Abstract
Gas-phase conjugation to unprotonated arginine side-chains via N-hydroxysuccinimide (NHS) esters is demonstrated through both charge reduction and charge inversion ion/ion reactions. The unprotonated guanidino group of arginine can serve as a strong nucleophile, resulting in the facile displacement of NHS from NHS esters with concomitant covalent modification of the arginine residue. This reactivity is analogous to that observed with unprotonated primary amines such as the N-terminus or ε-amino group of lysine. In solution, however, the arginine residues tend to be protonated at pH values low enough to prevent hydrolysis of NHS esters, which would render them relatively unreactive with NHS esters. This work demonstrates novel means for gas-phase conjugation to arginine side-chains in polypeptide ions.
Keywords: Gas-phase bioconjugation, ion/ion reactions, N-hydroxysuccinimide esters and arginine, gas-phase cross-linking
Bioconjugation reactions are widely used in modern biology research.1 Many common approaches involve targeted conjugation, such as the use of NHS esters to react with primary amines to form amide bonds or the use of maleimide derivatives to react with sulfhydryl groups to form thioether linkages.1 Some uses of these conjugation approaches include labeling with chromophores,2 elucidating structural information via cross-linking,3–5 providing more complete sequence information for peptides6 and proteins,7 and preparing reactive surfaces for gas-phase reactions.8 The ability to target native functional groups of biomolecules, such as those associated with the nucleobases of RNA or DNA or the amino acid residues of proteins, allows the development of site-specific methods to extract information.
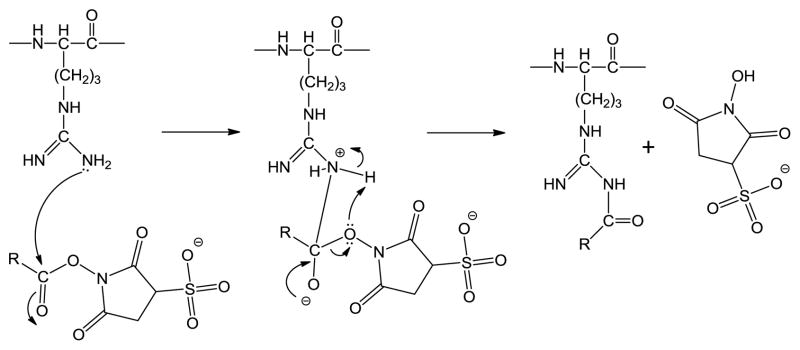
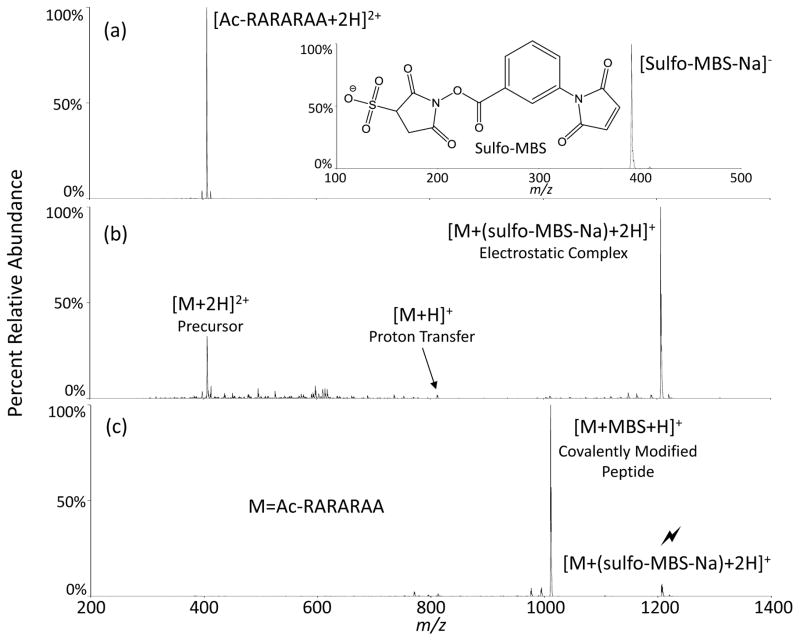
For example, primary amines such as the N-terminus or ε-amino group of lysine are used to react with NHS esters to produce a covalent linkage. These reactions proceed rapidly under physiological conditions, resulting in the formation of amide bonds through the acylation of the nucleophilic amines. With a pKA > 12,1 the guanidino group of arginine is protonated, and is therefore not nucleophilic, in the pH range within which NHS esters are stable to hydrolysis in aqueous solution. Arginine is therefore not regarded as a reactive site for NHS esters in solution. The gas-phase provides an environment in which both unprotonated arginine residues and NHS esters can coexist. A schematic for the general reaction involving the formation of an amide bond between arginine and the sulfo-NHS ester is illustrated in Scheme 1. The reactivity of arginine with NHS esters can be explored in straightforward fashion via gas-phase ion/ion reactions.9–11 NHS esters have previously been shown to be reactive with the N-terminus12 and lysine side chains12,13 in the gas-phase via ion/ion reactions. Here, we focus on the reactivity of unprotonated arginine toward sulfo-NHS esters in the gas-phase. Sulfo-NHS esters have been shown to have essentially the same specificity and reactivity as NHS esters14 in solution and are more water soluble. They are particularly useful gas-phase anionic reagents due to the strong interaction of the sulfonate group with protonated sites on polypeptide cations, which leads to the formation of long-lived complexes that facilitate covalent reactions.15,16 The general process by which ions are reacted and subsequently fragmented in the gas-phase is illustrated in Figure 1. The di-cationic precursor peptide ion, N-terminally acetylated RARARAA is mass selected (Figure 1a) in the first quadrupole of a hybrid linear ion trap based on a triple quadrupole design17 and stored in the reaction cell. Two of the arginine residues are expected to sequester the two protons, leaving the third arginine unprotonated and therefore free to react. The m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester (sulfo-MBS) anion is then mass selected (Figure 1a, inset) and transferred to the reaction cell. Following a defined “mutual storage” reaction period, the dominant reaction channel observed in Figure 1b is the formation of a long-lived electrostatic complex [Ac-RARARAA+(sulfo-MBS-Na)+2H]+. In this case, competition from ion/ion proton transfer to yield the singly protonated peptide is minimal.18 The strong interaction between the sulfonate and guanidinium groups likely underlies the stability of this complex.19 Collision-induced dissociation (CID) of the complex, indicated by the lightning bolt above the peak in Figure 1c, results in essentially exclusive loss of neutral sulfo-NHS. This loss is highly suggestive of a nucleophilic displacement of the sulfo-NHS leaving group and concomitant covalent modification of the polypeptide. This modification was confirmed by subsequent fragmentation (Figure S1).
Scheme 1.
General reaction schematic for arginine reacting with sulfo-NHS esters.
Figure 1.
Process of reacting arginine with a sulfo-NHS ester: (a) Precursor ions individually mass-selected. (b) Post-ion/ion reaction spectrum. (c) Collisional activation and fragmentation of the electrostatic complex, rendering a covalent modification. (Lightning bolt indicates ion subjected to activation).
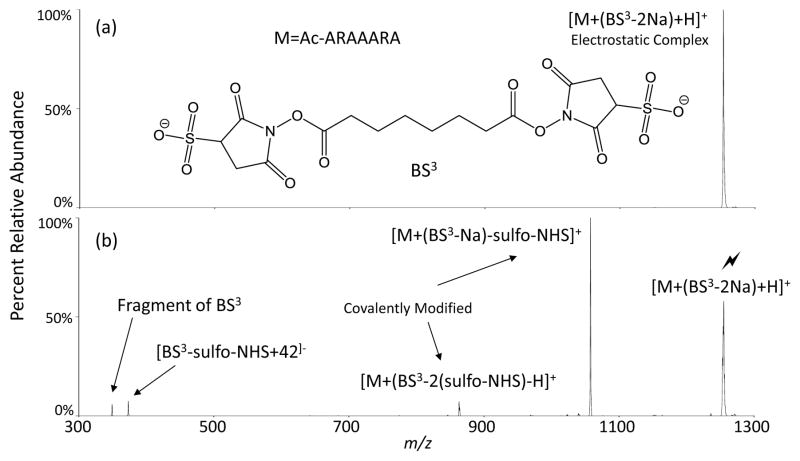
The reaction presented in Figure 1 results in the neutralization of a charge from a doubly-charged cation, thereby maintaining an overall positive charge. The reactivity of arginine with NHS esters was also investigated under charge inversion conditions. This was effected by reacting the singly-protonated, N-terminally acetylated peptide Ac-ARAAARA with the doubly-charged anionic homobifunctional crosslinker bis(sulfosuccinimidyl) suberate (BS3). Figure 2a shows the isolation of the electrostatic complex formed between the two ions to produce [Ac-ARAAARA+(BS3-2Na)+H]−. Figure 2b shows the CID of the complex to produce mainly a single sulfo-NHS neutral loss, corresponding to a single covalent modification. The presence of a second sulfo-NHS neutral loss represents a “do-si-do” type mechanism, involving multiple steps, which ultimately result in a second covalent modification on a different arginine, leaving the peptide intra-molecularly cross-linked. Briefly, once the first covalent bond is formed and the initial neutral sulfo-NHS is lost, a proton is transferred from the protonated arginine to the remaining sulfonate, neutralizing the charge and creating an unprotonated arginine, which then reacts with the sulfo-NHS ester to produce a second covalent bond and loss of the second neutral sulfo-NHS. (Note that the singly charged ion generated from the solution modification of YGGFLR to yield a fixed charge site at the N-terminus showed only loss of one sulfo-NHS group upon charge inversion with [BS3-2Na]2− in the gas-phase (see Figure S2). This result suggests that the arginine side-chain does not readily react twice with NHS-esters.) Two small peaks in the spectrum of Figure 2b are related to the cross-linker, with the first peak at m/z 350 corresponding to cleavage of one of the ester bonds (Figure S3a). The peak at m/z 374 corresponds to one end of the cross-linker being covalently attached to arginine followed by cleavage of the δN-εC bond of the arginine side chain, resulting in a loss of HN=C=N− from the arginine side-chain and its addition to the cross-linker (Figure S3b). The m/z 374 fragment is a signature for arginine reacting with this particular cross-linker based on the characteristic loss of HN=C=N−, which cannot occur with lysine or the N-terminal amine. Loss of 42 Da from arginine side-chains upon CID is well-known20 and a similar process in which the covalent modification was lost in conjunction with an additional 41 Da piece from the polypeptide was noted in the CID data for the [Ac-RARARAA+(sulfo-MBS-Na)+2H]+ system (see Figure S1). Studies with other NHS ester reagents have shown a variety of dissociation behaviors associated with covalently modified arginine side-chains, with transfer of the CN2H moiety to the reagent being one of the prominent processes. The net result of the process is the transformation of an arginine residue to an ornithine residue.
FIGURE 2.
Charge inversion reaction showing reactivity of arginine with a crosslinker, BS3, using an RF-DC isolation, resulting in (a) the mass-selected electrostatic complex. (b) CID of this complex renders one and two covalent bond formations. (Lightning bolt indicates ion subjected to activation.)
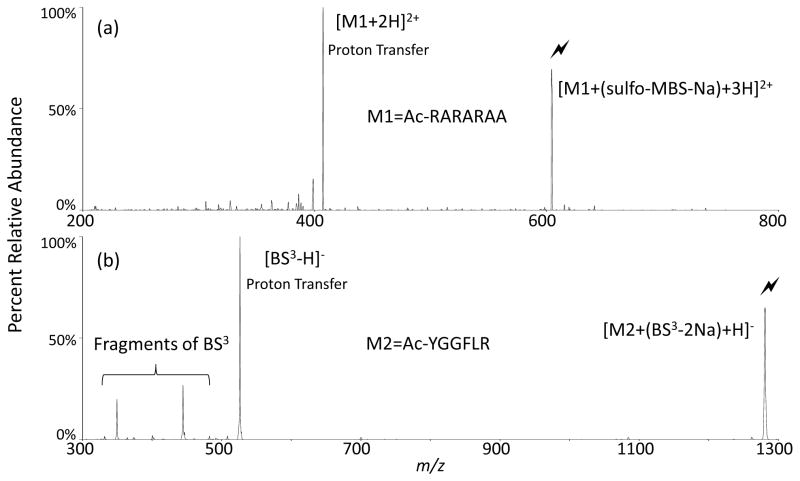
The reaction of arginine with NHS esters in aqueous solution is, at most, a minor process21 probably because the arginine is typically protonated in solution. To simulate these conditions in the gas-phase, peptides that have all of the arginines protonated were reacted with reagent anions. If the protonated arginines are reactive toward the sulfo-NHS esters, activation of this complex will result in neutral losses of sulfo-NHS, producing covalently modified ions, as has been shown in Figures 1 and 2. In the absence of a covalent reaction, the most facile CID process associated with the peptide/sulfo-NHS ester complex is simply the loss of the intact sulfo-NHS ester as a neutral species (i.e., a net proton transfer reaction). Figure 3a shows the CID spectrum of the charge-reduced complex of [Ac-RARARAA+(sulfo-MBS-Na)+3H]2+, which produces the proton transfer product [Ac-RARARAA+2H]2+. Likewise, Figure 3b shows the CID spectrum of the charge-inversion complex [Ac-YGGFLR+(BS3-2Na)+H]−, which produces the proton transfer product [BS3-H]−. Thus, as has also been noted for primary amines in the gas-phase,13 protonation of the arginine side-chain renders it non-nucleophilic and unreactive towards NHS esters. Note that in these cases, the sulfonate group is expected to undergo a strong electrostatic interaction with one of the protonated arginine residues. Hence, all of the arginine residues are nominally protonated with one undergoing a strong electrostatic interaction with the sulfonate group.
FIGURE 3.
Activation of electrostatic complexes containing only protonated arginine residues; (a) CID of [Ac-RARARAA+(sulfo-MBS-Na)+3H]2+; (b) CID of [Ac-YGGFLR+(BS3-2Na)+H]−. (Lightning bolt indicates ion subjected to activation.)
The present results show that NHS ester chemistry can be used to modify arginine side-chains in polypeptides in the gas-phase, provided they are unprotonated. The latter condition is difficult to achieve in solution under pH conditions that avoid hydrolysis of NHS esters. This constitutes an example of distinct behaviors in the gas-phase and aqueous solution. In the gas-phase, this condition is generally met with negative ions and polypeptide cations in which the total charge is exceeded by the number of arginine side-chains. NHS ester chemistry is expected to be specific for primary amines (i.e., N-termini and lysine ε-amino groups) when the total charge of the cation exceeds the number of arginine residues but does not exceed the sum of the arginine residues, lysine residues, and N-termini. When a mixture of unprotonated primary amines and arginine residues are present, both sites are expected to be reactive with NHS esters. (Control experiments have shown that NHS esters are unreactive towards unprotonated histidine residues. We have also found that unprotonated arginine side-chains are unreactive with aldehyde groups, unlike primary amines, which have been shown to form Schiff bases.16) These results add to the growing array of specific covalent chemistries that can be used to modify polypeptide ions in the gas-phase in a selective fashion.6,12,13,16
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health under Grant GM 45372.
ABBREVIATIONS
- CID
collision-induced dissociation
- NHS
N-hydroxysuccinimide
- Sulfo-MBS
m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester
- BS3
(bis[sulfosuccinimidyl] suberate
- R
arginine
- A
alanine
- Y
tyrosine
- G
glycine
- F
phenylalanine
- L
leucine
Footnotes
Supporting Information. Materials and experimental procedures, CID spectra of modified Ac-RARARAA+H and Pr-RARARAA (where Pr- indicates N-terminal propionylation), a CID spectrum of the electrostatic complex [TMAB-YGGFLR+(BS3-2Na)]− and likely structures of small peaks present in Figure 2b. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hermanson GT. Bioconjugate Techniques. 2. Academic Press; Amsterdam: 1998. [Google Scholar]
- 2.Vasicek L, Brodbelt JS. Anal Chem. 2010;82:9441–9446. doi: 10.1021/ac102126s. [DOI] [PubMed] [Google Scholar]
- 3.Tubb MR, Silva RAGD, Fang J, Tso P, Davidson WS. J Biol Chem. 2008;283:17314–17323. doi: 10.1074/jbc.M800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh P, Panchaud A, Goodlett DR. Anal Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowsk Willis JC, Nilges M, Cramer P, Rappsilber J. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stutzman JR, Hassell KM, McLuckey SA. Int J Mass Spectrom. 2012;312:195–200. doi: 10.1016/j.ijms.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsumoto H, Murata C, Miyata N, Kohda K, Taguchi R. Rapid Commun Mass Spectrom. 2007;21:3815–3824. doi: 10.1002/rcm.3279. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Hadjar O, Gassman P, Laskin J. Phys Chem Chem Phys. 2008;10:1512–1522. doi: 10.1039/b717617a. [DOI] [PubMed] [Google Scholar]
- 9.Ogorzalek Loo RR, Udseth HR, Smith RD. J Am Chem Soc. 1992;114:695–705. doi: 10.1016/1044-0305(92)87082-A. [DOI] [PubMed] [Google Scholar]
- 10.Pitteri SJ, McLuckey SA. Mass Spectrom Rev. 2005;24:931–958. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 11.McLuckey SA, Stephenson JL., Jr Mass Spectrom Rev. 1998;17:369–407. doi: 10.1002/(SICI)1098-2787(1998)17:6<369::AID-MAS1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 12.Mentinova M, Barefoot NZ, McLuckey SA. J Am Soc Mass Spectrom. 2011;23:282–289. doi: 10.1007/s13361-011-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mentinova M, McLuckey SA. J Am Chem Soc. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staros JV. Biochemistry. 1982;21:3950–3955. doi: 10.1021/bi00260a008. [DOI] [PubMed] [Google Scholar]
- 15.He M, Emory JF, McLuckey SA. Anal Chem. 2005;77:3173–3182. doi: 10.1021/ac0482312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han H, McLuckey SA. J Am Chem Soc. 2009;131:12884–12885. doi: 10.1021/ja904812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hager JW. Rapid Commun Mass Spectrom. 2002;16:512–526. [Google Scholar]
- 18.McLuckey SA. Eur J Mass Spectrom. 2010;16:429–439. doi: 10.1255/ejms.1031. [DOI] [PubMed] [Google Scholar]
- 19.Schug KA, Lindner W. Chem Rev. 2005;105:67–113. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- 20.Summerfeld SG, Dale VCM, Despyroux DD, Jennings KR. Eur J Mass Spectrom. 1995;1:183–194. [Google Scholar]
- 21.Miller BT, Collins TJ, Rogers ME, Kurosky A. Peptides. 1997;18:1585–1595. doi: 10.1016/s0196-9781(97)00225-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.