Abstract
Proteasomes are ubiquitously expressed multicatalytic complexes that serve as key regulators of protein homeostasis. There are several lines of evidence indicating that proteasomes exist in heterogeneous subpopulations in cardiac muscle, differentiated, in part, by post-translational modifications (PTMs). PTMs regulate numerous facets of proteasome function, including catalytic activities, complex assembly, interactions with associating partners, subcellular localization, substrate preference, and complex turnover. Classical technologies used to identify PTMs on proteasomes have lacked the ability to determine site specificity, quantify stoichiometry, and perform large-scale, multi-PTM analysis. Recent advancements in proteomic technologies have largely overcome these limitations. We present here a discussion on the importance of PTMs in modulating proteasome function in cardiac physiology and pathophysiology, followed by the presentation of a state-of-the-art proteomic workflow for identifying and quantifying PTMs of cardiac proteasomes.
Keywords: bioinformatics, protein degradation
this article is part of a collection on Post-translational Modification of Proteases in Cardiac Muscle Physiological Function and Characterization Using Proteomics. Other articles appearing in this collection, as well as a full archive of all collections, can be found online at http://ajpheart.physiology.org/.
Introduction
Proteasomes are abundant, multicatalytic complexes expressed in all mammalian cells, including cardiac muscle, which serve as key regulators of protein homeostasis (28, 30). Alterations in proteasome function have been observed in ischemic (52), hypertrophic (22, 56), atrophic (58), desmin-related (42), and diabetic (84) cardiomyopathies, thus indicating an important role for proteasomes in the remodeling of cardiac phenotypes.
The period of functional impact for most cardiac proteins is controlled by proteasome-dependent degradation (53, 55, 72), therefore proteasomes degrade a diverse mosaic of substrates. These include transcriptional factors [e.g., p53 (60)], signaling kinases [e.g., PKCδ (15)], sarcomeric proteins [e.g., α-actinin (41)], and apoptotic factors [e.g., Bid (9)], which may be marked for degradation in a variety of ways, including polyubiquitin tags or denaturation/unfolding due to stress. Cardiac proteasomes exhibit heterogeneity (23, 54, 71) and discrete subcellular distribution (28, 30, 63), which specializes subpopulations of proteasomes to degrade distinct substrate repertories (63). Post-translational modifications (PTMs) are recognized to be an important underlying mechanism for functional diversification of proteasomes (80, 81). There are several lines of evidence demonstrating that PTMs modulate numerous facets of proteasome function (Table 1). To date, significant progress has been made in identifying PTMs on cardiac proteasome complexes; however, quantitative information regarding the endogenous stoichiometric abundance of individual PTMs is lacking. The advancement of proteomic technologies has offered unique solutions for systematically characterizing the molecular basis of proteasomal regulation by PTMs. Moreover, the implementation of innovative bioinformatics platforms has aided in the dissemination of PTM discoveries and enabled synergistic efforts across the scientific community in this research arena. As proteasomes influence a broad spectrum of cell biology, the significance of understanding how PTMs modulate proteasomes in cardiovascular physiology and medicine is paramount.
Table 1.
PTMs identified in proteasome complexes
| PTM | Subunit | Site | Tissue | Technique | Biological Function | Reference |
|---|---|---|---|---|---|---|
| Phosphorylation | α2 | Y121 | CWSV1 | Mutagenesis | Trafficking | (5) |
| Phosphorylation | α2 | S198 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α2 | T204 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α2 | ND | HEK293 | 32P | Catalytic activity | (26) |
| Phosphorylation | α3 | T33 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α3 | S75 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α3 | S81 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α3 | S153 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α3 | Y156 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α3 | S173 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α3 | ND | HEK293 | 32P | Catalytic activity | (26) |
| Phosphorylation | α4 | T97 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α4 | Y153 | HEK293 | 2DE, mutagenesis | Catalytic activity | (44) |
| Phosphorylation | α5 | S134 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α5 | T219 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α5 | T230 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | α7 | S13 | Liver | LC-MS/MS | ND | (46) |
| Phosphorylation | α7 | T186 | Liver | LC-MS/MS | ND | (46) |
| Phosphorylation | α7 | S250 | COS-7 | LC-MS/MS | 26S assembly | (7) |
| Phosphorylation | α7 | His | Molt-4 | 32P | NDP kinase | (74) |
| Phosphorylation | β1 | S157 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β1 | T181 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β2 | T273 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β2 | S277 | Liver | LC-MS/MS | ND | (46) |
| Phosphorylation | β3 | Y85 | Liver | LC-MS/MS | ND | (46) |
| Phosphorylation | β3 | T86 | Liver | LC-MS/MS | ND | (46) |
| Phosphorylation | β3 | His | Molt-4 | 32P | ND | (74) |
| Phosphorylation | β4 | S39 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β5 | T48 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β5 | S192 | Liver | LC-MS/MS | ND | (46) |
| Phosphorylation | β5 | S204 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β6 | T38 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β6 | S48 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β6 | S167 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β6 | S169 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | β7 | S93 | Heart | LC-MS/MS | ND | (46) |
| Phosphorylation | Rpt5 | ND | Hela | 32P | Catalytic activity | (68) |
| Phosphorylation | Rpt6 | S120 | MDA 468 | 32P, mutagenesis | Catalytic activity | (76) |
| Phosphorylation | Rpt6 | ND | Heart | 32P | 26S assembly | (61) |
| Phosphorylation | 11S | ND | Reticulocyte | 32P | Catalytic activity | (43) |
| Acetylation | α1 | K102 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α1 | K104 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α2 | K70 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α2 | K171 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α3 | K127 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α3 | K176 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α3 | K238 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α4 | K227 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α7 | K57 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α7 | K110 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α7 | K206 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α7 | K230 | NS | LC-MS/MS | ND | (14) |
| Acetylation | α7 | K238 | NS | LC-MS/MS | ND | (14) |
| Acetylation | β1i | K53 | NS | LC-MS/MS | ND | (14) |
| Acetylation | β1i | K109 | NS | LC-MS/MS | ND | (14) |
| Acetylation | β3 | K77 | NS | LC-MS/MS | ND | (14) |
| Acetylation | β4 | K68 | NS | LC-MS/MS | ND | (14) |
| Acetylation | β4 | K185 | NS | LC-MS/MS | ND | (14) |
| Acetylation | β6 | K204 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt1 | K116 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt1 | K422 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt2 | K238 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt3 | K397 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt3 | K401 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt3 | K258 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt4 | K20 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt4 | K72 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt4 | K206 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpt6 | K222 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn2 | K310 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn5 | K221 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn5 | K368 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn5 | K448 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn6 | K417 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn8 | K204 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn8 | K214 | NS | LC-MS/MS | ND | (14) |
| Acetylation | Rpn9 | K298 | NS | LC-MS/MS | ND | (14) |
| Acetylation | 11Sγ | K195 | NS | LC-MS/MS | ND | (14) |
| Oxidation | Rpt3 | ND | Heart | 2DE | Catalytic activity | (21) |
| Oxidation | Rpt5 | ND | Heart | 2DE | Catalytic activity | (21, 56) |
| HNE modification | α1 | ND | Heart | 2DE | Catalytic activity | (12) |
| HNE modification | α2 | ND | Heart | 2DE | Catalytic activity | (12) |
| HNE modification | α4 | ND | Heart | 2DE | Catalytic activity | (12) |
| Poly-ADP ribosylation | ND | ND | K562 | 14C-ADP ribose | Catalytic activity | (67) |
| O-GlcNAc modification | Rpt2 | ND | NRK | 2DE | Catalytic activity | (78) |
| Ubiquitination | α1 | K59 | MCF-7 | LC-MS/MS | ND | (20) |
| Ubiquitination | α6 | K115 | Hela | LC-MS/MS | ND | (48) |
| Ubiquitination | α6 | K208 | Hela | LC-MS/MS | ND | (48) |
| Ubiquitination | β3 | ND | C2C12 | 2DE | ND | (69) |
| Ubiquitination | Rpt2 | K237 | HEK293 | LC-MS/MS | ND | (47) |
| Ubiquitination | Rpn10 | K84 | Yeast | Mutagenesis | Substrate specificity | (35) |
| Glycosylation | β3 | ND | MDCK | LC-MS/MS | ND | (13) |
| Glycosylation | Rpt11 | N241 | MDCK | LC-MS/MS | ND | (13) |
| Sumoylation | β2 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpt2 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpn1 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpn2 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpn5 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpn8 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpn10 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpn12 | ND | Hela | LC-MS/MS | ND | (65) |
| Sumoylation | Rpt1 | ND | HEK293 | LC-MS/MS | ND | (31) |
| Sumoylation | Rpt6 | ND | HEK293 | LC-MS/MS | ND | (31) |
| Sumoylation | α3 | ND | Yeast | LC-MS/MS | ND | (51) |
| Sumoylation | Rpn1 | ND | Yeast | LC-MS/MS | ND | (51) |
| Sumoylation | Rpn7 | ND | Yeast | LC-MS/MS | ND | (51) |
| Sumoylation | Rpn12 | ND | Yeast | LC-MS/MS | ND | (51) |
| Nitrosylation | α2 | Y228 | Pituitary | MALDI/MS/MS | ND | (75) |
Post-translational modifications (PTMs) and the specific site (if known) on proteasome subunits in various tissues. The techniques used in the discovery of PTMs and the biological function (if known) are included. In summary, 10 distinct PTMs have been identified on proteasome subunits. From those studies in which site-specific information was obtained, there were 33 phosphorylation sites on 12 20S, 2 19S, and 1 11S subunits; 38 acetylation sites on 9 20S, 10 19S, and 1 11S subunit; 5 ubiquitination sites on 3 20S and 2 19S subunits; 1 glycosylation site on 1 19S subunit; and 1 nitrosylation site on 1 20S subunit.
LC-MS/MS, liquid chromatography-tandem mass spectometry; ND, not determined; 2DE, two-dimensional electrophoresis; NDP, nucleoside diphosphate; HNE, 4-hydroxy-2-nonenal; O-GlcNAc, O-linked N-acetylglucosamine; NS, not specified; NRK, normal rat kidney cells; MDCK, Madin-Darby canine kidney cells; MALDI, matrix-assisted laser desorption/ionization.
PTMs Regulate Diverse Functions of Cardiac Proteasomes
As terminally differentiated cells, cardiomyocytes are charged with maintaining a healthy proteome for survival. Cardiomyocytes undergo rhythmic perturbations, including fluctuations in ionic strength, oxidoreductive potential, and mechanical strain. As proteasomes exhibit relatively long half-lives, ranging from 1 day (33) to 1 wk (19), PTMs serve as an essential mechanism for the instantaneous regulation of proteasome function to adapt to a dynamic intracellular environment. As multiple amino acid residues in proteasome proteins house a myriad of different PTMs, an essential first step in understanding the physiological function is the reliable and site-specific identification. To date, a diverse array of PTMs has been successfully identified on proteasomes (Table 1). As a result, studies aimed at elucidating the function of site-specific PTMs in physiology and disease are on the rise, which presents an exciting opportunity in the field. A summary of findings to date is presented below.
Irreversible PTMs.
NH2-terminal modifications are the most frequently observed PTMs on proteasome subunits. Subunits of 20S proteasomes are members of the NH2-terminal nucleophile hydrolase family, which have active centers located at their NH2-termini (β1, β2, and β5). In addition to enabling targeted initiation of hydrolase activity within the chamber of 20S, propeptides ensure that the catalytic centers of active subunits are protected from deactivating modifications, such as Nα-acetylation (1). Catalytically inactive β6- and β7-subunits are also processed by neighboring active subunits. Propeptides affect the recruitment of assembly partners. Processing procedures for the active subunits are conducted cooperatively during the process of proteasome assembly, which is an essential rate-limiting step in proteasome maturation. Overexpression of proteasome subunit β5 elevated the gross level of proteasome complexes as well as their proteolytic activities (45), which imparted enhanced resistance toward oxidative stress (40). The effect of β5 overexpression on propeptide processing efficiency and maturation of proteasome complexes remains to be delineated.
NH2-terminal acetylation and the removal of NH2-terminal methionine residues of proteasome subunits proceed according to the specificity of N-acetyl transferases and methionine aminopeptidases, which are primarily determined by the penultimate residue of protein substrates. Traditionally, NH2-terminal modifications have been linked to protein stability, which has been recently challenged by large-scale proteomics and bioinformatics studies. Current findings suggest that NH2-terminal acetylation may affect protein complex assembly (62), an intriguing mechanism that has not been tested in proteasome complexes. While it is uncertain how NH2-terminal modifications may change in cardiac disease, enhanced NH2-terminal acetylation of 20S α7 was reported in brain lysates from Alzheimer's patients, suggesting that NH2-terminal acetylation may be linked to neurodegenerative diseases (27).
N-myristoylation is catalyzed by N-myristoyltransferases. The cardiac 19S subunit Rpt2 contains a highly conserved motif and is indeed modified by this form of irreversible modification (30). N-myristoylation is thought to have a role in signal transduction and membrane targeting of proteins. Proteasome complexes have been identified in membranous regions (10, 11); however, the specific function of N-myristoylation in proteasome biology remains to be fully delineated.
Proteasomes are also regulated through irreversible PTMs in the form of cleavage by other proteases. Caspases and proteasomes can degrade one another (37, 66, 73), which establishes an essential checkpoint for cell survival. Apoptosis is initiated when caspases dominate this balance. In parallel, the lysosomal system also targets proteasome complexes (19). As proteasomes and lysosomes coordinate the degradation of the intracellular protein pool, perturbations of this dynamic relationship may affect a large number of protein substrates (39). The dynamics and regulation of this paradigm remain largely unknown.
Reversible PTMs.
Phosphorylation is the most studied form of reversible modification on proteasome complexes. Protein kinase A (PKA), an integral effector of the β-adrenergic pathway, is a functional associating partner of cardiac proteasomes (80). The in vitro or in vivo activation of PKA resulted in the phosphorylation of proteasomes, which enhanced proteolytic activities (2, 76, 80) and 26S assembly (2). Phosphorylation coordinates a wide array of proteasome function, including complex assembly, trafficking, and proteolytic activity, as well as recruitment of regulatory complexes (6, 7, 26, 38, 44, 61, 64, 80). Several kinases and phosphatases orchestrate the functional dynamics of proteasome complexes in the heart (7, 26, 80). Among them, PKA and protein phosphatase 2A are known to be important players in cardiac pathogenesis. In vivo inhibition of PKA eliminated the beneficial effects of ischemic preconditioning on proteasome assembly and activity (2), and activation of endogenous PKA rescued depressed 20S catalytic activities in isoproterenol-induced cardiac hypertrophy (22). In contrast, hyperphosphorylation of hepatic 20S in ethanol-induced liver disease has been correlated with decreased proteasome activities (4). It is likely that the divergent effects of phosphorylation on catalytic activities are due to differences in phosphorylation sites being targeted, modulated by distinct panels of kinases/phosphatases.
Lysine residues are subject to different forms of PTMs; acetylation and ubiquitination (Ub) have been identified on proteasomes, and many more remain to be discovered. Recent reports identified lysine acetylation on cardiac proteasome subunits (79). Histone acetyltransferases (HATs) and histone deacetylases (HDACs) determine the occupancy of these motifs. Studies also indicate that application of HDAC inhibitors may lead to cardiac benefits under stress (16). HATs and HDACs modulate gene transcription in the nucleus and metabolism in mitochondria, which serve as two important mechanisms for cell protection. However, the integration of protein degradation in these pathways remains largely unknown. A recent report demonstrated that HDAC inhibition led to upregulation of lysine acetylation on cardiac proteasome subunits, which resulted in enhanced proteolytic activities. These data suggest that lysine acetylation may modulate degradation capacities in health and disease (79).
Proteasome subunits are regulated by Ub as well. It has been reported that polyUb regulates both the proteolytic capacity and substrate specificity of proteasome complexes (32, 35, 69). The subcellular targeting of proteasome complexes or recruitment of the ubiquitin-associated domain containing regulators may be affected by polyUb as well. The synergistic or antagonistic interplay between lysine acetylation and Ub is an intriguing topic to study.
Proteasomes can be modified by O-linked N-acetylglucosamine, which is regulated by O-GlcNAc transferases and O-GlcNAcases. This PTM serves as a sensor for both metabolic dynamics and stress (8). It has been reported that the 19S ATPase subunit Rpt2 can be modified by O-GlcNAc, leading to an inhibition of proteolytic activity (78). However, the specific site of modification remains to be defined. Interplay between O-GlcNAc and phosphorylation has been reported, as both target Ser and Thr residues (34). This regulatory paradigm has yet to be investigated on proteasome complexes.
Proteasome complexes are essential for turning over proteins misfolded by oxidative stress. In fact, the extent of myocardial injury correlates to the functional potency of proteasome complexes after an ischemic event (55). Poly-ADP ribosylation of 20S proteasomes, catalyzed by poly(ADP-ribosyl) transferase, selectively elevates proteolytic activities against oxidized substrate repertoires (67). However, the oxidative modification of proteasome subunits directly is generally detrimental to proteasome function. Oxidative PTMs represent a broad array of modifications that involve the removal of a hydrogen atom or incorporation of an oxygen atom in protein substrates, which is largely nonenzymatical. Increased levels of oxidative PTMs on proteasome subunits have been identified in ischemic (12, 25) and hypertrophic cardiomyopathies (56), which may provide a mechanistic explanation for the observed impairment in degradation capacities.
Proteomics as an Essential Tool to Identify and Quantify PTMs in Proteasome Complexes
A significant portion of PTMs on proteasome complexes have been identified using classic biochemical approaches. Though these efforts were pivotal for establishing a basic understanding of PTM dynamics in cardiac muscle, recent developments in functional proteomics have offered major improvements in the depth and breadth with which we can understand PTMs in cardiac physiology (Fig. 1). First, proteomics removes the need for radioactive isotopes, which have limited clinical utility, unwanted transformative effects on the biological sample, and safety hazards. Second, proteomic analysis offers unbiased, site-specific analysis of PTMs (i.e., multiple sites within the same peptide), which has been a major shortcoming of PTM-directed antibodies. Third, proteomics supports large-scale analyses in that hundreds/thousands of distinct PTM events can be simultaneously probed in hundreds/thousands of proteins, allowing for the elucidation of PTM interplay within and among proteins. This feature is not practically feasible using classical biochemical approaches, which have typically involved the analysis of one or a few proteins at one time. Fourth, recent developments in bioinformatics have fostered the construction of spectral libraries, which provide superior sensitivity, portability, and user cumulativeness for PTM analysis, thus increasing the efficiency in data analysis. Finally, targeted proteomic approaches offer elucidation of multisite PTM stoichiometry through absolute quantitative approaches, which are linear over several orders of magnitude. With quantitative data capabilities for a panel of PTMs, biologists can begin to unravel how PTM dynamics tune the function of entire organelles, such as the proteasome, in cardiovascular physiology and pathology. A state-of-the-art proteomic workflow for identifying and quantifying PTMs on proteasomes is presented below.
Fig. 1.
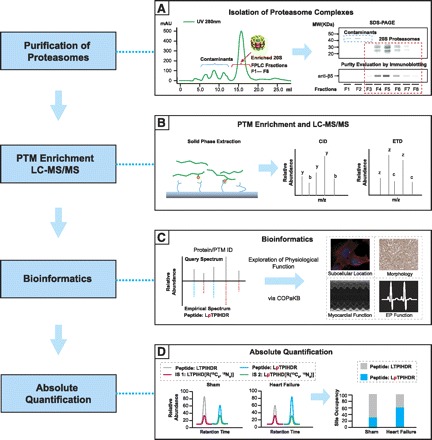
A state-of-the-art proteome workflow for characterizing post-translational modifications (PTMs) on proteasomes. A: an essential first step of PTM analysis of proteasomes is to fractionate cardiomyocytes into subcellular regions and/or to purify proteasome complexes to overcome issues imposed by the large dynamic range. A, left: chromatographic purification of 20S complexes from contaminants. A, right: validation of proteasome content in individual fractions using SDS-PAGE and immunoblotting. MW, molecular weight; FPLC, fast-performance liquid chromatography. B: following digestion of proteasome proteins, peptides containing PTMs can be selectively sequestered using solid-phase extraction techniques with chemistries specific for the PTM of interest (left). B, right: during liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, multiple complimentary fragmentation technologies can be used to both preserve labile PTM moieties and increase the likelihood of PTM detection. Upon collision-induced dissociation (CID), peptide fragments possessing peptide NH2-termini are labeled as the b signal series and fragments possessing the COOH-termini are labeled as the y signal series. Upon electron transfer dissociation (ETD), peptide fragments possessing peptide NH2-termini are labeled as the c signal series and fragments possessing the COOH-termini are labeled as the z signal series. C: peptide and PTM identities can be reliably decoded using an empirical spectral library. C, left: query spectrum matched against an experimental spectrum in the database for the phosphopeptide LpTPIHDR. pT, phosphothreonine. C, right: cardiovascular function of the proteins and PTMs identified using the spectral library can then be explored using the cardiac organellar protein atlas knowledge base (COPakB; www.heartproteome.org), a premier resource aimed at disseminating proteomic findings throughout the cardiovascular community. EP, electrophysiology. D: the final step in characterization of PTMs on proteasomes is to obtain stoichiometric measurements of individual PTMs sites using heavy-labeled internal standard (IS) peptides. IS peptides (green, pink) with sequences identical to the endogenous modified peptide (blue, LpTPIHDR) and unmodified peptide (gray, LTPIHDR) are spiked into the endogenous sample in known quantities, allowing for the accurate calculation of site-specific PTM stoichiometry. D, left and middle: extracted ion chromatograms of signal intensity vs. chromatographic retention time. D, right: quantification of phosphorylation site occupancies. Note that the quantified levels of modified peptide and unmodified peptide are inversely correlated.
Subcellular fractionation and enrichment of proteasome PTMs in cardiomyocytes.
PTMs on proteasomes exist at substoichiometric levels, therefore an essential first step is to enrich effective concentrations of PTMs by reducing the complexity of the cardiac sample. While proteasomes are relatively abundant within cells, the dynamic range of proteins in cardiac muscle is vast compared with other tissue proteomes. Proteins of the sarcomeric apparatus along with the rich population of mitochondria constitute the majority of cellular protein, which would be dominant in a shotgun proteomic approach. This necessitates the use of protein fractionation techniques to isolate proteasome complexes. Differential centrifugation is commonly used to separate subcellular organelles (3). Subsequently, 20S proteasomes or regulatory particles can be enriched through multidimensional purification strategies (e.g., chromatography, ultracentrifugation) to increase the effective concentration of proteasome complexes and their acquired PTMs. Purification strategies have been developed for three distinct proteasome complexes 20S (80), 19S (71), and 26S (30). Importantly, these strategies have resulted in purified proteasome complexes that are functionally viable, allowing for the downstream assessment of enzymatic activities. Proteolytic digestion is necessary to prepare proteasome complexes for shotgun proteomics analyses. In contrast to protein identification, where approximately two distinct peptides from any region of the protein are required for a confident identification, PTMs are localized to specific peptides. The capture of PTM-containing peptides is therefore mandatory and often requires the careful selection of endopeptidases other than trypsin. For large-scale studies, the application of a panel of endopeptidases significantly expands the overall coverage of PTM detection (80). A subsequent enrichment step with solid-phase extraction selectively sequesters peptides containing a substoichiometric PTM to enhance detection by mass spectrometry. Solid-phase extraction platforms involve an immobilized chemistry (e.g., antibodies) with specific affinity for a desired PTM. Successful strategies have been titanium dioxide (TiO2) and immobilized metal affinity columns (phosphorylation), lectin chromatography (glycosylation), immune affinity chromatography, etc. Our laboratory recently acquired the first comprehensive map of site-specific cardiac 20S phosphorylation using a combination of the methods above: subcellular fractionation to obtain a cytosolic fraction, 20S proteasome purification to obtain functionally viable proteasome complexes, complementary proteolytic digestion, and TiO2 enrichment of phosphorylated peptides. Using this workflow, we were able to effectively enrich low-abundant proteasome PTMs, enabling the identification of 52 phosphorylation sites in cardiac and liver proteasomes (46).
Characterization of PTMs using mass spectrometry.
The potential for identification of proteasome PTMs by mass spectrometry has vastly improved over the past decade with the development of mass spectrometry instruments, which is now the state-of-the-art platform enabling large-scale, site-specific PTM analysis. Historically, the main obstacles in identifying PTMs with mass spectrometry have been 1) achieving adequate sensitivity to capture the low-abundant posttranslationally modified peptides and 2) preserving labile PTM moieties during fragmentation of peptide precursor ions. The sensitivity of detection has been significantly enhanced through application of nanoscale chromatography in-line with new generation linear ion trap mass spectrometers with high ion capacity. Multidimensional chromatography, such as a miniaturized TiO2-reverse phase columns (e.g., PhosphoChip), or single dimension chromatography has been employed for the detection of PTMs. Ultraperformance liquid chromatography is fast becoming routine in proteomics. Ultraperformance liquid chromatography capitalizes on the use of solid-phase resin of miniscule (<2 μm) particle size to increase the speed, sensitivity, and resolution of chromatography. High-powered mass spectrometers (e.g., Thermo LTQ-Orbitrap Elite) with superior mass resolution and speed are continually being engineered. This translates into enhanced confidence and sensitivity for PTM identifications. The preservation of PTMs in the mass spec during fragmentation has been greatly aided by the development of electron transfer dissociation (ETD). ETD uses radical anions to fragment peptide backbones in a manner that leaves PTM moieties intact. This differs from the commonly employed collision-induced dissociation method, which fragments peptides after repeated collisions with an inert gas, resulting in loss of labile PTMs. A comprehensive mapping of 20S phosphorylation by our laboratory exploited the complementarity of the two fragmentation methods (46). Identified phosphopeptides could be classified into three groups: those identified by collision-induced dissociation only (34 phosphopeptides), those identified by ETD only (10 phosphopeptides), and those identified by both collision-induced dissociation and ETD (8 phosphopeptides). This study represents the most comprehensive mapping of cardiac proteasome phosphorylation available to date and was the direct result of methods described above aimed at enhancing sensitivity of detection and preservation of PTMs on peptides.
Bioinformatics are essential for PTM analyses.
Sophisticated bioinformatics platforms are required to accurately decode PTM dynamics in mass spectra. Commonly employed database search engines (e.g., SEQUEST, Mascot) identify PTMs by comparing query spectra with simulated, theoretical spectra based on protein sequences (24). This approach enables us to interrogate a series of selected PTMs within the context of a targeted organism. However, this workflow also presents the following limitations: 1) simulated spectra lack features for spectral pattern recognition (e.g., ion intensity), 2) the number of PTMs queried is limited by the power of computational platforms, and 3) protein sequence databases for an organism of interest may be incomplete. As an alternative approach, de novo sequencing algorithms directly analyze the raw spectra and define the peptide sequence based solely on fragment ions. For this approach to be successful, peptide spectra need to present sufficient fragmentation signatures for unequivocal conclusions. This removes the prerequisite for a protein database or a list of PTMs of interest. However, de novo sequencing algorithms calculate the probability of all possible combinations of peptide sequences, requiring an enormous search space that imposes a significant burden on the computational platform.
Identifying PTMs by comparing query spectra against an annotated library of experimentally derived spectra offers a solution for both the specificity and efficiency of data analysis. We have developed a cardiac organellar protein atlas knowledge base (COPaKB, www.heartproteome.org), including specific spectral library modules on human and murine cardiac proteasomes (83). COPaKB capitalizes on the use of both existing and newly generated high-quality experimental PTM spectra for the continuous enhancement of a superior reference against which query spectra are compared. The major advantages of this approach for PTM identification lie in the inclusion of descriptive features of fragmentation spectra that cannot be reliably predicted and therefore are absent from theoretical spectra. Searching query spectra against experimental spectra in a spectral library would provide a highly confident match in that the experimental library spectra would include gas-phase chemistries (both m/z shifts and intensities) unique to post-translationally modified precursor ions.
Quantitative analyses.
The final goal in the characterization of proteasome PTMs is to understand how a particular PTM influences the biological function of proteasomes in the endogenous cellular environment. To accomplish this, stoichiometry of identified PTMs at baseline and in a perturbed state (e.g., disease) must be quantified in a site-specific and reproducible fashion. Multiple reaction monitoring (MRM) is a customizable mass spectrometric technique that allows for sensitive detection and quantification of a target set of peptides. This technique typically employs the use of a triple quadrupole mass spectrometer in which the first quadrupole (Q1) gates for the precursor ion, the second (q2) fragments the precursor ion with an inert gas into daughter ions, and the third (Q3) specifically gates for selected q2-generated daughter ions. A precursor ion detected in Q1 and a daughter ion detected in Q3 are collectively referred to as a transition. Superior sensitivity of MRM comes from targeting only selected transitions that produces a high signal-to-noise ratio, and enhanced reproducibility comes from the continuous monitoring of these transitions that results in the acquisition of a high number of data points per transition. MRM-based quantification of PTMs is also linear over several orders of magnitude (17), making it ideally suited for quantifying PTMs exhibiting a large dynamic range. For example, the phosphorylation of Ser250 on murine α7 is significantly more abundant than the phosphorylation of other 20S subunits (81); MRM-based quantification would thus be highly suitable for the global quantification of 20S phosphorylation. These features in combination underscore the great promise that this technology holds for quantifying PTMs on proteasome complexes. The nature of MRM also allows for quantification of specific sites of phosphorylation in peptides where multiple potential phosphorylation sites are present. This requires the careful design of diagnostic transitions. For example, if phosphorylation of the peptide AGsAASPNVK (s = phosphorylated Ser) was being quantified using MRM, diagnostic transitions would include b3, b4, b5, y5, y6, and y7 ions, since they are representative of cleavages between the two potential phosphoserines (Ser3 and Ser6) that would unequivocally demarcate the site of phosphorylation. This feature can be challenging when the diagnostic fragmentation of peptides is not favored in the gas phase; thus it is critical that parameters affecting the sensitivity of detection (i.e., collision energy) be carefully optimized. In conclusion, MRM offers unique advantages for the global, unbiased, sensitive, and site-specific quantification of PTMs over traditional approaches.
Future Directions
Discovery of novel PTMs with functional relevance using tailored bioinformatics.
PTM databases, including PSI-MOD ontology (49) (http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MOD) and Unimod (18) (http://www.unimod.org), contain ∼1,000 distinct forms of PTMs. This number is still increasing with the discovery of novel PTMs. The evolving nature of PTM discovery presents a challenge for bioinformatics platforms to characterize proteasome PTMs in a systematic manner. Taking the perspective that spectra of PTM peptides share common signatures with their unmodified counterpart, a tailored database search using cardiac spectral libraries can be employed to identify novel PTMs. The measured mass difference between a post-translationally modified peptide and unmodified peptide can aid in correlating fragment ions between them to decipher the molecular nature of the potential PTM. With state-of-the-art mass spectrometers that can achieve beyond a mass resolution of 2 parts/million (50), it is highly likely that PTMs can be deduced from observed shifts in mass. The delineation of biological function should follow through designing gain- or loss-of-function models and through comparison of the abundance of such PTMs in diseased versus normal phenotypes.
Signaling kinetics to proteasomes and PTM interplay in the endogenous cellular environment.
To understand how proteasomes integrate into cardiac signaling pathways, PTMs of proteasomes must be analyzed within the intricate context of the endogenous cellular environment (Fig. 2), both in acute signaling (e.g., β-adrenergic receptor stimulation) and in disease. Moreover, it is highly likely that the functional dynamics of the proteasome complexes are tuned by various PTMs in coordination. Previous reports have suggested that PTMs on proteasomes interact to modulate proteasome function. Interferon-γ triggers a decrease in phosphorylation of both 20S and 19S subunits, leading to the disassembly of 26S proteasomes in response to stress (59). PKA induced the phosphorylation of Rpt6 and O-GlcNAc modification of Rpt2 coordinate glucose metabolism (77). In addition, PKA and protein phosphatase 2A play antagonistic roles in regulating proteasomal function (80). While an initial step in elucidating the functional importance of PTMs on proteasomes is to study the effect of an enzyme (e.g., kinase, acetyltransferase) on proteasome function in an isolated system, this provides limited information regarding the endogenous kinetics of PTM modifications and the coordinated interplay of PTMs on proteasomes. Progress in this area has been attenuated in part by the lack of information regarding signaling to modulating enzymes (e.g., N-myristoyltransferases) but also by the lack of a global and site-specific method for quantifying PTMs on proteasome subunits. Quantifying PTM stoichiometries is essential for delineating the interwoven functions of PTMs in modulating proteasome dynamics in cardiomyocytes. Furthermore, to grasp the dynamics of individual PTMs on proteasomes, multiple time points should be analyzed following a stimulus to gauge whether PTMs change with the time course of agonist treatment or with disease progression. Now that we have sensitive, site-specific and unbiased technologies (e.g., MRM) with which to quantify PTMs, these studies are possible. This approach will allow us to understand the nature of proteasome modulation by PTMs, which will help us to understand how to manipulate specific functions that go awry in disease.
Fig. 2.
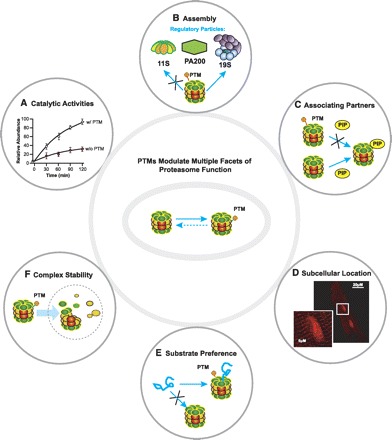
PTMs of proteasomes modulate several facets of proteasome function. PTMs act as a mechanism by which proteasomes can achieve instantaneous and/or chronic regulation to respond to a dynamic cellular environment. The following functions of proteasomes are known to be regulated by PTMs: caspase-like, trypsin-like, and chymotrypsin-like catalytic activities (A), assembly of 20S with regulatory particles (e.g., 11S, 19S, and PA200; B), association of proteasome complexes with proteasome interacting partners (e.g., PKA; C), discrete subcellular locations (D), substrate preference [e.g., ubiquitin tagged vs. misfolded/denatured proteins (E) and proteasome complex stability (F)]. “X” in B, C, and E conceptually illustrates inhibition of a particular pathway or process governed by the presence or absence of PTMs on proteasomes. PIP, proteasome-interacting proteins.
In conclusion, proteasomes have a widespread impact on cardiac physiology, affecting virtually all cellular processes. Understanding the dynamics of PTM modulation of proteasomes will aid in the development of inhibitors or activators that can tune discrete functions of proteasome subpopulations. This will ultimately lead to tightened and more refined protein quality control in cardiac muscle, which will ultimately provide targeted avenues for combating cardiac disease.
GRANTS
This work is supported in part by the National Heart, Lung, and Blood Institute Proteomics Center Award HHSN268201000035C (to P. Ping), Grant R01-HL-098954 (to P. Ping), Grant R01-HL-088640 (to E. Stefani), and NRSA Grant F32-HL-099029 (to S. B. Scruggs).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
S.B.S., N.C.Z., E.S., and P.P. conception and design of research; S.B.S., N.C.Z., and D.W. prepared figures; S.B.S. and N.C.Z. drafted manuscript; S.B.S., N.C.Z., and P.P. edited and revised manuscript; S.B.S., N.C.Z., D.W., E.S., and P.P. approved final version of manuscript.
REFERENCES
- 1. Arendt CS, Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J 18: 3575–3585, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, Takahama H, Sasaki H, Higo S, Asakura M, Takashima S, Hori M, Kitakaze M. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol 46: 452–462, 2009 [DOI] [PubMed] [Google Scholar]
- 3. Baines CP, Zhang J, Wang GW, Zheng YT, Xiu JX, Cardwell EM, Bolli R, Ping P. Mitochondrial PKCepsilon and MAPK form signaling modules in the murine heart: enhanced mitochondrial PKCepsilon-MAPK interactions and differential MAPK activation in PKCepsilon-induced cardioprotection. Circ Res 90: 390–397, 2002 [DOI] [PubMed] [Google Scholar]
- 4. Bardag-Gorce F, Venkatesh R, Li J, French BA, French SW. Hyperphosphorylation of rat liver proteasome subunits: the effects of ethanol and okadaic acid are compared. Life Sci 75: 585–597, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Benedict CM, Clawson GA. Nuclear multicatalytic proteinase subunit RRC3 is important for growth regulation in hepatocytes. Biochemistry 35: 11612–11621, 1996 [DOI] [PubMed] [Google Scholar]
- 6. Benedict CM, Ren L, Clawson GA. Nuclear multicatalytic proteinase alpha subunit RRC3: differential size, tyrosine phosphorylation, and susceptibility to antisense oligonucleotide treatment. Biochemistry 34: 9587–9598, 1995 [DOI] [PubMed] [Google Scholar]
- 7. Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J 378: 177–184, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bowe DB, Sadlonova A, Toleman CA, Novak Z, Hu Y, Huang P, Mukherjee S, Whitsett T, Frost AR, Paterson AJ, Kudlow JE. O-GlcNAc integrates the proteasome and transcriptome to regulate nuclear hormone receptors. Mol Cell Biol 26: 8539–8550, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem 275: 21648–21652, 2000 [DOI] [PubMed] [Google Scholar]
- 10. Brooks P, Fuertes G, Murray RZ, Bose S, Knecht E, Rechsteiner MC, Hendil KB, Tanaka K, Dyson J, Rivett J. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J 346: 155–161, 2000 [PMC free article] [PubMed] [Google Scholar]
- 11. Brooks P, Murray RZ, Mason GG, Hendil KB, Rivett AJ. Association of immunoproteasomes with the endoplasmic reticulum. Biochem J 352: 611–615, 2000 [PMC free article] [PubMed] [Google Scholar]
- 12. Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem 276: 30057–30063, 2001 [DOI] [PubMed] [Google Scholar]
- 13. Chiangjong W, Sinchaikul S, Chen ST, Thongboonkerd V. Calcium oxalate dihydrate crystal induced changes in glycoproteome of distal renal tubular epithelial cells. Mol Biosyst 7: 1917–1925, 2011 [DOI] [PubMed] [Google Scholar]
- 14. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840, 2009 [DOI] [PubMed] [Google Scholar]
- 15. Churchill EN, Ferreira JC, Brum PC, Szweda LI, Mochly-Rosen D. Ischaemic preconditioning improves proteasomal activity and increases the degradation of deltaPKC during reperfusion. Cardiovasc Res 85: 385–394, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Colussi C, Illi B, Rosati J, Spallotta F, Farsetti A, Grasselli A, Mai A, Capogrossi MC, Gaetano C. Histone deacetylase inhibitors: keeping momentum for neuromuscular and cardiovascular diseases treatment. Pharmacol Res 62: 3–10, 2010 [DOI] [PubMed] [Google Scholar]
- 17. Cox DM, Zhong F, Du M, Duchoslav E, Sakuma T, McDermott JC. Multiple reaction monitoring as a method for identifying protein posttranslational modifications. J Biomol Tech 16: 83–90, 2005 [PMC free article] [PubMed] [Google Scholar]
- 18. Creasy DM, Cottrell JS. Unimod: Protein modifications for mass spectrometry. Proteomics 4: 1534–1536, 2004 [DOI] [PubMed] [Google Scholar]
- 19. Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem 227: 792–800, 1995 [DOI] [PubMed] [Google Scholar]
- 20. Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics 7: 868–874, 2007 [DOI] [PubMed] [Google Scholar]
- 21. Divald A, Kivity S, Wang P, Hochhauser E, Roberts B, Teichberg S, Gomes AV, Powell SR. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circ Res 106: 1829–1838, 2010 [DOI] [PubMed] [Google Scholar]
- 22. Drews O, Tsukamoto O, Liem D, Streicher J, Wang Y, Ping P. Differential regulation of proteasome function in isoproterenol-induced cardiac hypertrophy. Circ Res 107: 1094–1101, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drews O, Wildgruber R, Zong C, Sukop U, Nissum M, Weber G, Gomes AV, Ping P. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol Cell Proteomics 6: 2021–2031, 2007 [DOI] [PubMed] [Google Scholar]
- 24. Eng JK, McCormack A, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989, 1994 [DOI] [PubMed] [Google Scholar]
- 25. Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch Biochem Biophys 453: 135–142, 2006 [DOI] [PubMed] [Google Scholar]
- 26. Feng Y, Longo DL, Ferris DK. Polo-like kinase interacts with proteasomes and regulates their activity. Cell Growth Differ 12: 29–37, 2001 [PubMed] [Google Scholar]
- 27. Gillardon F, Kloss A, Berg M, Neumann M, Mechtler K, Hengerer B, Dahlmann B. The 20S proteasome isolated from Alzheimer's disease brain shows post-translational modifications but unchanged proteolytic activity. J Neurochem 101: 1483–1490, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, Lu H, Stefani E, Ping P. Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Mol Cell Proteomics 8: 302–315, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, Bardag-Gorce F, Ping P. Mapping the murine cardiac 26S proteasome complexes. Circ Res 99: 362–371, 2006 [DOI] [PubMed] [Google Scholar]
- 31. Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, Eizirik DL, She JX, Wang CY. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun 337: 1308–1318, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Haas AL. Regulating the regulator: Rsp5 ubiquitinates the proteasome. Mol Cell 38: 623–624, 2010 [DOI] [PubMed] [Google Scholar]
- 33. Heink S, Ludwig D, Kloetzel PM, Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci USA 102: 9241–9246, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett 584: 2526–2538, 2010 [DOI] [PubMed] [Google Scholar]
- 35. Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell 38: 733–745, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jang M, Park BC, Lee AY, Na KS, Kang S, Bae KH, Myung PK, Chung BC, Cho S, Lee do H, Park SG. Caspase-7 mediated cleavage of proteasome subunits during apoptosis. Biochem Biophys Res Commun 363: 388–394, 2007 [DOI] [PubMed] [Google Scholar]
- 38. Knuehl C, Seelig A, Brecht B, Henklein P, Kloetzel PM. Functional analysis of eukaryotic 20S proteasome nuclear localization signal. Exp Cell Res 225: 67–74, 1996 [DOI] [PubMed] [Google Scholar]
- 39. Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584: 1393–1398, 2010 [DOI] [PubMed] [Google Scholar]
- 40. Kwak MK, Cho JM, Huang B, Shin S, Kensler TW. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free Radic Biol Med 43: 809–817, 2007 [DOI] [PubMed] [Google Scholar]
- 41. Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest 114: 1058–1071, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest 121: 3689–3700, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li N, Lerea KM, Etlinger JD. Phosphorylation of the proteasome activator PA28 is required for proteasome activation. Biochem Biophys Res Commun 225: 855–860, 1996 [DOI] [PubMed] [Google Scholar]
- 44. Liu X, Huang W, Li C, Li P, Yuan J, Li X, Qiu XB, Ma Q, Cao C. Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol Cell 22: 317–327, 2006 [DOI] [PubMed] [Google Scholar]
- 45. Liu Y, Liu X, Zhang T, Luna C, Liton PB, Gonzalez P. Cytoprotective effects of proteasome beta5 subunit overexpression in lens epithelial cells. Mol Vis 13: 31–38, 2007 [PMC free article] [PubMed] [Google Scholar]
- 46. Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, Li X, Whitelegge J, Drews O, Yang PY, Ping P. Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Mol Cell Proteomics 7: 2073–2089, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics 5: 4145–4151, 2005 [DOI] [PubMed] [Google Scholar]
- 48. Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res 7: 4566–4576, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Montecchi-Palazzi L, Beavis R, Binz PA, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat Biotechnol 26: 864–866, 2008 [DOI] [PubMed] [Google Scholar]
- 50. Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4: 2010–2021, 2005 [DOI] [PubMed] [Google Scholar]
- 51. Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279: 41346–41351, 2004 [DOI] [PubMed] [Google Scholar]
- 52. Powell SR, Divald A. The ubiquitin-proteasome system in myocardial ischaemia and preconditioning. Cardiovasc Res 85: 303–311, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Powell SR, Gurzenda EM, Teichberg S, Mantell LL, Maulik D. Association of increased ubiquitinated proteins with cardiac apoptosis. Antioxid Redox Signal 2: 103–112, 2000 [DOI] [PubMed] [Google Scholar]
- 54. Powell SR, Samuel SM, Wang P, Divald A, Thirunavukkarasu M, Koneru S, Wang X, Maulik N. Upregulation of myocardial 11S-activated proteasome in experimental hyperglycemia. J Mol Cell Cardiol 44: 618–621, 2008 [DOI] [PubMed] [Google Scholar]
- 55. Powell SR, Wang P, Katzeff H, Shringarpure R, Teoh C, Khaliulin I, Das DK, Davies KJ, Schwalb H. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: essential role of the proteasome. Antioxid Redox Signal 7: 538–546, 2005 [DOI] [PubMed] [Google Scholar]
- 56. Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, Pagani F, Powell SR, Day SM. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 121: 997–1004, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Razeghi P, Baskin KK, Sharma S, Young ME, Stepkowski S, Essop MF, Taegtmeyer H. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem Biophys Res Commun 342: 361–364, 2006 [DOI] [PubMed] [Google Scholar]
- 59. Rivett AJ, Bose S, Brooks P, Broadfoot KI. Regulation of proteasome complexes by gamma-interferon and phosphorylation. Biochimie 83: 363–366, 2001 [DOI] [PubMed] [Google Scholar]
- 60. Saito A, Hayashi T, Okuno S, Nishi T, Chan PH. Modulation of p53 degradation via MDM2-mediated ubiquitylation and the ubiquitin-proteasome system during reperfusion after stroke: role of oxidative stress. J Cereb Blood Flow Metab 25: 267–280, 2005 [DOI] [PubMed] [Google Scholar]
- 61. Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry 40: 314–319, 2001 [DOI] [PubMed] [Google Scholar]
- 62. Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scruggs SB, Ping P, Zong C. Heterogeneous cardiac proteasomes: mandated by diverse substrates? Physiology (Bethesda) 26: 106–114, 2011 [DOI] [PubMed] [Google Scholar]
- 64. Tanaka K, Yoshimura T, Tamura T, Fujiwara T, Kumatori A, Ichihara A. Possible mechanism of nuclear translocation of proteasomes. FEBS Lett 271: 41–46, 1990 [DOI] [PubMed] [Google Scholar]
- 65. Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal 4: rs4, 2011 [DOI] [PubMed] [Google Scholar]
- 66. Thorpe JA, Christian PA, Schwarze SR. Proteasome inhibition blocks caspase-8 degradation and sensitizes prostate cancer cells to death receptor-mediated apoptosis. Prostate 68: 200–209, 2008 [DOI] [PubMed] [Google Scholar]
- 67. Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci USA 96: 6223–6228, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Um JW, Im E, Park J, Oh Y, Min B, Lee HJ, Yoon JB, Chung KC. ASK1 negatively regulates the 26 S proteasome. J Biol Chem 285: 36434–36446, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ventadour S, Jarzaguet M, Wing SS, Chambon C, Combaret L, Bechet D, Attaix D, Taillandier D. A new method of purification of proteasome substrates reveals polyubiquitination of 20 S proteasome subunits. J Biol Chem 282: 5302–5309, 2007 [DOI] [PubMed] [Google Scholar]
- 71. Wang D, Zong C, Koag MC, Wang Y, Drews O, Fang C, Scruggs SB, Ping P. Proteome dynamics and proteome function of cardiac 19S proteasomes. Mol Cell Proteomics 10: 1–10, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol 301: H2207–H2219, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang XH, Zhang L, Mitch WE, Ledoux JM, Hu J, Du J. Caspase-3 cleaves specific 19S proteasome subunits in skeletal muscle stimulating proteasome activity. J Biol Chem 285: 21249–21257, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yano M, Mori S, Kido H. Intrinsic nucleoside diphosphate kinase-like activity is a novel function of the 20 S proteasome. J Biol Chem 274: 34375–34382, 1999 [DOI] [PubMed] [Google Scholar]
- 75. Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem 354: 279–289, 2006 [DOI] [PubMed] [Google Scholar]
- 76. Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem 282: 22460–22471, 2007 [DOI] [PubMed] [Google Scholar]
- 77. Zhang F, Paterson AJ, Huang P, Wang K, Kudlow JE. Metabolic control of proteasome function. Physiology (Bethesda) 22: 373–379, 2007 [DOI] [PubMed] [Google Scholar]
- 78. Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115: 715–725, 2003 [DOI] [PubMed] [Google Scholar]
- 79. Zong C, Fang C, Wang D, Yu H, Zhang J, Yang P, Ping P. Acetylation affords enhanced proteolytic function in normal and diseased myocardium. Circ Res 109: AP286, 2011 [Google Scholar]
- 80. Zong C, Gomes AV, Drews O, Li X, Young GW, Berhane B, Qiao X, French SW, Bardag-Gorce F, Ping P. Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ Res 99: 372–380, 2006 [DOI] [PubMed] [Google Scholar]
- 81. Zong C, Young GW, Wang Y, Lu H, Deng N, Drews O, Ping P. Two-dimensional electrophoresis-based characterization of post-translational modifications of mammalian 20S proteasome complexes. Proteomics 8: 5025–5037, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zong NC, Deng N, Chen L, Wang Y, Fang C, Jimenez R, Xu T, Hermjakob H, Duan H, Yates J, Apweiler R, Ping P. COPa library: a proteomic knowledge base for cardiovascular biology and medicine. Circ Res 109: AP327, 2011 [Google Scholar]
- 84. Zu L, Bedja D, Fox-Talbot K, Gabrielson KL, Van Kaer L, Becker LC, Cai ZP. Evidence for a role of immunoproteasomes in regulating cardiac muscle mass in diabetic mice. J Mol Cell Cardiol 49: 5–15, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]


