Quantitative polymerase chain reaction using serum rather than whole blood showed a wider window of positivity from day 2 to day 15 of fever and higher sensitivity than single sample serology for diagnosing leptospirosis. Quantitative leptospiremia did not predict severe clinical manifestations.
Abstract
(See the Editorial Commentary by Katz, on pages 1256–8.)
Background. Quantitative polymerase chain reaction (qPCR), despite cost and logistical challenges, has the potential to provide accurate and timely diagnosis for leptospirosis at the point-of-care in endemic areas. We studied optimal sample types for qPCR, timing of sampling, and clinical manifestations in relation to quantitative leptospiremia.
Methods. A new qPCR assay using pathogenic Leptospira-specific 16S ribosomal RNA (rRNA) gene Taqman primers and an optimized temperature stepdown protocol was used to analyze patient blood samples. Serum was compared with whole blood as sample source. Quantitative leptospiremia was compared with clinical manifestations of leptospirosis and outcome.
Results. The diagnostic sensitivity of qPCR of whole blood and serum was 18.4% (95% confidence interval [CI]: 9.97%–31.4%) and 51.0% (95% CI: 37.5%–64.4%) respectively. The qPCR on suspected cases confirmed infection in 58 of 381 cases (15.2%). Of these, 6 cases confirmed by nested polymerase chain reaction (PCR) and sequencing were serologically negative using a standard but not regionally optimized microscopic agglutination test panel. The bacterial load in serum/blood ranged from 102 to 106 Leptospira/mL. Median leptospiral load for uncomplicated, renal failure, myocarditis, and multi-organ failure patients were 8616, 11007, 36100, and 15882 Leptospira/mL respectively. The qPCR window of positivity ranged from day 2 to day 15; sensitivity of qPCR was not affected by the length of the interval between the onset of symptoms and sample collection (P = .328).
Conclusions. Quantitative PCR shows potential as a valid diagnostic test with a wider window of positivity than previously thought. Quantitative leptospiremia in serum/whole blood samples did not directly correlate with clinical manifestations of outcome in this patient population.
Leptospirosis affects primarily poor and marginalized populations in tropical and subtropical countries. The median incidence of human leptospirosis in highly endemic countries is as high as 975 per 100000 population. Severe life-threatening complications due to acute renal and acute lung injury are common with mortality rates as high as 67% and 87%, respectively [1].
The microscopic agglutination test (MAT) and culture used to diagnosis leptospirosis are only available in reference laboratories. MAT is limited by its subjectivity, requires the maintenance of live Leptospira, and requires a convalescent sample for conclusive results. Culture is insensitive and requires prolonged incubation (up to 3 months). The lack of diagnostic test result availability in relation to patient care impairs the clinical management of leptospirosis and makes assessment of the burden of this disease difficult.
Molecular-based diagnostic testing is being used increasingly for leptospirosis diagnosis [2]. Quantitative polymerase chain reaction (qPCR)-based assays are more sensitive than conventional PCR assays and offers the ability to measure the bacterial load in clinical specimens. A recent analysis of SYBR green and Taqman based qPCR assays used in leptospirosis diagnosis showed that published methods can detect between 102 and 103 bacteria/mL in whole blood. Yet, diagnostic sensitivity of qPCR in clinical samples is reported to be lower than MAT [3]. In clinical application, urine, whole blood, serum, and plasma have been used as sample types for analysis, but published studies showed mixed results on the detection of antigen in different blood fraction [4, 5]. For clinical application of qPCR methods, data are still needed to determine optimal sample types.
Few studies (none from Asia) have attempted to associate leptospiral bacterial load with disease outcome or severity. A critical threshold of 104 bacteria/mL was reported based on 12 confirmed cases [6]. In Peru, a similar threshold level was associated with severe pulmonary manifestations based on only 7 cases [7]. Extensive studies of other viral and bacterial diseases indicate that pathogen load in blood is a main predictor of disease severity and outcome [8–14]. Quantification of pathogen load in these bacterial diseases could provide an early indication of severe disease with important prognostic implications.
Using a well-characterized cohort of laboratory-confirmed leptospirosis patients, this study compares the diagnostic utility of qPCR analysis of serum and whole blood specimens, investigates the association between leptospiral load and clinical outcome and determines the window of positivity for qPCR.
METHODS
Study Units and Samples
Samples for the present study were obtained from a systematically sampled cohort of patients during a 2008 outbreak of leptospirosis in Sri Lanka [15, 16]. This cohort of patients included samples from 381 acute fever patients (fever <15 days) with possible leptospirosis recruited from 3 major hospitals in Kegalle, Kandy, and Matale districts of central Sri Lanka. From these samples we selected 49 MAT-positive confirmed leptospirosis cases and 56 MAT-negative controls. Of the available samples, only these 105 patients had the first (acute) whole blood/serum sample collected within the first 10 days of the onset of symptoms and their second (convalescent) sample collected after day 14, with all specimens having a minimum of 7 days between acute and convalescent sampling. Standard leptospirosis diagnosis was based on MAT as described elsewhere [15].
DNA Extraction
DNA was extracted from 100 μL of serum or whole blood using the DNeasy Blood and Tissue Kit (QIAGEN) according to manufacturer’s directions. For use in preparing standard curves (with spiked bacteria for extraction) and negative controls, venous blood was collected from a healthy individual. Exponential-phase Leptospira interrogans serovar Copenhageni, strain L1-130 [17] cultured in liquid EMJH (Ellinghausen-McCullogh-Johnson-Harris) media was inactivated in 10% formalin for 15 minutes and then counted in a Petroff-Hausser counting chamber. Known numbers of live Leptospira were then spiked into whole blood/serum and diluted to give final concentrations of 1 × 100 to 1 × 108 Leptospira/mL. DNA was extracted as described above. For negative controls, unspiked whole blood/serum from the same healthy individual was extracted as described.
Quantitative Polymerase Chain Reaction Primers and Probes
Quantitative PCR was performed using primer pair lepto16S620f and lepto16S730r as described elsewhere [17] and internal probe (5′-CAAGTCGCCTTCGCCACTGGTGTTCCTCCAGA-3′) “16S Taqman probe1” labeled with the fluorescent reporter dye FAM (6-carboxyfluorescein) at the 5′ end, and Black Hole quencher one (BHQ-1) at the 3′ end.
Polymerase Chain Reaction Conditions and Identification of Positive Samples
PCR reactions (in triplicate) used iQ Supermix (Bio-Rad) with final primer and probe concentrations of 0.5 and 0.2 μmol/L, respectively, and 5 μL DNA (samples/standard curves and controls) in 20 μL reaction volume using a DNA Engine Opticon 2 (MJ Research) according to the following protocol: 95°C for 3 minutes, 15 cycles of 10 seconds at 94°C, 45 seconds at 80°C; annealing temperature was decreased by 1°C per cycle to 65°C; amplification continued for another 24 cycles of 10 seconds at 94°C and 45 seconds at 65°C. All positives had a quantitative signal within the linear part of the standard curve greater than the lowest positive control.
Quantification of Leptospira in Human Blood
All qPCR-positive samples were reamplified using a single standard curve, and quantification was carried out using Opticon 2 software. Quantification was not carried out during the initial screening stage to minimize variability between measurements.
Single Tube Nested Polymerase Chain Reaction and Sequencing
The qPCR positive samples were amplified using a previously published nested PCR protocol [15]. PCR products were purified using Wizard SV Gel and PCR Clean-Up System (Promega). Purified PCR products were cloned into Topo cloning vector pCR2.1 (Invitrogen) and transformed into Escherichia coli TOP10F cells. The plasmid construct was isolated and purified using QIAprep Miniprep Kit (QIAGEN) before sequencing.
Data Analysis
We used proportions and 95% confidence interval (CI) to analyze sensitivity of qPCR in serum and whole blood samples. Specificity was not calculated because of the lack of a true gold standard; controls were probable fever patients in whom leptospirosis was excluded using paired sample MAT. All categorical data are presented as proportions and percentages. The Mann-Whitney U test was used to compare duration of illness among qPCR positive and negative samples.
Human Subjects Protections
This study was approved by the Ethical Review Committee, Faculty of Medicine, University of Peradeniya, Sri Lanka. Human blood was obtained for experimental work under a protocol approved by the UCSD Human Subjects Protection Program with verbal assent.
RESULTS
Comparison of Diagnostic Sensitivity of Quantitative Polymerase Chain Reaction for Serum and Whole Blood
Compared with MAT, the diagnostic sensitivity of qPCR of whole blood was 18.4% (95% CI: 9.97%–31.4%) and 51.0% (95% CI: 37.5%–64.4%) for serum (Table 1). Observed difference of positive proportions between serum and whole blood was highly significant (P = .0007). Two MAT-negative patients had positive qPCR results; sequencing confirmed that both were false-negative by the MAT. Both patients were admitted to hospital before 7th day of illness, with fever, headache, and prostration. On examination, both patients had mild conjunctival suffusion and mild muscle tenderness. Urinalysis detected albumin and red cells in the first patient, whereas liver and renal functions were normal. The second patient had normal blood and urine biochemistry. Both patients were discharge from hospitals without any complications. Single tube nested PCR (STNPCR) amplification was positive, and the sequencing confirmed infection with L. interrogans without further identification.
Table 1.
Comparison of Diagnostic Sensitivity of Quantitative Polymerase Chain Reaction for Serum and Whole Blood for Leptospirosis
| MAT Positive |
MAT Negative |
||||
| n | % | n | % | Total | |
| Whole blood | |||||
| Positive | 9 | 18.4 | 1 | 1.8 | 10 |
| Not detected | 40 | 81.6 | 55 | 98.2 | 95 |
| Total | 49 | 100.0 | 56 | 100.0 | 105 |
| Serum | |||||
| Positive | 25 | 51.0 | 1 | 1.8 | 26 |
| Not detected | 24 | 49.0 | 55 | 98.2 | 79 |
| Total | 49 | 100.0 | 56 | 100.0 | 105 |
Abbreviation: MAT, microscopic agglutination test.
Case Confirmation and Species Identification
Quantitative PCR was used to confirm the diagnosis of leptospirosis among 381 fever patients comprising the same patient population [15], including the 105 patients analyzed above. A total of 58 (15.2%) cases were detected using qPCR. Of these, 38 were confirmed previously, whereas 7 cases previously classified as “presumptive” were confirmed by qPCR, as well as 13 patients who were previously considered as negative for leptospirosis (Table 2). Of these 20 newly confirmed cases, 6 patients had negative MAT results (these include 2 patients used in serum-whole blood comparison), another 2 patients had negative MAT but the second sample was taken on day 8 and 9, and the other 12 patient have not had MAT results due to lack of paired samples. All 6 patients with MAT negative results in paired samples were positive in STNPCR, and sequencing was compatible with L. interrogans. Altogether, 26 cases were positive in STNPCR (except the 6 STNPCR positive patients that were previously confirmed). The deduced Leptospira species were L. interrogans (25) and L. weilli (1). There were no differences either in clinical manifestation or outcome between qPCR-positive vs qPCR-negative patients.
Table 2.
Retrospective Diagnosis of Probable Cases of Leptospirosis Using Quantitative Polymerase Chain Reaction From 2008 Leptospirosis Outbreak in Sri Lanka
| Availability (n) | Results | n | % |
| Whole blood and serum (192) | Both positive | 12 | 6.2 |
| Serum positive | 24 | 12.5 | |
| Whole blood positive | 2 | 1.0 | |
| Not detected | 154 | 80.2 | |
| Whole blood only (170) | Positive | 13 | 7.6 |
| Not detected | 157 | 92.4 | |
| Serum only (19) | Positive | 7 | 36.8 |
| Not detected | 12 | 63.2 | |
| Total (381) | Positive | 58 | 15.2 |
| Not detected | 323 | 84.8 |
Window of Positivity
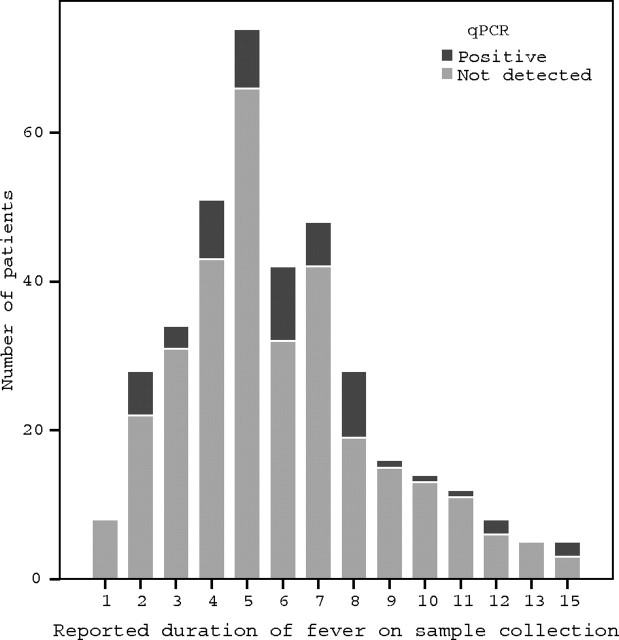
First we compared the window of positivity among qPCR-positive and -negative patients among 49 MAT-positive cases to determine whether the sensitivity of qPCR was affected by the timing of sample collection. The median duration of fever was 6 (interquartile range [IQR] 4–8) and 5 (IQR 5–6) days for qPCR-positive and -negative patients, respectively. Sensitivity of qPCR was not affected by the interval between onset of symptoms and sample collection in patients with fever <10 days (Mann-Whitney U test, P = .33). Among the 381 cases studied, qPCR was positive up to day 15 after onset of fever (Figure 1). Among 30 patients presenting to hospital between days 11 and 15, 5 (17%) were positive in qPCR compared with 53 (15%) patients who presented within first 10 days of fever.
Figure 1.
Window of positivity for qPCR among 381 suspected leptospirosis cases. Abbreviation: qPCR, quantitative polymerase chain reaction.
Comparison of Acute Phase Microscopic Agglutination Test and Quantitative Polymerase Chain Reaction
We compared the qPCR and acute sample MAT results (acute blood sample obtained within 15 days of fever) to determine the utility of these tests. Of the 87 confirmed leptospirosis patients (previously demonstrated by paired sample MAT), 73 samples were available for qPCR testing in the present analysis. Of acute serum samples available for these 73 patients, 36 (49%) were qPCR positive, whereas only 13 (18%) had a diagnostic MAT titer (≥1/400).
Quantification of Bacterial Load
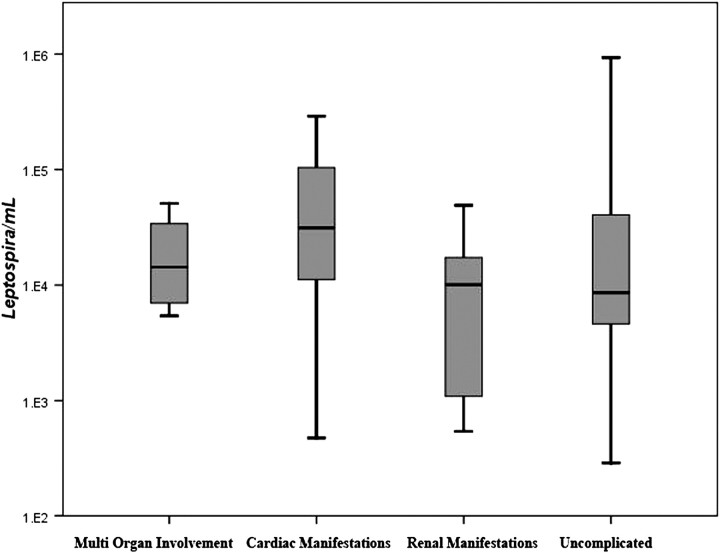
The bacterial load in serum/blood ranged from 102 to 106 Leptospira/mL among 58 positive cases. Median bacterial load was 9577 Leptospira/mL (IQR, 4623–49 580/mL). Of 58 qPCR-positive patients, 40 had uncomplicated disease, 8 had acute kidney injury (serum creatinine >1.5 mmol/L), 6 had myocarditis (ECG with T-wave inversions and/or diffuse ST elevations) with or without heart failure, and 4 patients had multi-organ failure including renal failure and myocarditis. No patient had pulmonary hemorrhage. Patients with positive qPCR results were compared to identify any association between bacterial load and clinical manifestations (Figure 2). Median leptospiral load for uncomplicated, renal failure, myocarditis, and multi-organ failure patients were 8616 (IQR, 4611–41727), 11007 (IQR, 1102–17417), 36100 (IQR, 11162–104067), and 15882 (IQR, 7216–36871) Leptospira/mL, respectively. Although the median bacterial load was lowest in uncomplicated group, bacterial load was not statistically different from other outcome categories (Mann-Whitney U test, P = .59). Among 40 male patients positive for qPCR, median leptospiral load was 15640 Leptospira/mL (IQR, 6053–53980 Leptospira/mL), and this was significantly (Mann-Whitney U test, P = .022) higher than that of female patients (median, 5611, IQR, 3891–14 383 Leptospira/mL).
Figure 2.
Distribution of leptospiral bacteremia among 58 quantitative polymerase chain reaction positive patients, grouped by complications.
DISCUSSION
We describe the direct clinical application of molecular based techniques to the diagnosis of acute leptospirosis in the resource-poor setting of rural Sri Lanka. The data and analysis here allow us to describe several important advances: (1) that a new qPCR protocol to diagnose acute leptospirosis is sensitive (often more than acute or retrospective gold-standard MAT serology); (2) that the window of positivity for qPCR in human clinical samples is longer than previously known (up to 15 days); and (3) that serum may be better than whole blood as a specimen type for the molecular diagnosis of acute human leptospirosis. These results have important implications not only for the diagnosis of acute leptospirosis but also for making new inferences into the pathogenesis of this disease.
Evidence suggests that although the MAT is considered to be the gold standard diagnostic test for leptospirosis, MAT sensitivity and specificity are often <100% because of limitations of knowing the regionally specific serovars [18, 19]. Our initial comparison data have confirmed these findings; we show through sequencing that 2 patients with a negative MAT and detected by qPCR and STNPCR were confirmed to have had leptospirosis despite negative MAT. Indeed, 6 MAT-negative serum samples later proved positive by qPCR and STNPCR and were identified as L. interrogans. A likely explanation could be that local serovars or serogroups in Sri Lanka are not represented in the panel used in the diagnosis, although it contained the strains isolated from Sri Lanka as well as common strains to represent a broad panel of serovars. Alternatively, some patients may simply be unable to make detectable antibody responses to leptospiral lipopolysaccharide. Nonetheless, regional differences in leptospiral biodiversity will always present a problem for serological diagnosis in geographically disparate regions. On the other hand, by their nature structural RNAs and essential core genes are less variable and are therefore more amenable to broad-range strain detection. Indeed Weisburg et al [20] have described several generic 16S primers that amplify several bacterial genera. Because MAT has been the standard reference laboratory confirmation procedure, our study findings raise the question about actual sensitivity of MAT [21–23]. This observation further confirms the need for more sensitive diagnostic tests that are available at point of care [22]. We found, during the acute phase of leptospirosis (up to 15 days in this study), the diagnostic sensitivity of qPCR was nearly 3 times higher than acute phase MAT test, considering 400 as the cutoff. This finding strengthens the validity of qPCR as a valid diagnostic tool over serology-based methods. However, the sensitivity and specificity of qPCR are still suboptimal, and further work is needed to improve the application of this technique to the diagnosis of leptospirosis.
Contrary to the previous reports that the leptospiral load is a main predictive factor of severity of illness, our study showed that high levels of leptospiremia—comparable with previous reports of severe/fatal leptospirosis—could occur without such complications. Previous studies [24, 25] observed that >104 organisms in bloodstream was a critical factor associated with severe pulmonary manifestations and death. In our study, 17 patients had bacterial load of >104 organisms/mL in whole blood/serum yet did not have severe complications. The difference in our observation from 2 previously published studies may be due to variability of virulence among different leptospiral species/serovars/strains. Even within our study sample, complications with low bacterial load and not having complications with high level of bacterial load could be due to different serovars that manifest in different ways. On the other hand, complications with a bacterial load of 103 per mL is difficult to interpret because the timing of sample might not been at peak leptospiremia. However, the number of patients in each complication category was small, and further studies are needed to strengthen this observation.
In this study, serum was a better specimen type than whole blood for obtaining DNA for diagnostic qPCR. However, rather than reflecting intrinsic differences between samples, this observation could reflect the fact that that there could be 2–3 times as much leptospiral DNA per microliter of serum compared with whole blood, assuming Leptospira are only found in serum. However, this is an unlikely explanation because phagocytosed Leptospira appear to be concentrated in the buffy coat [5]. Moreover, most serum-positive/whole blood-negative samples were well above the theoretical limit of detection of the qPCR (3 × 102 per mL under the conditions used). Thus, it is more likely that PCR inhibitors in blood such as heme/heme derivatives [26], host leukocyte DNA [27], or added anticoagulants [28] present in whole blood samples reduced the efficiency of qPCR amplification of leptospiral DNA. In this comparison we used only sensitivity but not specificity due to the limitation of study design and leptospirosis diagnostics we used for comparison. First, the control group was not ideal “controls” but febrile patients who could be “possible” cases of leptospirosis. Although these patients showed negative results in paired sample MAT, it is difficult to exclude the diagnosis because MAT is not 100% sensitive, as confirmed in this study. Further, the ideal test for assessing sensitivity of any diagnostic test would be a direct demonstration of the presence of Leptospira in acute blood samples such as by a positive blood culture. However, in this study, because of limitations of resources, we were unable to perform blood cultures on patient samples; isolation of Leptospira is nonetheless recognized to be insensitive itself because of the difficulty in isolating fastidious Leptospira.
A notable observation made during this work was significantly high leptospiremia among male patients. Epidemiological evidence has consistently reported that males are more often affected by leptospirosis than females; whether there is a biological explanation of this observation or males are simply more often exposed to Leptospira remains unclear. However, in animal populations where the exposure bias is minimal, males have shown higher seropositivity rate [29, 30]. Our finding of higher leptospiremia in human males provides evidence for a probable higher biological susceptibility of men for leptospirosis. This preliminary observation needs further investigation.
Limitations of the data from this study should be recognized. First, culture isolation of infecting serovars was not performed. Quantification and clinical correlation of leptospiremia should ideally be performed with systematic serial sampling to determine the peak leptospiremia and correlate it with clinical outcome. It is difficult to do this because it would require withholding of antibiotic treatment. Nonetheless, the diagnostic utility of qPCR was done using acute phase blood sample taken on admission, which reflects clinical practice. The molecular testing we performed did not differentiate infecting Leptospira beyond the species level. We did attempt to carry out multilocus sequence typing (MLST) on the qPCR-positive specimens in this study, 12 of which preliminarily suggested the presence of a limited number of sequence types (ST) similar to ST 1 (n = 11) and ST44 (n = 1) (unpublished observations). Finally, the samples for this study were obtained in 2008–2009, whereas the qPCR was done in 2011. During this period, samples were thawed at least 3–4 times, which might have reduced the sensitivity of qPCR.
The most important conclusion that we draw from this study is that qPCR is promising as a rapid diagnostic tool in the diagnosis of acute leptospirosis with a wide window of positivity. However, the technical expertise and cost needed for qPCR still impede its use in resource-poor settings.
Notes
Acknowledgments.
We would like to thank Jason S. B. Lehmann and Anne Spichler, MD, PhD, for their important advice and contributions, particularly in support of qPCR work. We also thank Paula Maguina for her expert scientific, administrative and logistical support of this work.
Financial support.
This work was supported by US Public Health Service grants, 1RO1TW05860, 1U01AI075420, 1K24AI068903, 5R25GM083275 and 1D43TW007120.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Report of the second meeting of leptospirosis burden epidemiology reference group. Geneva, Switzerland: World Health Organization; 2011. [Google Scholar]
- 2.Bourhy P, Bremont S, Zinini F, Giry C, Picardeau M. Comparison of real-time PCR assays for detection of pathogenic Leptospira spp. in blood and identification of variations in target sequences. J Clin Microbiol. 2011;49:2154–60. doi: 10.1128/JCM.02452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smythe LD, Smith G, Dohnt M, Symonds M, Barnett L, McKay L. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis. 2002;2:13. doi: 10.1186/1471-2334-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009;64:247–55. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Kositanont U, Rugsasuk S, Leelaporn A, Phulsuksombati D, Tantitanawat S, Naigowit P. Detection and differentiation between pathogenic and saprophytic Leptospira spp by multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2007;57:117–22. doi: 10.1016/j.diagmicrobio.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Truccolo J, Charavay F, Merien F, Perolat P. Quantitative PCR assay to evaluate ampicillin, ofloxacin, and doxycycline for treatment of experimental leptospirosis. Antimicrob Agents Chemother. 2002;46:848–53. doi: 10.1128/AAC.46.3.848-853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segura ER, Ganoza CA, Campos K, et al. Clinical spectrum of pulmonary involvement in leptospirosis in a region of endemicity, with quantification of leptospiral burden. Clin Infect Dis. 2005;40:343–51. doi: 10.1086/427110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang YC, Chang SC, Wang WK. High and increasing Oxa-51 DNA load predict mortality in Acinetobacter baumannii bacteremia: implication for pathogenesis and evaluation of therapy. PLoS One. 2010;5:e14133. doi: 10.1371/journal.pone.0014133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darton T, Guiver M, Naylor S, et al. Severity of meningococcal disease associated with genomic bacterial load. Clin Infect Dis. 2009;48:587–94. doi: 10.1086/596707. [DOI] [PubMed] [Google Scholar]
- 10.Hackett SJ, Guiver M, Marsh J, et al. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch Dis Child. 2002;86:44–6. doi: 10.1136/adc.86.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrol ED, Guiver M, Nkhoma S, et al. High pneumococcal DNA loads are associated with mortality in Malawian children with invasive pneumococcal disease. Pediatr Infect Dis J. 2007;26:416–22. doi: 10.1097/01.inf.0000260253.22994.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rello J, Lisboa T, Lujan M, et al. Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest. 2009;136:832–40. doi: 10.1378/chest.09-0258. [DOI] [PubMed] [Google Scholar]
- 13.Margolis DA, Burns J, Reed SL, Ginsberg MM, O'Grady TC, Vinetz JM. Septicemic plague in a community hospital in California. Am J Trop Med Hyg. 2008;78:868–71. [PMC free article] [PubMed] [Google Scholar]
- 14.Sonthayanon P, Chierakul W, Wuthiekanun V, et al. Association of high Orientia tsutsugamushi DNA loads with disease of greater severity in adults with scrub typhus. J Clin Microbiol. 2009;47:430–4. doi: 10.1128/JCM.01927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agampodi SB, Peacock SJ, Thevanesam V, et al. Leptospirosis outbreak in Sri Lanka in 2008: lessons for assessing the global burden of disease. Am J Trop Med Hyg. 2011;85:471–8. doi: 10.4269/ajtmh.2011.11-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agampodi SB, Nugegoda DB, Thevanesam V. Determinants of leptospirosis in Sri Lanka: study protocol. BMC Infect Dis Nov. 2010;10:332. doi: 10.1186/1471-2334-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, Vinetz JM. Asymptomatic renal colonization of humans in the Peruvian Amazon by Leptospira. PLoS Negl Trop Dis. 2010;4:e612. doi: 10.1371/journal.pntd.0000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthias MA, Ricaldi JN, Cespedes M, et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis. 2008;2:e213. doi: 10.1371/journal.pntd.0000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyokawa T, Ohnishi M, Koizumi N. Diagnosis of acute leptospirosis. Expert Rev Anti Infect Ther. 2011;9:111–21. doi: 10.1586/eri.10.151. [DOI] [PubMed] [Google Scholar]
- 20.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thaipadunpanit J, Chierakul W, Wuthiekanun V, et al. Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipl32 genes for human leptospirosis in Thailand: a case-control study. PLoS One. 2011;6:e16236. doi: 10.1371/journal.pone.0016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7:736–47. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merien F, Baranton G, Perolat P. Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J Infect Dis. 1995;172:281–5. doi: 10.1093/infdis/172.1.281. [DOI] [PubMed] [Google Scholar]
- 24.Truccolo J, Serais O, Merien F, Perolat P. Following the course of human leptospirosis: evidence of a critical threshold for the vital prognosis using a quantitative PCR assay. FEMS Microbiol Lett. 2001;204:317–21. doi: 10.1111/j.1574-6968.2001.tb10904.x. [DOI] [PubMed] [Google Scholar]
- 25.Al-Soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–93. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morata P, Queipo-Ortuno MI, de Dios Colmenero J. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for diagnosis of human brucellosis. J Clin Microbiol. 1998;36:2443–6. doi: 10.1128/jcm.36.9.2443-2446.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JT, Wang TH, Sheu JC, Lin SM, Lin JT, Chen DS. Effects of anticoagulants and storage of blood samples on efficacy of the polymerase chain reaction assay for hepatitis C virus. J Clin Microbiol. 1992;30:750–3. doi: 10.1128/jcm.30.3.750-753.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colagross-Schouten AM, Mazet JA, Gulland FM, Miller MA, Hietala S. Diagnosis and seroprevalence of leptospirosis in California sea lions from coastal California. J Wildl Dis. 2002;38:7–17. doi: 10.7589/0090-3558-38.1.7. [DOI] [PubMed] [Google Scholar]
- 29.Trainer DO, Hanson RP, Pope EP, Carbrey EA. The role of deer in the epizootiology of leptospirosis in Wisconsin. Am J Vet Res. 1963;24:159–67. [PubMed] [Google Scholar]