanson and Hanson, 1996; Chistoserdova et al., 2009). The canonical formaldehyde oxidation pathway requires folate as a cofactor for C1 transfer and formate dehydrogenase complexes (see below). The classical gene folA involved in the last step of folate-biosynthesis (encoding dihydrofolate reductase, FolA) is absent in “Ca. Methylacidiphilum” strains V4 and SolV. Hou et al. (2008) suggested that the role of this enzyme could be taken over by an alternative dihydropteroate synthase (FolP). The gene encoding for this enzyme was also present in strain SolV and was constitutively expressed at RPKM values of 133–236. The presence of the folD gene in the “Ca. Methylacidiphilum” strains (V4 and SolV) and expression data of this gene in strain SolV (Table 2), indeed suggest that conversion of formaldehyde is tetrahydrofolate-dependent. The “archaeal” tetrahydromethanopterin cofactor-based pathway for C1 transfer found in other methylotrophs is not present in the genomes of the “Ca. Methylacidiphilum” strains (Hou et al., 2008; Khadem et al., 2012).
Formaldehyde can also directly be oxidized by a formaldehyde dehydrogenase. The genome data of stain SolV reveal several candidates for formaldehyde oxidation: a NADPH:quinone reductase (or related Zn-dependent oxidoreductases), Zn-dependent alcohol dehydrogenases or the NAD-dependent aldehyde dehydrogenases. The genes encoding for these enzymes were expressed under all conditions tested (Table 2). A role for these enzymes should be further supported by enzyme purification and characterization studies. Genes encoding for soluble and membrane bound NAD-dependent formate dehydrogenases were also predicted from the draft genome of strain SolV and there expression levels were not significantly different the same under all experimental conditions (Table 2). This enzyme performs the last step of CH4 oxidation, converting formate into CO2.
Carbon metabolism (carbon fixation)
The genome data of the verrucomicrobial methanotrophs (Hou et al., 2008; Khadem et al., 2012) showed differences in carbon assimilation compared to proteobacterial methanotrophs (Chistoserdova et al., 2009). Analyses of the draft genome of “Ca. M. fumariolicum” strain SolV revealed that the key genes needed for an operational RuMP pathway, hexulose-6-P synthase and hexulose-6-P isomerase were absent. In addition, the crucial genes encoding key enzymes of the serine pathway, malyl coenzyme A lyase, and glycerate kinase, were not found (Khadem et al., 2011). However, all genes required for an active CBB cycle could be identified in the SolV genome. These genes were highly expressed in both chemostat cultures (Table A1 in Appendix), to levels identical to those of cells in batch cultures growing at μmax (Khadem et al., 2011). The constitutive expression in all cell cultures was expected, assuming biomass carbon in strain SolV growing on methane can only be derived from fixation of CO2 via the CBB cycle (Khadem et al., 2011). Our transcriptome data of the chemostat cultures and batch cultures showed low expression of the cbbR gene, encoding a possible RuBisCO operon transcriptional regulator. The cbbR gene product is a LysR-type transcriptional regulator and the key activator protein of cbb operons in facultative autotrophs (Bowien and Kusian, 2002). As an autotroph, strain SolV may not need much regulation of the CBB cycle genes. For strain V4 a coupling of this cbbR gene to nitrate reduction and transport was suggested (Hou et al., 2008).
Although, the genes encoding for the ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), the key enzyme of the CBB cycle was found in the genome of some proteobacterial methanotrophs like M. capsulatus Bath (Ward et al., 2004) and Methylocella silvestris BL2 (Chen et al., 2010) and the non-proteobacterial methanotroph “Candidatus Methylomirabilis oxyfera” (Ettwig et al., 2010), autotrophic growth in liquid cultures has not been reported for these methanotrophs yet.
Carbohydrate metabolism
The presence and transcription of genes involved in the pentose phosphate pathway suggested the possibility of gluconeogenesis in strain SolV (Table A1 in Appendix). In M. capsulatus Bath, gluconeogenesis was suggested as follows: a putative phosphoketolase, condenses pyruvate, and glyceraldehyde-3-phosphate into xylulose-5-phosphate, which in turn is fed into the ribulose-5-phosphate pool for formation of glucose-6-phosphate through the pentose phosphate pathway (Ward et al., 2004). Since a putative phosphoketolase is also present and expressed in strain SolV, gluconeogenesis might take place in the same way. Another possibility for the production of glucose-6-phosphate from glyceraldehyde-3-phosphate would be the consecutive action of triose-P-isomerase, fructose-1,6-bisphosphate aldolase, fructose-1,6-bisphosphate phosphatase, and glucose-6-P isomerase. All genes encoding these enzymes are expressed under the growth conditions tested (Table A1 in Appendix).
In many gammaproteobacterial methanotrophs, the tricarboxylic acid (TCA) cycle is believed to be incomplete, because they lack the α-ketoglutarate dehydrogenase activity (Hanson and Hanson, 1996). However in the M. capsulatus genome homologs of this enzyme were indentified, suggesting that the TCA cycle might operate in this microorganism (Ward et al., 2004). Alphaproteobacterial methanotrophs are known to have a complete TCA cycle (Hanson and Hanson, 1996; Dedysh et al., 2002; Chen et al., 2010). The genes encoding for the TCA cycle enzymes were predicted from the genomes of strains V4 and SolV (Hou et al., 2008; Khadem et al., 2012). Our transcriptome analysis showed that these genes were expressed under the conditions applied, with slightly lower expression levels under N2 fixing conditions (Table A2 in Appendix). The presence of an operational TCA cycle in strain SolV suggests that growth on two carbon compounds like acetate should be possible. The presence and transcription of a gene encoding acetyl-coenzyme A synthetase (acs), allows acetate to be activated and fed into the TCA cycle (Table A2 in Appendix). Three alphaproteobacterial genera Methylocella, Methylocapsa, and Methylocystis, which were shown to be able to grow or survive on acetate, also posses a TCA cycle (Dedysh et al., 2005; Dunfield et al., 2010; Belova et al., 2011; Semrau et al., 2011).
Potential carbon and energy storage
Many bacteria start to accumulate reserve polymers when enough supply of suitable carbon is available, but nitrogen is limited (Wanner and Egli, 1990). This phenomenon is also known for methanotrophs (Pieja et al., 2011). Recently it was shown that type II methanotrophs contained the gene phaC, which encodes for the poly-3-hydroxybutyrate (PHB) synthase enable them to produce PHB (Pieja et al., 2011). At least three genes (phaC, phaA, phaB) were considered to be crucial for PHB synthesis. These genes are absent in type I methanotrophs and in the “Ca. Methylacidiphilum” strains (Hou et al., 2008; Khadem et al., 2012). However, genes encoding for glycogen synthesis, degradation, and transport (glycogen synthase, glycogen debranching enzyme, and ADP-glucose pyrophosphorylase) were predicted based on the draft genome of strain SolV. These genes were expressed under all conditions tested (Table A3 in Appendix). This supports the ability of carbon storage by strain SolV, but further physiological studies with cells growing under excess of carbon and nitrogen limitation are needed. Thus far literature on glycogen synthesis in methanotrophs is sparse, but several of the publicly available genomes of proteobacterial methanotrophs contain glycogen synthesis genes (M. capsulatus str. Bath; Methylomonas methanica MC09; Methylomicrobium alcaliphilum; Methylocystis sp. ATCC 49242; http://www.ncbi.nlm.nih.gov/genomes).
The presence and constitutive expression of genes involved in phosphate transport, polyphosphate synthesis, and utilization (ABC-type phosphate transport system, polyphosphate kinase, adenylate kinase, and exopolyphosphatase; Table A3 in Appendix) suggest that strain SolV is able to store polyphosphate as energy and phosphorus reserve.
Nitrogen metabolism: Ammonium, nitrate, and amino acid metabolism
Based on the genome and supported by the transcriptome data the main route for ammonium assimilation in “Ca. M. fumariolicum” occurs via glutamine synthase (glnA)/glutamate synthase (gltB) and/or the alanine and glutamate dehydrogenases (ald, gdh). Expression values of ald and gdh were about three- to fivefold lower compared to glnA and gltB under the conditions tested (Table 3). Also the genes encoding the glutamine-hydrolyzing carbamoyl-phosphate synthase (carA and carB) were constitutively expressed. This enzyme coverts glutamine and carbon dioxide into glutamate and carbamoyl-phosphate. The latter substrate can be fed into the urea cycle. Except for the gene encoding arginase all other genes (argDHFG) encoding enzymes of the urea cycle were present and constitutively expressed. The most likely function of this partial cycle will be arginine synthesis. For strain V4 it was suggested that the ornithine needed can be supplied by 4-aminobutyrate aminotransferase through a part of the TCA cycle and glutamate synthesis (Hou et al., 2008). In strain SolV, the gene encoding 4-aminobutyrate aminotransferase is also present and expressed. Other methylotrophs possess neither arginase nor ArgD (Hou et al., 2008).
Table 3.
Transcription of genes involved in nitrogen metabolism in “Ca. M. fumariolicum” strain SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM) |
||
|---|---|---|---|---|---|
| Cells at μmax | N2 fixing cells | O2 limited cells | |||
| Glutamine synthetase | glnA | Mfum_90015 | 1542 | 1052 | 725 |
| Glutamine synthetase regulatory protein PII | glnB | Mfum_90016 | 1039 | 1701 | 914 |
| UTP:GlnB (Protein PII) uridylyltransferase | glnD | Mfum_230007 | 169 | 111 | 145 |
| Nitrogen regulatory protein PII | glnK | Mfum_140026 | 166 | 60 | 203 |
| Alanine dehydrogenase | ald | Mfum_290047 | 248 | 279 | 256 |
| Glutamate dehydrogenase | gdhA | Mfum_810044 | 436 | 182 | 500 |
| Glutamate synthase alpha chain | gltB | Mfum_940063 | 1343 | 1360 | 1355 |
| Glutamate synthase beta chain | gltD | Mfum_270076 | 133 | 114 | 125 |
| Ornithine/acetylornithine aminotransferase | argD1 | Mfum_190040 | 736 | 736 | 410 |
| argD2 | Mfum_1010035 | 383 | 421 | 410 | |
| Argininosuccinate lyase | argH | Mfum_970020 | 226 | 186 | 107 |
| Ornithine carbamoyltransferase | argF | Mfum_1010036 | 286 | 267 | 375 |
| Argininosuccinate synthase | argG | Mfum_270005 | 725 | 527 | 678 |
| Carbamoyl-phosphate synthase small chain | carA | Mfum_270024 | 450 | 340 | 479 |
| Carbamoyl-phosphate synthase large chain | carB | Mfum_700048 | 395 | 267 | 504 |
| Ammonium transporter | amtB | Mfum_430001_160001a | 343 | 1143 | 420 |
| Nitrate ABC transporter, nitrate-binding protein | tauA | Mfum_140012 | 28 | 117 | 33 |
| Nitrate transporter | nasA | Mfum_140017 | 22 | 26 | 16 |
| Assimilatory nitrate reductase large subunit | bisC | Mfum_140014 | 13 | 14 | 8 |
| Assimilatory nitrite reductase | nirB | Mfum_140015 | 14 | 40 | 13 |
| Ferredoxin subunit of nitrite reductase | nirD | Mfum_140016 | 32 | 18 | 16 |
| Signal transduction histidine kinase with PAS domain | ntrB | Mfum_920004 | 283 | 294 | 250 |
| Signal transduction response regulator, NtrC family | ntrC1 | Mfum_110018 | 90 | 76 | 67 |
| ntrC2 | Mfum_170043 | 116 | 110 | 111 | |
| ntrC3 | Mfum_260013 | 623 | 361 | 525 | |
| ntrC4 | Mfum_920003 | 225 | 154 | 216 | |
| Hydroxylamine oxidoreductase | haoA | Mfum_970027 | 357 | 124 | 568 |
| Nitric oxide reductase B subunit | norB | Mfum_970100 | 261 | 120 | 342 |
| Nitric oxide reductase subunit C | norC | Mfum_970099 | 192 | 93 | 139 |
| Nitrite reductase (NO-forming) | nirK | Mfum_270071 | 72 | 63 | 200 |
| DNA-binding response regulator, NarL family | mxaB | Mfum_1030004 | 232 | 74 | 142 |
| DNA-binding response regulator, LuxR family | citB1 | Mfum_790006 | 1760 | 5611 | 4679 |
| DNA-binding response regulator, LuxR family | citB2 | Mfum_580001 | 394 | 200 | 239 |
The mRNA expression is expressed as RPKM according to Mortazavi et al. (2008). Changes in expression in the chemostat cultures (N2 fixing cells or O2 limited cells) compared to batch culture cells growing at μmax are indicated by shading: up regulation >2 times (dark gray), down regulation <0.5 (light gray). aMfum_430001 and Mfum_160001 encode the N- and C-terminal part, respectively.
The genes encoding nitrate/nitrite transporters and the assimilatory nitrite and nitrate reductases showed very low expression levels (8–117), probably due to the absence of nitrate in the growth media used. The ammonium transporter gene (amtB type) is three- to fourfold upregulated in N2 fixing cells, which reflects increased ammonium scavenging under nitrogen limited conditions.
Nitrogen fixation
The genomes of strain SolV and strain V4 show a complete set of genes necessary for N2 fixation (Hou et al., 2008; Khadem et al., 2012). Most of these genes and their organization in putative operons resemble those of M. capsulatus Bath (Ward et al., 2004), a gammaproteobacterial methanotroph that has been shown to fix atmospheric N2 (Oakley and Murrell, 1991). N2 fixation is widely distributed among methanotrophs as shown by the presence of both nifH gene fragments and acetylene reduction activity in a variety of alpha- and gammaproteobacterial methanotroph strains (Auman et al., 2001). Also the deep-sea anaerobic methane-oxidizing Archaea were shown to fix N2 and share the products with their sulfate-reducing bacterial symbionts (Dekas et al., 2009).
Gene expression data of strain SolV showed that all the genes involved in nitrogen fixation were upregulated only in absence of ammonium and nitrate indicating the effect of nitrogen availability on the expression of these genes (Table 4). The genes encoding for the nitrogenase (nifH, nifD, nifK) were 100- to 325-fold upregulated, while the gene involved in regulation (nifA) and the Fe/Mo cofactor biosynthesis genes showed 30- to 235-fold increased expression levels. Our previous physiological studies already confirmed that nitrogenase was active in N2 fixing chemostat cultures (Khadem et al., 2010).
Table 4.
Transcription of genes involved in nitrogen fixation in “Ca. M. fumariolicum” strain SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM) |
||
|---|---|---|---|---|---|
| Cells at μmax | N2 fixing cells | O2 limited cells | |||
| Nitrogenase iron protein | nifH | Mfum_690040 | 69 | 22651 | 80 |
| Nitrogenase molybdenum-iron protein alpha chain | nifD | Mfum_690039 | 87 | 14359 | 65 |
| Nitrogenase molybdenum-iron protein beta chain | nifK | Mfum_690038 | 35 | 3348 | 34 |
| Nitrogenase Mo/Fe cofactor biosynthesis protein NifE | nifE | Mfum_690037 | 77 | 5370 | 67 |
| Nitrogenase Mo/Fe cofactor biosynthesis protein NifN | nifN | Mfum_690036 | 64 | 3677 | 52 |
| Protein NifX | nifX | Mfum_690035 | 100 | 2831 | 26 |
| Nif-specific regulatory protein | nifA | Mfum_690018 | 123 | 1080 | 109 |
| Nitrogenase Mo/Fe cofactor biosynthesis protein NifB | nifB | Mfum_690029 | 19 | 3316 | 14 |
| Pyruvate-flavodoxin oxidoreductase | nifJ | Mfum_940083 | 149 | 133 | 185 |
| NifQ family protein | nifQ | Mfum_690020 | 65 | 270 | 22 |
| Cysteine desulfurase | nifS1 | Mfum_690022 | 93 | 801 | 87 |
| nifS2 | Mfum_90010 | 159 | 133 | 116 | |
| nifS3 | Mfum_190023 | 681 | 491 | 486 | |
| nifS4 | Mfum_970062 | 162 | 275 | 145 | |
| nifS5 | Mfum_310028 | 226 | 151 | 264 | |
| NifU-like protein involved in FeS cluster formation | nifU | Mfum_310029 | 54 | 16 | 79 |
| Nitrogenase-stabilizing/protective protein NifW | nifW | Mfum_690011 | 62 | 2260 | 43 |
| NifZ domain protein | nifZ | Mfum_690023 | 158 | 1373 | 50 |
| Electron transfer flavoprotein beta chain | fixA | Mfum_690010 | 60 | 3311 | 73 |
| Electron transfer flavoprotein alpha chain | fixB | Mfum_690009 | 81 | 3043 | 69 |
| Flavoprotein-ubiquinone oxidoreductase | fixC | Mfum_690008 | 101 | 3632 | 100 |
| Ferredoxin-like protein | fixX | Mfum_690007 | 126 | 2917 | 135 |
| Nitrogen fixation protein FixU | fixU | Mfum_690015 | 164 | 2265 | 91 |
| FeS cluster assembly scaffold protein, HesB/SufA family | sufA1 | Mfum_690026 | 40 | 2301 | 22 |
| sufA2 | Mfum_810040 | 149 | 43 | 71 | |
| sufA3 | Mfum_1020116 | 214 | 143 | 183 | |
| FeS cluster assembly protein SufB | sufB | Mfum_970056 | 1099 | 2409 | 622 |
| FeS cluster assembly protein SufD | sufD | Mfum_970057 | 663 | 2169 | 279 |
| FeS cluster assembly protein SufE family | sufE | Mfum_110022 | 375 | 209 | 219 |
| FeS4 cluster protein with leucine rich repeats | Mfum_690024 | 88 | 1884 | 61 | |
| Ferredoxin-like protein in nif region | frxA | Mfum_690027 | 112 | 7892 | 46 |
| Uptake hydrogenase large subunit | hupB | Mfum_50004 | 92 | 372 | 1840 |
| Uptake hydrogenase small subunit | hupS | Mfum_50003 | 186 | 997 | 2955 |
| Nickel/iron-hydrogenase I, small subunit | hyaA | Mfum_870019 | 2380 | 4157 | 4280 |
| Nickel/iron-hydrogenase I, large subunit | hyaB | Mfum_870018 | 2566 | 1879 | 3595 |
| Ni,Fe-hydrogenase I cytochrome b subunit | hyaC | Mfum_50005 | 122 | 593 | 984 |
| [NiFe] hydrogenase Ni incorporation protein HypA | hypA | Mfum_730023 | 119 | 110 | 95 |
| [NiFe] hydrogenase Ni incorporation-associated protein HypB | hypB | Mfum_870009 | 421 | 335 | 692 |
| [NiFe] hydrogenase metallocenter assembly protein HypC | hypC | Mfum_870006 | 830 | 600 | 321 |
| [NiFe] hydrogenase expression/formation protein HypD | hypD | Mfum_870005 | 1159 | 893 | 1246 |
| [NiFe] hydrogenase metallocenter assembly protein HypE | hypE | Mfum_870004 | 424 | 476 | 759 |
| [NiFe] hydrogenase metallocenter assembly protein HypF | hypF | Mfum_870007 | 92 | 81 | 167 |
| Hydrogenase expression protein HupH | hupH | Mfum_50006 | 160 | 631 | 879 |
| Hydrogenase expression/formation protein HoxQ | hoxQ | Mfum_730017 | 177 | 169 | 136 |
| Hydrogenase maturation protease | hycI | Mfum_730022 | 69 | 44 | 89 |
The mRNA expression is expressed as RPKM according to Mortazavi et al. (2008). Changes in expression in the chemostat cultures (N2 fixing cells or O2 limited cells) compared to batch culture cells growing at μmax are indicated by shading: up regulation >2 times (dark gray), down regulation <0.5 (light gray).
Growth on atmospheric nitrogen in the chemostat was only observed when the dO2 was below 0.5% oxygen saturation. Our previous batch incubations in the presence of ammonium and 0.5% O2 saturation resulted in doubling time of 10 h (Khadem et al., 2010). This indicates that in N2 fixing chemostat cultures, this low oxygen was not growth limiting. Maintaining a low oxygen concentration in both batch and chemostat is required for an active nitrogenase, since this enzyme is irreversibly damaged by O2 (Robson and Postgate, 1980). Low oxygen requirement for N2 fixation was also demonstrated for other proteobacterial methanotrophs (Murrell and Dalton, 1983; Takeda, 1988; Dedysh et al., 2004). The effect of high oxygen concentration on the expression of genes encoding N2 fixing enzymes, in absence of ammonium/nitrate still needs to be addressed in strain SolV.
Methanotrophic hydrogenases are considered to have a role in N2 fixation or CH4 oxidation. The role of hydrogenase as a source of reducing power for CH4 oxidation was demonstrated in M. capsulatus Bath (Hanczar et al., 2002). Hydrogen uptake and evolution activities during N2 fixation were reported for Methylosinus trichosporium (De Bont and Mulder, 1976) and Methylocystis T-1 (Takeda, 1988), respectively. However, knock out studies of ΔhupSL encoding for the large and small subunit of the Ni/Fe-dependent hydrogenase in M. capsulatus Bath, did not show differences in viability under nitrogen fixing and non-nitrogen fixing condition in comparison to the wild strain (Csaki et al., 2001). Based on these results, the authors suggested that the hydrogenase is probably regulated by oxygen availability rather than by the hydrogen generated by the nitrogenase enzyme complex. Our expression data also show an increased expression under both nitrogen fixing and oxygen limited conditions (Table 4). Since under oxygen limitation the nitrogen fixing genes were not expressed, while the hydrogenase encoding genes were expressed to even higher levels, oxygen seems to be the regulatory factor for the latter set of genes.
The PII signal transduction proteins (encoded by glnB and glnK) are used to transduce the nitrogen status of the cell to the NtrB–NtrC two-component regulatory system and the σ54-dependent amtB promoters to tune nif gene transcription (for a detailed overview see Dixon and Kahn, 2004). The glnB gene of strain SolV was highly expressed under all conditions and slightly upregulated (1.5-fold) under N2 fixing conditions. Expression of glnK was overall about fivefold lower and threefold downregulated under nitrogen fixing conditions. In addition genes encoding for uridylyltransferase (glnD), NtrB, and NtrC showed expression levels under three conditions tested which did not significantly differ (Table 3). This suggests that in strain SolV the PII proteins are involved in sensing and regulating the status of fixed nitrogen in the cell. Transcription of the nif genes is regulated by nifA and nifL genes (Dixon and Kahn, 2004). The expression of nifA is regulated by oxygen and/or fixed nitrogen and nifL gene is involved in oxygen sensing. We could not identify a nifL gene in the genome of strain SolV (Khadem et al., 2012). However, nifA is present and was 30-fold upregulated under N2 fixing conditions.
Nitrogenase is believed to be sensitive for reactive oxygen species (ROS), and during nitrogen fixation the level of ROS is reduced by up regulation of ROS-detoxifying genes. In Gluconacetobacter diazotrophicus upregulation of these genes was observed during nitrogen fixation (Alquéres et al., 2010). Although, in the genome of strain SolV two sodA genes encoding for superoxide dismutases can be identified, they both are highly expressed under all conditions tested (RPKM values: Mfum_810007, 797 ± 163; Mfum_980001, 961 ± 669), but expression seems to be 1.5- to 3-fold lower under N2 fixing and O2 limited conditions.
Nitrosative stress
The pMMO enzyme involved in the first step of CH4 oxidation in methanotrophs, also oxidizes ammonium which results in the formation of the intermediate hydroxylamine (NH2OH; Hanson and Hanson, 1996; Nyerges and Stein, 2009 and reference therein; Stein and Klotz, 2011). Ammonia-oxidizers can relay electrons from hydroxylamine oxidation to the quinone pool to drive energy production and cellular growth (Klotz and Stein, 2008), but methanotrophs lack this relay and can not produce energy from this oxidation. Since hydroxylamine is a highly toxic intermediate, methanotrophs rely on mechanisms to quickly remove it. In their natural habitat the “Ca. M. fumariolicum” cells are confronted with varying nitrogen levels (1–28 mM, Khadem et al., 2010) which means that the cells have to balance assimilation and tolerance to reactive-N. Detoxification can be achieved by conversion of hydroxylamine back to ammonium or to nitrite through the use of a hydroxylamine reductase enzyme (HAO). The nitrite in turn can be converted to N2O via NO by putative denitrifying enzymes (nitrite reductase and NO-reductase, Campbell et al., 2011). Genes involved in these conversions may include hao, cytL, cytS, nirBD, nirS or nirK, and norCB. The genes hao, norCB were shown to be present in the genomes of the verrucomicrobial methanotrophs (Hou et al., 2008; Khadem et al., 2012), while a nirK homolog was only found in strain SolV. The gene inventory in methanotrophic bacteria for handling hydroxylamine or other toxic nitrosating intermediates and for those encoding putative denitrifying enzymes is diverse and unpredictable by phylotype or taxon (Stein and Klotz, 2011).
In our study we found that although expressed under all conditions tested, expression of hao and norBC were 1.5- to 4.5-fold lower under nitrogen fixing conditions (Table 3), which makes sense in view of the expected lower ammonium levels in the cells. However, for nirK expression was low (RPKM = 63–72) except for the cells grown under oxygen limitation (RPKM = 200).
Conclusion
In this study we analyzed the genome wide changes in expression during three different growth conditions which helped very much to understand the physiology of “Ca. M. fumariolicum” strain SolV. The analysis indicated that the two of the three pmoCAB operons are probably regulated by oxygen, although the effect of other factors like growth rate, cell density can not be excluded. Further, the hydrogen produced during N2 fixation can be recycled, and that nitrosative stress is counter acted. the results point to a regulation of the pmoCAB1/pmoCAB2 genes by the oxygen concentration.
The obtained information will be a guide to design future physiological and biochemical studies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Evolutionary_and_Genomic_Microbiology/10.3389/fmicb.2012.00266/abstract
RNA-Seq analysis of “Ca. Methylacidiphilum fumariolicum” SolV grown under different conditions.
Housekeeping genes to test robustness of transcriptome data.
Acknowledgments
This work was supported by Mosaic grant 62000583 from the Dutch Organization for Scientific Research-NWO. Mike S. M. Jetten is supported by ERC grant number 232937.
Appendix
Table A1.
Transcription of genes involved in carbon fixation and the pentose phosphate pathway in “Ca. M. fumariolicum” strain SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM) |
||
|---|---|---|---|---|---|
| Cells at μmax | N2 fixing cells | O2 limited cells | |||
| 6-Phosphogluconolactonase | nagB | Mfum_380003 | 166 | 70 | 285 |
| Glucose-6-phosphate isomerase | pgi | Mfum_180006 | 218 | 156 | 112 |
| Ribose-5-phosphate-3-epimerase | rpe | Mfum_830014 | 238 | 227 | 335 |
| Transaldolase | mipB | Mfum_70005 | 279 | 301 | 256 |
| Phosphoketolase | xfp | Mfum_200044 | 326 | 253 | 302 |
| Transaldolase | mipB | Mfum_120006 | 380 | 205 | 187 |
| Glucose-6-phosphate 1-dehydrogenase | zwf | Mfum_380002 | 569 | 478 | 469 |
| Triosephosphate isomerase | tpiA | Mfum_170033 | 587 | 428 | 522 |
| Fructose-1,6-bisphosphatase II | glpX | Mfum_1080012 | 591 | 670 | 750 |
| 6-Phosphogluconate dehydrogenase | gnd | Mfum_1020039 | 610 | 334 | 411 |
| Glucose-6-phosphate 1-dehydrogenase | zwf | Mfum_1020106 | 686 | 302 | 794 |
| Ribose 5-phosphate isomerase B | rpiB | Mfum_1020038 | 752 | 335 | 1250 |
| Phosphoribulokinase, chromosomal | cfxP | Mfum_280006 | 829 | 682 | 662 |
| Phosphoglycerate kinase | pgk | Mfum_170032 | 891 | 1066 | 671 |
| Fructose-1,6-bisphosphatase class 1 | fbp | Mfum_280007 | 1174 | 974 | 1078 |
| Phosphoribulokinase | udk | Mfum_70009 | 1798 | 938 | 1300 |
| Glyceraldehyde-3-phosphate dehydrogenase | cbbG | Mfum_170031 | 1830 | 483 | 926 |
| Fructose-bisphosphate aldolase | fbaA | Mfum_310038 | 2016 | 1688 | 1917 |
| Transketolase (TK) | cbbT | Mfum_70010 | 2119 | 1527 | 2448 |
| Protein CbxX | CbxX | Mfum_70008 | 6882 | 2807 | 8188 |
| Ribulose bisphosphate carboxylase small subunit | cbbS | Mfum_70007 | 7200 | 3606 | 6693 |
| Ribulose bisphosphate carboxylase large subunit) | cbbL | Mfum_70006 | 10209 | 7348 | 8567 |
| RuBisCO operon transcriptional regulator CbbR | lysR | Mfum_140011 | 47 | 41 | 28 |
| Carbonic anhydrase | cynT | Mfum_890009 | 1832 | 2022 | 851 |
| Ribose-phosphate pyrophosphokinase | prs | Mfum_870060 | 1105 | 1091 | 1055 |
The mRNA expression is expressed as RPKM according to Mortazavi et al. (2008). Changes in expression in the chemostat cultures (N2 fixing cells or O2 limited cells) compared to batch culture cells growing at μmax are indicated by shading: up regulation >2 times (dark gray), down regulation <0.5 (light gray).
Table A2.
Transcription of genes involved the TCA cycle in “Ca. M. fumariolicum” strain SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM) |
||
|---|---|---|---|---|---|
| Cells at μmax | N2 fixing cells | O2 limited cells | |||
| Pyruvate 2-oxoglutarate dehydrogenase complex (E1), alpha subunit | acoA | Mfum_180024 | 1016 | 891 | 813 |
| Pyruvate 2-oxoglutarate dehydrogenase complex (E1), beta subunit | acoB | Mfum_180023 | 922 | 937 | 987 |
| Pyruvate 2-oxoglutarate dehydrogenase complex, dihydrolipoamide acyltransferase (E2) | aceF | Mfum_180020 | 877 | 813 | 297 |
| Pyruvate 2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) | lpd1 | Mfum_180019 | 478 | 241 | 322 |
| lpd 2 | Mfum_720053 | 121 | 42 | 103 | |
| Succinate dehydrogenase flavoprotein subunit | sdhA | Mfum_300009 | 419 | 359 | 256 |
| Succinate dehydrogenase catalytic subunit | sdhB | Mfum_300008 | 177 | 84 | 231 |
| Succinate dehydrogenase cytochrome b subunit | sdhC | Mfum_300010 | 185 | 72 | 59 |
| Succinyl-CoA synthetase subunit beta | sucC | Mfum_170019 | 279 | 227 | 214 |
| Succinyl-CoA ligase (ADP-forming) subunit alpha | sucD | Mfum_170018 | 831 | 1248 | 751 |
| Pyruvate kinase | pykF1 | Mfum_940089 | 155 | 86 | 130 |
| pykF2 | Mfum_990021 | 630 | 382 | 499 | |
| 6-Phosphofructokinase | pfkA | Mfum_920021 | 413 | 288 | 540 |
| Deoxyribose-phosphate aldolase | deoC | Mfum_850005 | 100 | 44 | 92 |
| Enolase | eno | Mfum_310014 | 1540 | 2423 | 1136 |
| Fumarate hydratase class II | fumC | Mfum_220008 | 192 | 146 | 156 |
| Phosphoenolpyruvate carboxykinase (ATP) | pckA | Mfum_200061 | 250 | 227 | 161 |
| Isocitrate dehydrogenase (NADP) | icd | Mfum_200008 | 242 | 154 | 122 |
| Aconitate hydratase | acn | Mfum_170037 | 343 | 170 | 156 |
| Phosphoglycerate mutase (PhoE family) | phoE | Mfum_70011 | 530 | 282 | 306 |
| Citrate synthase | gltA | Mfum_260030 | 830 | 404 | 368 |
| Acyl-CoA synthetase (AMP forming) | Acs | Mfum_1020005 | 448 | 297 | 414 |
| Acetate kinase | ackA | Mfum_310026 | 256 | 180 | 161 |
The mRNA expression is expressed as RPKM according to Mortazavi et al. (2008). Changes in expression in the chemostat cultures (N2 fixing cells or O2 limited cells) compared to batch culture cells growing at μmax are indicated by shading: up regulation >2 times (dark gray), down regulation <0.5 (light gray).
Table A3.
Transcription of genes involved carbon and energy storage in “Ca. M. fumariolicum” strain SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM) |
||
|---|---|---|---|---|---|
| Cells at μmax | N2 fixing cells | O2 limited cells | |||
| Glycogen synthase 2 | glgA | Mfum_1010040 | 306 | 242 | 346 |
| 1,4-Alpha-glucan-branching enzyme | glgB | Mfum_170041 | 154 | 91 | 110 |
| Malto-oligosyltrehalose trehalohydrolase | glgB | Mfum_170046 | 162 | 128 | 81 |
| Glucose-1-phosphate adenylyltransferase | glgC | Mfum_1020013 | 391 | 158 | 416 |
| Glucan phosphorylase | glgP1 | Mfum_1020098 | 656 | 447 | 843 |
| glgP2 | Mfum_220010 | 113 | 98 | 122 | |
| glgP3 | Mfum_880004 | 447 | 382 | 450 | |
| Glycogen operon protein homolog | glgX | Mfum_40003 | 381 | 218 | 263 |
| Glycogen debranching enzyme | gdb | Mfum_200059 | 256 | 246 | 106 |
| ABC-type phosphate transport system, permease | pstA | Mfum_300005 | 81 | 45 | 98 |
| Phosphate import ATP-binding protein | pstB | Mfum_300006 | 137 | 99 | 249 |
| ABC-type phosphate transport system, permease | pstC | Mfum_300004 | 72 | 35 | 83 |
| ABC-type phosphate transport system, periplasmic component | pstS | Mfum_300003 | 108 | 97 | 129 |
| Exopolyphosphatase | gppA1 | Mfum_1010048 | 581 | 430 | 486 |
| gppA2 | Mfum_1020105 | 101 | 63 | 101 | |
| Polyphosphate kinase | ppk | Mfum_1030014 | 266 | 206 | 197 |
| Exopolyphosphatase-related protein | Mfum_550017 | 506 | 283 | 404 | |
| Adenylate kinase | adk | Mfum_210014 | 247 | 88 | 170 |
The mRNA expression is expressed as RPKM according to Mortazavi et al. (2008). Changes in expression in the chemostat cultures (N2 fixing cells or O2 limited cells) compared to batch culture cells growing at μmax are indicated by shading: up regulation >2 times (dark gray), down regulation <0.5 (light gray).
Figure A1.
Plots of expression levels (log2(RPKM + 1)) of 394 genes (in total 443 kbp) involved in energy generation, ribosomes, carbon fixation (CBB cycle), Cl metabolism (except for pmo), amino acid synthesis, cell wall synthesis, translation, transcription, DNA replication, and tRNA synthesis.
References
- Alquéres S. M. C., Oliveira J. H. M., Nogueira E. M., Guedes H. V., Oliveira P. L., Camara F., Baldani J. I., Martins O. B. (2010). Antioxidant pathways are up-regulated during biological nitrogen fixation to prevent ROS-induced nitrogenase inhibition in Gluconacetobacter diazotrophicus. Arch. Microbiol. 192, 835–841 10.1007/s00203-010-0609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman A. J., Speake C. C., Lidstrom M. E. (2001). nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ. Microbiol. 67, 4009–4016 10.1128/AEM.67.9.4009-4016.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baani M., Liesack W. (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SCZ. Proc. Natl. Acad. Sci. U.S.A. 105, 10203–10208 10.1073/pnas.0702643105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belova S. E., Baani M., Suzina N. E., Bodelier P. L. E., Liesack W., Dedysh S. N. (2011). Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 3, 36–46 10.1111/j.1758-2229.2010.00180.x [DOI] [PubMed] [Google Scholar]
- Boetius A., Ravenschlag K., Schubert C. J., Rickert D., Widdel F., Gieseke A., Amann R., Jorgensen B. B., Witte U., Pfannkuche O. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626 10.1038/35036572 [DOI] [PubMed] [Google Scholar]
- Bowien B., Kusian B. (2002). Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha. Arch. Microbiol. 178, 85–93 10.1007/s00203-002-0441-3 [DOI] [PubMed] [Google Scholar]
- Campbell M. A., Nyerges G., Kozlowski J. A., Poret-Peterson A. T., Stein L. Y., Klotz M. G. (2011). Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 322, 82–89 10.1111/j.1574-6968.2011.02340.x [DOI] [PubMed] [Google Scholar]
- Chaudhuri R. R., Yu L., Kanji A., Perkins T. T., Gardner P. P., Choudhary J., Maskell D. J., Grant A. J. (2011). Quantitative RNA-seq analysis of the Campylobacter jejuni transcriptome. Microbiology 157, 2922–2932 10.1099/mic.0.050278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Crombie A., Rahman M. T., Dedysh S. N., Liesack W., Stott M. B., Alam M., Theisen A. R., Murrell J. C., Dunfield P. F. (2010). Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J. Bacteriol. 192, 3840–3841 10.1128/JB.00506-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L., Chen S. W., Lapidus A., Lidstrom M. E. (2003). Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185, 2980–2987 10.1128/JB.185.10.2980-2987.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L., Kalyuzhnaya M. G., Lidstrom M. E. (2009). The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63, 477–499 10.1146/annurev.micro.091208.073600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292 10.1111/j.1758-2229.2009.00038.x [DOI] [PubMed] [Google Scholar]
- Croucher N. J., Thomson N. R. (2010). Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 13, 619–624 10.1016/j.mib.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaki R., Hanczar T., Bodrossy L., Murrell J. C., Kovacs K. L. (2001). Molecular characterization of structural genes coding for a membrane bound hydrogenase in Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 205, 203–207 10.1111/j.1574-6968.2001.tb10948.x [DOI] [PubMed] [Google Scholar]
- De Bont J. A. M., Mulder E. G. (1976). Invalidity of acetylene reduction assay in alkane utilizing, nitrogen-fixing bacteria. Appl. Environ. Microbiol. 31, 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh S. N., Khmelenina V. N., Suzina N. E., Trotsenko Y. A., Semrau J. D., Liesack W., Tiedje J. M. (2002). Methylocapsa acidiphila gen. nov., sp nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52, 251–261 [DOI] [PubMed] [Google Scholar]
- Dedysh S. N., Knief C., Dunfield P. F. (2005). Methylocella species are facultatively methanotrophic. J. Bacteriol. 187, 4665–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh S. N., Ricke P., Liesack W. (2004). NifH and NifD phylogenies: an evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 150, 1301–1313 10.1099/mic.0.26585-0 [DOI] [PubMed] [Google Scholar]
- Dekas A. E., Poretsky R. S., Orphan V. J. (2009). Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426 10.1126/science.1178223 [DOI] [PubMed] [Google Scholar]
- Dixon R., Kahn D. (2004). Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631 10.1038/nrmicro954 [DOI] [PubMed] [Google Scholar]
- Dunfield P. F., Belova S. E., Vorob’Ev A. V., Cornish S. L., Dedysh S. N. (2010). Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int. J. Syst. Evol. Microbiol. 60, 2659–2664 10.1099/ijs.0.020149-0 [DOI] [PubMed] [Google Scholar]
- Dunfield P. F., Yuryev A., Senin P., Smirnova A. V., Stott M. B., Hou S. B., Ly B., Saw J. H., Zhou Z. M., Ren Y., Wang J. M., Mountain B. W., Crowe M. A., Weatherby T. M., Bodelier P. L. E., Liesack W., Feng L., Wang L., Alam M. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–883 10.1038/nature06411 [DOI] [PubMed] [Google Scholar]
- Erikstad H. A., Jensen S., Keen T. J., Birkeland N. K. (2012). Differential expression of particulate methane monooxygenase genes in the verrucomicrobial methanotroph “Methylacidiphilum kamchatkense” Kam1. Extremophiles 16, 405–409 10.1007/s00792-012-0439-y [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., Butler M. K., Le Paslier D., Pelletier E., Mangenot S., Kuypers M. M. M., Schreiber F., Dutilh B. E., Zedelius J., de Beer D., Gloerich J., Wessels H. J. C. T., van Alen T., Luesken F., Wu M. L., van de Pas-Schoonen K. T., Op den Camp H. J. M., Janssen-Megens E. M., Francoijs K. J., Stunnenberg H., Weissenbach J., Jetten M. S. M., Strous M. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–550 10.1038/nature08883 [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., Shima S., van de Pas-Schoonen K. T., Kahnt J., Medema M. H., Op den Camp H. J. M., Jetten M. S. M., Strous M. (2008). Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10, 3164–3173 10.1111/j.1462-2920.2008.01724.x [DOI] [PubMed] [Google Scholar]
- Hanczar T., Csaki R., Bodrossy L., Murrell J. C., Kovacs K. L. (2002). Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch. Microbiol. 177, 167–172 10.1007/s00203-001-0372-4 [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Hanson T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60, 439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S. B., Makarova K. S., Saw J. H. W., Senin P., Ly B. V., Zhou Z. M., Ren Y., Wang J. M., Galperin M. Y., Omelchenko M. V., Wolf Y. I., Yutin N., Koonin E. V., Stott M. B., Mountain B. W., Crowe M. A., Smirnova A. V., Dunfield P. F., Feng L., Wang L., Alam M. (2008). Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3, 1–25 10.1186/1745-6150-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T., Jensen S., Reigstad L. J., Larsen O., Birkeland N. K. (2008). Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A. 105, 300–304 10.1073/pnas.0704162105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartal B., Maalcke W. J., de Almeida N. M., Cirpus I., Gloerich J., Geerts W., Op den Camp H. J. M., Harhangi H. R., Janssen-Megens E. M., Francoijs K. J., Stunnenberg H. G., Keltjens J. T., Jetten M. S. M., Strous M. (2011). Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–130 10.1038/nature10453 [DOI] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Jetten M. S. M., Op den Camp H. J. M. (2010). Nitrogen fixation by the verrucomicrobial methanotroph ‘Methylacidiphilum fumariolicum’ SolV. Microbiology 156, 1052–1059 10.1099/mic.0.036061-0 [DOI] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Wieczorek A., Mohammadi S. S., Francoijs K. J., Stunnenberg H. G., Jetten M. S. M., Op den Camp H. J. M. (2011). Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193, 4438–4446 10.1128/JB.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem A. F., Wieczorek A. S., Pol A., Vuilleumier S., Harhangi H. R., Dunfield P. F., Kalyuzhnaya M. G., Murrell J. C., Francoijs K-J., Stunnenberg H. G., Stein L. Y., DiSpirito A. A., Semrau J. D., Lajus A., Médigue C., Klotz M. G., Jetten M. S. M., Op den Camp H. J. M. (2012). Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J. Bacteriol. 194, 3729–3730 10.1128/JB.00501-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz M. G., Stein L. Y. (2008). Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 278, 146–456 10.1111/j.1574-6968.2007.00970.x [DOI] [PubMed] [Google Scholar]
- Malone J. H., Oliver B. (2011). Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 9, 34. 10.1186/1741-7007-9-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Murrell J. C., Dalton H. (1983). Nitrogen fixation in obligate methanotrophs. J. Gen. Microbiol. 129, 3481–3486 [Google Scholar]
- Murrell J. C., Gilbert B., McDonald I. R. (2000). Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173, 325–332 10.1007/s002030000158 [DOI] [PubMed] [Google Scholar]
- Nyerges G., Stein L. Y. (2009). Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol. Lett. 297, 131–136 10.1111/j.1574-6968.2009.01674.x [DOI] [PubMed] [Google Scholar]
- Oakley C. J., Murrell J. C. (1991). Cloning of nitrogenase structural genes from the obligate methanotroph Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 78, 121–126 10.1016/0378-1097(91)90144-Y [DOI] [PubMed] [Google Scholar]
- Op den Camp H. J. M., Islam T., Stott M. B., Harhangi H. R., Hynes A., Schouten S., Jetten M. S. M., Birkeland N. K., Pol A., Dunfield P. F. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306 10.1111/j.1758-2229.2009.00022.x [DOI] [PubMed] [Google Scholar]
- Pieja A. J., Rostkowski K. H., Criddle C. S. (2011). Distribution and selection of poly-3-hydroxybutyrate production capacity in methanotrophic proteobacteria. Microb. Ecol. 62, 564–573 10.1007/s00248-011-9873-0 [DOI] [PubMed] [Google Scholar]
- Pol A., Heijmans K., Harhangi H. R., Tedesco D., Jetten M. S. M., Op den Camp H. J. M. (2007). Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450, 874–879 10.1038/nature06222 [DOI] [PubMed] [Google Scholar]
- Raghoebarsing A. A., Pol A., van de Pas-Schoonen K. T., Smolders A. J. P., Ettwig K. F., Rijpstra W. I. C., Schouten S., Damste J. S. S., Op den Camp H. J. M., Jetten M. S. M., Strous M. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921 10.1038/nature04617 [DOI] [PubMed] [Google Scholar]
- Robson R. L., Postgate J. R. (1980). Oxygen and hydrogen in biological nitrogen fixation. Annu. Rev. Microbiol. 34, 183–207 10.1146/annurev.mi.34.100180.001151 [DOI] [PubMed] [Google Scholar]
- Semrau J. D., Chistoserdov A., Lebron J., Costello A., Davagnino J., Kenna E., Holmes A. J., Finch R., Murrell J. C., Lidstrom M. E. (1995). Particulate methane monooxygenase genes in methanotrophs. J. Bacteriol. 177, 3071–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau J. D., DiSpirito A. A., Vuilleumier S. (2011). Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol. Lett. 323, 1–12 10.1111/j.1574-6968.2011.02315.x [DOI] [PubMed] [Google Scholar]
- Stein L. Y., Klotz M. G. (2011). Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem. Soc. Trans. 39, 1826–1831 10.1042/BST20110712 [DOI] [PubMed] [Google Scholar]
- Stolyar S., Costello A. M., Peeples T. L., Lidstrom M. E. (1999). Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology 145, 1235–1244 10.1099/13500872-145-5-1235 [DOI] [PubMed] [Google Scholar]
- Takeda K. (1988). Characteristics of a nitrogen fixing methanotroph, Methylocystis T-1. Antonie Van Leeuwenhoek 54, 521–534 10.1007/BF00588388 [DOI] [PubMed] [Google Scholar]
- Tavormina P. L., Orphan V. J., Kalyuzhnaya M. G., Jetten M. S. M., Klotz M. G. (2011). A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ. Microbiol. Rep. 3, 91–100 10.1111/j.1758-2229.2010.00192.x [DOI] [PubMed] [Google Scholar]
- Taylor S., Ninjoor V., Dowd D. M., Tappel A. L. (1974). Cathepsin b2 measurement by sensitive fluorometric ammonia analysis. Anal. Biochem. 60, 153–162 10.1016/0003-2697(74)90140-7 [DOI] [PubMed] [Google Scholar]
- van Passel M. W. J., Kant R., Zoetendal E. G., Plugge C. M., Derrien M., Malfatti S. A., Chain P. S. G., Woyke T., Palva A., de Vos W. M., Smidt H. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6, e16876. 10.1371/journal.pone.0016876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Horn M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249 10.1016/j.copbio.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 10.1038/nrm2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner U., Egli T. (1990). Dynamics of microbial-growth and cell composition in batch culture. FEMS Microbiol. Rev. 75, 19–44 10.1111/j.1574-6968.1990.tb04084.x [DOI] [PubMed] [Google Scholar]
- Ward N., Larsen O., Sakwa J., Bruseth L., Khouri H., Durkin A. S., Dimitrov G., Jiang L. X., Scanlan D., Kang K. H., Lewis M., Nelson K. E., Methe B., Wu M., Heidelberg J. F., Paulsen I. T., Fouts D., Ravel J., Tettelin H., Ren Q. H., Read T., DeBoy R. T., Seshadri R., Salzberg S. L., Jensen H. B., Birkeland N. K., Nelson W. C., Dodson R. J., Grindhaug S. H., Holt I., Eidhammer I., Jonasen I., Vanaken S., Utterback T., Feldblyum T. V., Fraser C. M., Lillehaug J. R., Eisen J. A. (2004). Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2, e303. 10.1371/journal.pbio.0020120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimga M. T., Dunfield P. F., Ricke P., Heyer J., Liesack W. (2003). Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69, 5593–5602 10.1128/AEM.69.9.5593-5602.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-Seq analysis of “Ca. Methylacidiphilum fumariolicum” SolV grown under different conditions.
Housekeeping genes to test robustness of transcriptome data.