Abstract
Previous studies have found that inappropriate elevation of matrix metalloproteinase-9 (MMP9) expression and activity is coincident with early onset of emphysema in Smad3-null mice. Herein, we further investigated the role of increased MMP9 in emphysema pathogenesis and the related molecular regulatory mechanisms of elevated MMP9 in Smad3-null lung. Genetic blockade of MMP9 in Smad3-null mice significantly attenuated emphysema pathology but not hypoalveolarization during early postnatal lung development. Furthermore, Smad3 was found to be a transcription factor to positively regulate a protein deacetylase sirtuin 1 (SIRT1) by binding to an AP-1 site of SIRT1 promoter. A synergistic regulatory effect on SIRT1 expression was also detected between Smad3 and c-Jun. Consistently, Smad3 knockout lung at P28 had reduced SIRT1 expression, which in turn resulted in increased acetylation of histone H3 at the transcription factor activator protein 1 (AP-1), NF-κB, and Pea3 binding sites of MMP9 promoter and increased acetylation of NF-κB. In addition, increased Pea3 expression and nuclear accumulation was also detected in Smad3-null lungs at P28. Consistently, bindings of acetylated NF-κB and Pea3 to the MMP9 promoter were elevated in Smad3-null lung. We thus propose that deficiency of Smad3 causes downregulation of SIRT1 and increased Pea3 expression/nuclear accumulation, respectively. Decreased SIRT1 activity resulted in increased acetylation of histone H3 and NF-κB. Subsequently, increased bindings of transcription factors including NF-κB and Pea3 to MMP9 promoter significantly upregulate MMP9 transcription, contributing to emphysema pathogenesis.
Keywords: matrix metalloproteinase-9, sirtuin 1, Pea3, NF-κB
pulmonary emphysema, a major component of chronic obstructive pulmonary disease (COPD), is defined as “abnormal and permanent enlargement of peripheral airspaces including respiratory bronchioles, alveolar ducts, and alveoli, accompanied by destruction of extracellular matrix without obvious fibrosis” (26). These pathological changes result in dramatic loss of functional gas-exchange surfaces and compliance of the lung, exacerbating respiratory failure in patients with COPD. Excessive degradation of lung extracellular matrix caused by proteinase-antiproteinase imbalance and ineffective repair of extracellular matrix components are considered to be the key pathogenic events of emphysema (12), but the related molecular mechanisms remain incompletely understood.
Transforming growth factor (TGF)-β signaling plays important roles in regulating lung development and remodeling, as well as lung injury repair and pulmonary inflammation. Upon TGF-β ligand binding, constitutively active TGF-β receptor II (TβRII) kinase phosphorylates TGF-β receptor I (TβRI), which subsequently phosphorylates Smad2 and Smad3 proteins. These phosphorylated Smad2/3 form complexes with Smad4 and translocate into the nucleus, acting as transcriptional comodulators to alter TGF-β target gene expression (16).
Numerous human and animal studies have shown that TGF-β signaling is involved in emphysema pathogenesis, associated with its effect on synthesis of specific matrix metalloproteinases (MMPs) that degrade the components of extracellular matrices (17, 29). In human, genetic polymorphisms of the TGF-β1, TβRI, or TβRII gene are associated with emphysema in smokers (4, 31). Decreased expression of TGF-β1 and TβRII has been reported in lung tissue specimens from patients with COPD (1). Loss of function mutation of TβRII was found in patients with Marfan's syndrome (22), who have enlargement of the distal airspaces with or without discrete bullae (15). In addition, a polymorphism in the MMP9 gene promoter has been reported to be associated with an increased incidence of emphysema among smokers in Japan (21). In mice, deficiency in the core fucosylation of TGF-β receptors in mice by abrogating α1, 6-fucosyltransferase resulted in emphysema with reduced TGF-β receptor activity (30). Smad3-null mice spontaneously developed pulmonary emphysema, associated with increased MMP9 expression and activity in the lung (3, 5). However, little is known whether this altered MMP9 level leads to subsequent emphysema development and how defective TGF-β-Smad3 signaling altered MMP9 expression.
Gene expression can be regulated by genetic and epigenetic approaches. Acetylation of core histones by histone acetyltransferases leads to unwinding of DNA, which subsequently allows transcription factors and RNA polymerase II to switch on gene transcription. Conversely, deacetylation of core histones by histone deacetylase is generally associated with transcriptional repression (2). Sirtuins (SIRTs) are enzymes that catalyze NAD+-dependent deacetylation and/or ADP-ribosylation of target proteins and have been implicated in aging, metabolism, and stress resistance (20). Seven human isoforms exist (SIRT1-SIRT7), which differ in their subcellular localization, tissue distribution, and protein substrates. In mammals, SIRT1 is primarily a nuclear protein in most cell types and has evolved to deacetylate transcription factors and cofactors, histones and other chromatin proteins, and components of DNA repair machinery, which govern many central metabolic pathways (14). Recently, Nakamura et al. (23) demonstrated that SIRT1, but not other isoforms, is a negative regulator of MMP9 in human monocytes. However, whether SIRT1 expression can be regulated by TGF-β signaling has never been studied.
To determine whether increased MMP9 expression in Smad3-null lung is responsible for the subsequent emphysema development, we herein show that abrogation of MMP9 in Smad3-null mice rescues the lung from an emphysema phenotype. In addition, we demonstrate that TGF-β-activated Smad3 cooperates with c-Jun to promote SIRT1 expression in the lung, and, conversely, blockade of Smad3 decreases SIRT1 expression. This amplifies MMP9 transcription by enhancing acetylation of histone H3 at the activator protein (AP)-1, NF-κB, and Pea3 binding sites of the MMP9 promoter. Furthermore, upregulation of nuclear Pea3, a transcriptional activator of MMP9, may also contribute to elevated MMP9 expression in Smad3 knockout emphysema lung.
MATERIALS AND METHODS
Mouse strains and breeding.
Smad3 heterozygous mice were provided by Dr. Xiao-Fan Wang at Duke University (9), and MMP9 heterozygous mice were obtained from the Jackson Laboratory (Bar Harbor, ME) (28). All of them were backcrossed to the C57BL/6J genetic background. Smad3/MMP9 double heterozygous mice generated by crossing Smad3 heterozygous mice with MMP9 heterozygous mice survived with an expected Mendelian ratio. Smad3 knockout mice and Smad3/MMP9 double knockout mice were obtained from the intercrosses of Smad3/MMP9 double heterozygous mice. All these mice were genotyped by genomic DNA PCR (5, 28). The protocol for animal experiments was approved by the IACUC at Children's Hospital Los Angeles.
Histology and morphometric analysis.
Mouse lungs were inflated under 25 cmH2O pressure and fixed in 4% paraformaldehyde for histology and morphometric analysis. Five-micrometer tissue sections were stained with hematoxylin and eosin (H&E) for histological examination. Five H&E-stained lung sections randomly chosen from each lobe at ∼250-μm intervals were used for mean linear intercept (MLI) measurement according to established methods, as described previously (5, 10, 27).
RNA isolation and real-time RT-PCR.
Total RNA was isolated from lung tissue using Trizol reagent following the manufacturer's protocol (Invitrogen, Carlsbad, CA). Two micrograms of total RNA were used for cDNA synthesis using an iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA). Real-time PCR reactions were carried out with diluted reverse transcription products using an iQ SYBR Green Supermix PCR kit (Bio-Rad). Transcripts of β-actin and GAPDH were used as internal controls for normalization (32, 33).
Protein extraction and Western blot.
Total cellular proteins were isolated from lung tissue using RIPA buffer. Cytoplasmic and nuclear proteins were isolated from lung tissue by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). Specific proteins were detected by Western blots as previously described (32), using the following antibodies: SIRT1 (D60E1; Cell Signaling Technology, Danvers, MA), Pea3 (16; Santa Cruz Biotechnology, Santa Cruz, CA), NF-κB (p65; Millipore, Temecula, CA), acetylated NF-κB (Lys310; Cell Signaling Technology), c-Jun (60A8; Cell Signaling Technology), Sp1 (Millipore), RNA polymerase II (8WG16; Covance, Emeryville, CA), Smad3 (C67H9; Cell Signaling Technology), phosphorylated Smad3 (pS423 and pS425; Rockland, Gilbertsville, PA), phosphorylated epidermal growth factor receptor (EGFR) (Tyr1068, D7A5; Cell Signaling Technology), EGFR (D38B1; Cell Signaling Technology), GAPDH (6C5; Fitzgerald Industries International, Concord, MA), and β-actin (C4; MP Biomedicals, Solon, OH). MMP9 activity was measured by SDS-PAGE zymography as described previously (5).
Pulmonary macrophage isolation.
Isolated lung tissues were finely minced into pieces and digested in Hank's Balanced Salt Solution (HBSS) containing 0.1% trypsin and 0.001% DNase, followed by a second digestion in HBSS containing 0.1% collagenase and 0.001% DNase. Macrophages were labeled with PE-conjugated anti-mouse F4/80 antibody (Biolegend, San Diego, CA), incubated with anti-PE magnetic beads, and isolated by loading onto MACS magnetic separation columns (Miltenyi Biotec, Auburn, CA).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays using lung tissue were performed as previously described (32, 33), using antibodies against acetylated histone H3 (Lys9 and Lys14, Millipore), acetylated histone H4 (Lys5, Cell Signaling Technology), Pea3 (16, Santa Cruz Biotechnology), acetylated NF-κB (Lys310, Cell Signaling Technology), Smad3 (Abcam, Cambridge, MA), and c-Jun (60A8, Cell Signaling Technology). Immunoprecipitated samples were analyzed by PCR with primers spanning potential binding sites.
Plasmids, DNA transfection, and luciferase reporter assay.
A segment of the mouse SIRT1 gene promoter from nucleotides −3,363 to −304 containing ScaI and MluI restriction sites was amplified from mouse genomic DNA and subcloned into SmaI and MluI restriction sites of the pGL2-Basic luciferase reporter vector from Promega (Sirt1-luc; Madison, WI). The sequence was verified by DNA sequencing. The potential AP-1 binding element in the Sirt1-luc was mutated using GeneTailor Site-Directed Mutagenesis System (Invitrogen) according to the manufacturer's protocol. pRK5-Flag-Smad3 is a gift from Dr. Rick Derynck (University of California, San Francisco). Mouse c-Jun cDNA from ATCC was subcloned into pcDNA3.1(-). F9 cells were transfected using FuGENE HD Transfection Reagent (Roche Diagnostics, Indianapolis, IN) following the manufacturer's instructions. Luciferase activity in cell lysates was detected by Dual-luciferase Reporter Assay System (Promega) and normalized according to Renilla luciferase activity of cotransfected pRL-cytomegalovirus (CMV). Empty pCR-CMV vector was used to adjust the total amount of vector DNA for transfection.
Cell culture.
Mouse lung epithelial cell line 12 (MLE12), human lung fibroblast cell line 1 (HFL1), and RAW264.7 (mouse monocyte/macrophage cell line) cells were cultured in RPMI 1640, F-12K, and DMEM, respectively, containing 10% FBS, 1% penicillin/streptomycin, and 2 mM l-glutamine. The cells were starved with 1% FBS and 2 mM l-glutamine 1 day before TGF-β1 treatment (5 ng/ml).
Data presentation and statistical analysis.
All experiments were repeated at least three times. Data are presented as means ± SE. Statistical difference between two different groups were assessed by independent-samples t-test and were considered to be significant if P < 0.05.
RESULTS
Blockade of MMP9 in Smad3-null mice significantly attenuated emphysema pathology.
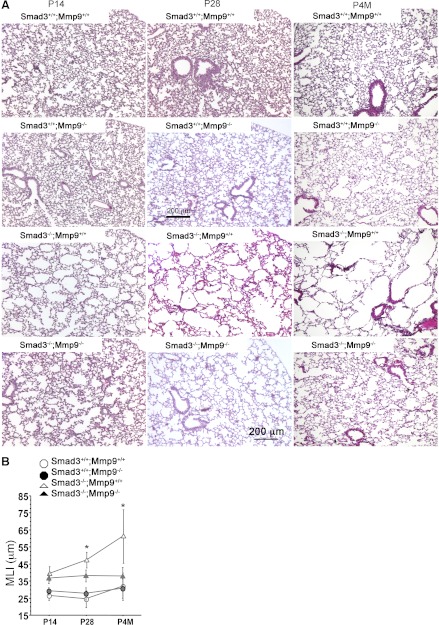
We have reported that increases of MMP9 expression and activity in Smad3-null lung at postnatal day (P) 28 were coincident with its early onset of emphysema (3, 5). To determine whether the elevated MMP9 expression in Smad3-null lung is responsible for the emphysema development, blockade of MMP9 in vivo was achieved by crossing conventional MMP9-null mice with Smad3-null mice, and the lung morphology of Smad3/MMP9 double knockout mice was compared with their wild-type, Smad3 single knockout, and MMP9 single knockout littermates at P14, P28, and postnatal 4 mo (P4M). Consistent with our previous report (5), retarded alveolarization in Smad3-null lungs was detected at P14, as characterized by reduced secondary septa formation and uniformly larger terminal air spaces (Fig. 1A). This phenotype was also seen in Smad3/MMP9 double knockout lungs at P14 (Fig. 1A). From P28 to P4M, a subsequent emphysema phenotype characterized by accelerated uneven enlargement of airspaces in the peripheral lung was observed in Smad3 single knockout mice but was not present in Smad3/MMP9 double knockout lungs, in which the alveolar size remained similar to that at P14 (Fig. 1A). MMP9-null lungs displayed normal alveolar structures similar to those seen in wild-type littermate controls (Fig. 1A). Changes in peripheral alveolar size at P14 and P28 as well as P4M were further quantified by MLI measurement (Fig. 1B). Significant reductions of MLI (23 and 42%) were detected in P28 and P4M Smad3/MMP9 double knockout lungs vs. Smad3 single knockout lungs, respectively. These data suggest that the increase of MMP9 expression observed in Smad3-null mouse lungs is an important factor contributing to their emphysema.
Fig. 1.
Blockade of matrix metalloproteinase 9 (MMP9) in Smad3-null mice significantly attenuated emphysema pathology. A: hematoxylin and eosin-stained lung sections of wild-type, MMP9 single knockout, Smad3 single knockout, and Smad3/MMP9 double knockout mice at postnatal day 14 (P14), P28, and P4M (n ≥ 3, each). B: morphometric analyses of pulmonary alveolar sizes of those mouse lungs at P14, P28, and P4M, as measured by mean linear intercept (MLI). *P < 0.05.
Increased MMP9 transcriptional expression in Smad3-null lung was associated with decreased SIRT1-mediated histone H3 deacetylation of the MMP9 promoter.
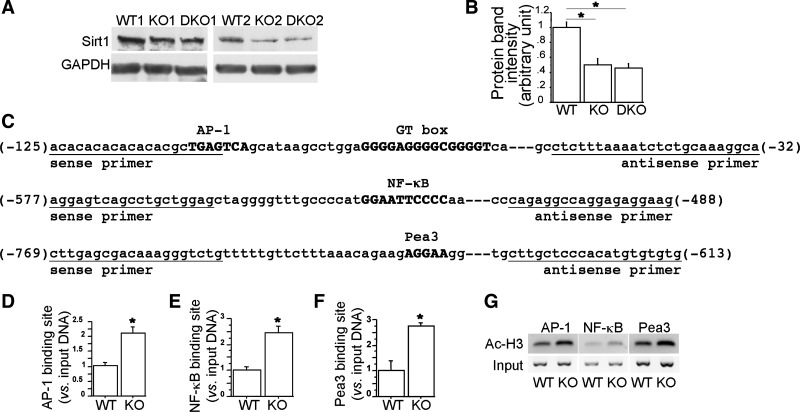
AP-1, NF-κB, and Pea3 are known as the key transcription factors involved in regulating MMP9 expression in a variety of cells (7, 18, 24, 25). Recently, Nakamura et al. (23) have demonstrated that decreased expression of SIRT1 under oxidative stress in human monocyte U937 cells causes MMP9 elevation by reducing deacetylation of core histones at the AP-1 and NF-κB binding sites of the MMP9 promoter (23). We therefore examined SIRT1 expression in Smad3-null lungs. Western blots revealed a 50% reduction of SIRT1 protein levels in both Smad3 single knockout and Smad3/MMP9 double knockout lungs at P28 compared with wild-type littermate control lungs (Fig. 2A), which was quantified by densitometry (Fig. 2B). Thus, blockade of MMP9 expression in Smad3 knockout lung had no effect on SIRT1 expression. Reduced expression of histone deacetylase SIRT1 may result in accumulated acetylation of core histones at the target gene promoter. We therefore compared acetylated histone H3 and H4 levels at the transcription factor-binding sites of the MMP9 promoter between wild-type and Smad3-null lungs at P28. ChIP assays using specific antibodies against acetylated histone H3 (Lys9 and Lys14) and H4 (Lys5) followed by real-time PCR demonstrated that reduction of SIRT1 expression in Smad3-null lungs significantly enhanced acetylation of histone H3, but not H4 (Lys5, data not shown), at the AP-1, NF-κB, and Pea3 binding sites of the MMP9 promoter (Fig. 2, C–G). These data may suggest a molecular mechanism underlying inappropriate elevation of MMP9 expression in Smad3-null lungs.
Fig. 2.
Decreased sirtuin 1 (SIRT1) expression enhanced histone H3 acetylation at the activator protein-1 (AP-1), NF-κB, and Pea3 binding sites of the MMP9 promoter in Smad3-null lungs. SIRT1 protein levels in wild-type (WT), Smad3 single knockout (KO), and Smad3/MMP9 double knockout (DKO) lungs at P28 (n ≥ 3, each) were measured by Western blots (A) and quantified by densitometry (B). GAPDH was a loading control. WT and KO lungs at P28 (n = 3, each) were collected for chromatin immunoprecipitation (ChIP) assay using anti-acetylated histone H3 antibody. The relative amount of the coimmunoprecipitated DNA at the AP-1, NF-κB, and Pea3 binding sites of the MMP9 promoter (C, sequences shown in bold uppercase), which was bound to acetylated histone H3, were quantified by real-time PCR (D–F) and also illustrated on 2% agarose gel (G). *P < 0.01.
TGF-β1 regulated expression of SIRT1 and MMP9 in a variety of lung cells in vitro and in vivo.
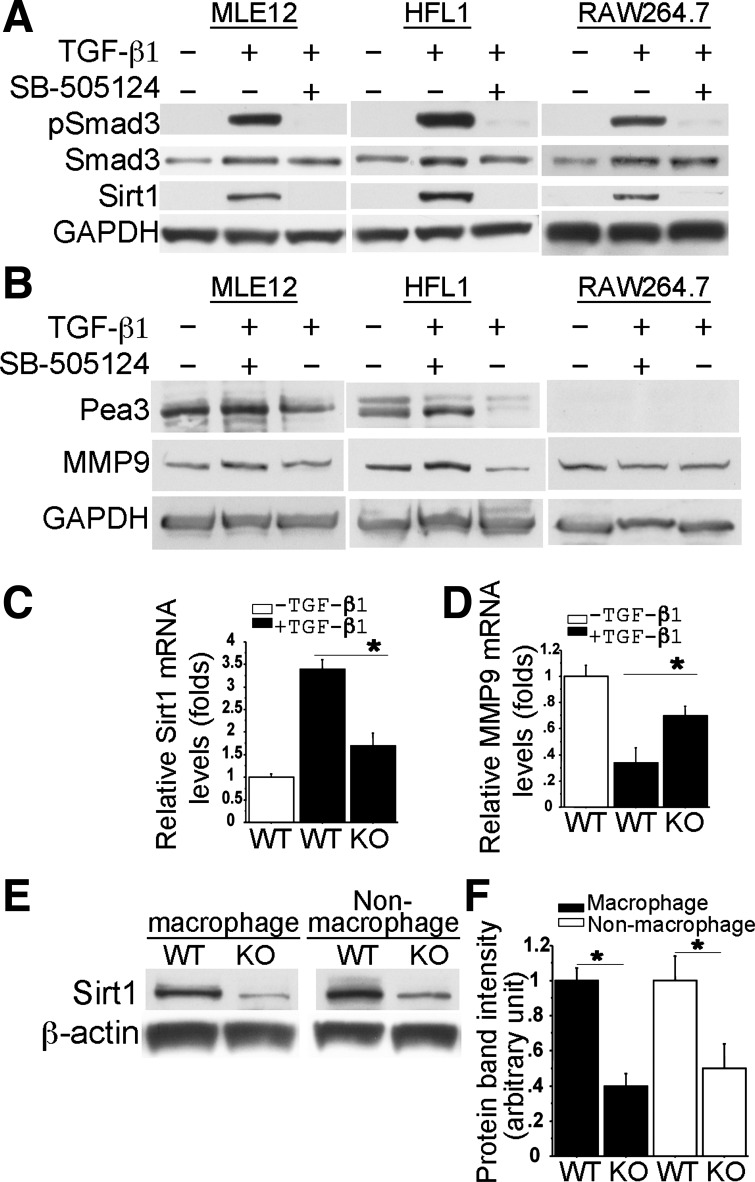
To test whether activated TGF-β signaling is able to upregulate SIRT1 expression directly, we studied TGF-β signaling activation and SIRT1 expression in a variety of cells in vitro. In cultured MLE12 cells, HFL1 cells, and mouse monocyte/macrophage cells (RAW264.7), activation of TGF-β signaling was achieved by addition of exogenous TGF-β1 (5 ng/ml), as determined by increased Smad3 phosphorylation (pSmad3) after 1-h treatment. Increased SIRT1 protein expression was then detected in all tested cells 48 h later (Fig. 3A). The TGF-β-specificity in SIRT1 regulation was further verified by the result that blockade of Smad3 phosphorylation with pretreatment of 1 μM SB-505124, a TβRI inhibitor (8), abolished TGF-β-induced SIRT1 expression. These suggest that TGF-β-mediated SIRT1 induction is an ubiquitous transcriptional regulation in vitro. In addition, we also examined whether TGF-β activation could alter MMP9 and its transcriptional regulators in these cell lines. After 48-h TGF-β1 treatment (5 ng/ml), MMP9 and Pea3 protein levels were significantly reduced in both MLE12 and HFL1 cells. These changes were also abolished by SB-505124 pretreatment (Fig. 3B). Pea3 expression in macrophages was not detected, which was consistent with another report (6). Interestingly, TGF-β1 treatment did not significantly suppress MMP9 expression in RAW264.7 cells (Fig. 3B).
Fig. 3.
Expression of SIRT1, Pea3, and MMP9 was regulated by TGF-β1 in a variety of cells in vitro and in vivo. Western blots showed that Smad3 phosphorylation was induced 1 h after TGF-β (5 ng/ml) stimulation in mouse lung epithelial cell line 12 (MLE12), human lung fibroblast cell line 1 (HFL1), and RAW264.7 cells (A). 48 h later, induction of Pea3, SIRT1, and MMP9 in these cells was further examined by Western blot (A and B). Both pSmad3 and SIRT1 induction were abolished by pretreatment with SB-505124 (1 μM). GAPDH was a loading control (A and B). The specific induction of Sirt1 and MMP9 expression by TGF-β-Smad3 in primary macrophages, which were isolated from either WT or Smad3 KO mouse lungs, was also examined by real-time PCR after 18h TGF-β (5 ng/ml) treatment (C and D, n = 3). Alteration of endogenous SIRT1 protein levels in F4/80 antibody FACS-sorted macrophages and nonmacrophages from WT and Smad3 KO lungs were also examined by Western blot (E) and quantified by densitometry (F, n = 3) *P < 0.01.
Because gene expression and its response to growth factor regulation may be different between immortalized cell lines and related primary cells in vitro, we then examined whether altered expression of SIRT1 and MMP9 could be induced by exogenous TGF-β stimulation in primary lung macrophages isolated from P28 mouse lung tissues. TGF-β1 treatment (5 ng/ml for 18 h) resulted in significant increases (3.4-fold) of Sirt1 expression and decreases (2.9-fold) of MMP9 expression at the mRNA level in the wild-type pulmonary macrophages (Fig. 3, C and D). However, these TGF-β-induced changes of Sirt1 and MMP9 expression were significantly diminished in the lung macrophages isolated from Smad3 knockout mice (Fig. 3, C and D). Consistent with the in vitro data, SIRT1 protein levels were significantly decreased in both macrophages and nonmacrophages of Smad3-null lungs compared with the wild-type controls (Fig. 3, E and F). Therefore, SIRT1 downregulation caused by deficient TGF-β-Smad3 signaling occurred ubiquitously in emphysema lung. This may be one of the molecular mechanisms by which abrogation of Smad3 results in abnormal expression of the genes involved in emphysema pathogenesis, such as MMP9.
Smad3 and c-Jun interacted with and synergistically activated the SIRT1 promoter.
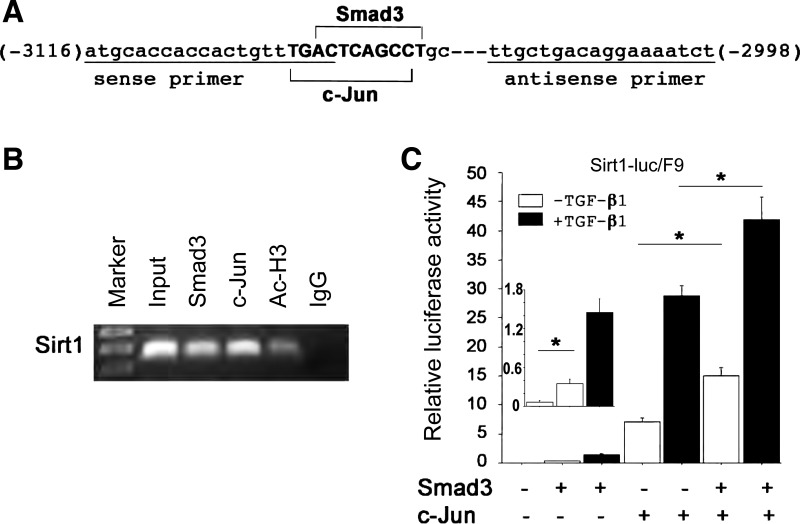
To determine how TGF-β-Smad3 signaling can directly regulate SIRT1 expression, we analyzed mouse SIRT1 promoter DNA sequences for potential Smad3 binding sites. Interestingly, a potential Smad3 binding site (actcagcc) was found to be overlapped with a potential c-Jun binding site (tgactcagcc) (Fig. 4A). As previously reported, in response to TGF-β, phosphorylated Smad3 interacts with the AP-1 complex at the AP-1-binding site to regulate target gene transcription (37). We therefore tested whether endogenous Smad3 and c-Jun physically interact with the overlapping Smad3 and c-Jun binding site of the SIRT1 promoter by ChIP assays. Using wild-type lungs at P28, specific ChIP signals with antibodies to Smad3, c-Jun, and acetylated histone H3 were reproducibly detected with the same primers spanning this overlapping binding site (Fig. 4B). In Smad3-null lung tissue at P28, no ChIP signals were detected from immunoprecipitation with Smad3 antibody becase there was no Smad3 protein, whereas ChIP signals was still detected from immunoprecipitation with c-Jun antibody, similar to that in wild-type lung (data not shown). These data confirmed that Smad3 and c-Jun specifically and independently interact with this particular overlapping binding site of the SIRT1 promoter in the lung.
Fig. 4.
Smad3 and c-Jun interacted with and synergistically activated the SIRT1 promoter. A: Smad3 (actcagcc) and c-Jun (tgactcagcc) overlapping binding sequences are present at the mouse SIRT1 promoter, and a primer pair (underlined) was used to span the binding site. B: Smad3 and c-Jun were present on the mouse SIRT1 promoter examined by ChIP assays using WT lungs at P28. Acetylated histone H3 (Ac-H3) and mouse IgG were positive and negative controls, respectively. C: with or without TGF-β treatment (5 ng/ml), Sirt1-luc was co-transfected with Smad3 and/or c-Jun in different combinations into F9 cells, followed by luciferase assays 48 h after transfection. *P < 0.01.
To determine whether Smad3 and c-Jun could synergistically activate the SIRT1 promoter, the SIRT1 promoter/reporter plasmid (Sirt1-luc) was transiently transfected into F9 cells, which lack c-Jun and c-Fos but have endogenous Smad3 and Smad4. As shown in Fig. 4C, transcription activity from this reporter reached 7-fold induction when c-Jun was overexpressed compared with the base level and 15-fold induction when both Smad3 and c-Jun were overexpressed. Moreover, in the presence of TGF-β1 stimulation, transcription from this reporter reached 29-fold induction by c-Jun cotransfection and 42-fold induction by Smad3 and c-Jun cotransfection. The reporter activity was slightly induced in the absence of c-Jun (Fig. 4C) and was totally abolished when the Smad3/c-Jun overlapping binding site was deleted (data not shown). These data suggest that Smad3 and c-Jun synergistically activate the SIRT1 gene promoter.
Increased Pea3 expression and NF-κB acetylation also contributed to elevated MMP9 transcription in Smad3-null lungs.
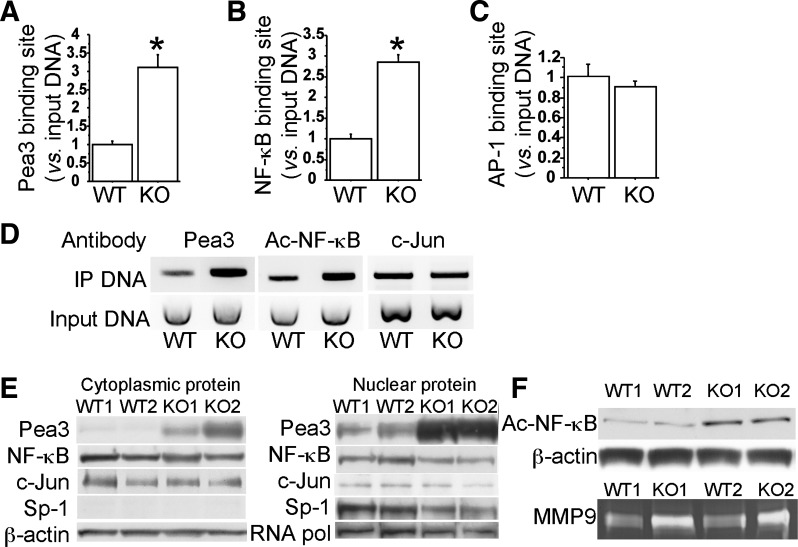
In addition to enhanced histone H3 acetylation on the MMP9 promoter by reduced SIRT1, elevated MMP9 transcription in Smad3-null lungs could also be mediated by altered binding activity and/or expression levels of transcription factors AP-1, Pea3, and NF-κB. We therefore compared the levels of the transcription factors that were present at the AP-1, Pea3, and NF-κB binding sites of the MMP9 promoter between wild-type and Smad3-null lungs at P28. ChIP assays revealed significant increases of Pea3 and NF-κB that were bound to the specific sites of MMP9 promoter in Smad3-null lungs compared with wild-type controls (Fig. 5, A, B, and D), whereas binding of c-Jun to the AP-1 site of the MMP9 promoter was not altered in Smad3-null lungs (Fig. 5, C and D).
Fig. 5.
Increased binding on MMP9 promoter and expression of Pea3 and acetylated NF-κB in Smad3-null lungs (KO). WT and Smad3 KO lungs at P28 (n = 3, each) were collected for ChIP assays using Pea3 or acetylated NF-κB or c-Jun antibody. The relative amount of the coimmunoprecipitated DNA at Pea3, NF-κB, and AP-1 binding sites were detected by real-time PCR (A–C, *P < 0.01), and illustrated on 2% of agarose gel (D). E: cytoplasmic and nuclear Pea3, NF-κB, and c-Jun protein levels were compared between WT and Smad3 KO at P28 by Western blot. β-Actin was a loading control for cytoplasmic proteins, whereas Sp-1 and RNA polymerase II (RNA pol.) were used for nuclear protein-loading controls. F: acetylated NF-κB protein levels and MMP9 activities in lung macrophages of WT and Smad3 KO at P28 were examined by Western blot and zymography, respectively.
Because increased transcription factor and promoter interaction may be due to elevated expression of transcription factors, we then examined proteins levels of Pea3, NF-κB, and c-Jun in wild-type and Smad3-null lungs at P28. Western blots indicated that both cytoplasmic and nuclear protein levels of Pea3, which is expressed in both lung epithelial and mesenchymal cells (6), were dramatically increased in Smad3-null lungs (Fig. 5E), whereas protein levels of NF-κB and c-Jun were not altered between wild-type and Smad3-null lungs (Fig. 5E).
It has been reported that acetylation of NF-κB is required for its transcriptional regulatory activity and that SIRT1 is able to deacetylate RelA/p65 Lys310 residue of NF-κB (36). We therefore further determined the level of acetylated NF-κB (Lys310). As shown in Fig. 5F, acetylated NF-κB (Lys310) level was elevated in macrophages of Smad3-null lungs at P28, compared with wild-type controls, suggesting that increased NF-κB and MMP9 promoter interaction in Smad3-null lungs (Fig. 5B) is likely due to reduced SIRT1-mediated deacetylation of RelA/p65 but not due to change in protein level. Moreover, increased MMP9 levels in pulmonary macrophages of Smad3 knockout mice were verified by MMP9 zymography (Fig. 5F). Taken together, elevated MMP9 transcription in Smad3-null lungs can also be explained by increased Pea3 expression and NF-κB acetylation, in addition to altered histone H3 acetylation.
DISCUSSION
Imbalance between proteinase and antiproteinase is one of the important mechanisms underlying lung tissue destruction in emphysema/COPD through excessively degrading extracellular matrix components including type I collagen and elastin. Our previous studies have shown that early onset of emphysema occurs in Smad3-null mice accompanied by increased MMP9 expression and activity (3, 5). Whether increased MMP9 in these Smad3-null lungs contributes to emphysema pathology has not been determined. In addition, although a number of studies have linked the TGF-β signaling pathway to emphysema through its effect on synthesis of specific MMPs, the related molecular mechanisms are not fully understood. In the present study, we have shown that early onset of emphysema in Smad3-null lung could be rescued by blocking MMP9 activity through a genetic deletion of MMP9 (Fig. 1). Furthermore, we have demonstrated that the molecular mechanisms that increased MMP9 expression in Smad3-null lung were mediated by upregulation of its transcription factor Pea3 and downregulation of TGF-β-Smad3 target gene SIRT1-mediated deacetylation of histone H3 and NF-κB.
Elevated expression and nuclear accumulation of Pea3 and subsequently more binding of Pea3 to the MMP9 promoter are considered to be one of the mechanisms for altered MMP9 expression in Smad3-null lungs (Fig. 5, A and D). Pea3, a transcription factor of the ETS family, is known to be downstream of EGFR signaling. Elevated EGFR activation affects tumor progression in part by promoting tumor invasion through nuclear accumulation of Pea3 and directly binding of Pea3 to the endogenous MMP9 and MMP14 promoters (7). Moreover, Xu et al. (34) have recently shown that TGF-β-Smad3 signaling upregulates receptor type protein tyrosine phosphatase-κ (RPTP-κ) at the levels of both mRNA and protein in human keratinocytes and that RPTP-κ induction decreases basal and EGF-stimulated EGFR-tyrosine (1680) phosphorylation to reduce EGFR signaling activity. Interestingly, we found that Smad3 knockout lung had significant decrease of RPTP-κ expression and increase of EGFR-tyrosine (1680) phosphorylation (data not included). The detailed mechanism by which TGF-β signaling regulates RPTP-κ expression remains to be investigated. Nevertheless, it is reasonable to assume that blockade of Smad3 may relieve the suppressive effect of TGF-β on EGFR activation in the lung, resulting in EGFR-mediated nuclear accumulation of Pea3 and direct binding of Pea3 to the MMP9 promoter.
Downregulation of TGF-β-Smad3 target gene SIRT1 is considered to be another mechanism for altered MMP9 expression in Smad3-null lungs (Fig. 2, A and B). SIRT1 has evolved to deacetylate transcription factors, cofactors, histones, and other chromatin proteins that govern many central metabolic pathways (14). SIRT1 has been shown to act as a negative regulator for MMP9 expression in human monocytes by deacetylating histone at the AP-1 and NF-κB binding sites of the MMP9 promoter (23). Moreover, anti-inflammatory property of SIRT1 has been shown to be associated with decreased NF-κB transcriptional activity by deacetylating RelA/p65 at Lys310 residue (36). Given the fact that a significant reduction of SIRT1 occurs in Smad3-null lungs, it is likely that downregulation of SIRT1 contributes to MMP9 upregulation, not only by enhancing histone H3 acetylation at the AP-1, NF-κB, and Pea3 binding sites of the MMP9 promoter (Fig. 2, C–F), but also probably by reducing deacetylation of RelA/p65 to increase NF-κB transcriptional activity (Fig. 5, B and F).
In mammals, SIRT1 is primarily a nuclear protein in most cell types. SIRT1 expression could be induced by TGF-β1 in mouse lung epithelial cells, human lung fibroblast cells, and mouse monocyte/macrophage cells (Fig. 3A). In addition, SIRT1 protein levels were significantly decreased in both macrophages and nonmacrophages (lung parenchymal cells, including epithelial and mesenchymal cells) of Smad3-null lungs (Fig. 3, E and F). MMP9, a downstream target of SIRT1, is also expressed in both lung parenchymal and infiltrated inflammatory cells (11, 19, 35). Although a previous study has reported that overexpression of human MMP9 in mouse macrophages is sufficient to induce emphysema in mice, accompanied by loss of alveolar elastin (13), we think that downregulated SIRT1-mediated MMP9 overexpression occurs in a variety of cell types in Smad3-null mouse lung and contributes to emphysema pathogenesis in this disease model.
In summary, we have shown that blockade of MMP9 in Smad3-null mice attenuated subsequent airspace enlargement, indicating that MMP9 overexpression is primarily responsible for the subsequent emphysema observed in Smad3-null lung. In addition, we have discovered the molecular mechanisms by which abnormal MMP9 overexpression in Smad3-deficient emphysema lung can be mediated by 1) Pea3 upregulation and nuclear accumulation, and 2) downregulation of a protein deacetylase SIRT1, which is a direct target of TGF-β-Smad3 signaling. Reduced SIRT1 results in increased protein acetylation of histone H3 around MMP9 promoter and NF-κB, which are all key regulators for MMP9 transcription.
GRANTS
This work was supported by National Institute of Health grants HL109932 and HL068597 (W. Shi), American Heart Association grant 10BGIA4150155 (B. Xu), and a California Institute of Regenerative Medicine training grant (W. Xu).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: B.X., D.W., M.K., J.G., and W.S. conception and design of research; B.X., H.C., W.X., W.Z., S.B., S.-G.Z., and W.S. performed experiments; B.X., S.-G.Z., and W.S. analyzed data; B.X., D.W., M.K., J.G., and W.S. interpreted results of experiments; B.X. and W.S. prepared figures; B.X. drafted manuscript; S.B. and W.S. edited and revised manuscript; W.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Xiao-Fan Wang at Duke University for providing Smad3 knockout mouse line.
REFERENCES
- 1. Baraldo S, Bazzan E, Turato G, Calabrese F, Beghe B, Papi A, Maestrelli P, Fabbri LM, Zuin R, Saetta M. Decreased expression of TGF-beta type II receptor in bronchial glands of smokers with COPD. Thorax 60: 998– 1002, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J 25: 552– 563, 2005 [DOI] [PubMed] [Google Scholar]
- 3. Bonniaud P, Kolb M, Galt T, Robertson J, Robbins C, Stampfli M, Lavery C, Margetts PJ, Roberts AB, Gauldie J. Smad3 null mice develop airspace enlargement and are resistant to TGF-beta-mediated pulmonary fibrosis. J Immunol 173: 2099– 2108, 2004 [DOI] [PubMed] [Google Scholar]
- 4. Celedon JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, Demeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, Sylvia JS, Hernandez M, Speizer FE, Weiss ST, Silverman EK. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 13: 1649– 1656, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Chen H, Sun J, Buckley S, Chen C, Warburton D, Wang XF, Shi W. Abnormal mouse lung alveolarization caused by Smad3 deficiency is a developmental antecedent of centrilobular emphysema. Am J Physiol Lung Cell Mol Physiol 288: L683– L691, 2005 [DOI] [PubMed] [Google Scholar]
- 6. Chotteau-Lelièvre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene 15: 937– 952, 1997 [DOI] [PubMed] [Google Scholar]
- 7. Cowden Dahl KD, Zeineldin R, Hudson LG. PEA3 is necessary for optimal epidermal growth factor receptor-stimulated matrix metalloproteinase expression and invasion of ovarian tumor cells. Mol Cancer Res 5: 413– 421, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DaCosta BS, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol 65: 744– 752, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol 19: 2495– 2504, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunnill MS. Quantitative methods in the study of pulmonary pathology. Thorax 17: 320– 328, 1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunsmore SE, Saarialho-Kere UK, Roby JD, Wilson CL, Matrisian LM, Welgus HG, Parks WC. Matrilysin expression and function in airway epithelium. J Clin Invest 102: 1321– 1331, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkington PT, Friedland JS. Matrix metalloproteinases in destructive pulmonary pathology. Thorax 61: 259– 266, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foronjy R, Nkyimbeng T, Wallace A, Thankachen J, Okada Y, Lemaitre V, D'Armiento J. Transgenic expression of matrix metalloproteinase-9 causes adult-onset emphysema in mice associated with the loss of alveolar elastin. Am J Physiol Lung Cell Mol Physiol 294: L1149– L1157, 2008 [DOI] [PubMed] [Google Scholar]
- 14. Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp Quant Biol. In press [DOI] [PubMed] [Google Scholar]
- 15. Hall JR, Pyeritz RE, Dudgeon DL, Haller JA., Jr Pneumothorax in the Marfan syndrome: prevalence and therapy. Ann Thorac Surg 37: 500– 504, 1984 [DOI] [PubMed] [Google Scholar]
- 16. Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev 14: 627– 644, 2000 [PubMed] [Google Scholar]
- 17. Matrisian LM. The matrix-degrading metalloproteinases. Bioessays 14: 455– 463, 1992 [DOI] [PubMed] [Google Scholar]
- 18. Mauviel A, Chung KY, Agarwal A, Tamai K, Uitto J. Cell-specific induction of distinct oncogenes of the Jun family is responsible for differential regulation of collagenase gene expression by transforming growth factor-beta in fibroblasts and keratinocytes. J Biol Chem 271: 10917– 10923, 1996 [DOI] [PubMed] [Google Scholar]
- 19. Mercer BA, Kolesnikova N, Sonett J, D'Armiento J. Extracellular regulated kinase/mitogen activated protein kinase is up-regulated in pulmonary emphysema and mediates matrix metalloproteinase-1 induction by cigarette smoke. J Biol Chem 279: 17690– 17696, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J 404: 1– 13, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minematsu N, Nakamura H, Tateno H, Nakajima T, Yamaguchi K. Genetic polymorphism in matrix metalloproteinase-9 and pulmonary emphysema. Biochem Biophys Res Commun 289: 116– 119, 2001 [DOI] [PubMed] [Google Scholar]
- 22. Mizuguchi T, Collod-Beroud G, Akiyama T, Abifadel M, Harada N, Morisaki T, Allard D, Varret M, Claustres M, Morisaki H, Ihara M, Kinoshita A, Yoshiura K, Junien C, Kajii T, Jondeau G, Ohta T, Kishino T, Furukawa Y, Nakamura Y, Niikawa N, Boileau C, Matsumoto N. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet 36: 855– 860, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamaru Y, Vuppusetty C, Wada H, Milne JC, Ito M, Rossios C, Elliot M, Hogg J, Kharitonov S, Goto H, Bemis JE, Elliott P, Barnes PJ, Ito K. A protein deacetylase SIRT1 is a negative regulator of metalloproteinase-9. FASEB J 23: 2810– 2819, 2009 [DOI] [PubMed] [Google Scholar]
- 24. Ogawa K, Chen F, Kuang C, Chen Y. Suppression of matrix metalloproteinase-9 transcription by transforming growth factor-beta is mediated by a nuclear factor-kappaB site. Biochem J 381: 413– 422, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O'Malley BW, Xu J. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol 28: 5937– 5950, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Snider GL, Kleinerman J, Thurlbeck WM, Bengali ZH. The definition of emphysema. Report of a National Heart, Lung, and Blood Institute, Division of Lung Diseases Workshop. Am Rev Respir Dis 132: 182– 185, 1985 [DOI] [PubMed] [Google Scholar]
- 27. Thurlbeck WM. Internal surface area and other measurements in emphysema. Thorax 22: 483– 496, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vu TH, Shipley JM, Bergers G, Berger JE, Helms JA, Hanahan D, Shapiro SD, Senior RM, Werb Z. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell 93: 411– 422, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev 14: 2123– 2133, 2000 [DOI] [PubMed] [Google Scholar]
- 30. Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, Uozumi N, Ihara S, Lee SH, Ikeda Y, Yamaguchi Y, Aze Y, Tomiyama Y, Fujii J, Suzuki K, Kondo A, Shapiro SD, Lopez-Otin C, Kuwaki T, Okabe M, Honke K, Taniguchi N. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci USA 102: 15791– 15796, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu L, Chau J, Young RP, Pokorny V, Mills GD, Hopkins R, McLean L, Black PN. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax 59: 126– 129, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu B, Chen C, Chen H, Zheng SG, Bringas P, Jr, Xu M, Zhou X, Chen D, Umans L, Zwijsen A, Shi W. Smad1 and its target gene Wif1 coordinate BMP and Wnt signaling activities to regulate fetal lung development. Development 138: 925– 935, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu B, Qu X, Gu S, Doughman YQ, Watanabe M, Dunwoodie SL, Yang YC. Cited2 is required for fetal lung maturation. Dev Biol 317: 95– 105, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Y, Baker D, Quan T, Baldassare JJ, Voorhees JJ, Fisher GJ. Receptor type protein tyrosine phosphatase-kappa mediates cross-talk between transforming growth factor-beta and epidermal growth factor receptor signaling pathways in human keratinocytes. Mol Biol Cell 21: 29– 35, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao PM, Buhler JM, d'Ortho MP, Lebargy F, Delclaux C, Harf A, Lafuma C. Expression of matrix metalloproteinase gelatinases A and B by cultured epithelial cells from human bronchial explants. J Biol Chem 271: 15580– 15589, 1996 [DOI] [PubMed] [Google Scholar]
- 36. Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23: 2369– 2380, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 394: 909– 913, 1998 [DOI] [PubMed] [Google Scholar]