Abstract
Transcripts originating from the SV40 late promoter of pSV2-neo or pSV2-cat contain pBR322 sequences and are polyadenylated at the SV40 late poly(A) site, resulting in an RNA of 3500 nt. If the SV40(L) poly(A) signal is destroyed, late orientation transcripts are polyadenylated at a site within pBR322 sequences, yielding in an RNA of 2500 nt. This cryptic poly(A) site is located 42-46 nucleotides downstream from an AAUAAA. Utilization of the pBR322 poly(A) signal is undetectable in late orientation transcripts from pSV2-neo or pSV2-cat, although it is located 966 nucleotides upstream from the SV40(L) poly(A) signal. The pBR322 site is not utilized when the spacing between the two poly(A) signals is varied from 209 to 1913 nucleotides. The pBR322 poly(A) site was utilized only in constructs in which all or portions of the SV40(L) poly(A) signal were deleted, such as in a construct with a 7 bp deletion into the SV40(L) AATAAA and adjacent sequences.
Full text
PDF



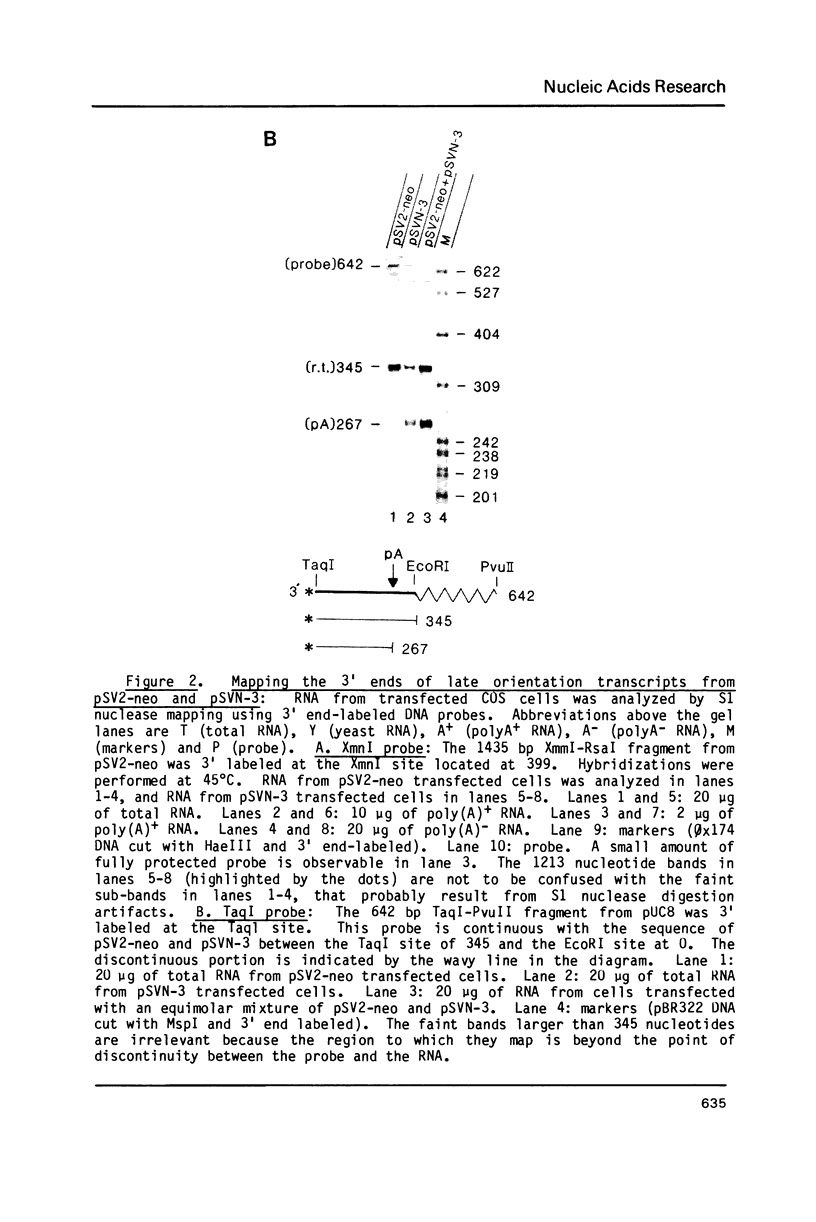

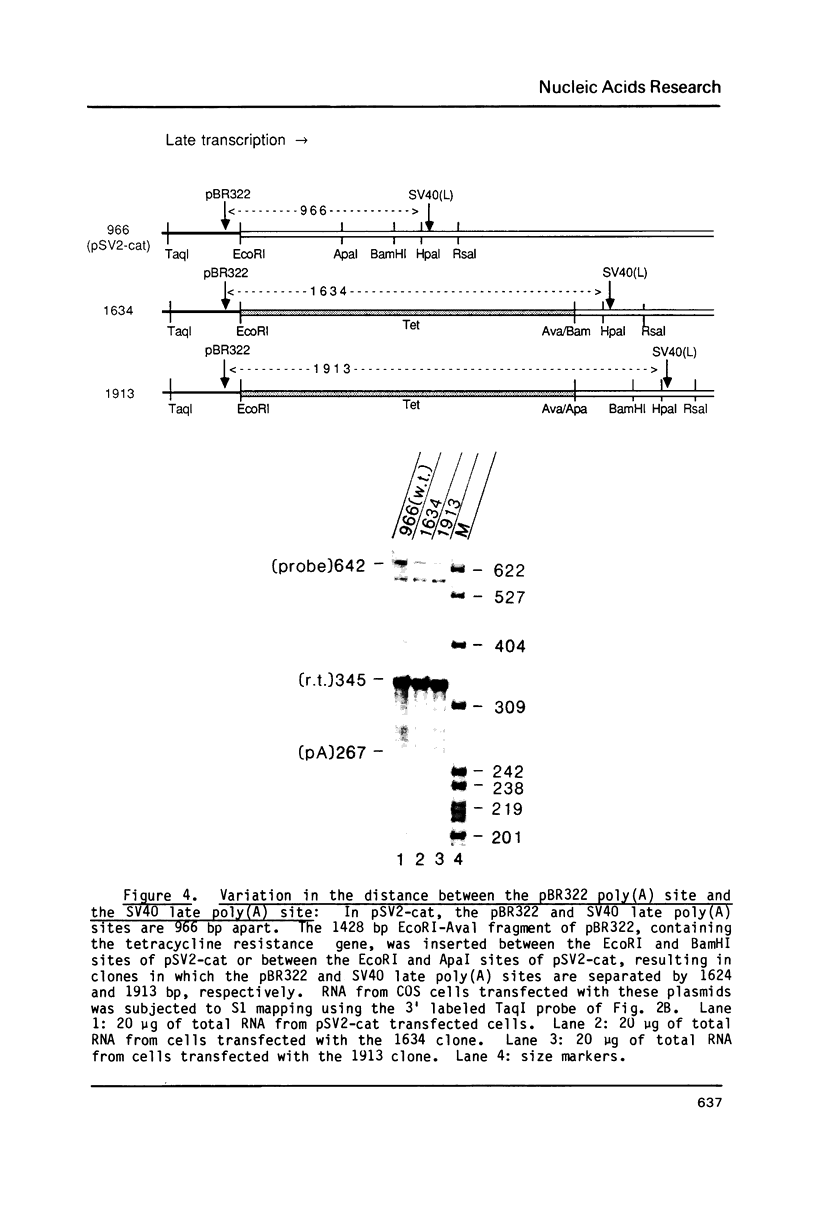





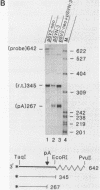
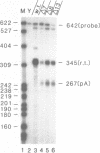
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Capetanaki Y. G., Ngai J., Flytzanis C. N., Lazarides E. Tissue-specific expression of two mRNA species transcribed from a single vimentin gene. Cell. 1983 Dec;35(2 Pt 1):411–420. doi: 10.1016/0092-8674(83)90174-5. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Stacy T. P. Identification of sequences in the herpes simplex virus thymidine kinase gene required for efficient processing and polyadenylation. Mol Cell Biol. 1985 Aug;5(8):2104–2113. doi: 10.1128/mcb.5.8.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gerlinger P., Krust A., LeMeur M., Perrin F., Cochet M., Gannon F., Dupret D., Chambon P. Multiple initiation and polyadenylation sites for the chicken ovomucoid transcription unit. J Mol Biol. 1982 Dec 5;162(2):345–364. doi: 10.1016/0022-2836(82)90531-9. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Ali H., Nevins J. R. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol Cell Biol. 1985 Nov;5(11):2975–2983. doi: 10.1128/mcb.5.11.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M. M., Beckendorf R. C., Westhafer M. A., Nordstrom J. L. Requirement of A-A-U-A-A-A and adjacent downstream sequences for SV40 early polyadenylation. Nucleic Acids Res. 1986 Jun 25;14(12):4939–4952. doi: 10.1093/nar/14.12.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J., Tseng R. W., Acheson N. H. Duplication of functional polyadenylation signals in polyomavirus DNA does not alter efficiency of polyadenylation or transcription termination. J Virol. 1986 Jun;58(3):733–742. doi: 10.1128/jvi.58.3.733-742.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983 Mar 11;11(5):1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- Nelson K. J., Haimovich J., Perry R. P. Characterization of productive and sterile transcripts from the immunoglobulin heavy-chain locus: processing of micron and muS mRNA. Mol Cell Biol. 1983 Jul;3(7):1317–1332. doi: 10.1128/mcb.3.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R. Definition and mapping of adenovirus 2 nuclear transcription. Methods Enzymol. 1980;65(1):768–785. doi: 10.1016/s0076-6879(80)65072-1. [DOI] [PubMed] [Google Scholar]
- Nordstrom J. L., Hall S. L., Kessler M. M. Polyadenylylation of sea urchin histone RNA sequences in transfected COS cells. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1094–1098. doi: 10.1073/pnas.82.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom J. L., Westhafer M. A. Splicing and polyadenylylation at cryptic sites in RNA transcribed from pSV2-neo. Biochim Biophys Acta. 1986 Jun 20;867(3):152–162. doi: 10.1016/0167-4781(86)90075-8. [DOI] [PubMed] [Google Scholar]
- Parnes J. R., Robinson R. R., Seidman J. G. Multiple mRNA species with distinct 3' termini are transcribed from the beta 2-microglobulin gene. 1983 Mar 31-Apr 6Nature. 302(5907):449–452. doi: 10.1038/302449a0. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Mermod J. J., Amara S. G., Swanson L. W., Sawchenko P. E., Rivier J., Vale W. W., Evans R. M. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983 Jul 14;304(5922):129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tosi M., Young R. A., Hagenbüchle O., Schibler U. Multiple polyadenylation sites in a mouse alpha-amylase gene. Nucleic Acids Res. 1981 May 25;9(10):2313–2323. doi: 10.1093/nar/9.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik R. P., Lyons R. H., Post L., Rottman F. M. Requirement for the 3' flanking region of the bovine growth hormone gene for accurate polyadenylylation. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]