Abstract
Cholesterol 7α-hydroxylase (CYP7A1) is the initiating and rate-limiting enzyme in the neutral pathway that coverts cholesterol to primary bile acids (BA). CYP7A1-deficient (Cyp7a1−/−) mice have a depleted BA pool, diminished intestinal cholesterol absorption, accelerated fecal sterol loss, and increased intestinal cholesterol synthesis. To determine the molecular and physiological effects of restoring the BA pool in this model, adult female Cyp7a1−/− mice and matching Cyp7a1+/+ controls were fed diets containing cholic acid (CA) at modest levels [0.015, 0.030, and 0.060% (wt/wt)] for 15–18 days. A level of just 0.03% provided a CA intake of ∼12 μmol (4.8 mg) per day per 100 g body wt and was sufficient in the Cyp7a1−/− mice to normalize BA pool size, fecal BA excretion, fractional cholesterol absorption, and fecal sterol excretion but caused a significant rise in the cholesterol concentration in the small intestine and liver, as well as a marked inhibition of cholesterol synthesis in these organs. In parallel with these metabolic changes, there were marked shifts in intestinal and hepatic expression levels for many target genes of the BA sensor farnesoid X receptor, as well as genes involved in cholesterol transport, especially ATP-binding cassette (ABC) transporter A1 (ABCA1) and ABCG8. In Cyp7a1+/+ mice, this level of CA supplementation did not significantly disrupt BA or cholesterol metabolism, except for an increase in fecal BA excretion and marginal changes in mRNA expression for some BA synthetic enzymes. These findings underscore the importance of using moderate dietary BA levels in studies with animal models.
Keywords: bile acid synthesis and transport, bile acid excretion, cholesterol synthesis and transport, small intestine, liver
the formation of bile acids from cholesterol in the liver and their eventual excretion in the stools represents a major route for the elimination of cholesterol from the body (16, 28, 45, 46). Bile acid synthesis occurs in the liver via several pathways, the predominant one usually being the classic or neutral pathway that is initiated by cholesterol 7α-hydroxylase (CYP7A1). There are also alternate pathways that involve an initial hydroxylation of the side chain via sterol 27-hydroxylase (CYP27A1) or cholesterol 25-hydroxylase (CH25H). Both 27-hydroxycholesterol and 25-hydroxycholesterol are substrates for CYP7B1, an oxysterol 7α-hydroxylase. In another pathway, 24-hydroxycholesterol, generated in the brain via cholesterol 24-hydroxylase (CYP46A1), is transported to the liver, where it serves as a substrate for CYP39A1, another oxysterol 7α-hydroxylase. Each of these reactions, as well as all the subsequent steps in the bile acid synthetic pathway and the mechanisms that ultimately control the amount of cholesterol converted to bile acids, is discussed in recent reviews (9, 32, 33).
Bile acids, in conjugated form, are secreted via the bile into the lumen of the small bowel, where they enter an intestinal pool, the preservation of which is essential for the absorption of sterols and various other classes of lipids, including essential fatty acids and fat-soluble vitamins (7). Indeed, it has long been known that key factors influencing the amount of cholesterol that is absorbed from the small intestine and transported to the liver are the size and composition of the bile acid pool (37, 48, 51). In the steady state, the fraction of bile acid in this pool that is excreted from the body each day is ordinarily replaced by an equal amount of newly synthesized bile acid. The bile acid pool undergoes continuous enterohepatic flux, a process that is facilitated through the action of multiple transporters (1, 11). Specifically, these proteins are responsible for pumping bile acids across the canalicular membrane into the bile [bile salt export pump (BSEP)], reclaiming them from the ileum [apical sodium-dependent bile acid transporter (ASBT) and organic solute transporter (OSTα-OSTβ)], or extracting them from the portal blood [sodium-taurocholate cotransporter polypeptide (NCTP)]. Long before these proteins were identified and their specific roles in bile acid handling delineated, the pharmacological interruption of the enterohepatic circulation of bile acids was used as an effective strategy for lowering the plasma LDL-cholesterol concentration in the treatment of atherosclerosis (15, 20).
In recent years, new areas of research point to a more global role for bile acids in metabolic regulation beyond their critical involvement in the solubilization of lipids and the maintenance of cholesterol balance across the liver and the body as a whole (2, 19). This development stems largely from major advances in our knowledge of how bile acids serve as activators of several nuclear receptors, such as farnesoid X receptor and pregnane X receptor, and also of cell signaling pathways in the liver and gastrointestinal tract (14, 19, 30). One of the most clinically significant findings centers around the apparent relationship between bile acid metabolism, hepatic insulin resistance, and glucose homeostasis, which may be mediated through farnesoid X receptor-dependent and -independent mechanisms (22, 26, 38, 49, 56). To the contrary, a more recent study suggests that glucose and insulin may be regulators of bile acid synthesis (25). These and other findings are driving a wave of research into the association between bile acid metabolism and various disorders, such as type 2 diabetes and obesity. Many of these new areas of investigation utilize animal models with genetically altered bile acid metabolism and/or experimental diets enriched with various bile acids. In most cases, the level of bile acid incorporated into such diets is 0.5% (wt/wt), but in some studies, the amount of supplementation far exceeds this level (4, 8, 10, 18, 29, 48, 50, 53, 55). Depending on the design of the study, such high levels of supplementation could be considered unphysiological, because the daily oral intake of bile acid is manyfold greater than the size of the animal's bile acid pool and vastly greater than the basal rate of bile acid synthesis in that animal, as measured by its daily excretion rate of acidic sterols. This practice arises in part because of the lack of published data defining the impact of graded increases in dietary bile acid supplementation on bile acid and cholesterol metabolism in the mouse and other species.
Mice deficient in CYP7A1 manifest a marked reduction in bile acid pool size and excretion, diminished intestinal cholesterol absorption, accelerated fecal neutral sterol excretion, and increased intestinal cholesterol synthesis (12, 36). In the present study, we established a dietary cholic acid level that was sufficient to normalize these various parameters in this model. We then systematically quantitated, at a molecular and biochemical level, the changes in multiple components of intestinal and hepatic bile acid and sterol metabolism in CYP7A1-deficient (Cyp7a1−/−) mice and also in their matching Cyp7a1+/+ controls at this level, as well as lower and higher levels, of cholic acid intake. Together, the data show that decisive shifts in bile acid and cholesterol metabolism occur in response to dietary bile acid levels far lower than those traditionally used to perturb the enterohepatic circulation of bile acids in animal models.
MATERIALS AND METHODS
Animals and diets.
Cyp7a1−/− mice were generated by crossing homozygous carriers (21). The colony was maintained on a mixed-strain background (C57BL/6:129/SvEv), and C57BL/6:129/SvEv hybrids (Cyp7a1+/+) served as control animals. To increase the survival rate of homozygous pups, the diet of nursing females was routinely supplemented with cholic acid [0.25% (wt/wt)] until the pups were weaned at 23–25 days of age. The age of the mice was 7–10 mo at the time of study in all experiments, except those involving the measurement of cholesterol absorption and neutral sterol excretion, where the mice were 3–4 mo old. All studies were done with female mice fed the meal from a cereal-based rodent diet (Wayne Lab Blox, No. 8604, Harland Teklad, Madison, WI), which contained 0.02% (wt/wt) cholesterol and ∼5% (wt/wt) total lipid. This regimen, referred to as the basal diet, was made to contain varying levels of cholic acid (see below). Depending on the metabolic parameter being measured, the mice were housed individually or in groups of three or four in plastic colony cages with wood shavings in a light-cycled room. All animals were studied in the fed state toward the end of the dark phase of their light cycle. Many parameters of bile acid and cholesterol metabolism are sensitive to the level of caloric intake, as demonstrated by recent studies describing diurnal changes in multiple parameters of bile acid metabolism in mice (39, 54). All experiments were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical School.
Establishment of dietary cholic acid levels to be tested in Cyp7a1−/− and Cyp7a1+/+ mice.
Mature female mice lacking CYP7A1 were documented to excrete ∼5 μmol of bile acid per day per 100 g body wt, which represented only ∼36% of the output in matching Cyp7a1+/+ females (∼14 μmol·day−1·100 g body wt−1) (37). These mice, irrespective of genotype, consumed ∼16 g of the basal diet per day per 100 g body wt (data not shown), which corresponds to the food intake we reported previously for other mouse strains (41). From these data, we thus determined that the addition of 0.03% (wt/wt) cholic acid (mol wt 408.6) to the diet ought to provide a daily intake of bile acid (exclusive of that taken in through coprophagy) of ∼11.7 μmol·day−1·100 g body wt−1. This supplementation, in addition to the animal's basal rate of bile acid excretion of 5 μmol·day−1·100 g body wt−1, would raise the excretion rate in the Cyp7a1−/− mice to a level not very different from that typically found in their Cyp7a1+/+ counterparts. Nevertheless, it was decided that, in addition to 0.03% (wt/wt) cholic acid, we would also test the response of mice of both genotypes to half [0.015% (wt/wt)] and twice [0.060% (wt/wt)] this level of supplementation. Irrespective of the level of cholic acid supplementation, all mice maintained their weights over the feeding period. For all groups, the final body weights were 24–28 g.
Bile acid pool size and composition and rate of fecal bile acid excretion.
Pool size was determined as the total bile acid content of the small intestine, gallbladder, and liver combined. The bile acids were extracted in ethanol in the presence of [24-14C]taurocholic acid (PerkinElmer, Waltham, MA) and analyzed by HPLC (36). Bile acids were detected by measurement of the refractive index and identified by comparison with authentic standards. For muricholic acids, no attempt was made to determine whether the major peak identified as β-muricholic acid might have also represented unknown amounts of α- and ω-muricholic acid. In all animals, cholic and muricholic acid together represented >90% of the bile acids in the pool, although the ratio of cholic to muricholic acid varied widely, depending largely on the level of dietary cholic acid supplementation. Essentially all bile acids detected in the extracted pools were taurine-conjugated. Pool size was expressed as micromoles per 100 g body wt. For the fecal bile acid measurements, stools collected over 3 days from individually housed mice were dried, weighed, and ground to a fine powder. A 1-g aliquot of this material was used to determine total bile acid content by an enzymatic method previously described (36). The excretion rate of bile acids was expressed as micromoles per day per 100 g body wt.
Intestinal cholesterol absorption and rate of fecal neutral sterol excretion.
Fractional cholesterol absorption was measured by a dual-isotope method. A mixture of 2 μCi of [5,6-3H]sitostanol (American Radiolabeled Chemicals, St. Louis, MO) and 1 μCi of [4-14C]cholesterol (PerkinElmer) in medium-chain triglyceride oil (Mead Johnson, Evansville, IN) was administered intragastrically by gavage. Mice were housed individually, and stools were collected over the next 3 days. Aliquots of stool and of the original dosing mixture were extracted, and fractional cholesterol absorption (%) was calculated from the ratio of 14C to 3H, as previously described (36). This method is an adaptation of a method that we originally used in hamsters and has been validated by direct comparison with the dual-isotope plasma ratio technique in that species (40). The dual-isotope plasma and fecal ratio methods were originally established in the rat (5, 57). These and other techniques have been systematically evaluated for measurement of intestinal cholesterol absorption in the mouse (47). A second aliquot of stool was used to quantitate the amounts of cholesterol, coprostanol, epicoprostanol, and cholestanone by gas chromatography, and these data were used to calculate the rate of fecal neutral sterol excretion as micromoles per day per 100 g body wt.
Concentrations of total, unesterified, and esterified cholesterol in liver and small intestine.
Aliquots of liver and the whole small intestine were saponified and extracted, and their total cholesterol concentrations were measured by gas chromatography using stigmastanol as an internal standard (36). In one study, additional aliquots of liver were extracted in chloroform-methanol [2:1 (vol/vol)] for the measurement of unesterified and esterified cholesterol concentrations, as described elsewhere (43).
Rates of cholesterol synthesis in liver and small intestine.
Rates of cholesterol synthesis in liver and small intestine were measured in vivo using [3H]water, as described elsewhere (36). At 1 h after intraperitoneal administration of ∼40 mCi of [3H]water to the mice, the liver and whole small intestine were removed, rinsed, blotted, and weighed. The organs were then saponified, and the labeled sterols were extracted and quantitated as described elsewhere (36). The rate of cholesterol synthesis in each organ was calculated as nanomoles of [3H]water incorporated into sterols per hour per gram of tissue.
Relative mRNA expression analysis.
Small intestines were removed, flushed with ice-cold phosphate-buffered saline, and cut into three sections of similar length. The proximal and distal sections were opened longitudinally, and the mucosae were removed by gentle scraping. These scrapings, along with aliquots of liver, were quickly frozen in liquid nitrogen. mRNA levels were measured using a quantitative real-time PCR assay (23). All analyses were performed using the comparative cycle number at threshold method (User Bulletin No. 2, Perkin-Elmer Life Sciences) with cyclophilin as the internal control. The primer sequences used to measure RNA levels are given in Table 1. Relative mRNA levels in individual animals were determined by expression of the amount of mRNA found relative to that obtained for Cyp7a1+/+ mice fed the basal diet alone, which in each case was arbitrarily set at 1.0. For RNA blotting analyses, total RNA was prepared from various tissues of 4–10 mice (all male, except for ovary and uterus), equal amounts per animal were pooled, and poly(A)+-enriched mRNA was purified. Five micrograms of this RNA were loaded per lane, separated by electrophoresis, transferred from gel to membrane, and hybridized with [32P]cDNA probes, as previously described (14).
Table 1.
Quantitative real-time PCR primer sequences for bile acid- and cholesterol-related genes in the mouse
| Gene | Gene Name | NCBI Accession No. | Sequence of Primers (5′–3′) | Amplicon Nucleotide Nos. |
|---|---|---|---|---|
| Bile acid synthetic enzymes | ||||
| Cyp7a1 | Cholesterol 7α-hydroxylase | NM_007824 | F: AGCAACTAAACAACCTGCCAGTACTA | 1073–1154 |
| R: GTCCGGATATTCAAGGATGCA | ||||
| Ch25h | Cholesterol 25-hydroxylase | NM_009890 | F: CACAAGGTGCATCACCAGAACT | 478–565 |
| R: CGAAGAAGGTCAGCGAAAGC | ||||
| Cyp27a1 | Sterol 27-hydroxylase | NM_024264 | F: GCCTCACCTATGGGATCTTCA | 485–547 |
| R: TCAAAGCCTGACGCAGATG | ||||
| Cyp39a1 | Oxysterol 7α-hydroxylase | NM_018887 | F: GGTAGTGAAGCCAGTTAAAATTCTGA | 1228–1308 |
| R: CTGTGTAGCCAGAATGGAGACAA | ||||
| Cyp7b1 | Oxysterol 7α-hydroxylase | NM_007825 | F: TAGCCCTCTTTCCTCCACTCATA | 1298–1374 |
| R: GAACCGATCGAACCTAAATTCCT | ||||
| Hsd3b7 | 3β-Hydroxy-Δ5-C27 steroid oxidoreductase | NM_133943 | F: CCAGGTAGTTGGCAAAACTGATT | 1464–1542 |
| R: TGTTCCTCCCTGCTTGAGAAA | ||||
| Cyp8b1 | Sterol 12α-hydroxylase | NM_010012 | F: GCCTTCAAGTATGATCGGTTCCT | 1252–1326 |
| R: GATCTTCTTGCCCGACTTGTAGA | ||||
| Akr1d1 | Δ4-3-Oxosteroid 5β-reductase | NM_145364 | F: GAACGTCTCTTCTCCACCCTTGT | 762–871 |
| R: TCCCTCGCTGGATGTTGAA | ||||
| Akr1c4 | 3α-Hydroxysteroid dehydrogenase | NM_030611 | F: TGCCTTGTGCCAGATGTCA | 1037–1129 |
| R: CAGAAGCTTGGATTAGGGTGATATG | ||||
| Slc27a5 | Bile acid-CoA ligase | NM_009512 | F: GACCACTGGACTCCCAAAGC | 976–1044 |
| R: GACAGCACGTTGCTCACTTGT | ||||
| Amacr | 2-Methylacyl-CoA racemase | NM_008537 | F: GTGGATGAACAGCAATGAAGTCA | 1279–1356 |
| R: CGCAATCGTTATCGTGTAACCA | ||||
| Acox2 | Branched-chain acyl-CoA oxidase | NM_053115 | F: GACGGTCCTGAACGCATTT | 484–562 |
| R: CATTCATGGCAATACCATGTAAGTT | ||||
| Hsd17b4 | D-Bifunctional protein | NM_008292 | F: CACTGTGTGCTGTTAAAGGAGTCA | 2240–2319 |
| R: CACTCCTGTGGATCGCAGAA | ||||
| Acaa1 | Peroxisomal thiolase | NM_130864 | F: GCAGAAGCAGGATGACTTTGC | 809–891 |
| R: CAATCTCAGCACGGAGCAT | ||||
| Baat | Bile acid-CoA:amino acid N-acyltransferase | NM_007519 | F: AGCACCACTCCTCACTTCCATAG | 1517–1605 |
| R: TCCATCCTCCTGTATTTTCTTGTG | ||||
| Genes involved in enterohepatic flux of cholesterol and bile acid | ||||
| Abca1 | ATP-binding cassette member A1 | NM_013454 | F: CGTTTCCGGGAAGTGTCCTA | 6805–6883 |
| R: GCTAGAGATGACAAGGAGGATGGA | ||||
| Abcb11 | Bile salt export pump (BSEP) | NM_021022 | F: AAGCTACATCTGCCTTAGACACAGAAA | 3858–3941 |
| R: CAATACAGGTCCGACCCTCTCT | ||||
| Abcg8 | ATP-binding cassette member G8 | NM_026180 | F: TGCCCACCTTCCACATGTC | 1759–1831 |
| R: ATGAAGCCGGCAGTAAGGTAGA | ||||
| Fabp6 | Ileal bile acid-binding protein (IBABP) | NM_008375 | F: CAAGGCTACCGTGAAGATGGA | 240–317 |
| R: CCCACGACCTCCGAAGTCA | ||||
| Fgf15 | Fibroblast growth factor 15 | NM_008003 | F: ACGGGCTGATTCGCTACTC | 497–562 |
| R: TGTAGCCTAAACAGTCCATTTCCT | ||||
| Flot1 | Flotillin 1 | NM_008027 | F: CAAGAGTGAAAAGGTTTATACCCG | 309–394 |
| R: CCTTGTTCTGGCCCTGGATTT | ||||
| Flot2 | Flotillin 2 | NM_008028 | F: AGGTGACATCAGAAGTAAACCG | 1383–1468 |
| R: GGGTATCTTTGAAAGGTCCACACC | ||||
| Hmgcs1 | 3-Hydroxy-3-methylglutaryl-CoA synthase 1 | NM_145942 | F: GCCGTGAACTGGGTCGAA | 536–612 |
| R: GCATATATAGCAATGTCTCCTGCAA | ||||
| Npc1l1 | Niemann-Pick type C1-like protein 1 | NM_207242 | F: TGGACTGGAAGGACCATTTCC | 1547–1650 |
| R: GACAGGTGCCCCGTAGTCA | ||||
| Nr0b2 | Short heterodimer partner (SHP) | NM_011850 | F: CAGCGCTGCCTGGAGTCT | 492–565 |
| R: AGGATCGTGCCCTTCAGGTA | ||||
| Nr2a1 | Hepatic nuclear factor 4α (HNF4α) | NM_008261 | F: CCAACCTCAATTCATCCAACA | 254–316 |
| R: CCCGGTCGCCACAGAT | ||||
| Nr5a2 | Liver receptor homolog 1 (LRH-1) | NM_030676 | F: TGGGAAGGAAGGGACAATCTT | 1406–1506 |
| R: CGAGACTCAGGAGGTTGTTGAA | ||||
| Ostα | Organic solute transporter-α | NM_145932 | F: AACAGAACATGGGATCCAAGTTT | 1103–1161 |
| R: CAGGGCGGTCAGGATGA | ||||
| Slc10a1 | Sodium-taurocholate cotransporter polypeptide (NTCP) | NM_011387 | F: GAAGTCCAAAAGGCCACACTATGT | 568–645 |
| R: ACAGCCACAGAGAGGGAGAAAG | ||||
| Slc10a2 | Ileal bile acid transporter (IBAT or ASBT) | NM_011388 | F: TGGGTTTCTTCCTGGCTAGACT | 768–848 |
| R: TGTTCTGCATTCCAGTTTCCAA | ||||
| ScarbI | Scavenger receptor type BI (SRBI) | NM_016741 | F: TCCCCATGAACTGTTCTGTGAA | 1331–1397 |
| R: TGCCCGATGCCCTTGA | ||||
NCBI, National Center for Biotechnology Information; F, forward; R, reverse.
Analysis of data.
Values are means ± SE for the specified number of animals. Prism 5 software (GraphPad, San Diego, CA) was used for all statistical analyses. Differences between means were tested for statistical significance (P < 0.05) by one- or two-way analysis of variance with genotype and diet as factors. The Newman-Keuls multiple-comparison test for statistical significance was used for all one-way analyses of variance. Transformed data were used if unequal variance among the groups was evident by Bartlett's test.
RESULTS
Level of expression of mRNA for CYP7A1 and most other enzymes in the bile acid biosynthetic pathway is highly localized in liver.
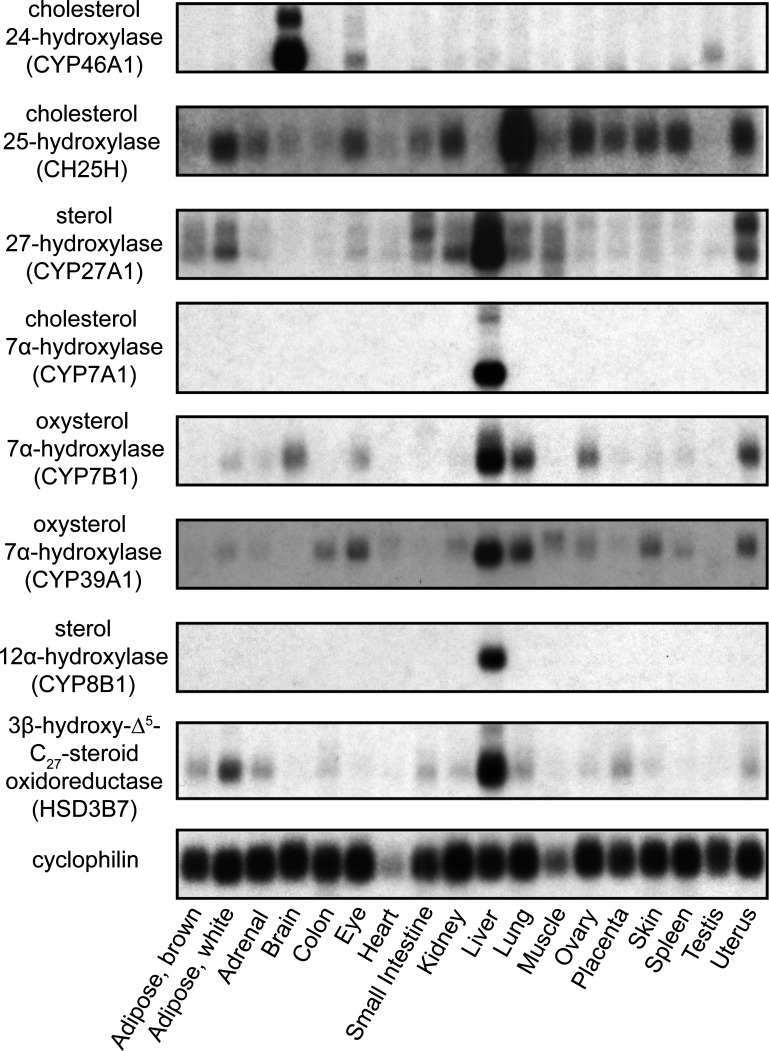
The tissue distribution of eight of the key enzymes involved in bile acid biosynthesis, as measured by RNA blotting, is shown in Fig. 1. With the exception of CH25H and CYP46A1, all enzymes were primarily localized to the liver. In the case of CH25H, the expression level was highest in the lung, and CYP46A1 was found almost exclusively in the brain.
Fig. 1.
Tissue distribution of key enzymes of bile acid biosynthesis in the mouse. Poly(A)+-enriched RNA from various mouse tissues (all male, except for ovary and uterus) were resolved by electrophoresis, transferred to a membrane, and detected by [32P]cDNA probes. Membrane was stripped and reprobed to detect RNAs encoding key enzymes of bile acid synthesis and the housekeeping gene cyclophilin.
Loss of Cyp7a1 function has a differential effect on the expression levels of mRNAs encoding other enzymes involved in bile acid biosynthesis.
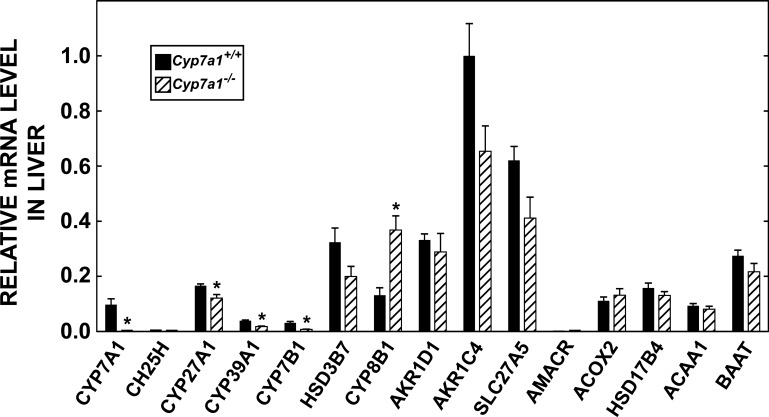
Although the Cyp7a1−/− mouse has been widely studied, there is no documented information regarding how the expression levels of mRNA for other enzymes involved in the primary and alternate pathways of bile acid synthesis shift in the absence of CYP7A1. Given that one of the objectives of this study was to determine the impact of modest cholic acid supplementation on the level of mRNA expression for enzymes in the hepatic bile acid biosynthetic pathways of adult Cyp7a1−/− and matching Cyp7a1+/+ mice, we carried out a preliminary study to measure the mRNA levels for these enzymes in mice fed a basal diet. As shown in Fig. 2, the enzyme with the highest abundance of mRNA was 3α-hydroxysteroid dehydrogenase (AKR1C4). Thus this level was arbitrarily set at 1.0, and the mRNA levels for other enzymes were expressed relative to this baseline. Surprisingly, for the majority of enzymes, there was no significant difference in the relative mRNA level between the Cyp7a1−/− and Cyp7a1+/+ mice. The clearest exception was the mRNA level for sterol 12α-hydroxylase (CYP8B1), which showed a decisive increase in the Cyp7a1−/− mice. In contrast, there were modest reductions in the mRNA levels for CYP27A1 and both of the oxysterol 7α-hydroxylases (CYP39A1 and CYP7B1).
Fig. 2.
Relative levels of expression of mRNA for enzymes involved in conversion of cholesterol to bile acids in control (Cyp7a1+/+) and CYP7A1-deficient (Cyp7a1−/−) mice fed a basal rodent chow diet. Quantitative real-time PCR was used to measure the mRNA level of each enzyme in liver of adult mice of both genotypes. Order in which enzymes are listed follows that in a recent review (33). Full name, gene accession number, and primer sequence for each enzyme are given in Table 1. mRNA level for individual enzymes was established by the cycle number at threshold method, with cyclophilin used as the invariant housekeeping gene. Values were adjusted arithmetically to convey 3α-hydroxysteroid dehydrogenase (AKR1C4) as the most abundant RNA species, the expression level of which was set arbitrarily at 1.0. CYP46A1 was omitted, because this mRNA is expressed at very low levels in liver (27). In the case of CH25H and 2-methylacyl-CoA-racemase (AMACR), only trace amounts of mRNA were detected in Cyp7a1+/+ and Cyp7a1−/− mice. Values are means ± SE of determinations in 5 mice of each genotype. *Significantly different from matching Cyp7a1+/+ control (P < 0.05).
Low levels of dietary cholic acid supplementation cause dramatic changes in bile acid metabolism in Cyp7a1−/− mice with comparatively little effect in Cyp7a1+/+ controls.
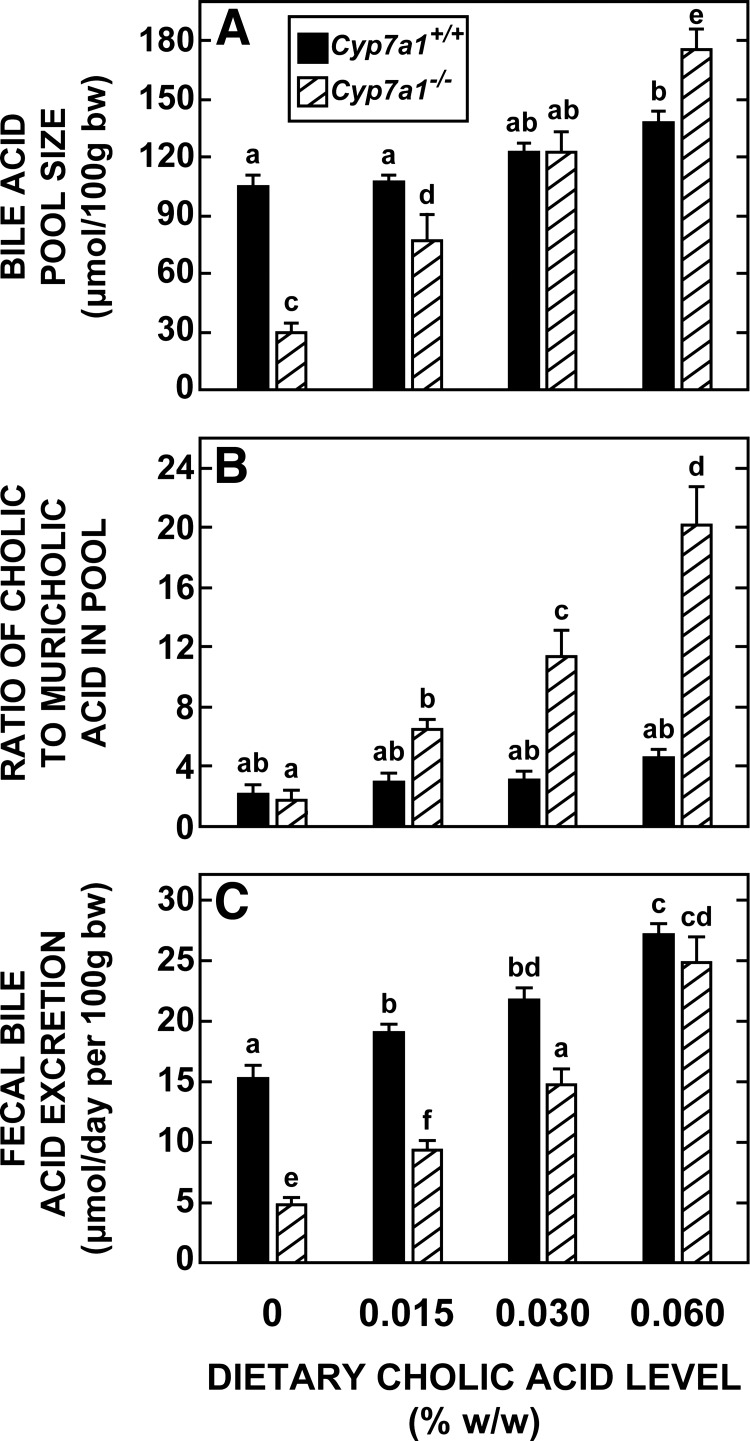
The data in Fig. 3 reveal two notable findings with respect to how the major parameters of bile acid metabolism in Cyp7a1−/− mice change when small amounts of cholic acid are added to their diet. First, at a dietary cholic acid level of 0.030% (wt/wt), bile acid pool size (Fig. 3A) and fecal bile acid excretion (Fig. 3C) were restored to values very close to those found for Cyp7a1+/+ mice fed the basal diet alone; however, at this level of supplementation, there was a 6.4-fold increase in the ratio of cholic to muricholic acid in the pool of the Cyp7a1−/− mice (Fig. 3B). This rise, which reflected an increase in the mass of cholic acid in the pool, far exceeded that in the matching Cyp7a1+/+ mice, where the ratio increased from 2.16 ± 0.14 in mice fed the basal diet alone to 3.05 ± 0.21 in the matching group fed the diet containing 0.030% (wt/wt) cholic acid. Second, in Cyp7a1+/+ mice, cholic acid supplementation, while having comparatively little impact on pool size and composition, consistently raised the rate of fecal bile acid excretion, but the magnitude of change at each level of supplementation was never as pronounced as it was for matching Cyp7a1−/− mice (Fig. 3C).
Fig. 3.
Bile acid pool size and composition and fecal bile acid excretion rate in Cyp7a1−/− and Cyp7a1+/+ mice fed graded levels of cholic acid in their diet. Groups of adult female Cyp7a1−/− and matching Cyp7a1+/+ mice were fed a basal rodent chow diet containing 0–0.060% (wt/wt) cholic acid for 15–18 days. Values are means ± SE of data from 8 mice in each group. Because 2-way ANOVA revealed significant interaction between diet and genotype, 1-way ANOVA was performed; different letters (a–f) denote statistically different values (P < 0.05).
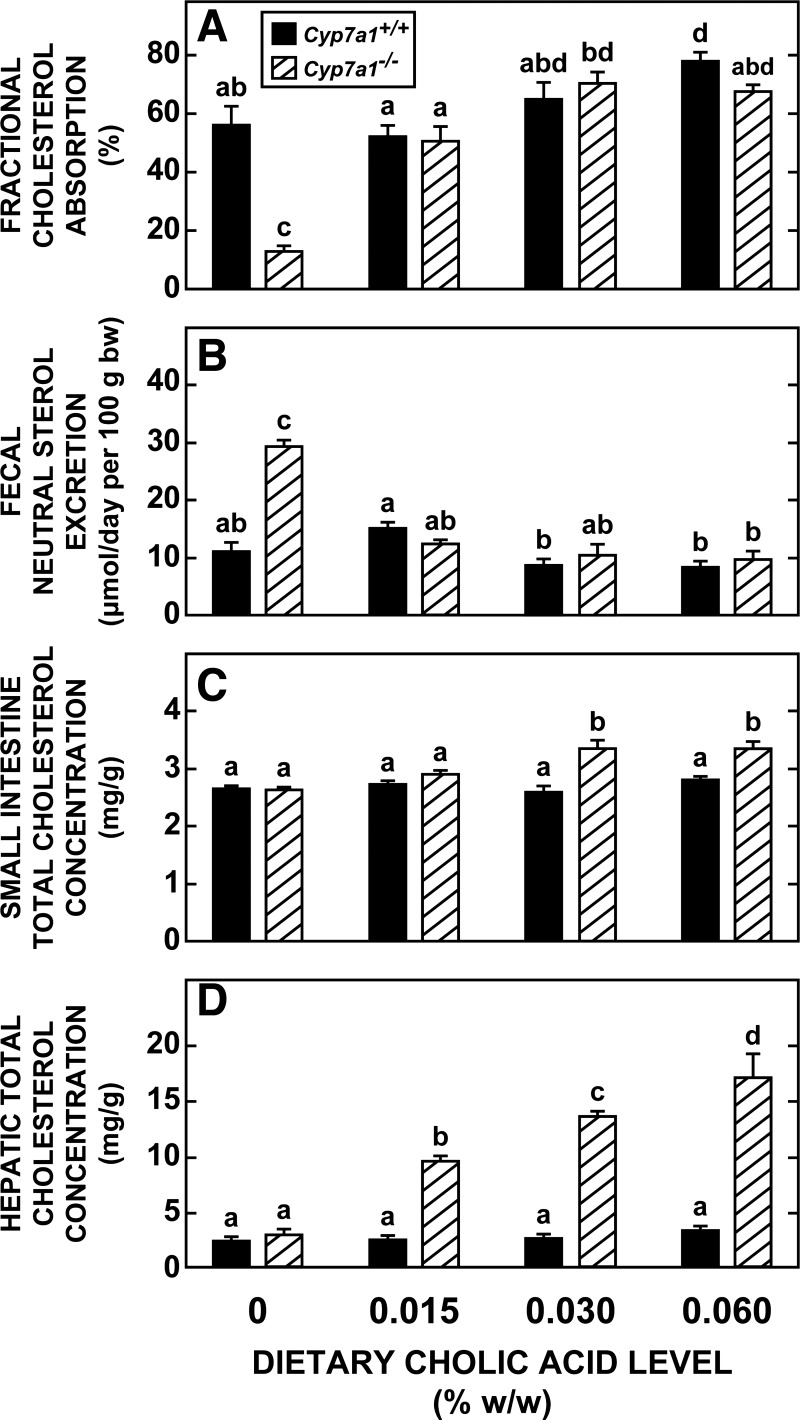
Normalization of bile acid pool size in Cyp7a1−/− mice by cholic acid feeding restores cholesterol absorption and rate of cholesterol excretion and is accompanied by a marked increase in hepatic cholesterol concentration.
As shown in Fig. 4, the characteristically low level of cholesterol absorption and accelerated rate of fecal neutral sterol excretion evident in the Cyp7a1−/− mice fed the basal diet alone were restored by cholic acid [0.030% (wt/wt)] feeding to values seen in matching Cyp7a1+/+ controls fed the basal diet (Fig. 4, A and B). This restoration was almost fully achieved at a cholic acid level of only 0.015% (wt/wt). Doubling the cholic acid to 0.060% (wt/wt) did not have an additional effect on cholesterol absorption and excretion in the Cyp7a1−/− mice, but it did marginally raise the fractional cholesterol absorption in the Cyp7a1+/+ controls. At the two higher levels of cholic acid, there was a modest, but significant, increase in the total cholesterol concentration in the small intestine in Cyp7a1−/− mice, but not in their corresponding controls (Fig. 4C). One of the most striking effects of these low levels of cholic acid supplementation in the Cyp7a1−/− mice was on their total cholesterol concentrations in the liver, which increased 5.9-fold from 2.86 ± 0.15 mg/g in the group fed the basal diet alone to 17.01 ± 2.17 mg/g in the group fed the diet containing 0.060% (wt/wt) cholic acid (Fig. 4D). The weight of the liver, expressed relative to body weight in this latter group of mice, was 6.0 ± 0.2%, which was just above the range for all other mice in this study (5.1–5.6%; data not shown).
Fig. 4.
Intestinal cholesterol absorption, fecal neutral sterol excretion, and total cholesterol concentrations in small intestine and liver of Cyp7a1−/− and Cyp7a1+/+ mice fed graded levels of cholic acid in their diet. Groups of adult female Cyp7a1−/− and matching Cyp7a1+/+ mice were fed diets described in Fig. 3 legend. At 3 days before the end of the experiment, mice were dosed with radiolabeled sterols, and their stools were collected over the following 3 days for measurement of fractional cholesterol absorption and fecal neutral sterol excretion. Values are means ± SE of data for 4 or 5 mice in each group in A–C and 8 mice per group in D. Because 2-way ANOVA revealed significant interaction between diet and genotype, 1-way ANOVA was performed; different letters (a–d) denote statistically significant values (P < 0.05).
Compensatory inhibition of hepatic and intestinal cholesterol synthesis accompanies increase in tissue cholesterol concentrations in Cyp7a1−/− mice given cholic acid supplementation.
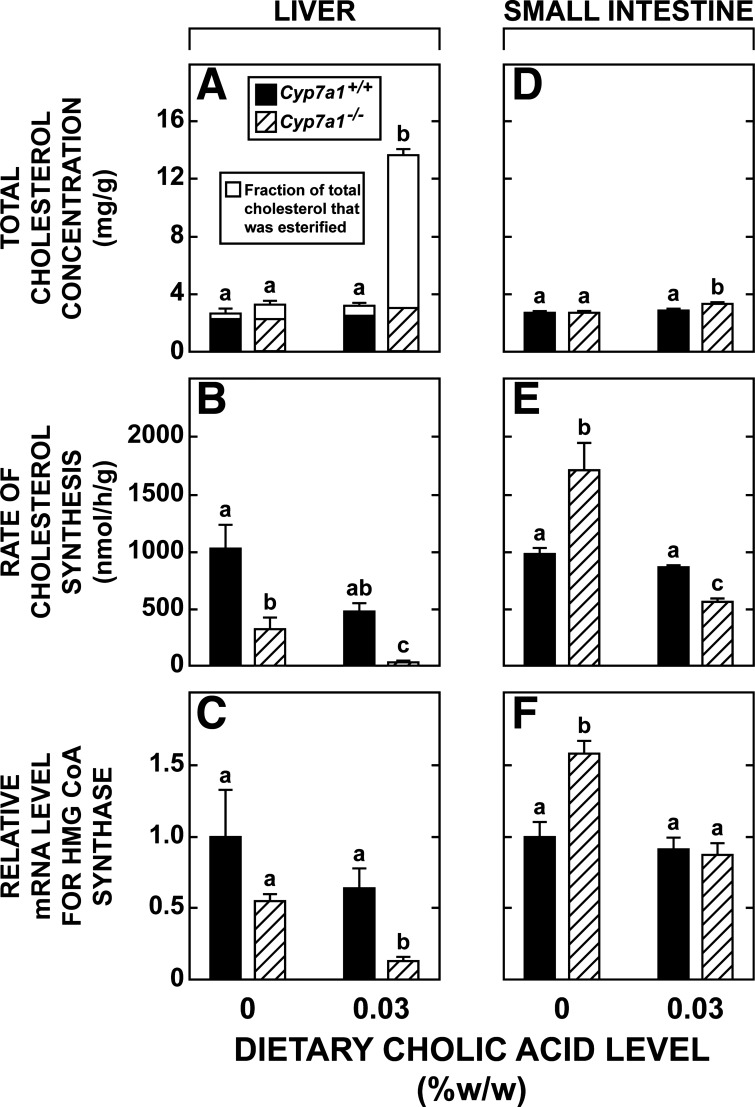
For the remaining studies, on the basis of the findings described in Figs. 3 and 4, it was decided to use only a single level of cholic acid supplementation [0.030% (wt/wt)]. The primary objective of these additional experiments was to determine how cholic acid supplementation affected the rate of hepatic and intestinal cholesterol synthesis, as well as the relative level of mRNA expression for a constellation of enzymes, transcription factors, and transporters involved in the synthesis and handling of bile acids and of other proteins that regulate the intestinal synthesis and movement of cholesterol. The data in Fig. 5A show that essentially all the increase in hepatic cholesterol levels in Cyp7a1−/− mice given cholic acid (Fig. 4D) reflected a rise in the esterified fraction. Other groups of matching mice manifested a near-complete inhibition of hepatic cholesterol synthesis (Fig. 5B) and a significantly lower level of hepatic mRNA expression for 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase (Fig. 5C) in parallel with the markedly expanded pool of cholesteryl ester. In the case of the Cyp7a1+/+ mice fed the diet supplemented with cholic acid, there was a clear trend toward lower rates of hepatic cholesterol synthesis (Fig. 5B) and levels of mRNA expression for HMG-CoA synthase (Fig. 5C), although these were not statistically significant.
Fig. 5.
Concentrations of unesterified and esterified cholesterol in liver and rates of cholesterol synthesis in liver and small intestine of Cyp7a1−/− and Cyp7a1+/+ mice fed a single level of cholic acid in their diet. Level of dietary cholic acid supplementation was set at 0 or 0.030% (wt/wt). All measurements were made in adult female mice fed their respective diets for 15–18 days. Data are derived from several experiments. Data in A are from the same animals used for measurements in Fig. 4D. Other groups of matching mice were used for measurement of total cholesterol concentration in intestine (D), rates of hepatic and intestinal cholesterol synthesis (B and E), and mRNA level for 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase (HMG)-CoA synthase (C and F). mRNA data for the small intestine are for the distal third of the organ, whereas cholesterol synthesis data are for the entire small intestine. Liver and small intestine (distal third only) taken for these mRNA analyses are from the same animals used in experiments described in legends of Figs. 6 and 7. Values are means ± SE of data from 5–8 mice in each group. In A, height of each bar defines total cholesterol concentration, and relevant SE is shown. SE for the esterified fraction is not indicated. Because 2-way ANOVA of data in A, D, and E revealed significant interaction between diet and genotype, 1-way ANOVA was performed; different letters (a–c) denote statistically different values (P < 0.05).
Cholic acid feeding had comparatively little effect on intestinal cholesterol concentrations; however, as shown in Fig. 5D, in Cyp7a1−/− mice fed the diet with 0.03% (wt/wt) cholic acid, the total cholesterol concentration in the small intestine (3.36 ± 0.11 mg/g) was significantly higher than in matching Cyp7a1−/− and Cyp7a1+/+ mice fed the basal diet alone (2.70 ± 0.06 and 2.75 ± 0.10 mg/g, respectively). Although cholic acid feeding did not significantly affect the rate of cholesterol synthesis or the mRNA level for HMG-CoA synthase in the small intestine of the Cyp7a1+/+ mice (Fig. 5, E and F, respectively), this treatment did lower both of these parameters in the Cyp7a1−/− mice.
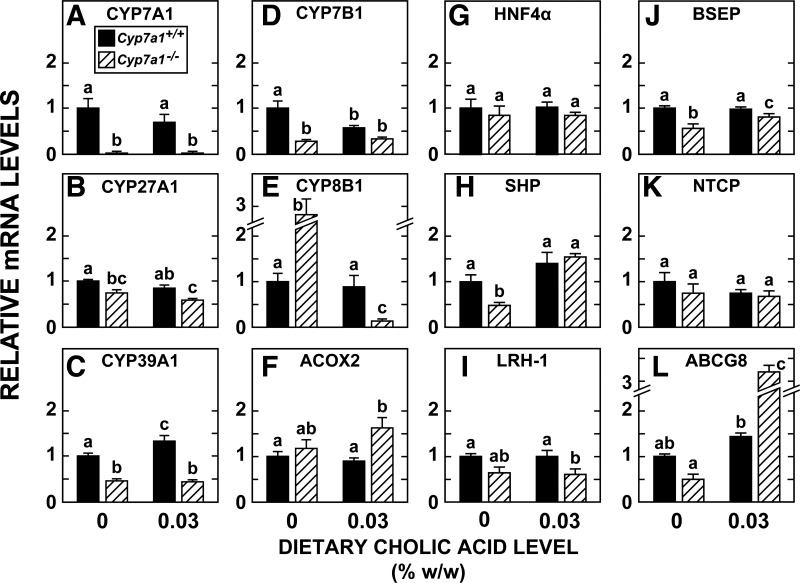
Marked elevation in mRNA expression for CYP8B1 in liver of Cyp7a1−/− mice is abolished by cholic acid feeding.
Although the relative mRNA level for multiple proteins involved in the conversion of cholesterol to bile acids was measured, for the majority of these, there was no change as a function of genotype or of cholic acid intake. Therefore, in Fig. 6, A–F, the mRNA expression levels of just 6 of the 15 enzymes listed in Fig. 1 and Table 1 are shown. The most striking changes associated with CYP7A1 deficiency were a 73% reduction in the relative mRNA level for CYP7B1 (Fig. 6D) and a 2.8-fold increase in the mRNA level for CYP8B1 (Fig. 6E). While this level of mRNA expression for CYP7B1 in the Cyp7a1−/− mice was unaltered by cholic acid feeding, in the case of CYP8B1, the supplementation lowered the mRNA expression level to just a fraction of that in the matching and unsupplemented Cyp7a1−/− mice and also to levels well below those in the Cyp7a1+/+ mice fed the basal diet alone or the basal diet containing cholic acid. In parallel with the findings for CYP7B1, the mRNA level for CYP39A1 (the other oxysterol 7α-hydroxylase) was lower in the Cyp7a1−/− mice than in the matching Cyp7a1+/+ controls, especially after cholic acid feeding (Fig. 6C). The dietary level of cholic acid that produced these changes in the Cyp7a1−/− mice had comparatively little effect on the matching Cyp7a1+/+ controls, with the exception of a twofold increase in the relative mRNA level for CYP39A1 (Fig. 6C) and a 43% reduction in the relative mRNA level for CYP7B1 (Fig. 6D).
Fig. 6.
Relative mRNA levels for various enzymes, transcription factors, and transporters involved in bile acid metabolism in livers of Cyp7a1−/− and Cyp7a1+/+ mice fed a single level of cholic acid in their diet. Groups of adult female mice were fed the same diets used in experiments described in Fig. 5 legend. Liver and small intestine were removed for RNA measurements. A–F: enzymes involved in conversion of cholesterol to bile acids. ACOX2, acyl-CoA oxidase 2. G–L: proteins that play a role in bile acid synthesis and transport within the liver or, in the case of ATP-binding cassette (ABC) transporter G8 (ABCG8), in facilitating hepatic cholesterol efflux. HNF-4α, hepatocyte nuclear factor 4α; SHP, small heterodimer partner; LRH-1, liver receptor homolog 1; BSEP, bile salt export pump; NTCP, sodium-taurocholate cotransporting polypeptide. Values are means ± SE of data for 5 or 6 mice in each group. Different letters denote statistically different values (P < 0.05), as determined by 1-way ANOVA.
The remaining data in Fig. 6 show the relative mRNA levels in liver for various regulatory transcription factors in bile acid metabolism [hepatocyte nuclear factor-4α, small heterodimer partner (SHP), and liver receptor homolog 1; Fig. 6, G-I], bile acid transporters (Fig. 6, J and K), and a canalicular sterol transporter, ABCG8 (Fig. 6L). The most notable changes associated with CYP7A1 deficiency alone were reductions of ∼40–50% in the mRNA levels for SHP (Fig. 6H), BSEP (Fig. 6J), and ABCG8 (Fig. 6L), all of which were reversed to varying degrees by cholic acid supplementation. The most striking change in this regard was for ABCG8: the mRNA level in the Cyp7a1−/− mice given cholic acid was 5.2-fold greater than in the unsupplemented Cyp7a1−/− controls and 3.2-fold greater than in the unsupplemented Cyp7a1+/+ mice (Fig. 6L).
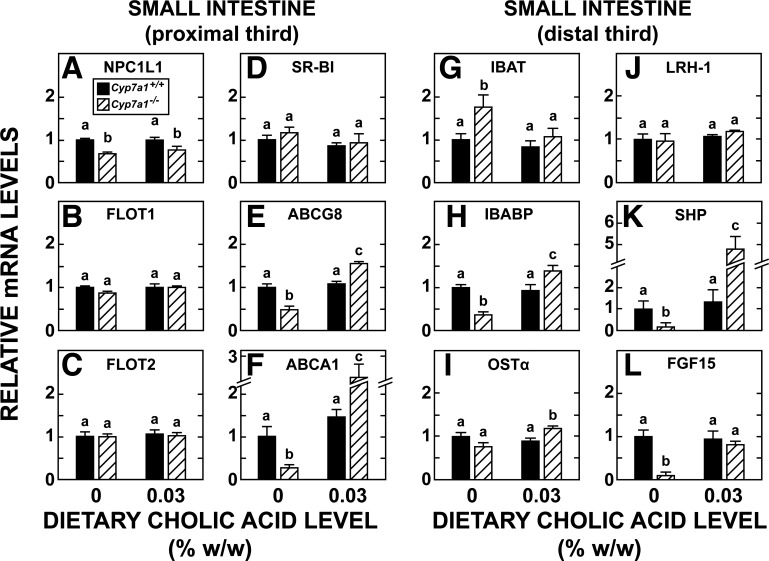
Diminished intestinal expression of mRNA for various sterol and bile acid transporters and related proteins resulting from loss of CYP7A1 function is reversed to a variable degree by cholic acid feeding.
One of the main objectives of these studies was to define the impact of CYP7A1 deficiency and bile acid pool restoration on the expression of a constellation of genes involved in cholesterol and bile acid handling in the small intestine. The data in Fig. 7 show mRNA levels for multiple genes in the proximal and distal small intestine of the same mice used for mRNA analyses in the liver. The focus in the proximal region (Fig. 7, A–F) was on proteins that facilitate sterol movement across the enterocyte. For Niemann-Pick type C-like 1 (NPC1L1; Fig. 7A), the expression level fell (32%) in the Cyp7a1−/− mice, and this outcome changed little with cholic acid feeding. This was also the case in the Cyp7a1+/+ controls given cholic acid. Although flotillin-1 and flotillin-2 are believed to play a key role in NPC1L1-mediated cholesterol uptake (13), mRNA levels for these proteins remained unchanged in the Cyp7a1−/− animals and were unresponsive to cholic acid supplementation (Fig. 7, B and C). This was also the case for scavenger receptor class B member 1 (SR-B1; Fig. 7D). The pattern of mRNA expression for ABCG8 (Fig. 7E) followed that for liver (Fig. 6L), although the changes were not as large. In the case of ABCA1, the expression level in the Cyp7a1−/− mice fed the basal diet without supplementation was only 27% of that in the corresponding Cyp7a1+/+ controls. Although cholic acid feeding did not significantly change the mRNA level for ABCA1 in the Cyp7a1+/+ mice, in the case of Cyp7a1−/− mice, it caused a substantial increase.
Fig. 7.
Relative mRNA levels for various proteins involved in movement of cholesterol or synthesis and transport of bile acids in small intestine of Cyp7a1−/− and Cyp7a1+/+ mice fed a single level of cholic acid in their diet. Analyses were carried out in mucosal scrapings from proximal (A–F) and distal (G–L) sections of small intestine of adult female Cyp7a1−/− and Cyp7a1+/+ mice fed diets described in legends to Figs. 5 and 6. NPC1L1, Niemann-Pick C1-like 1; FLOT1 and FLOT2, flotillin-1 and -2; SR-B1, scavenger receptor class B1; IBAT, ileal bile acid transporter; IBABP, ileal bile acid-binding protein; OSTα, organic solute transporter-α; FGF15, fibroblast growth factor 15. Values are means ± SE of data for 5 or 6 mice per group. Different letters (a–c) denote statistically different values (P < 0.05), as determined by 1-way ANOVA.
In the distal small intestine, the most pronounced change in mRNA expression levels resulting from CYP7A1 deficiency alone was seen for SHP (Fig. 7K) and fibroblast growth factor 15 (FGF15; Fig. 7L), with the relative mRNA level for both proteins being 80–90% lower than in matching Cyp7a1+/+ mice fed the basal diet, and also for ileal bile acid-binding protein (IBABP; Fig. 7H), where a reduction of 64% was seen. In the Cyp7a1−/− mice, cholic acid feeding essentially normalized the expression levels for IBABP and FGF15. However, for SHP, it dramatically increased the relative mRNA in Cyp7a1−/− mice to a level that was 27-fold greater than in their counterparts fed the basal diet alone and 4.8-fold more than for the Cyp7a1+/+ controls fed the basal diet. The mRNA levels for OSTα (Fig. 7I) and liver receptor homolog 1 (Fig. 7J) showed little change with either genotype or diet. In the case of the mRNA level for ileal bile acid transporter (Fig. 7G), there was a significant increase in the Cyp7a1−/− mice, but this change was normalized with cholic acid feeding.
DISCUSSION
Several points relating to the objectives and design of these studies warrant elaboration. The first relates to the experiments being run in normal, as well as Cyp7a1−/−, mice. This design allowed us to make a side-by-side comparison of the impact of administration of small, graded increases of bile acid via the diet, not only to mice with normal bile acid pool sizes and intestinal cholesterol metabolism but, moreover, to mice with inherently small bile acid pools, altered rates of intestinal cholesterol absorption and synthesis, and enhanced fecal sterol loss, all stemming from their lack of CYP7A1. The Cyp7a1−/− mouse was selected mainly because this model has been defined more completely with respect to sterol metabolism than others with genetically altered bile acid metabolism (12, 36, 37, 50). Another point concerns the selection of the bile acid species used for supplementation. While it would have been interesting to compare cholic acid and chenodeoxycholic acid, as has been done previously in a short-term feeding study with mice lacking CYP8B1 (24), we focused specifically on cholic acid, because it is a predominant bile acid in the pool of the mouse and also because it has a key role in the feedback regulation of bile acid biosynthesis (3, 24). We gauged the response of the mice to cholic acid supplementation through the measurement of a constellation of parameters that encompass the major steps involved in the synthesis, enterohepatic flux, and disposition of bile acids and cholesterol. Together, the data reveal informative findings about the widely differing sensitivity of Cyp7a1−/− mice and their Cyp7a1+/+ counterparts to the delivery of physiological amounts of cholic acid from the diet into their intestinal pools.
Two major sets of conclusions can be drawn from these data: one relates to the changes in the synthesis, storage, transport, and disposition of bile acids, while the other centers on the impact of these changes in bile acid metabolism on the handling of cholesterol by the small intestine and liver. Although we had projected that a dietary cholic acid level as low as 0.03% (wt/wt) should be sufficient to restore bile acid pool size in Cyp7a1−/− mice to that in Cyp7a1+/+ mice fed the basal diet alone, our initial experiments nevertheless also tested levels of cholic acid supplementation equal to half [0.015% (wt/wt)] and twice [0.060% (wt/wt)] this dose. While the intermediate level of supplementation, which provided an intake of cholic acid of just 4.8 mg (11.7 μmol) per day per 100 g body weight, did normalize the pool size in the Cyp7a1−/− mice (Fig. 3A), it also resulted in marked cholic acid enrichment of the pool (Fig. 3B). This enrichment had no discernible impact on the expression of mRNA for any of the three enzymes involved in the initiating steps of the alternate pathways of bile acid synthesis: CYP27A1 (Fig. 6B), CYP39A1 (Fig. 6C), and CYP7B1 (Fig. 6D). However, there was a large reduction in the level of mRNA expression of CYP8B1 (Fig. 6E), as well as a greater than twofold increase in the relative level of mRNA for SHP (Fig. 6H). There was also a marginal rise in the mRNA level for BSEP (Fig. 6J) but no discernible change in mRNA for NTCP (Fig. 6K). In the distal small intestine, the impact of restoring the bile acid pool on the level of mRNA expression for several proteins that act as sensors of changes in bile acid flux or that facilitate the transport of bile acids at various points in the enterohepatic cycle was particularly striking. Thus, with the deficiency of bile acids now corrected, the mRNA level for ileal bile acid transporter fell (Fig. 7G), while the mRNA level for the cytosolic IBABP (Fig. 7H) and Ostα (Fig. 7I) increased significantly. In the case of SHP (Fig. 7K) and FGF15 (Fig. 7L), the expression level of mRNA was manyfold greater in the Cyp7a1−/− mice fed the diet containing cholic acid than in those fed the basal diet alone. While many of the changes accompanying pool restoration are directionally what might have been predicted for this model, what should be emphasized is the low level of dietary cholic acid content that precipitated these changes.
With respect to bile acid metabolism in the matching Cyp7a1+/+ mice, their response to the intermediate level of cholic acid intake was more subtle. The relative mRNA level for CYP7A1 was only marginally lower (P > 0.05) in the cholic acid-fed Cyp7a1+/+ mice (Fig. 6A), and there was also no consistent impact on the mRNA levels for CYP27A1 (Fig. 6B), CYP39A1 (Fig. 6C), or CYP7B1 (Fig. 6D). A suppression was nevertheless evident in the case of CYP7B1. Although mRNA was not measured in the liver of any of the mice fed the diet supplemented with 0.06% (wt/wt) cholic acid, a more conspicuous change in the mRNA level for CYP7A1, and perhaps for other genes that regulate bile acid synthesis and handling, might have been manifest at this level of supplementation, because there were discernible changes in the size of the bile acid pool (Fig. 3A) and its composition (Fig. 3B). This possibility seems likely on the basis of findings in other studies in which there was a marked-to-complete suppression of expression of mRNA for CYP7A1 (24, 34, 50) and up to more than a twofold increase in the level of mRNA for SHP (24) in Cyp7a1+/+ mice fed diets containing significantly more cholic acid [0.10, 0.25, or 0.5% (wt/wt)]. At a dietary cholic acid level of 0.5% (wt/wt), the mRNA level for CYP7B1 was reduced by ∼70% (34), and there was also a clear reduction in the mRNA level of the bile acid transporter NCTP (50).
The data in Fig. 6, B–D, raise the general question: Why were the mRNA levels for CYP27A1, CYP39A1, and CYP7B1 consistently lower in the Cyp7a1−/− mice than in their matching Cyp7a1+/+ controls, irrespective of whether they were given any cholic acid in their diet? This question cannot be answered from other measurements described here, but it is clear from earlier studies in Cyp7a1−/− mice that the amount of bile acid generated by the alternate pathways, while equal to about only one-third of that found when CYP7A1 is present, is sufficient to sustain the animals throughout life after ∼3 wk of age (12, 21, 35, 37). It is also known that the amount of bile acid made via these alternate pathways does not vary with gender and remains unchanged in the face of a marked increase in dietary cholesterol intake or the disruption of the enterohepatic circulation of bile acids by feeding of cholestyramine (37). All these various findings were based on the use of fecal bile acid excretion rates as a surrogate measure of the rate of bile acid synthesis in the intact animal. While this technique has been applied in many animal models and also in humans under a diverse set of experimental conditions (15, 17, 31, 42, 52), its application to the measurement of bile acid synthesis rates in animals supplemented with even small levels of dietary bile acid is not valid, because, with rare exception, there is no way to determine the fraction of the acidic sterols appearing in the stool that comes from new synthesis vs. the diet. Given this caveat, a particular comment on what can be concluded from the fecal bile acid excretion rates in the present studies (Fig. 3C) is warranted. For the Cyp7a1+/+ and Cyp7a1−/− mice, the rate of fecal bile acid excretion increased significantly with almost every step-wise increment in the dietary cholic acid level starting at 0.015% (wt/wt). However, the magnitude of increase from one step to another was always higher in the Cyp7a1−/− mice than in their corresponding Cyp7a1+/+ controls. One interpretation of this outcome is simply that, with progressively greater degrees of inhibition of the CYP7A1 activity paralleling cholic acid intake, the quantity of bile acid newly synthesized via the neutral pathway appearing in the stool of the Cyp7a1+/+ mice falls proportionately but never to zero, as it does in the Cyp7a1−/− mice.
The more important point to be taken from the data in Fig. 3C relates to the practice of coprophagy by mice in general. In the early stages of maturity, mice reingest ≥10% of their daily stool output (6). Clearly, the higher the level of dietary bile acid supplementation, the more bile acid will be taken in through consumption of stool, thus adding to the exogenous bile acid load with which the small bowel and liver must deal. Moreover, the species of bile acid in the stool may be quite variable and include the secondary bile acids deoxycholic and lithocholic acid (12, 35). Thus, when these points are considered together, the first major conclusion from these studies is that, depending partly on the genetic makeup of the animal, the feeding of exaggerated amounts of bile acid in the diet might well induce changes in one or more aspects of bile acid metabolism that do not represent a true physiological response.
The second major conclusion from this work relates to the marked sensitivity of many parameters of cholesterol metabolism to changes in bile acid pool size and composition resulting from dietary bile acid supplementation. The magnitude of these changes is dictated largely by the extent to which cholesterol absorption and the amount of chylomicron-cholesterol delivered to the liver change as a consequence of shifts in bile acid pool size and composition. In the case of the Cyp7a1+/+ mice, the only discernible change in cholesterol metabolism in response to the intermediate level of cholic acid supplementation was a trend toward a lower rate of hepatic cholesterol synthesis (P > 0.05; Fig. 5, B and C). Clearly, more decisive changes in this and other parameters of cholesterol metabolism might have been found in Cyp7a1+/+ mice at twice the level of cholic acid supplementation [i.e., 0.06% (wt/wt)], because the subtle changes in bile acid pool size and composition at this dose (Fig. 3, A and B) were accompanied by an increase in fractional cholesterol absorption from 56.5 ± 6.3% to 78.6 ± 3.0% (Fig. 4A). Irrespective of whether this is the case, in the Cyp7a1−/− mice given cholic acid at the intermediate level, there were striking changes in virtually every aspect of cholesterol metabolism measured. Normalization of bile acid pool size and the accompanying cholic acid enrichment of the pool (Fig. 3, A and B) culminated in a major increase in hepatic cholesterol content, mainly in the esterified fraction (Figs. 4D and 5A). This outcome was likely due to the inability of the liver to convert cholesterol of intestinal origin to bile acids, not only because of the absence of CYP7A1, but also because of the unresponsiveness of the alternate pathways of bile acid synthesis to an expanding intrahepatic pool of surplus cholesterol (37). In addition to esterifying much of this cholesterol, the liver exhibited the classic responses of completely repressing de novo synthesis (Fig. 5, B and C) and accelerating cholesterol efflux via ABCG5/ABCG8 (Fig. 6L). One unexpected finding was the impact of the intermediate dietary cholic acid level on cholesterol metabolism in the small intestine of the Cyp7a1−/− mice. There was a modest, but significant, rise in the total cholesterol concentration (Fig. 4C), a marked inhibition of cholesterol synthesis (Fig. 5, E and F), and an increase in the expression levels of mRNA for ABCG8 and ABCA1 in the proximal region of the small intestine (Fig. 7, E and F).
Taken together, the results from these studies point to the importance of using very low levels of bile acid in diets, especially when they are given to mouse models with genetic alterations in some aspect of their bile acid metabolism. Ideally, this level of bile acid supplementation should only be large enough to restore the pool size and activity of relevant metabolic pathways to physiological levels and should not be so high as to induce pharmacological or, possibly, artifactual changes in a biological process of interest. The current studies illustrate how small the supplements should be to delineate with greater certainty the role that specific bile acid species might play in the pathogenesis and management of metabolic diseases such as obesity and type 2 diabetes (22, 26, 44, 49, 56).
GRANTS
This research was supported by National Institutes of Health Grants HL-09610 (J. M. Dietschy and S. D. Turley), HL-20948 (D. W. Russell), and DK-078592 (J. J. Repa) and the Moss Heart Fund (J. M. Dietschy).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.D.J., J.J.R., J.M.D., and S.D.T. are responsible for conception and design of the research; R.D.J., J.J.R., and S.D.T. performed the experiments; R.D.J., J.J.R., and S.D.T. analyzed the data; R.D.J., J.J.R., D.W.R., J.M.D., and S.D.T. interpreted the results of the experiments; R.D.J. and S.D.T. prepared the figures; R.D.J., J.J.R., and S.D.T. drafted the manuscript; R.D.J., J.J.R., D.W.R., J.M.D., and S.D.T. edited and revised the manuscript; R.D.J., J.J.R., D.W.R., J.M.D., and S.D.T. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Mario Saucedo, Carolyn Crumpton, Adam Lopez, Lam Le, Stephen Ostermann, and Monti Schneiderman for excellent technical assistance.
REFERENCES
- 1. Alrefai WA, Gill RK. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm Res 24: 1803–1823, 2007 [DOI] [PubMed] [Google Scholar]
- 2. Amaral JD, Viana RJS, Ramalho RM, Steer CJ, Rodrigues CMP. Bile acids: regulation of apoptosis by ursodeoxycholic acid. J Lipid Res 50: 1721–1734, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beher WT, Filus AM, Rao B, Beher ME. A comparative study of bile acid metabolism in the rat, mouse, hamster, and gerbil. Proc Soc Exp Biol Med 130: 1067–1074, 1969 [DOI] [PubMed] [Google Scholar]
- 4. Biddinger SB, Haas JT, Yu BB, Bezy O, Jing E, Zhang W, Unterman TG, Carey MC, Kahn CR. Hepatic insulin resistance directly promotes formation of cholesterol gallstones. Nat Med 14: 778–782, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borgström B. Quantitative aspects of the intestinal absorption and metabolism of cholesterol and β-sitosterol in the rat. J Lipid Res 9: 473–481, 1968 [PubMed] [Google Scholar]
- 6. Brown CJ, Donnelly TM. Rodent husbandry and care. Vet Clin North Am Exot Anim Pract 7: 201–225, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Carey MC, Cahalane MJ. Enterohepatic circulation. In: The Liver: Biology and Pathobiology, edited by Arias IM, Jakoby WB, Popper H, Schachter D, Shafritz DA. New York: Raven, 1988, p. 573–616 [Google Scholar]
- 8. Cheng X, Buckley D, Klaassen CD. Regulation of hepatic bile acid transporters Ntcp and Bsep expression. Biochem Pharmacol 74: 1665–1676, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiang JYL. Bile acids: regulation of synthesis. J Lipid Res 50: 1955–1966, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho JY, Matsubara T, Kang DW, Ahn SH, Krausz KW, Idle JR, Luecke H, Gonzalez FJ. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J Lipid Res 51: 1063–1074, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res 50: 2340–2357, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erickson SK, Lear SR, Deane S, Dubrac S, Huling SL, Nguyen L, Bollineni JS, Shefer S, Hyogo H, Cohen DE, Shneider B, Sehayek E, Ananthanarayanan M, Balasubramaniyan N, Suchy FJ, Batta AK, Salen G. Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. J Lipid Res 44: 1001–1009, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Ge L, Qi W, Wang LJ, Miao HH, Qu YX, Li BL, Song BL. Flotillins play an essential role in Niemann-Pick C1-like 1-mediated cholesterol uptake. Proc Natl Acad Sci USA 108: 551–556, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodwin B, Gauthier KC, Umetani M, Watson MA, Lochansky MI, Collins JL, Leitersdorf E, Mangelsdorf DJ, Kliewer SA, Repa JJ. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci USA 100: 223–228, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grundy SM, Ahrens EH, Jr, Salen G. Interruption of the enterohepatic circulation of bile acids in man: comparative effects of cholestyramine and ileal exclusion on cholesterol metabolism. J Lab Clin Med 78: 94–121, 1971 [PubMed] [Google Scholar]
- 16. Grundy SM, Bilheimer DW. Inhibition of 3-hydroxy-3-methlyglutaryl-CoA reductase by mevinolin in familial hypercholesterolemia heterozygotes: effects on cholesterol balance. Proc Natl Acad Sci USA 81: 2538–2542, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hahn C, Reichel C, von Bergmann K. Serum concentration of 7α-hydroxycholesterol as an indication of bile acid synthesis in humans. J Lipid Res 36: 2059–2066, 1995 [PubMed] [Google Scholar]
- 18. Huang W, Bansode RR, Xie Y, Rowland L, Mehta M, Davidson NO, Mehta KD. Disruption of the murine protein kinase Cβ gene promotes gallstone formation and alters biliary lipid and hepatic cholesterol metabolism. J Biol Chem 286: 22795–22805, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res 50: 1509–1520, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Insull W., Jr Clinical utility of bile acid sequestrants in the treatment of dyslipidemia: a scientific review. South Med J 99: 257–273, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW. Disruption of cholesterol 7α-hydroxylase gene in mice. I. Postnatal lethality reversed by bile acid and vitamin supplementation. J Biol Chem 271: 18017–18023, 1996 [DOI] [PubMed] [Google Scholar]
- 22. Kobayashi M, Ikegami H, Fujisawa T, Nojima K, Kawabata Y, Noso S, Babaya N, Itoi-Babaya M, Yamaji K, Hiromine Y, Shibata M, Ogihara T. Prevention and treatment of obesity, insulin resistance, and diabetes by bile acid-binding resin. Diabetes 56: 239–247, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Methods Enzymol 389: 3–15, 2004 [DOI] [PubMed] [Google Scholar]
- 24. Li-Hawkins J, Gåfvels M, Olin M, Lund EG, Andersson U, Schuster G, Björkhem I, Russell DW, Eggertsen G. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest 110: 1191–1200, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li T, Franci JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, Chiang JYL. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem 287: 1861–1873, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JYL. Transgenic expression of cholesterol 7α-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 52: 678–690, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lund EG, Guileyardo JM, Russell DW. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci USA 96: 7238–7243, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McMurry MP, Connor WE, Lin DS, Cerqueira MT, Connor SL. The absorption of cholesterol and the sterol balance in the Tarahumara Indians of Mexico fed cholesterol-free and high cholesterol diets. Am J Clin Nutr 41: 1289–1298, 1985 [DOI] [PubMed] [Google Scholar]
- 29. Miyata M, Matsuda Y, Nomoto M, Takamatsu Y, Sato N, Hamatsu M, Dawson PA, Gonzalez FJ, Yamazoe Y. Cholesterol feeding prevents hepatic accumulation of bile acids in cholic acid-fed farnesoid X receptor (FXR)-null mice: FXR-independent suppression of intestinal bile acid absorption. Drug Metab Dispos 37: 338–344, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal 8: e005, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mott GE, Jackson EM, McMahan CA, McGill HC., Jr Dietary cholesterol and type of fat differentially affect cholesterol metabolism and atherosclerosis in baboons. J Nutr 122: 1397–1406, 1992 [DOI] [PubMed] [Google Scholar]
- 32. Norlin M, Wikvall K. Enzymes in the conversion of cholesterol into bile acids. Curr Mol Med 7: 199–218, 2007 [DOI] [PubMed] [Google Scholar]
- 33. Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res 50: S120–S125, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwarz M, Lund EG, Lathe R, Björkhem I, Russell DW. Identification and characterization of a mouse oxysterol 7α-hydroxylase cDNA. J Biol Chem 272: 23995–24001, 1997 [DOI] [PubMed] [Google Scholar]
- 35. Schwarz M, Lund EG, Setchell KDR, Kayden HJ, Zerwekh JE, Björkhem I, Herz J, Russell DW. Disruption of cholesterol 7α-hydroxylase gene in mice. II. Bile acid deficiency overcome by induction of oxysterol 7α-hydroxylase. J Biol Chem 271: 18024–18031, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7α-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res 39: 1833–1843, 1998 [PubMed] [Google Scholar]
- 37. Schwarz M, Russell DW, Dietschy JM, Turley SD. Alternate pathways of bile acid synthesis in the cholesterol 7α-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding. J Lipid Res 42: 1594–1603, 2001 [PubMed] [Google Scholar]
- 38. Staels B, Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care 32: S237–S245, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stroeve JHM, Brufau G, Stellaard F, Gonzalez FJ, Staels B, Kuipers F. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab Invest 90: 1457–1467, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turley SD, Daggy BP, Dietschy JM. Psyllium augments the cholesterol-lowering action of cholestyramine in hamsters by enhancing sterol loss from the liver. Gastroenterology 107: 444–452, 1994 [DOI] [PubMed] [Google Scholar]
- 41. Turley SD, Schwarz M, Spady DK, Dietschy JM. Gender-related differences in bile acid and sterol metabolism in outbred CD-1 mice fed low- and high-cholesterol diets. Hepatology 28: 1088–1094, 1998 [DOI] [PubMed] [Google Scholar]
- 42. Turley SD, Spady DK, Dietschy JM. Regulation of fecal bile acid excretion in male Golden Syrian hamsters fed a cereal-based diet with and without added cholesterol. Hepatology 25: 797–803, 1997 [DOI] [PubMed] [Google Scholar]
- 43. Turley SD, Valasek MA, Repa JJ, Dietschy JM. Multiple mechanisms limit the accumulation of unesterified cholesterol in the small intestine of mice deficient in both ACAT2 and ABCA1. Am J Physiol Gastrointest Liver Physiol 299: G1012–G1022, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uchida K, Makino S, Akiyoshi T. Altered bile acid metabolism in nonobese, spontaneously diabetic (NOD) mice. Diabetes 34: 79–83, 1985 [DOI] [PubMed] [Google Scholar]
- 45. Vanhanen H, Kesäniemi YA, Miettinen TA. Pravastatin lowers serum cholesterol, cholesterol-precursor sterols, fecal steroids, and cholesterol absorption in man. Metabolism 41: 588–595, 1992 [DOI] [PubMed] [Google Scholar]
- 46. von Bergmann K, Mok HY, Hardison WGM, Grundy SM. Cholesterol and bile acid metabolism in moderately advanced, stable cirrhosis of the liver. Gastroenterology 77: 1183–1192, 1979 [PubMed] [Google Scholar]
- 47. Wang DQH, Carey MC. Measurement of intestinal cholesterol absorption by plasma and fecal dual-isotope ratio, mass balance, and lymph fistula methods in the mouse: an analysis of direct versus indirect methodologies. J Lipid Res 44: 1042–1059, 2003 [DOI] [PubMed] [Google Scholar]
- 48. Wang DQH, Tazuma S, Cohen DE, Carey MC. Feeding natural hydrophilic bile acids inhibits intestinal cholesterol absorption: studies in the gallstone-susceptible mouse. Am J Physiol Gastrointest Liver Physiol 285: G494–G502, 2003 [DOI] [PubMed] [Google Scholar]
- 49. Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, Mataki C, Sato H, Tanigawara Y, Schoonjans K, Itoh H, Auwerx J. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem 286: 26913–26920, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolters H, Elzinga BM, Baller JFW, Boverhof R, Schwarz M, Stieger B, Verkade HJ, Kuipers F. Effects of bile salt flux variations on the expression of hepatic bile salt transporters in vivo in mice. J Hepatol 37: 556–563, 2002 [DOI] [PubMed] [Google Scholar]
- 51. Woollett LA, Wang Y, Buckley DD, Yao L, Chin S, Granholm N, Jones PJH, Setchell KDR, Tso P, Heubi JE. Micellar solubilisation of cholesterol is essential for absorption in humans. Gut 55: 197–204, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie C, Turley SD, Dietschy JM. Centripetal cholesterol flow from the extrahepatic organs through the liver is normal in mice with mutated Niemann-Pick type C protein (NPC1). J Lipid Res 41: 1278–1289, 2000 [PubMed] [Google Scholar]
- 53. Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem 275: 15482–15489, 2000 [DOI] [PubMed] [Google Scholar]
- 54. Zhang YKJ, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLos One 6: e16683, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Y, Klaassen CD. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J Lipid Res 51: 3230–3242, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zieve FJ, Kalin MF, Schwartz SL, Jones MR, Bailey WL. Results of the glucose-lowering effect of WelChol study (GLOWS): a randomized, double-blind, placebo-controlled pilot study evaluating the effect of colesevelam hydrochloride on glycemic control in subjects with type 2 diabetes. Clin Ther 29: 74–83, 2007 [DOI] [PubMed] [Google Scholar]
- 57. Zilversmit DB, Hughes LB. Validation of a dual-isotope plasma ratio method for measurement of cholesterol absorption in rats. J Lipid Res 15: 465–473, 1974 [PubMed] [Google Scholar]